Summary
Membrane contact sites (MCS) between the endoplasmic reticulum (ER) and the plasma membrane (PM) play fundamental roles in all eukaryotic cells. ER-PM MCS are particularly abundant in Saccharomyces cerevisiae, where approximately half of the PM surface is covered by cortical ER (cER). Several proteins, including Ist2, Scs2/22, and Tcb1/2/3 are implicated in cER formation, but the specific roles of these molecules are poorly understood. Here, we use cryo-electron tomography to show that ER-PM tethers are key determinants of cER morphology. Notably, Tcb proteins (tricalbins) form peaks of extreme curvature on the cER membrane facing the PM. Combined modeling and functional assays suggest that Tcb-mediated cER peaks facilitate the transport of lipids between the cER and the PM, which is necessary to maintain PM integrity under heat stress. ER peaks were also present at other MCS, implying that membrane curvature enforcement may be a widespread mechanism to regulate MCS function.
Keywords: membrane contact site, endoplasmic reticulum, plasma membrane, tricalbins, membrane curvature, cryo-electron tomography, cryo-ET, cryo-focused ion beam milling, cryo-FIB
Graphical Abstract
Highlights
-
•
Tethers of ER-plasma membrane (PM) contact sites shape cortical ER (cER) morphology
-
•
Tricalbins create peaks of extreme curvature on the cER membrane facing the PM
-
•
cER peaks are important to maintain PM integrity under heat stress
-
•
cER peaks may facilitate ER-to-PM lipid transport
Using cryo-electron tomography, Collado et al. show that tricalbins generate peaks of extreme curvature on the cortical ER (cER) membrane at ER-plasma membrane (PM) contact sites. Functional assays and theoretical modeling indicate that cER peaks are important to maintain PM integrity under heat stress, possibly by facilitating cER-to-PM lipid transport.
Introduction
Endoplasmic reticulum (ER)-plasma membrane (PM) membrane contact sites (MCS) are critical modulators of Ca2+ and lipid homeostasis in eukaryotic cells (Balla, 2018, Chang et al., 2017, Cockcroft and Raghu, 2018, Saheki and De Camilli, 2017a, Stefan, 2018). These structures, where the ER and the PM come into close apposition (10 – 30 nm), mediate store-operated Ca2+ entry (Carrasco and Meyer, 2011), insulin secretion by pancreatic beta cells (Lees et al., 2017), and excitation-contraction coupling in striated muscle (Bers, 2002). Consequently, dysregulation of ER-PM MCS is linked to multiple human diseases (Lacruz and Feske, 2015, Landstrom et al., 2014, Ríos et al., 2015).
ER-PM MCS are particularly abundant in the yeast Saccharomyces cerevisiae, where nearly half of the PM surface area is covered by cortical ER (cER) (Manford et al., 2012, Pichler et al., 2001, Quon et al., 2018, Toulmay and Prinz, 2012, West et al., 2011). The loss of six proteins (Ist2, Scs2/22, and Tcb1/2/3; “Δtether” cells) dramatically reduces the extent of ER-PM association, indicating that these proteins are important ER-PM tethers (Manford et al., 2012). Additional proteins, including Ice2 and the yeast StARkin orthologs, are also implicated in cER-PM function in S. cerevisiae (Gatta et al., 2015, Quon et al., 2018). Loss of cER triggers PM lipid imbalance (Manford et al., 2012, Quon et al., 2018), highlighting the physiological importance of these membrane junctions.
Ist2 is a member of the anoctamin/TMEM16 protein family (Whitlock and Hartzell, 2017). Ist2 resides on the ER membrane and consists of eight transmembrane domains plus a long C-terminal cytoplasmic tail that binds PM lipids (Figure 2A), thereby tethering the ER and the PM (Fischer et al., 2009, Jüschke et al., 2005, Maass et al., 2009, Manford et al., 2012). Deletion of Ist2 results in reduced cER levels, whereas Ist2 overexpression leads to increased ER-PM MCS (Manford et al., 2012, Wolf et al., 2012).
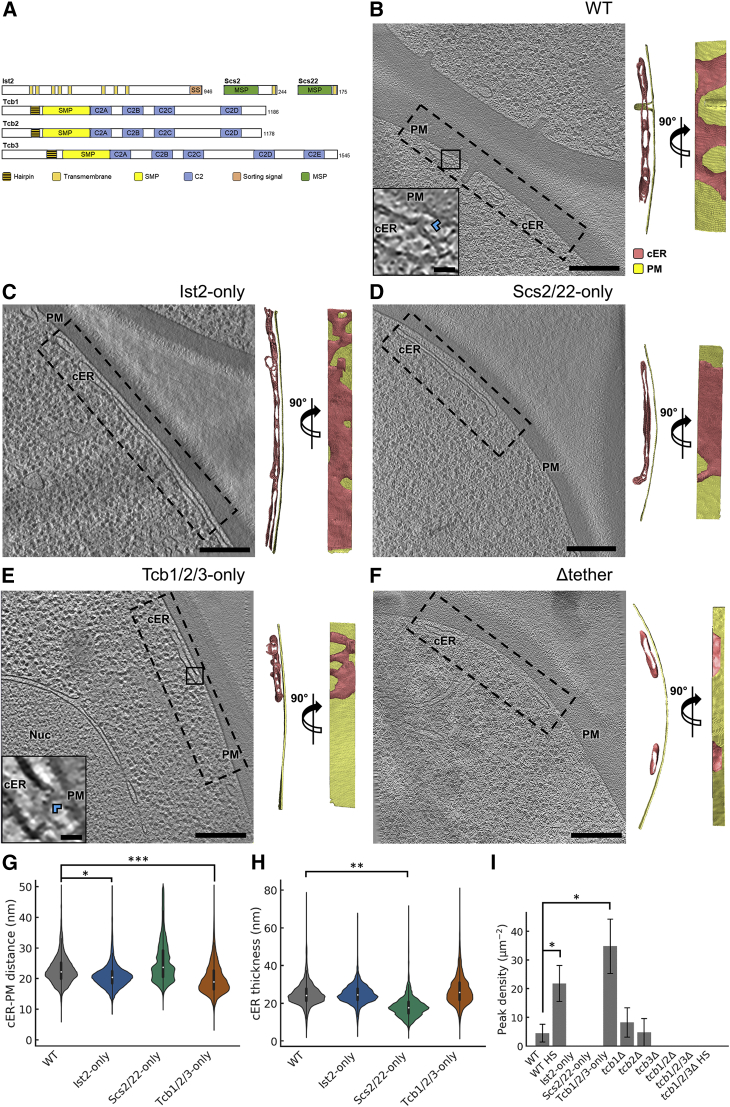
Figure 2.
cER Morphology in ER-PM MCS Tether Mutants
(A) Domain structure of the main ER-PM tethers. Ist2 is an ER multipass transmembrane protein with a long and presumably unstructured cytosolic tail. The C-terminal sorting signal (SS) binds the PM. Scs2 and Scs22 are ER transmembrane proteins containing an N-terminal MSP domain. Tcb proteins are anchored to the ER membrane by a hairpin sequence. In their cytoplasmic C-terminus, Tcbs contain an SMP domain and a variable number of C2 domains. Panels B through F show 1.4-nm-thick tomographic slices of cER in the indicated strains (left) and 3D renderings in two perpendicular orientations upon a 90° rotation along an axis parallel to the PM (right). cER: cortical ER (pink); Nuc: nucleus; PM: plasma membrane (gold). (B) WT cell, (C) Ist2-only cell, (D) Scs2/22-only cell, (E) Tcb1/2/3-only cell, (F) Δtether cell. Insets in (B) and (E) show cER peaks (blue arrowheads). Scale bars: 300 nm (main panels); 25 nm (insets). Panels G, H, and I show quantifications of cER-PM distance (G), cER thickness (H) and cER peak density per μm2 of cER membrane area (I). In G and H the violin plots show the complete distribution of values for all MCS analyzed. A white dot represents the median, a black slab the interquartile range, and a black line 1.5 times the interquartile range. Panel I shows average values (gray bars) and SE (error bars). HS: heat shock (42°C for 10 min). ∗, ∗∗, and ∗∗∗ indicate, respectively, p < 0.05, p < 0.01 and p < 0.01 by unpaired t test (G, H) or Mann-Whitney U test (I). N = 6 (WT), 7 (WT HS), 5 (Ist2-only), 5(Scs2/22-only), 9 (Tcb1/2/3-only), 5 (tcb1Δ), 5 (tcb2Δ), 5 (tcb3Δ), 5 (tcb1/2Δ), 5 (tcb1/2/3Δ), and 5 (tcb1/2/3Δ HS) cER-PM MCS. See also Figures S1 and S2; Table S1.
Scs2/22 are orthologs of the mammalian VAMP-associated proteins (VAPs), a family of ER-resident proteins widely implicated in MCS formation (Murphy and Levine, 2016, Stefan et al., 2011). Both Scs2 and Scs22 are C-terminally anchored to the ER by a transmembrane segment and contain an N-terminal major sperm protein (MSP) domain (Figure 2A). Scs2/22 function as ER-PM tethers thanks to the binding of their MSP domain to PM proteins containing FFAT or FFAT-like motifs (Manford et al., 2012, Murphy and Levine, 2016). A strong reduction in cER levels is observed in Scs2/22 knockout (KO) cells (Loewen et al., 2007, Manford et al., 2012).
The tricalbin proteins (Tcb1/2/3) are orthologs of the mammalian extended-synaptotagmins (E-Syts) and the plant synaptotagmins (SYTs) (Pérez-Sancho et al., 2016, Saheki and De Camilli, 2017b). Tcbs are likely anchored to the ER membrane by a hairpin sequence (Giordano et al., 2013, Saheki and De Camilli, 2017b) (Figure 2A) similar to those found in ER morphogenetic proteins such as reticulons (Hu et al., 2011). Tcbs harbor a synaptotagmin-like, mitochondrial, and lipid-binding protein (SMP) domain that can bind and transport lipids (Lee and Hong, 2006, Saheki et al., 2016, Schauder et al., 2014, Toulmay and Prinz, 2012, Yu et al., 2016). SMP domains have been found in multiple MCS-resident proteins and likely play a key role in the intermembrane exchange of lipids at these sites (Reinisch and De Camilli, 2016). C-terminal to the SMP domain, Tcbs contain a variable number of C2 domains (four in Tcb1/2 and five in Tcb3), some of which can bind membrane phospholipids in a manner either dependent upon or independent of Ca2+ (Creutz et al., 2004, Rizo and Südhof, 1998, Schulz and Creutz, 2004). Both the SMP and the C2 domains are required for Tcb targeting to ER-PM MCS (Manford et al., 2012, Toulmay and Prinz, 2012), and tethering likely takes place via PM binding by C2 domains (Giordano et al., 2013).
Although Ist2, Scs2/22, and Tcb1/2/3 are involved in the appropriate formation of cER, the exact functions of these proteins at ER-PM MCS are poorly understood. First, whereas Ist2 and Scs2/22 are important ER-PM tethers, their relative contributions to cER generation remain unclear. The functions of Tcbs are even more mysterious: Tcbs are bona fide ER-PM tethers, because cER levels are significantly higher in mutants expressing Tcbs but lacking Ist2 and Scs2/22 than in Δtether cells (Manford et al., 2012). However, loss of Tcbs on their own does not result in a substantial reduction in the amount of cER (Manford et al., 2012, Toulmay and Prinz, 2012), suggesting that the main role of Tcbs is not the mechanical anchoring of the ER to the PM. More broadly, the physiological functions of the mammalian E-Syts remain similarly unclear (Sclip et al., 2016, Tremblay and Moss, 2016), although their capacity to shuttle lipids at ER-PM MCS has been demonstrated (Bian et al., 2018, Saheki et al., 2016, Yu et al., 2016).
Here, we aimed to dissect the functional roles of Ist2, Scs2/22, and Tcb1/2/3 at ER-PM MCS. To this end, we used cryo-electron tomography (cryo-ET) to study the fine structure of the cER within mutant cells lacking specific tethers. Thanks to the advent of cryo-focused ion beam (cryo-FIB) technology and direct electron detectors, cryo-ET allows high resolution 3D imaging of a virtually unperturbed cell interior at molecular resolution (Beck and Baumeister, 2016, Rigort et al., 2012, Wagner et al., 2017). Given the narrow intermembrane space of MCS, these structures are particularly sensitive to alterations introduced by classical EM procedures such as chemical fixation, dehydration, and heavy-metal staining, which can alter membrane morphology. Therefore, cryo-ET is especially suited for the high-resolution study of native MCS architecture (Collado and Fernández-Busnadiego, 2017, Fernández-Busnadiego et al., 2015). Our results show that, besides simply anchoring the ER to the PM, each family of tethers uniquely contributes to shaping the cER. In particular, Scs2/22 are associated with cER sheets, whereas Tcbs favor cER tubules. Notably, Tcbs are necessary for the generation of peaks of extreme curvature at the cER membrane that contribute to maintaining PM integrity, possibly by facilitating the transport of cER lipids to the PM.
Results
MCS Architecture in S. cerevisiae
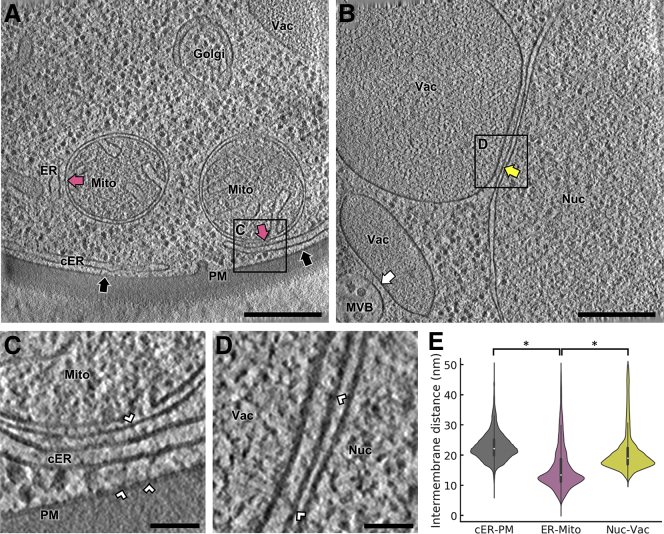
To study MCS architecture in situ by cryo-ET, S. cerevisiae cells were vitrified on EM grids and thinned down to 100–200-nm-thick lamellae using cryo-FIB. Lamellae were loaded into a cryo-TEM (Figure S1), and tomograms were acquired at suitable cellular locations. Cryo-tomograms of various MCS (Figures 1A and 1B) revealed abundant proteinaceous densities of diverse morphologies bridging the gap between the membranes (Figures 1C and 1D; Figure S2A). Interestingly, distance measurements showed a characteristic intermembrane spacing for different MCS. For example, while average nucleus-vacuole distance was 21 ± 7 nm (mean ± STD, N = 5 nucleus-vacuole MCS; Figure 1E), ER-mitochondria junctions were significantly narrower (16 ± 7 nm; mean ± STD, N = 5 ER-mitochondria MCS; p < 0.05 by unpaired t test; Figure 1E).
Figure 1.
Cryo-ET Imaging of MCS in WT S. cerevisiae
(A) 1.4 nm-thick tomographic slice showing cER-PM MCS (black arrows) and ER-mitochondria MCS (purple arrows). The boxed area is magnified in (C). ER: endoplasmic reticulum; cER: cortical ER; Golgi: Golgi apparatus; Mito: mitochondrion; PM: plasma membrane; Vac: vacuole.
(B) 1.4 nm-thick tomographic slice showing a nucleus-vacuole junction (yellow arrow) and a multivesicular body-vacuole MCS (white arrow). The boxed area is magnified in (D). MVB: multivesicular body; Nuc: nucleus.
(C) Magnification of the area boxed in (A). White arrowheads: intermembrane tethers.
(D) Magnification of the area boxed in (B).
(E) Violin plots showing the distribution of intermembrane distances of cER-PM, ER-mitochondrion and nucleus-vacuole MCS. The plots show the complete distribution of values including all MCS analyzed. A white dot represents the median, a black slab the interquartile range, and a black line 1.5x the interquartile range. ∗ indicates p < 0.05 by unpaired t test. N = 6 (cER-PM), 5 (ER-mitochondria) and 5 (nucleus-vacuole) MCS in WT cells. Scale bars: 300 nm (A, B), 50 nm (C, D). See also Figure S2; Table S1.
To gain further insights into the molecular determinants of MCS structure and function, we focused on ER-PM MCS, perhaps the most abundant MCS in S. cerevisiae (Manford et al., 2012, Pichler et al., 2001, Quon et al., 2018, West et al., 2011). Cryo-ET analysis showed that the cER of wild-type (WT) cells consisted of both membrane sheets and tubules (Figure 2B), with an average thickness of 24 ± 6 nm (mean ± STD, N = 6 cER-PM MCS; Figure 2H). For 95% of the MCS area, ER-PM distance ranged from 16 to 34 nm, with an average of 23 ± 5 nm (mean ± STD; Figures 1E and 2G). Thus, in WT cells the cER had a variable morphology and a relatively broad distribution of distances to the PM.
ER-PM Tethers Control cER Morphology
Because the simultaneous deletion of Ist2, Scs2/22, and Tcb1/2/3 largely abolishes ER-PM MCS (Manford et al., 2012), we sought to understand the individual contribution of each of these protein families to ER-PM tethering. To that end, we performed cryo-ET imaging of ER-PM MCS in mutant cells expressing only one family of tethers. These data confirmed previous observations (Loewen et al., 2007, Manford et al., 2012, Toulmay and Prinz, 2012, Wolf et al., 2012) that total levels of cER were still substantial in cells expressing only Ist2 (scs2/22Δ tcb1/2/3Δ; “Ist2-only” cells; Figure S1B) or the VAP orthologues Scs2 and Scs22 (ist2Δ tcb1/2/3Δ; “Scs2/22-only” cells; Figure S1C). However, cER levels in cells expressing only Tcb1/2/3 (ist2Δ scs2/22Δ; “Tcb1/2/3-only” cells; Figure S1D) were markedly lower than in WT (Figure S1A), although higher than in Δtether cells (Figure S1E), in agreement with previous results (Manford et al., 2012).
Next, we investigated the fine morphology of the cER in these mutants. In Ist2-only cells, the cER was a mixture of membrane sheets and tubules similar to WT cells (Figures 2B and 2C). Although average ER-PM distance was slightly shorter than WT (21 ± 4 nm, mean ± STD, N = 5 cER-PM MCS; p < 0.05 by unpaired t test; Figure 2G), cER thickness (25 ± 6 nm, mean ± STD; Figure 2H) was comparable to WT, suggesting that Ist2 is an important contributor to the morphology of the cER in WT cells.
Interestingly, cER morphology in Scs2/22-only cells was dramatically different from WT. In these mutants, cER tubules were rarely observed, as the cER consisted almost exclusively of extended sheets (Figure 2D). Average ER-PM distance in these cells spread across a wider range of values (26 ± 7 nm, mean ± STD, N = 5 cER-PM MCS; Figure 2G). On the other hand, the ER sheets observed in Scs2/22-only cells were significantly narrower than WT (18 ± 6 nm, mean ± STD; p < 0.01 by unpaired t test; Figure 2H). These data show that while Scs2/22 are not very effective in controlling ER-PM distance, they are important determinants of cER width.
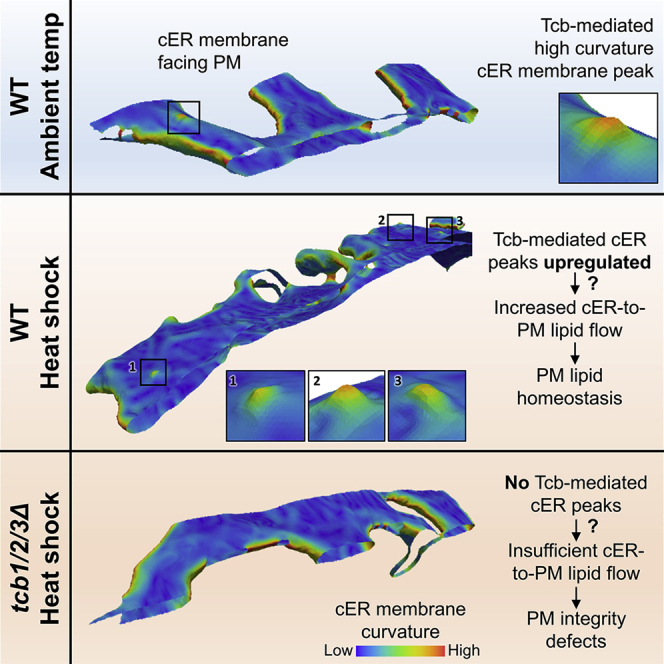
In contrast to Scs2/22-only cells, the cER was formed mainly by membrane tubules in Tcb1/2/3-only cells (Figure 2E). Interestingly, in these cells we also observed abundant peaks of very high curvature on the membrane of the cER facing the PM (Figure 2E, inset; Figure S2B). These peaks had a radius of ∼10 nm at their base and protruded ∼7 nm from the cER membrane (Figure S2C). The cER came into very close proximity of the PM at cER peaks, narrowing to distances of 7–8 nm (Figure S2C). Overall, the average cER-PM distance was significantly shorter in Tcb1/2/3-only (20 ± 5 nm, mean ± STD; N = 9 cER-PM MCS; p < 0.001 by unpaired t test; Figure 2G) than in WT cells (23 ± 5 nm, mean ± STD). Whereas cER peaks were not found in Ist2-only (Figures 2C and 2I), Scs2/22-only (Figures 2D and 2I), or ∆tether (Figure 2F) cells, they were also present in WT cells, albeit at a lower frequency (Figure 2B, inset; Figure 2I; p < 0.05 by Mann-Whitney U test; Figure S2B). The cER peaks in WT cells were morphologically indistinguishable from those found in Tcb1/2/3-only cells (Figure S2C). Therefore, the formation of high curvature peaks at the cER is likely controlled by the Tcb proteins. Because the peaks were only found on the part of the cER membrane opposed to the PM, these structures may be involved in intermembrane exchange.
Altogether, these data show that each of the ER-PM tethers play key and yet distinct roles in controlling cER morphology, especially in terms of membrane curvature.
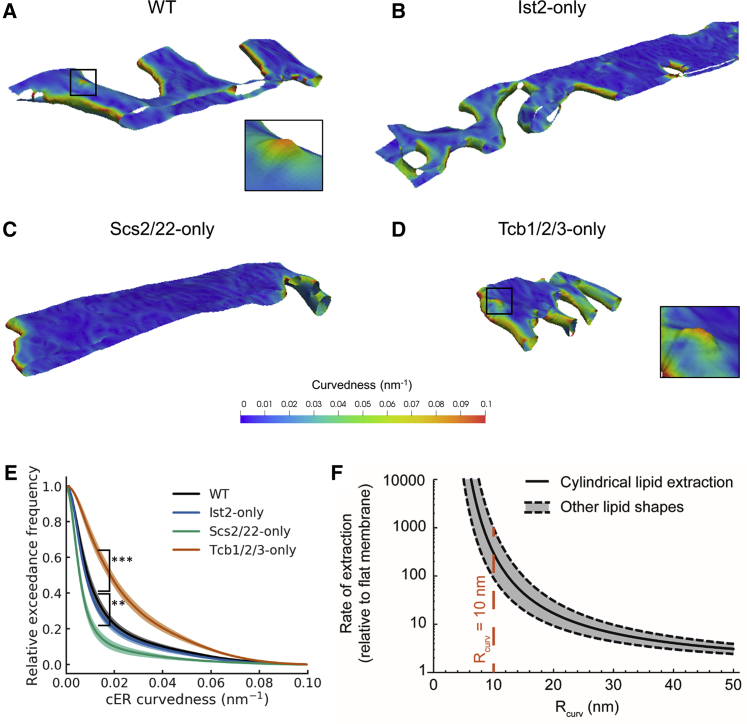
Quantitative Analysis of cER Membrane Curvature
Membrane curvature plays a major role in a wide variety of cellular processes (Kozlov et al., 2014) and is a fundamental determinant of ER morphology (Hu et al., 2011). Therefore, we further analyzed the cER membrane curvature alterations observed in the different ER-PM tether mutants. To that end, we implemented an algorithm allowing a quantitative determination of membrane curvature in cryo-ET data (Kalemanov et al., 2019). A global analysis was consistent with the visual impression that the average cER curvature observed in Scs2/22-only cells was lower than in WT (p < 0.01 by unpaired t test; Figures 3A, 3C, and 3E), indicating the higher prevalence of cER sheets. Conversely, the curvature of the cER membrane in Tcb1/2/3-only cells was significantly higher than WT (p < 0.001 by unpaired t test; Figures 3A, 3D, and 3E), reflecting the more tubular cER morphology in these cells. Local mapping of the curvature in cER membrane renderings highlighted the presence of peaks of extreme curvature (curvature radius ≤ 10 nm; Figure 3A, inset) in WT cells. These structures were enriched in Tcb1/2/3-only cells compared to WT (Figure 2I; Figure 3D, inset), and absent in tcb1/2/3Δ mutants (N = 5 cER-PM MCS; Figures 2I and 4E) and cells expressing only Scs2/22 or Ist2 (Figures 2I, 3B, and 3C). Therefore, this analysis reinforced the notion that Tcb1/2/3 are necessary for the generation of high curvature peaks on the side of the cER membrane directly facing the PM.
Figure 3.
Quantification of cER Curvature
(A–D) 3D visualizations of cER curvedness in the indicated strains. Insets in (A) and (D) show cER peaks. (A) WT cell, (B) Ist2-only cell, (C) Scs2/22-only cell, (D) Tcb1/2/3-only cell. (E) Quantification of cER curvedness, shown as an exceedance plot. The shaded lines represent the average across all MCS ± SE for each bin (1 nm-1.). ∗∗ and ∗∗∗ indicate, respectively, p < 0.01 and p < 0.001 by unpaired t test. N = 6 (WT), 7 (WT HS), 5 (Ist2-only), 5 (Scs2/22-only), and 9 (Tcb1/2/3-only) cER-PM MCS. (F) Enhancement of the rate of lipid extraction by membrane curvature according to a theoretical model. The plot shows the rate of extraction computed for a standard cylindrical lipid (black curve) as well as for lipids of other shapes, such as conical or inverted conical lipids (gray-shaded area between the dashed, black curves). The value of the radius of curvature of the experimentally observed cER peaks is denoted by the dashed red line. is equivalent to the curvedness for κ1=κ2. See also Figures S3 and S4; Table S1.
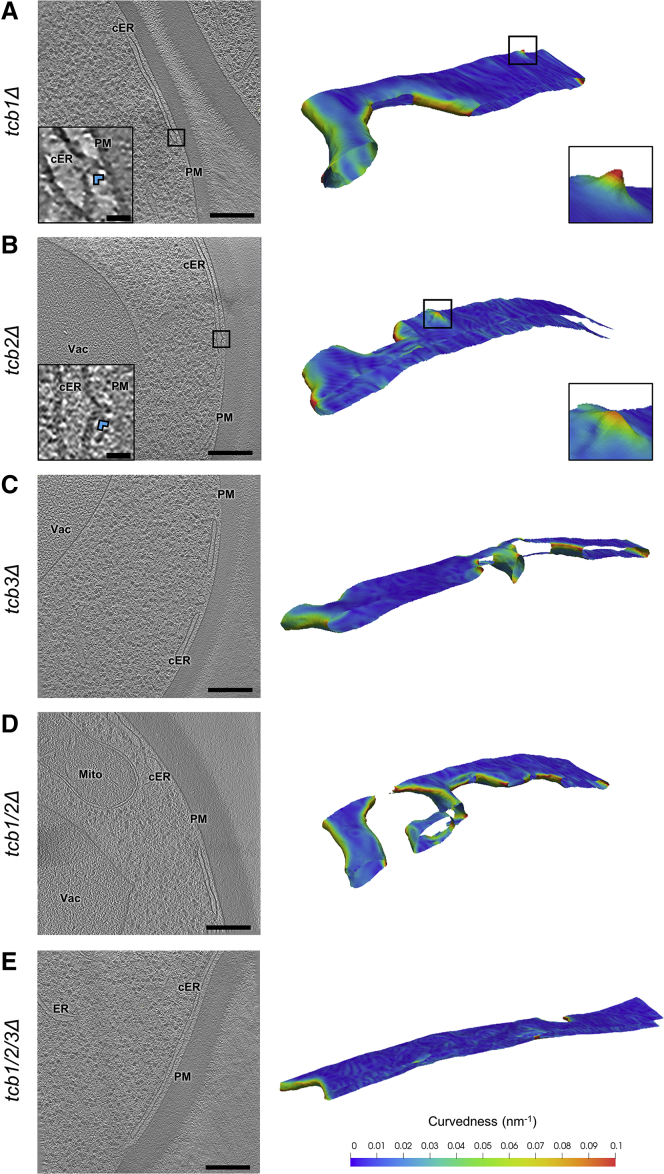
Figure 4.
cER Peaks in Tcb Mutants
(A–E) 1.4-nm-thick tomographic slices of cER in the indicated strains (left) and 3D renderings of cER curvature (right). (A) tcb1Δ, (B) tcb2Δ, (C) tcb3Δ, (D) tcb1/2Δ, (E) tcb1/2/3Δ cell. cER: cortical ER; Mito: mitochondrion; PM: plasma membrane; Vac: vacuole. Insets in (A) and (B) show cER peaks (blue arrowheads). Scale bars for tomographic slices: 300 nm (main panels), 25 nm (insets). See also Figure S2; Table S1.
To address the molecular basis of this phenomenon, we next investigated which Tcb proteins were required for cER peak formation by conducting cryo-ET of cells lacking specific Tcbs in the presence of all other ER-PM tethers. In tcb1Δ (N = 5 cER-PM MCS; Figures 4A and S2B) and tcb2Δ cells (N = 5 cER-PM MCS; Figures 4B and S2B), cER peaks were similar in frequency to WT cells (Figure 2I). cER peaks in these strains were also morphologically comparable to WT (Figure S2C). In contrast, no cER peaks were observed in tcb3Δ (N = 5 cER-PM MCS; Figures 2I and 4C) and tcb1/2Δ cells (N = 5 cER-PM MCS; Figures 2I and 4D). Therefore, the expression of Tcb3 and either Tcb1 or Tcb2 seems necessary for the efficient formation of cER peaks, consistent with reports that Tcbs (and E-Syts) can form heterodimers (Creutz et al., 2004, Giordano et al., 2013, Idevall-Hagren et al., 2015, Schulz and Creutz, 2004).
cER Peaks May Facilitate Lipid Transfer
Next, we investigated the biological function of Tcb-mediated cER peaks. Tcbs contain modules that can sense or induce membrane curvature (hairpin anchor, multiple C2 domains) and transport lipids (SMP domain) (Creutz et al., 2004, Lee and Hong, 2006, Manford et al., 2012, Martens et al., 2007, Schauder et al., 2014, Toulmay and Prinz, 2012). Tcbs may combine both their curvature-generation and lipid-transport properties by controlling the formation of cER peaks, which could facilitate cER-to-PM lipid transport by (1) reducing the physical distance between cER and PM, and/or (2) disturbing the cER lipid bilayer to facilitate lipid extraction and at the same time imposing cER-to-PM directionality on the transfer process.
To address this possibility, we used a semi-quantitative model (Campelo and Kozlov, 2014) to calculate how the induction of cER membrane curvature may facilitate the lipid-transfer process. We assume that this task is performed by a lipid-transport module such as the SMP domain of Tcbs. The total free energy required for lipid extraction by a lipid-transport protein (LTP) can be expressed as the sum of two components. The first one is independent from membrane geometry, incorporating electrostatic interactions and membrane-independent interactions between the lipid and the LTP. The second component is determined by the elastic stresses imposed on the membrane by its geometry prior to LTP binding, and by how these stresses change as a result of the lipid rearrangements caused by a partial insertion of the LTP into the membrane. In turn, membrane geometry can be determined by its lipid composition and/or by external factors such as curvature-generating proteins (Campelo et al., 2008). We focused on this last scenario, as it is unlikely that physiological lipid compositions result in the extreme membrane curvatures of cER peaks (Campelo et al., 2008, Sorre et al., 2009). With these premises, our calculations showed that the energy barrier for lipid extraction is reduced by ∼6 kBT when the radius of curvature of the membrane is 10 nm (Figure S3), as observed in Tcb-induced cER peaks. This is of similar magnitude to the facilitation of sterol extraction from a flat membrane by an LTP in comparison to its spontaneous desorption, estimated to be ∼2–3 kBT (Dittman and Menon, 2017), and would result in a ∼500-fold acceleration of the transfer reaction (Figure 3F). Therefore, our model predicts that cER peaks greatly facilitate lipid extraction by lipid transport modules.
cER Peaks Maintain PM Integrity
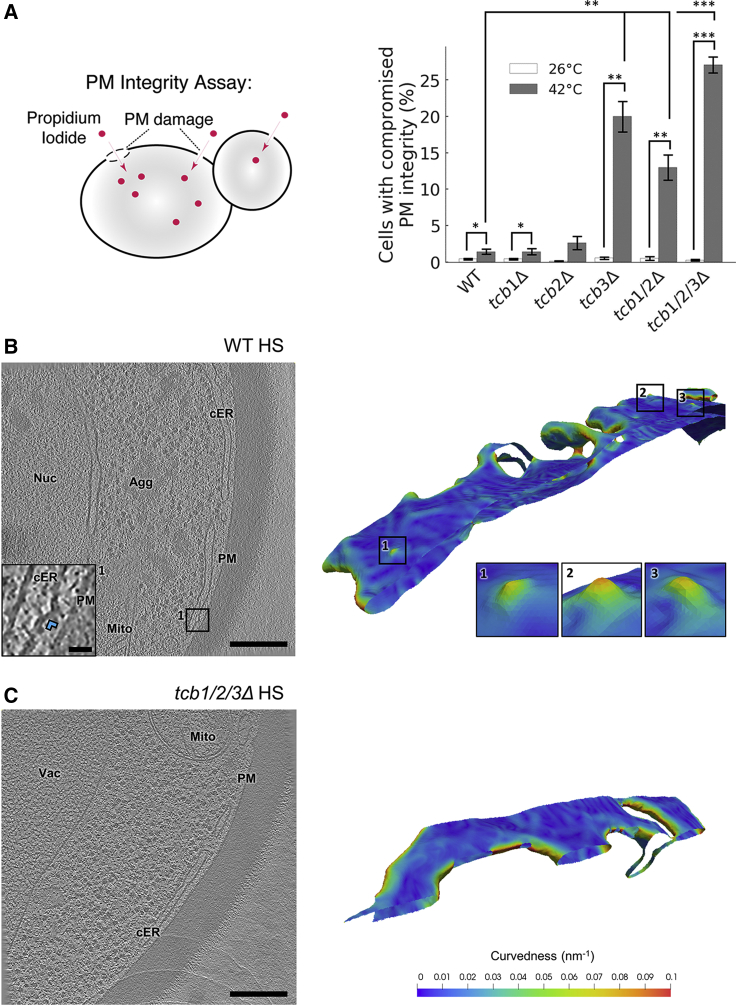
The synthesis of certain PM lipids—including phosphatidylinositol, phosphatidylserine, and sterols—is enhanced at the cER (Pichler et al., 2001). Interestingly, tcb1/2/3Δ cells show PM integrity defects upon heat stress (Omnus et al., 2016), a situation in which substantial traffic of lipids between the ER and the PM may be necessary to repair heat-induced alterations. Because Tcbs are required for the formation of cER peaks that may facilitate ER-PM lipid transfer, it is possible that cER peaks are important to maintain PM integrity under heat-stress conditions.
To test this hypothesis, we performed PM integrity assays in the different Tcb mutants. Cells were subjected to 42°C for 10 min, and PM integrity was monitored by measuring the entry of extracellular propidium iodide into cells using flow cytometry (Figure 5A). Because this dye is membrane impermeable, it only enters cells with compromised PM integrity (Zhao et al., 2013). Remarkably, these experiments revealed PM integrity defects for all conditions in which cER peaks were not observed (tcb3Δ, tcb1/2Δ cells, p < 0.01 by unpaired t test; tcb1/2/3Δ cells, p < 0.001 by unpaired t test; Figure 5A), whereas PM integrity in Tcb mutants showing cER peaks (tcb1Δ, tcb2Δ) was comparable to WT. Thus, there was a strong correlation between the absence of Tcb-induced cER peaks and PM integrity defects.
Figure 5.
PM Integrity and cER Curvature under Heat Stress
(A) Schematic of the propidium iodide assay to assess PM integrity (left) and PM integrity measurements of Tcb deletion mutants upon 10-min incubation at 42°C (right). The entry of propidium iodide in cells with compromised PM integrity was measured by flow cytometry. The plot shows average values (white/gray bars) for each condition ± SE (error bars). ∗, ∗∗, and ∗∗∗, respectively, indicate p < 0.05, p < 0.01, and p < 0.001 by Mann-Whitney U test (for tcb1Δ 26°C data, which was not normally distributed) or unpaired t test (for all other conditions). Four independent biological repeats were performed for all conditions. (B and C) 1.4-nm-thick tomographic slices of cER in the indicated strains (left) and 3D renderings of cER curvature (right). Agg: aggregate; cER: cortical ER; Mito: mitochondrion; Nuc: nucleus; PM: plasma membrane; Vac: vacuole. (B) WT cell under heat stress (HS). Insets show cER peaks (blue arrowhead in the tomographic slice inset). (C) tcb1/2/3Δ cell under heat stress. Scale bars: 300 nm (main panels), 25 nm (inset). See also Figures S2 and S4; Table S1.
However, the density of cER peaks was relatively low in WT cells under nonstress conditions (Figures 2B, 2I, and 3A); cER peak area was equivalent to 0.15% of the total cER area facing the PM in WT cells. Could these rare structures play an important role in maintaining PM integrity under heat-shock conditions? To address this question, we performed cryo-ET on heat-shocked cells. As expected, cells showed abundant amorphous aggregates in various cellular locations (Miller et al., 2015, Wagner et al., 2017) (Figure 5B). Also, heat-shocked WT cells showed a strong increase in the number of cER peaks (N = 7 cER-PM MCS; p < 0.05 by Mann-Whitney U test; Figures 2I and 5B), whereas these structures were absent in heat-shocked tcb1/2/3Δ cells (N = 5 cER-PM MCS; Figures 2I and 5C). Therefore, the Tcb-dependent formation of cER peaks is induced by conditions that challenge PM integrity, such as heat shock.
Of note, deletion of all Tcbs (tcb1/2/3Δ) did not result in integrity defects as pronounced as Δtether (Figure S4A), and even in the presence of Tcbs, tether mutants with severely impaired cER formation (scs2/22Δ, Tcb1/2/3-only; Manford et al., 2012) also suffered from PM integrity defects (Figure S4A). Therefore, non-Tcb enriched areas of the cER may also be important for PM integrity.
We investigated this issue combining theoretical modeling with our experimental measures of peak size and abundance. We assume that the cER membrane is completely flat outside of cER peaks, and that LTPs are homogenously distributed at the cER-PM interface. Taking into account the much higher rate of lipid transfer predicted at cER peaks, our model indicates that the total number of lipids extracted from cER peaks in WT cells (0.15% of total cER membrane area) is roughly equivalent to that of the flat parts (99.85% of total cER membrane area) (Figure S4B). Thus, lipid transfer at flat cER membranes may not be negligible. Nevertheless, in heat-shocked WT cells the observed ∼6-fold increase in cER peak density would translate into a ∼5-fold more lipids transferred from cER peaks than from the flat portions of the cER (Figure S4B). This analysis also shows that increasing cER peak density can only substantially increase lipid flows when the total levels of cER are high, possibly explaining the PM integrity defects observed in mutants with less cER such as scs2/22Δ or Tcb1/2/3-only (Figure S4A).
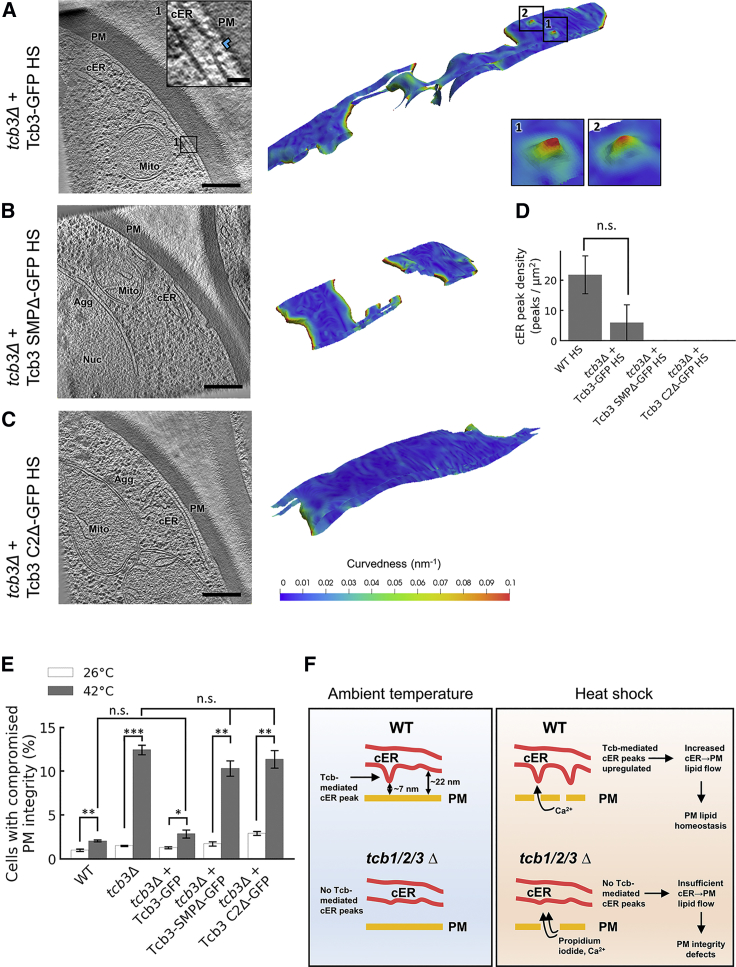
Tcb3 SMP and C2 Domains Are Necessary for cER Peak Formation
The higher density of cER peaks observed in heat-shocked WT cells provided a higher dynamic range to investigate by cryo-ET the specific roles of Tcb3 domains in cER peak formation at 42°C. We investigated GFP-tagged Tcb3 constructs, either full length (Tcb3-GFP), lacking the SMP domain (Tcb3 SMPΔ-GFP), or all C2 domains (Tcb3 C2Δ-GFP). In agreement with previous studies (Manford et al., 2012, Toulmay and Prinz, 2012), both truncations localized to the cER, although substantially higher fluorescence was detected in the nuclear ER compared to full-length Tcb3-GFP (Figure S4C).
We then inquired to what extent these Tcb3 constructs could rescue the defects observed in tcb3Δ cells in terms of cER peak formation (Figures 2I and 4C) and PM integrity (Figure 5A). Interestingly, cryo-ET imaging of heat-shocked cells and PM integrity assays revealed that neither truncation was able to complement tcb3Δ mutants (Figures 6B–6E), while substantial recovery was observed by expression of full-length Tcb3-GFP (Figures 6A, 6D, and 6E). These experiments indicate that the Tcb3 SMP and C2 domains are involved in cER peak formation and reinforce the correlation between PM integrity at elevated temperatures and the presence of Tcb-mediated cER peaks.
Figure 6.
cER Peaks and PM Integrity in Heat-Shocked Tcb3 Truncation Mutants
(A–C) 1.4-nm-thick tomographic slices of cER in the indicated strains (left) and 3D renderings of cER curvature (right). Agg: aggregate; cER: cortical ER; Mito: mitochondrion; Nuc: nucleus; PM: plasma membrane. (A) tcb3Δ + Tcb3-GFP HS, (B) tcb3Δ + Tcb3 SMPΔ- GFP HS, (C) tcb3Δ + Tcb3 C2Δ-GFP HS. Insets show cER peaks (blue arrowhead in the tomographic slice inset). Scale bars for tomographic slices: 300 nm (main panels); 25 nm (insets). The contrast of the tomographic slices in (A), (B), and (C) was enhanced using a deconvolution filter. (D) cER peak density per μm2 of cER membrane area showing average values (gray bars) ± SE (error bars). N = 7 (WT HS), 3 (tcb3Δ + Tcb3-GFP HS), 3 (tcb3Δ + Tcb3 SMPΔ-GFP HS), and 3 (tcb3Δ + Tcb3 C2Δ-GFP HS) ER-PM MCS. n.s. indicates p > 0.05 by Mann-Whitney U test. (E) PM integrity assay of tcb3Δ cells complemented with Tcb3 truncation mutants upon 10-min incubation at 42°C. The plot shows average values (white/gray bars) for each condition ± SE (error bars). n.s., ∗, ∗∗, and ∗∗∗, respectively, indicate p > 0.05, p < 0.05, p < 0.01, and p < 0.001 by unpaired t test. Four independent biological repeats were performed for all conditions. (F) Model for the function of cER peaks in maintaining PM integrity. In WT cells, Tcbs generate membrane peaks of extreme curvature on the cER membrane. This may facilitate the extraction of cER lipids and their delivery to the PM (top left). The generation of cER peaks is the main structural role of Tcbs at ER-PM MCS, as overall ER-PM tethering is not substantially affected by Tcb1/2/3 deletion. However, tcb1/2/3Δ cells lack cER peaks (bottom left). Under heat stress, influx of extracellular Ca2+ through a damaged PM drives the localized formation of additional Tcb-mediated cER peaks, which in turn facilitate sufficient delivery of cER lipids to the PM to maintain PM integrity (top right). Absence of cER peaks in heat stressed tcb1/2/3Δ cells leads to PM integrity defects allowing influx of propidium iodide (bottom right). See also Figure S4; Table S1.
Altogether, these data indicate that (1) Tcbs are necessary for the formation of cER peaks; (2) cER peaks may facilitate ER-to-PM lipid transfer; and (3) cER peaks are important to maintain PM integrity under heat stress, a condition in which the PM may require substantial lipid influx to restore its lipid homeostasis. Interestingly, similar high curvature peaks were observed at other ER-mediated MCS such as ER-mitochondria MCS (Figure S2D), suggesting that the induction of membrane curvature may be a general mechanism to facilitate intermembrane lipid exchange at various MCS.
Discussion
ER-PM Tethers Shape the cER
MCS exist between essentially all cellular membranes, and a great number of MCS-resident proteins and tethers have been identified. However, the functions of many of these molecules remain poorly understood (Bohnert and Schuldiner, 2018, Shai et al., 2018, Valm et al., 2017, Wu et al., 2018). For example, it is unclear why so many ER-PM tethers exist in S. cerevisiae (Gatta et al., 2015, Manford et al., 2012, Quon et al., 2018), as well as the possible functions of these proteins beyond the mechanical anchoring of the membranes. Here, we employed state-of-the-art in situ imaging by cryo-ET to reveal that ER-PM tethers are critical determinants of cER morphology and MCS function.
Our data confirm that Ist2 is an important ER-PM tether (Lavieu et al., 2010, Manford et al., 2012, Wolf et al., 2012). The distribution of ER-PM distances was particularly narrow in Ist2-only cells, indicating that Ist2 is very effective in maintaining an ER-PM separation of about 21 nm. This is surprising, because Ist2 bridges the ER and the PM by a 340-amino-acid-long linker that is predicted to be unstructured. This linker is probably not fully extended, as this would allow it to span up to 120 nm, but how it can precisely regulate intermembrane distance requires further investigation.
ER-PM distances were more broadly distributed in Scs2/22-only cells, possibly due to the promiscuous interactions of Scs2/22 with different FFAT/FFAT-like motif proteins at the PM (Murphy and Levine, 2016). Strikingly, the cER in these cells consisted almost exclusively of extended, narrow sheets. How such sheets are formed remains to be established, as direct interactions of Scs2/22 in trans across the ER lumen appear unlikely, given their short luminal sequences. In contrast to Scs2/22-only cells, the cER was mainly formed by tubules in Tcb1/2/3-only cells. This phenomenon may rely on the hairpin sequence that anchors Tcbs to the ER membrane. Tcb hairpin sequences could sense and/or generate membrane curvature as in reticulons and other ER morphogenetic proteins (Hu et al., 2011).
Alternatively, it is also possible that the cER morphologies observed in the different ER-PM tether mutants arise from the dysregulation of additional ER-PM MCS factors (Quon et al., 2018), other ER morphogenetic proteins, or lipids. However, WT cER also consists of a mixture of sheets and tubules, which could plausibly arise as a combination of the morphologies of the individual ER-PM tether mutants. Therefore, it is possible that the different families of tethers are enriched in partially segregated cER subdomains, consistent with their punctate localization observed by light microscopy (Creutz et al., 2004, Manford et al., 2012, Toulmay and Prinz, 2012, Wolf et al., 2012). Similarly, in plant and mammalian cells, different ER-PM tethers are known to co-exist at the same MCS but form separate subdomains (Giordano et al., 2013, Siao et al., 2016). Therefore, native ER-PM MCS may be established as a juxtaposition of molecular territories enriched in different tethers.
Tcbs Form Highly Curved Peaks at the cER Membrane
Besides being generally tubular, the cER in Tcb1/2/3-only cells was enriched in membrane peaks of extreme (< 10-nm radius) membrane curvature, one of the highest observed in a cell (Antonny, 2011). cER peaks were also present in WT cells at a lower frequency, suggesting that Tcbs generate these structures also in the WT case. In fact, our study of Tcb-deletion mutants indicated that the efficient formation of cER peaks requires expression of Tcb3 and Tcb1 or Tcb2, consistent with biochemical evidence that Tcbs/E-Syts form heterodimers (Creutz et al., 2004, Giordano et al., 2013, Idevall-Hagren et al., 2015, Schulz and Creutz, 2004). Given that the SMP domain is important for dimerization (Schauder et al., 2014), disruption of Tcb3-Tcb1/2 heterodimers may explain the lack of cER peaks in tcb3Δ cells expressing Tcb3 SMPΔ-GFP.
A Tcb3 truncation lacking all C2 domains also failed to rescue cER peak formation in tcb3Δ cells. On one hand, it is possible that C2 domains are also involved in Tcb dimerization or oligomerization (Zanetti et al., 2016). Additionally, binding of at least some C2 domains to the cER membrane may play an active role in curvature generation, by analogy with other multi-C2 domain proteins. For example, the C2 domains of mammalian SYT1 bind the PM in a Ca2+-dependent manner, generating curvature in the PM (Martens et al., 2007). Although C2 domains generally prefer negatively charged membranes such as the PM, binding of E-Syt C2 domains to the ER membrane has been speculated (Min et al., 2007, Schauder et al., 2014). The exact mechanisms by which Tcb SMP and C2 domains could generate cER peaks, perhaps synergistically with the hairpin sequence, require further investigation.
Bona fide membrane curvature generators such as reticulons have also been implicated in ER-PM MCS formation (Caldieri et al., 2017), and there is increasing evidence for important roles of curvature-sensing and/or generating proteins at other MCS (Ackema et al., 2016, de Saint-Jean et al., 2011, Ho and Stroupe, 2016, Moser von Filseck et al., 2015, Voss et al., 2012), consistent with our observations of high curvature peaks at, e.g., ER-mitochondria MCS. Thus, membrane curvature may be an important regulator of MCS function (Henne et al., 2015).
cER Peaks Are Important for PM Integrity under Stress
Most MCS harbor an intense nonvesicular exchange of lipids (Cockcroft and Raghu, 2018, Lees et al., 2017, Saheki et al., 2016), which is especially important at ER-PM MCS because most PM lipids are synthesized in the ER. Moreover, the E-Syts, mammalian orthologs of Tcbs, are directly implicated in ER-PM lipid transfer, as their SMP domain mediates lipid binding and transport (Bian et al., 2018, Saheki et al., 2016, Schauder et al., 2014, Yu et al., 2016). Because Tcb-induced cER peaks always faced the PM, we hypothesized that these peaks could play a role in an ER-PM lipid transfer.
Consistent with this idea, our semi-quantitative modeling indicated that cER peaks can dramatically enhance the rate of lipid extraction from the cER by facilitating the shallow insertion of lipid-transport modules into the lipid bilayer, in agreement with experimental studies (Machida and Ohnishi, 1980, Moser von Filseck et al., 2015). cER peaks also shorten cER-PM distance and can impose cER-to-PM directionality on the lipid transfer process. Thus, cER-to-PM lipid transfer may be greatly enhanced by Tcb-mediated cER peaks. It is also attractive to speculate that Tcb SMP domains may play an active role in such lipid transfer process.
The physiological roles of the Tcb protein family remain enigmatic. On one hand, these molecules are highly conserved and therefore likely to play important functions. However, no major alterations were discovered in yeast cells lacking all Tcbs (Manford et al., 2012, Toulmay and Prinz, 2012), nor in mammalian cells or mice lacking all three E-Syts (Saheki et al., 2016, Sclip et al., 2016, Tremblay and Moss, 2016). E-Syt triple knockout cells did display an accumulation of diacylglycerol at the PM upon phospholipase C activation (Saheki et al., 2016), suggesting that the main function of E-Syts/Tcbs is to respond to stimuli that perturb lipid homeostasis (Stefan, 2018).
Our data suggest that one of such stimulus is heat stress. Although Tcb1/2/3 deletion does not substantially reduce the levels of cER, heat-shocked tcb1/2/3Δ cells suffer from PM integrity defects (Omnus et al., 2016). Furthermore, our functional assays of Tcb mutants showed a strong correlation between the absence of cER peaks and PM integrity defects upon heat stress. Although the exact mechanisms by which heat stress compromises PM integrity remain to be established, heat alters PM protein and lipid homeostasis, as well as the physico-chemical properties of the bilayer (Fan and Evans, 2015, Verghese et al., 2012, Zhao et al., 2013). PM repair likely involves the addition of new lipids (Vaughan et al., 2014), and ER-PM MCS regulate phospholipid biogenesis (Pichler et al., 2001, Tavassoli et al., 2013). Thus, under conditions of PM damage, Tcb-mediated cER peaks could maintain PM integrity by ensuring sufficient flow of lipids synthesized in the cER toward the PM.
Consistently, we observed a substantial increase in the number of cER peaks in WT cells under heat stress. Since membrane binding of some Tcb C2 domains is regulated by Ca2+ (Schulz and Creutz, 2004), the formation of new cER peaks upon heat stress could be driven by the influx of extracellular Ca2+ through a damaged PM (Andrews and Corrotte, 2018, Jimenez and Perez, 2017). Ca2+ influx may trigger the binding of Tcb C2 domains to the cER membrane, inducing cER membrane curvature in a way similar to mammalian SYTs (Martens et al., 2007). At the same time, the cER is highly dynamic and explores most of the cellular PM over a few minutes, possibly monitoring PM status (Omnus et al., 2016). Thus, Ca2+ signals at sites of PM damage may trigger formation of Tcb-mediated cER peaks exactly where they are needed to locally enhance PM repair (Figure 6F). This mechanism may act in parallel or synergistically with other pathways to maintain PM homeostasis (Andrews and Corrotte, 2018, Jimenez and Perez, 2017, Omnus et al., 2016, Zhao et al., 2013).
In fact, our calculations show that besides cER peaks, high levels of cER may also be important for PM integrity, suggesting that optimal cER-PM lipid flow requires joint efforts by Tcbs and other tethers. Although our model indicates that the capacity to extract lipids from cER peaks is much higher than from flat membranes, lipid currents from cER peaks and flat areas appear comparable in WT cells, given the minute fraction of cER area occupied by peaks. However, lipid currents from cER peaks may dominate upon heat shock due to the increase in cER peak density, possibly explaining why the phenotypes of Tcb mutants only become apparent under stress conditions.
The important role of Tcbs in maintaining PM integrity upon stress agrees with findings on the plant SYTs, which also act as ER-PM tethers and are important factors protecting PM integrity from different stresses (Kawamura and Uemura, 2003, Lee et al., 2019, Pérez-Sancho et al., 2015, Schapire et al., 2008, Yamazaki et al., 2008). Other mammalian multi-C2 domain proteins like SYT7 and dysferlin are directly implicated in PM repair (Andrews and Corrotte, 2018, Jimenez and Perez, 2017). As with yeast Tcbs, membrane binding by some C2 domains of plant SYTs and mammalian E-Syts is regulated by Ca2+ (Giordano et al., 2013, Idevall-Hagren et al., 2015, Pérez-Sancho et al., 2015). Therefore, we propose that a crucial function of yeast Tcbs, plant SYTs, and possibly mammalian E-Syts is to respond to the influx of extracellular Ca2+ through a damaged PM by forming cER peaks, which may enhance the cER-to-PM lipid transfer necessary for PM repair. Further work should experimentally determine the extent and composition of these lipid flows.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Max Efficiency® DH5alpha | Invitrogen | Cat# 18258-012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phusion® HF DNA Polymerase | NEB | Cat# M0530 |
| DpnI | NEB | Cat# R0176 |
| Propidium Iodide | Invitrogen | Cat# P3566 |
| Critical Commercial Assays | ||
| QIAprep® Spin MiniPrep Kit | Qiagen | Cat# 27106 |
| Deposited Data | ||
| Cryo-Electron Tomogram of cER-PM MCS in a Heat-Shocked WT S. cerevisiae Cell | This study | EMDB ID: EMD-10378 |
| Cryo-Electron Tomogram of cER-PM MCS in a Heat-Shocked WT S. cerevisiae Cell (NAD Filtered) | This study | EMDB ID: EMD-10379 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: Strain Background SEY6210: MATαleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | (Robinson et al., 1988) | ATCC: 96099 |
| S. cerevisiae: Strain Background SEY6210.1: MATaleu2-3,112 ura3-52 his3-Δ200 trp1 Δ901 lys2 801 suc2-Δ9 | (Robinson et al., 1988) | SEY6210.1 |
| S. cerevisiae: Strain ANDY117: SEY6210 tcb1Δ::KANMX6 | (Manford et al., 2012) | ANDY117 |
| S. cerevisiae: Strain ANDY118: SEY6210.1 tcb1Δ::KANMX6 | (Manford et al., 2012) | ANDY118 |
| S. cerevisiae: Strain ANDY120: SEY6210.1 tcb2Δ::KANMX6 | (Manford et al., 2012) | ANDY120 |
| S. cerevisiae: Strain YCS2359: SEY6210.1 tcb3Δ::HISMX6 | This study | YCS2359 |
| S. cerevisiae: Strain YCS2430: SEY6210.1 tcb1Δ::KANMX6 tcb2Δ::KANMX6 | This study | YCS2429 |
| S. cerevisiae: Strain ANDY214: SEY6210.1 tcb1Δ::KANMX6 tcb2Δ::KANMX6 tcb3Δ::HISMX6 | (Manford et al., 2012) | ANDY214 |
| S. cerevisiae: Strain DBY356: SEY6210.1 scs2Δ::TRP1 scs22Δ:: HISMX6 | (Stefan et al., 2011) | DBY356 |
|
S. cerevisiae: Strain ANDY113 SEY6210.1 ist2Δ::HISMX6 |
(Manford et al., 2012) | ANDY113 |
| S. cerevisiae: Strain ANDY129: SEY6210.1 ist2Δ::HISMX6 scs2Δ::TRP1 scs22Δ::HISMX6 | (Manford et al., 2012) | ANDY129 |
| S. cerevisiae: Strain ANDY176: SEY6210.1 ist2Δ::HISMX6 tcb1Δ::KANMX6 tcb2Δ::KANMX6 tcb3Δ::HISMX6 | (Manford et al., 2012) | ANDY176 |
| S. cerevisiae: Strain ANDY196: SEY6210.1 scs2Δ::TRP1 scs22Δ::HISMX6 tcb1Δ::KANMX6 tcb2Δ::KANMX6 tcb3Δ::HISMX6 | (Manford et al., 2012) | ANDY196 |
| S. cerevisiae: Strain ANDY198: SEY6210.1 ist2Δ::HISMX6 scs2Δ::TRP1 scs22Δ::HISMX6 tcb1Δ::KANMX6 tcb2Δ::KANMX6 tcb3Δ::HISMX6 | (Manford et al., 2012) | ANDY198 |
| Oligonucleotides | ||
| Fwd deltaTCB3_SMP: AAGAAACCTTGTCG GATCGCGACATTATGGCTGCTCAATCAAAAGAAG |
metabion | N/A |
| Rv deltaTCB3_SMP: GCGATCCGACAAGGTTTCTT | metabion | N/A |
| Fwd TCB3-1: ATGACTGGCATCAAAGCTCAAGT | metabion | N/A |
| Recombinant DNA | ||
| Plasmid: pRS415-TCB3-GFP-TADH1 | (Manford et al., 2012) | pAM43 |
| Plasmid: pRS415-Tcb3 (1–491)-GFP-TADH1 | (Manford et al., 2012) | pAM44 |
| Plasmid: pRS415-TCB3 SMPΔ-GFP-TADH1 | This study | pCB2 |
| Software and Algorithms | ||
| Amira | Thermo Fisher Scientific |
https://www.fei.com/software/amira-3d-for-life-sciences/ RRID:SCR_014305 |
| BD FACSDiva | BD Biosciences |
https://www.bdbiosciences.com/en-us RRID:SCR_001456 |
| Fiji/ImageJ | NIH |
https://imagej.net/Fiji#Downloads RRID:SCR_002285 |
| Graph-Tool 2.2.44 | (Peixoto, 2017) | https://graph-tool.skewed.de |
| IMOD | (Kremer et al., 1996) |
http://bio3d.colorado.edu/imod/ RRID:SCR_003297 |
| K2Align | Dimitry Tegunov | https://github.com/dtegunov/k2align |
| Mathematica 9.0 | Wolfram Research, Inc. |
https://www.wolfram.com/mathematica RRID:SCR_014448 |
| MATLAB | MathWorks |
https://www.mathworks.com/ RRID:SCR_001622 |
| ParaView 5.5.2 | (Ahrens et al., 2005) | https://www.paraview.org RRID:SCR_002516 |
| Pyto | (Lučić et al., 2016) | N/A |
| Python 2.7.16 and 3.6.5 | N/A |
https://www.python.org RRID:SCR_008394 |
| Python 2.7 Packages Installed via Pip or Anaconda: matplotlib-1.5.1, networkx-1.11, nibabel-2.4.0, numpy-1.11.3, skimage-0.12.3, scipy-0.18.1, pandas-0.19.2, pathlib2-2.2.0, pathos-0.2.2.1, pytest-4.6.2 | N/A |
https://pypi.org https://www.anaconda.com RRID:SCR_008624 RRID:SCR_002498 RRID:SCR_008633 RRID:SCR_008058 |
| Python 3 Packages Installed via Anaconda: matplotlib-3.0.2, numpy-1.15.2, pandas-0.23.0, scipy-1.1.0, seaborn-0.8.1, Statsmodels-0.9.0 | N/A |
https://www.anaconda.com RRID:SCR_016074 |
| Python Software to Estimate Membrane Curvature and Calculate Distances between Membranes | This study and (Kalemanov et al., 2019) | https://github.com/kalemaria/pycurv |
| SerialEM | (Mastronarde, 2005) |
http://bio3d.colorado.edu/SerialEM/ RRID:SCR_017293 |
| TOM Toolbox | (Nickell et al., 2005) | https://www.biochem.mpg.de/tom |
| tom_deconv | Dimitry Tegunov | https://github.com/dtegunov/tom_deconv |
| TomSegMemTV | (Martinez-Sanchez et al., 2014) | https://sites.google.com/site/3demimageprocessing/tomosegmemtv |
| TOMOAND | (Fernández and Li, 2003) | https://sites.google.com/site/3demimageprocessing/tomoand |
| VTK 6.3.0 | (Schroeder et al., 2006) | http://www.vtk.org RRID: SCR_015013 |
| Other | ||
| Quantifoil grids 200 Mesh Copper R2/1 | Quantifoil MicroTools | N/A |
| Whatman Filter Paper 597 | Sigma Aldrich | Cat# WHA10311814 |
| Cover Glasses | Carl Zeiss | Cat# 474030-9000-000 |
Lead Contact and Material Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ruben Fernandez-Busnadiego (ruben.fernandezbusnadiego@med.uni-goettingen.de). Strains and plasmids are available from the authors upon request.
Experimental Model and Subject Details
Yeast Strains and Cell Culture
The yeast strains used in this study are listed in the Key Resources Table.
All strains analyzed were previously reported (Manford et al., 2012, Robinson et al., 1988, Stefan et al., 2011), except YCS2359 (tcb3Δ) and YCS2429 (tcb1Δ tcb2Δ), which were created for this study. Chromosomal deletion of the TCB3 gene was performed as previously described (Longtine et al., 1998). The tcb1Δ tcb2Δ double mutant strain (YCS2429) is a segregant from a genetic cross of tcb1Δ (ANDY117) and tcb2Δ (ANDY120) single mutant strains.
Yeast colonies grown on YPD plates were inoculated in liquid YPD and incubated at 30°C until reaching 0.6 OD600.
Plasmid Construction
The constructs used in this study are listed in the Key Resources Table.
Plasmids pAM43 and pAM44 were previously reported (Manford et al., 2012). For plasmid pCB2, the deletion of the SMP domain of TCB3 (272–479 aa) was created by linearizing the pAM43 vector using the Phusion® HF DNA polymerase (NEB). The sequences of the two oligos used were: 5′- AAGAAACCTTGTCGGATCGCGACATTATGGCTGCTCAATCAAAAGAAG - 3′ and 5′- GCGATCCGACAAGGTTTCTT -3′. The resulting PCR product was treated with DnpI restriction enzyme (NEB) and later purified using the phenol-chloroform protocol. The vector was re-circularized using Gibson strategy (Gibson et al., 2009) as described in https://openwetware.org/wiki/Gibson_Assembly. The complete reaction was transformed in Max Efficiency® DH5alpha (Invitrogen) and platted on an LB plate containing 100 μg/mL ampicillin. The plasmid was extracted using QIAprep® Spin Miniprep Kit (Qiagen) and sequenced by Microsynth with the oligo : 5′- ATGACTGGCATCAAAGCTCAAGT- 3′.Plasmids were transformed into the strains of interest.
Cells carrying plasmids were grown in complete synthetic media buffered at pH 6.25 with Soerensen buffer, lacking leucine to maintain plasmid selection and with 2% glucose as carbon source.
Method Details
Light Microscopy
Cells were grown in the appropriate liquid media and diluted the following day. Recovered cells reaching the exponential phase (OD600=0.6-0.8) were briefly spun down, and confocal Z-stack images were acquired with an LSM780 microscope (Zeiss) at 63× magnification. The images were analyzed using Fiji software (NIH).
Cell Vitrification
Cryo-EM grids (R2/1, Cu 200 mesh grid, Quantifoil microtools) were glow discharged using a plasma cleaner (PDC-3XG, Harrick) for 30 s and mounted on a Vitrobot Mark IV (FEI).
A 3.5 μl drop of yeast culture was deposited on the carbon side of the grid before being blotted from the back using filter paper (Whattman 597) at force setting 9 for 10 s. The grids were immediately plunged into a liquid ethane/propane mixture at liquid nitrogen temperature and stored in grid boxes submerged in liquid nitrogen until usage.
Cryo-Focused Ion Beam Milling
Vitrified grids were mounted into Autogrid carriers (FEI), held in place by a copper ring. They were subsequently inserted in a dual-beam Quanta 3D cryo-FIB / scanning electron microscope (SEM) (FEI) using a transfer shuttle and a cryo-transfer system (PP3000T, Quorum). Inside the microscope, the sample was kept at -180°C using a cryo-stage throughout the milling process.
To protect the sample from unwanted damage by the ion beam, a layer of organic platinum was deposited on top of the grid using a gas injection system from a 13.5 mm distance for 9 s.
Small groups of cells located near the center of the grid square were targeted for milling. Milling was done at a 20° tilt. Several sequential steps were taken, starting with the Ga2+ ion beam at 30 kV and 500 pA beam current for rough milling, down to 30 kV and 30 pA for fine milling.
The final lamellae were around 14 μm wide and 150–250 nm thick. SEM imaging at 5 kV and 13 pA was used to monitor the milling process. The final thickness was reached when the lamellae lacked contrast at 3 kV and 10 pA.
Cryo-Electron Tomography
Cryo-FIB lamellae were imaged at liquid nitrogen temperature in a Polara (tcb3Δ + Tcb3 SMPΔ-GFP, tcb3Δ + Tcb3 C2Δ-GFP, tcb3Δ + Tcb3-GFP cells, as well as one experiment of tcb1/2/3Δ, tcb3Δ and tcb1Δ cells) or Titan Krios (all other cases) cryo-electron microscopes (FEI) equipped with 300 kV field emission guns, post column energy filters (Gatan) and K2 Summit direct electron detectors (Gatan).
Low magnification (4500 ×, 27-Å pixel size in Polara; 3600 ×, 40-Å pixel size in Titan Krios; -100 μm defocus) images of the lamellae were taken to identify regions of interest. Tilt series were recorded using SerialEM software (Mastronarde, 2005) at higher magnification (34,000 ×, 3.509-Å pixel size in Polara; 42,000 ×, 3.42-Å pixel size in Titan Krios; -5 μm defocus), typically from -46° to +64° with increments of 2°. For one experiment of the tcb1/2/3Δ, tcb3Δ and tcb1Δ conditions, tilt series were recorded in Polara at 22,500 x (0.522-Å pixel size) using -5 μm defocus.
The cameras were operated in dose fractionation mode, producing frames every 0.2 s. 1/cos scheme was used to increase the exposure time at higher tilt angles, resulting in exposure times of 1–2 s per projection image. Tilt series acquired in the Titan Krios for the tcb1/2/3Δ, Δtether, WT heat shock and tcb1/2/3Δ heat shock conditions were recorded using a dose-symmetric scheme (Hagen et al., 2017). All other tilt series were acquired using a unidirectional scheme. In all cases, the total dose per tilt series was ∼120 e-/Å2.
K2 frames were aligned using in house software (K2Align) based on previous research (Li et al., 2013) and available at https://github.com/dtegunov/k2align. The tilt series were aligned using patch-tracking and reconstructed by weighted back projection in IMOD (Kremer et al., 1996). Tomograms were binned twice, to a final voxel size of ∼1.4 nm (2.1 nm for the tilt series recorded at 22,500 x). For visualization, the tomographic slices shown in all figures except Figures S2A, S2D, 6A, 6B, and 6C were denoised using a non-linear anisotropic filter (Fernández and Li, 2003). The contrast of Figures S2A, S2D, 6A, 6B, and 6C was enhanced using a deconvolution filter (https://github.com/dtegunov/tom_deconv) executed in MATLAB (MathWorks) using the functionalities of the TOM toolbox (Nickell et al., 2005).
Membrane Segmentation and Surface Generation
Membranes were automatically segmented along their middle line using TomoSegMemTV (Martinez-Sanchez et al., 2014), and refined manually using Amira (FEI). The intermembrane volumes were manually segmented. Segmentations consisted of binary voxel masks, which were smoothed using a Gaussian kernel with σ of 1 voxel. From these segmentations, isosurfaces were generated using the Marching Cubes algorithm (Lorensen and Cline, 1987) with an isosurface level of 0.7. Finally, isosurfaces were transformed into single-layer triangle mesh surfaces delineating the membranes. On average, surface triangles had an area of 0.6 ± 0. 4 nm2 (mean ± SD) for the PM and 0.7 ± 0. 4 nm2 (mean ± SD) for the cER membrane.
Membrane Curvature Determination
Curvature was estimated locally for each surface triangle using a novel algorithm (Kalemanov et al., 2019). Briefly, we used tensor voting with a neighborhood of triangles defined by the RadiusHit parameter to denoise the surface normal vectors and then estimate the principal directions and curvatures for each surface triangle center. The maximal (κ1) and the minimal (κ2) principal curvatures were combined into a single scalar value for each triangle by calculating curvedness (Koenderink and van Doorn, 1992):
A RadiusHit value of 10 nm was used, limiting the size of the smallest feature measured reliably to a radius of 10 nm, i.e. a curvedness of 0.1 nm-1. Higher values were excluded from the analysis. Values within 1 nm to the surface border were removed, as curvature estimation was not reliable in these areas. The total number of cER triangles where curvature was measured in each condition is shown in Table S1. Membrane curvature was visualized using ParaView (Ahrens et al., 2005).
Intermembrane Distance Measurements
To calculate distances between two membranes, surfaces following the cytosolic side of all membranes were generated. Denoised normal vectors from the first surface (PM in ER-PM MCS, mitochondria in ER-mitochondria MCS, vacuole in nucleus-vacuole MCS) were generated (see above) and extended until their intersection with the second surface. The Euclidean distance between the source triangle center and the intersection point was calculated as the intermembrane distance. These measurements were performed for all triangles of the first surface for which a normal vector intersected with the second surface within 50 nm (Table S1 shows the total number of measurements for each condition).
To calculate cER thickness, a surface following the luminal side of the cER membrane was generated. Denoised PM surface normal vectors were extended until their first and second intersections with the cER membrane surface. The Euclidean distance between the intersections was calculated as the cER thickness. These measurements were performed for all triangles of the PM for which cER thickness was within 80 nm (Table S1 shows the total number of measurements for each condition).
Intermembrane distances at ER-PM, ER-mitochondria and nucleus-vacuole MCS were defined as the distances between the cytosolic leaflets of the membranes. cER thickness was calculated as the distance between the luminal leaflets of the cER membrane. Given that the triangle-mesh surfaces go through the centers of the voxels on the edge of the segmentations, one voxel was added to all distances for correction.
cER Peak Morphology and Density Quantification
Measurements of cER peak height, radius and distance to the PM were done in tomographic slices using the measuring tool built in IMOD. The cER peak density was calculated per MCS by dividing the number of cER peaks by half of the cER surface area, since cER peaks were found only on the side of cER facing the PM. The cER surface area was calculated by summing up the areas of all triangles in the cER surface (the total cER surface area for each condition is shown in Table S1).
Membrane Modeling
To compute the change in the free energy barrier associated to the extraction of a lipid from a highly curved membrane as compared to the extraction from a flat membrane, we consider that the extraction is performed by a lipid transport protein (LTP). The lipid extraction reaction undergoes a series of steps, initiated by the binding and partial insertion of the LTP into the membrane (absorption), followed by the lipid extraction and detachment of the protein-lipid complex from the membrane (desorption) (Dittman and Menon, 2017, Wong et al., 2017). Hence, the total free energy required for lipid extraction, , corresponds to the change of the free energy of the system (including both the membrane and the LTP) resulting from the extraction of lipids by one lipid transfer reaction of a single LTP.
We can split this free energy of lipid extraction in two terms. The first one, denoted by , corresponds to contributions independent from membrane stress, such as hydrogen bonding and electrostatic interactions occurring during protein insertion, as well as LTP-lipid chemical interactions occurring both within the membrane and in solution. The second term, denoted by , corresponds to the elastic contribution dependent on membrane stress (Campelo and Kozlov, 2014). We denote by the change in the free energy of lipid extraction from a highly curved membrane (associated with a total curvature , where is the radius of curvature, and the local curvature at the tip of the peak is considered to be locally spherical; is equivalent to the curvedness for κ1=κ2) with respect to the extraction from a flat membrane (. Since is independent of the curvature or elastic stresses within the membrane, it follows that , where .
To calculate the elastic part of the free energy of lipid extraction as a function of the membrane curvature, , we consider that the main contributions to this energy arise from the shallow insertion of a domain of the LTP into the cytoplasmic leaflet of the cER membrane, , and from the elastic energy relaxation of the extracted lipids, .
We consider an elastic model of the lipid monolayer as a three-dimensional, anisotropic elastic material (Campelo et al., 2008) to compute the internal strains and stresses generated by the partial insertion of the LTP into the membrane, and hence the accumulated elastic energy, (Campelo and Kozlov, 2014, Campelo et al., 2008). One can define the curvature sensitivity parameter, , which accounts for the ability of a given protein domain to sense membrane curvature, and depends on the way the membrane curvature has been generated (Campelo and Kozlov, 2014). It has been computationally shown that the elastic energy of insertion can be written as , which allow us to write, , given that is the curvature-independent part of the protein insertion energy.
To compute the elastic energy relaxation of the lipid (or lipids) extracted by the LTP, we use the Helfrich model of membrane curvature energy (Helfrich, 1973). According to this model, we can express the change in the free energy relaxation of lipids, each of which having a cross-sectional surface area, , and an effective spontaneous curvature (Zimmerberg and Kozlov, 2006), , as , where is the bending rigidity of a single monolayer (Niggemann et al., 1995). kBT is the product of the Boltzmann constant and the absolute temperature. We do not consider here a possible dependence of the lipid free energy change on the lateral tension of the membrane, since we assume that there is no membrane tension gradient appearing as a result of membrane bending and therefore the membrane is under the same lateral tension regardless of its curvature.
In total, we can write down the free energy change for lipid extraction by an LTP as a function of the curvature of the donor membrane as
| (Equation 1) |
From the free energy change for lipid extraction, we can estimate the change in the rate of lipid extraction from a curved membrane relative to the flat membrane. Assuming Arrhenius kinetics, the rate of lipid extraction can be written as , where is the Arrhenius prefactor, which we consider to be curvature-independent (Dittman and Menon, 2017). Hence, the change in the rate of lipid extraction from a curved membrane relative to the flat membrane can be written as
| (Equation 2) |
The value of the curvature sensitivity parameter can be computationally calculated (Campelo and Kozlov, 2014), and depends on different parameters, in particular, on the size and depth of the insertion. Importantly, it depends on the way the membrane curvature has been generated. For membrane curvature generated by an externally applied torque (e.g. by protein scaffolds or by protein insertions), a cylindrical insertion of radius 0.5 nm, length 2 nm, and inserted 0.8 nm into the monolayer, the curvature sensitivity parameter has been calculated to be (Campelo and Kozlov, 2014). Depending on the geometrical parameters, the curvature sensitivity can range between (Campelo and Kozlov, 2014).
The relative dependence of the two contributions to the free energy change in Equation (1) can be quantitated by the ratio . For the characteristic parameters mentioned above (; ; ; ; ), the relative contribution to the extraction free energy of the protein insertion elastic energy is much higher than that of the lipid curvature stress for a wide range of lipid spontaneous curvatures (Figure S3A). The lipid bending stress only dominates for protein insertions with a relatively low curvature sensitivity extracting many lipids with a very large negative spontaneous curvature (conical lipids such as diacylglycerol, which has a spontaneous curvature (Szule et al., 2002)) (Figure S3B).
The plots of the calculated lipid extraction energy changes as a function of the membrane curvature, , and of the curvature sensitivity parameter, , for the extraction of both cylindrical () and highly conical () lipids are shown in Figure S3C. In addition, we present the computed values of the lipid extraction energy from a highly curved cER peak (radius of curvature ) as a function of the curvature sensitivity parameter for a cylindrical lipid (Figure S3D) and as a function of the lipid spontaneous curvature for (Figure S3E). Altogether, we can conclude that, for standard physiological conditions, the extraction of lipids by LTPs is more efficient when occurring from highly curved membranes because these proteins have a better insertion affinity into highly bent membranes associated with a large bending moment in the monolayer facing the PM.
Calculation of Lipid Extraction Currents
We define the lipid current as the total number of lipids extracted along the entire cER membrane per unit time, , where the local extraction rate, , is integrated along the entire cER membrane, which has a total surface area . We consider that the cER membrane is formed by (i) highly curved peaks of constant curvature, , with an area , and (ii) a flat region of zero curvature, with an area . Hence, the total lipid current is the sum of the lipid currents coming from the peaks and the flat part, , where the peak current is , and the flat current, .
We can relate to the experimentally measured peak number density, , (Figure 2I) as , where is the average surface area of a single cER peak, which can be estimated from our cryo-tomograms to be (by modeling the peak as a conic structure of base radius and height of , and hence average total curvature, ). Moreover, if we define the standard lipid current as the lipid current for a flat cER having a total surface area and with no peaks, , we can express the peak, flat, and total lipid currents, respectively, as
| (Equation 3a) |
| (Equation 3b) |
| (Equation 3c) |
In Figure S4B we plot the total lipid current relative to the standard lipid current, ,as a function of the peak density, , and of the relative cER surface area , for the computed value of the lipid extraction rate at the peaks, . We can see that the total amount of lipids being extracted from the cER depends not only on the number density of peaks per unit area, (horizontal axis in Figure S4B), but also on the total amount of cER surface area (plotted relative to WT levels in Figure S4B, vertical axis). When heat shocking WT cells, peak density increases from to (Figure 2I), and the total lipid current concomitantly increases about 3-fold.
It is also informative to evaluate the relative contribution to the lipid current between the peaks and the flat part, which, from Equations (3a) and (3b), is
| (Equation 4) |
Using the experimentally determined density of cER peaks, , whereas . Therefore, in WT cells the total number of lipids extracted from cER peaks is roughly equivalent to that of the flat parts, but lipid extraction from cER peaks becomes dominant upon heat shock due to the observed increase in cER peak density (Figure S4B).
PM Integrity Assays
Cells were grown at 26°C to mid-log phase in rich media (YPD) or selective drop out media (YND) as required. Cells were shifted to 42°C for 10 min and (1OD600 equivalents) were collected, resuspended in PBS, and incubated with propidium iodide (Invitrogen) for 10 min. Cells were then washed twice with ddH2O and analyzed by flow cytometry (BD LSR II). For each condition, 10,000 cells were measured. Background was determined by analyzing each of the cell strains prior to staining with propidium iodide. Four independent biological repeats were performed for all conditions. We noted that PM integrity defects were enhanced in mutant cells grown in YPD compared to YND, consistent previous results (Omnus et al., 2016).
Quantification and Statistical Analysis
For intermembrane distances (Figures 1E and 2G), cER thickness (Figure 2H) and cER curvedness (Figure 3E), a large number of measurements (Table S1) was performed automatically for surface triangles (see above). The figures show the complete distribution of values as violin plots (Figures 1E, 2G, and 2H) or histograms (Figure 3E), including all measurements for all MCS analyzed. For violin plots, a white dot represents the median, a black slab the interquartile range, and a black line 1.5x the interquartile range. The histogram in Figure 3E shows the relative frequency of exceedance (calculated as 1 – cumulative relative frequency) of cER curvedness averaged across MCS for each bin (solid line) ± SE (shaded area). Bin width was 1 nm-1. The frequency was weighted by triangle area and normalized to the total surface area.
Because measurements of neighboring triangles are not statistically independent from each other, statistical comparisons between conditions were performed using the mean of the measurements for each MCS. Consequently, in the text we report N as the number of MCS analyzed. In all cases, normality was assessed using Shapiro-Wilks test. To estimate statistical significance we used t-test for normally distributed samples and Mann-Whitney-U test for non-normally distributed samples.
Figure 2I shows mean peak density per μm2 cER surface area for each condition ± SE. cER peak morphology measurements (Figure S2C) were displayed by box plots. For PM integrity assays (Figures 5A, 6E, and S4A), average values from four independent experiments were analyzed. Bar plots show mean values for each condition ± SE.
These data were statistically analyzed as reported above. The number of independent biological experiments, MCS studied and measurements performed are reported in Table S1.
Data and Code Availability
The tomogram displayed in Figure 5A showing cER-PM MCS in a heat-shocked WT cell has been deposited at the Electron Microscopy Data Base (EMDB) with accession numbers EMD-10378 (raw tomogram) and EMD-10379 (tomogram filtered by non-linear anisotropic diffusion, as in Figure 5A).
The Python software used to estimate membrane curvature and calculate distances between membranes is available at https://github.com/kalemaria/pycurv. The software depends on the following external packages: Pyto (Lučić et al., 2016), Graph-tool (Peixoto, 2017) and VTK (Schroeder et al., 2006). Graph-tool and VTK are respectively available at https://graph-tool.skewed.de/ and https://vtk.org/.
Acknowledgments
We thank Philipp Erdmann, Günter Pfeifer, Jürgen Plitzko, and Miroslava Schaffer for electron microscopy support, and the Imaging Facility of the Max Planck Institute of Biochemistry for light microscopy support. We are grateful to Scott Emr for strains as well as Markus Hohle, Vladan Lucic, Eri Sakata, and Florian Wilfling for helpful discussions. We also thank Patrick C. Hoffmann and Wanda Kukulski for sharing unpublished results. J.C. and M.K. were supported by the Graduate School of Quantitative Biosciences Munich. J.C., M.K., A.M.-S., W.B. and R.F.-B. have received funding from the European Commission (FP7 GA ERC-2012-SyG_318987–ToPAG). R.F.-B. acknowledges funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through Germany’s Excellence Strategy - EXC 2067/1- 390729940 and SFB1190/P22. F.C. acknowledges financial support from the Spanish Ministry of Economy and Competitiveness (“Severo Ochoa” program for Centres of Excellence in R&D (SEV-2015-0522, FIS2015-63550-R, FIS2017-89560-R, BFU2015-73288-JIN, RYC-2017-22227, and AEI/FEDER/UE), Fundació Privada Cellex, and from the Generalitat de Catalunya through the CERCA program. R.L. acknowledges funding from the European Commission (ERC AdG TENDO - 834394), the Swiss National Science Foundation (31003A_179517), the Swiss National Centre for Competence in Research in Chemical Biology (51NF40-185898), and the Canton of Geneva. C.J.S. is supported by MRC funding to the MRC LMCB University Unit at UCL, award code MC_UU_00012/6.
Author Contributions
J.C. performed electron microscopy experiments and contributed to computational data analysis. M.K. and A.M.S. developed software procedures for data analysis. M.K. performed computational and statistical analysis of the data. F.C. performed theoretical modeling. C.B. carried out molecular biology and light microscopy experiments. F.T. and C.J.S. constructed strains, plasmids, and performed plasma membrane integrity assays. R.F.-B. supervised electron microscopy experiments and data analysis. J.C., F.C., C.B., R.L., W.B., C.J.S., and R.F.-B. designed the research. R.F.-B. wrote the manuscript with contributions from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: November 18, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2019.10.018.
Supplemental Information
References
- Ackema K.B., Prescianotto-Baschong C., Hench J., Wang S.C., Chia Z.H., Mergentaler H., Bard F., Frank S., Spang A. Sar1, a novel regulator of ER-mitochondrial contact sites- sar1. PLoS One. 2016;11:e0154280. doi: 10.1371/journal.pone.0154280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens J., Geveci B., Law C. Visualization Handbook. Elsevier; 2005. ParaView: an end-user tool for large data visualization. [Google Scholar]
- Andrews N.W., Corrotte M. Plasma membrane repair. Curr. Biol. 2018;28:R392–R397. doi: 10.1016/j.cub.2017.12.034. [DOI] [PubMed] [Google Scholar]
- Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Balla T. Ca(2+) and lipid signals hold hands at endoplasmic reticulum-plasma membrane contact sites. J. Physiol. (Lond.) 2018;596:2709–2716. doi: 10.1113/JP274957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Baumeister W. Cryo-electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Bers D.M. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- Bian X., Saheki Y., De Camilli P. Ca(2+) releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J. 2018;37:219–234. doi: 10.15252/embj.201797359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M., Schuldiner M. Stepping outside the comfort zone of membrane contact site research. Nat. Rev. Mol. Cell Biol. 2018;19:483–484. doi: 10.1038/s41580-018-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldieri G., Barbieri E., Nappo G., Raimondi A., Bonora M., Conte A., Verhoef L.G.G.C., Confalonieri S., Malabarba M.G., Bianchi F. Reticulon 3-dependent ER-PM contact sites control EGFR nonclathrin endocytosis. Science. 2017;356:617–624. doi: 10.1126/science.aah6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F., Kozlov M.M. Sensing membrane stresses by protein insertions. PLoS Comput. Biol. 2014;10:e1003556. doi: 10.1371/journal.pcbi.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F., McMahon H.T., Kozlov M.M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco S., Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu. Rev. Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.L., Chen Y.J., Liou J. ER-plasma membrane junctions: why and how do we study them? Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1494–1506. doi: 10.1016/j.bbamcr.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S., Raghu P. Phospholipid transport protein function at organelle contact sites. Curr. Opin. Cell Biol. 2018;53:52–60. doi: 10.1016/j.ceb.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado J., Fernández-Busnadiego R. Deciphering the molecular architecture of membrane contact sites by cryo-electron tomography. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1507–1512. doi: 10.1016/j.bbamcr.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Creutz C.E., Snyder S.L., Schulz T.A. Characterization of the yeast tricalbins: membrane-bound multi-C2-domain proteins that form complexes involved in membrane trafficking. Cell. Mol. Life Sci. 2004;61:1208–1220. doi: 10.1007/s00018-004-4029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittman J.S., Menon A.K. Speed limits for nonvesicular intracellular sterol transport. Trends Biochem. Sci. 2017;42:90–97. doi: 10.1016/j.tibs.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Evans R.M. Turning up the heat on membrane fluidity. Cell. 2015;161:962–963. doi: 10.1016/j.cell.2015.04.046. [DOI] [PubMed] [Google Scholar]
- Fernández J.J., Li S. An improved algorithm for anisotropic nonlinear diffusion for denoising cryo-tomograms. J. Struct. Biol. 2003;144:152–161. doi: 10.1016/j.jsb.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Fernández-Busnadiego R., Saheki Y., De Camilli P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc. Natl. Acad. Sci. USA. 2015;112:E2004–E2013. doi: 10.1073/pnas.1503191112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M.A., Temmerman K., Ercan E., Nickel W., Seedorf M. Binding of plasma membrane lipids recruits the yeast integral membrane protein Ist2 to the cortical ER. Traffic. 2009;10:1084–1097. doi: 10.1111/j.1600-0854.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- Gatta A.T., Wong L.H., Sere Y.Y., Calderón-Noreña D.M., Cockcroft S., Menon A.K., Levine T.P. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. ELife. 2015;4 doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Giordano F., Saheki Y., Idevall-Hagren O., Colombo S.F., Pirruccello M., Milosevic I., Gracheva E.O., Bagriantsev S.N., Borgese N., De Camilli P. PI(4,5)P2-dependent and Ca2+-regulated ER-PM interactions mediated by the Extended Synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen W.J.H., Wan W., Briggs J.A.G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C Biochem. Biophys. Biol. Virol. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Henne W.M., Liou J., Emr S.D. Molecular mechanisms of inter-organelle ER-PM contact sites. Curr. Opin. Cell Biol. 2015;35:123–130. doi: 10.1016/j.ceb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Ho R., Stroupe C. The HOPS/class C Vps complex tethers high-curvature membranes via a direct protein-membrane interaction. Traffic. 2016;17:1078–1090. doi: 10.1111/tra.12421. [DOI] [PubMed] [Google Scholar]
- Hu J., Prinz W.A., Rapoport T.A. Weaving the web of ER tubules. Cell. 2011;147:1226–1231. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall-Hagren O., Lü A., Xie B., De Camilli P. Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J. 2015;34:2291–2305. doi: 10.15252/embj.201591565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A.J., Perez F. Plasma membrane repair: the adaptable cell life-insurance. Curr. Opin. Cell Biol. 2017;47:99–107. doi: 10.1016/j.ceb.2017.03.011. [DOI] [PubMed] [Google Scholar]
- Jüschke C., Wächter A., Schwappach B., Seedorf M. SEC18/NSF-independent, protein-sorting pathway from the yeast cortical ER to the plasma membrane. J. Cell Biol. 2005;169:613–622. doi: 10.1083/jcb.200503033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalemanov M., Collado J.F., Baumeister W., Fernández-Busnadiego R., Martínez-Sánchez A. Estimation of membrane curvature for cryo-electron tomography. bioRxiv. 2019:579060. doi: 10.1371/journal.pcbi.1007962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y., Uemura M. Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J. 2003;36:141–154. doi: 10.1046/j.1365-313x.2003.01864.x. [DOI] [PubMed] [Google Scholar]
- Koenderink J.J., van Doorn A.J. Surface shape and curvature scales. Image Vis. Comput. 1992;10:557–564. [Google Scholar]
- Kozlov M.M., Campelo F., Liska N., Chernomordik L.V., Marrink S.J., McMahon H.T. Mechanisms shaping cell membranes. Curr. Opin. Cell Biol. 2014;29:53–60. doi: 10.1016/j.ceb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lacruz R.S., Feske S. Diseases caused by mutations in ORAI1 and STIM1. Ann. N. Y. Acad. Sci. 2015;1356:45–79. doi: 10.1111/nyas.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom A.P., Beavers D.L., Wehrens X.H. The junctophilin family of proteins: from bench to bedside. Trends Mol. Med. 2014;20:353–362. doi: 10.1016/j.molmed.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G., Orci L., Shi L., Geiling M., Ravazzola M., Wieland F., Cosson P., Rothman J.E. Induction of cortical endoplasmic reticulum by dimerization of a coatomer-binding peptide anchored to endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 2010;107:6876–6881. doi: 10.1073/pnas.1002536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Vanneste S., Pérez-Sancho J., Benitez-Fuente F., Strelau M., Macho A.P., Botella M.A., Friml J., Rosado A. Ionic stress enhances ER-PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2019;116:1420–1429. doi: 10.1073/pnas.1818099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Hong W. Diverse membrane-associated proteins contain a novel SMP domain. FASEB J. 2006;20:202–206. doi: 10.1096/fj.05-4581hyp. [DOI] [PubMed] [Google Scholar]
- Lees J.A., Messa M., Sun E.W., Wheeler H., Torta F., Wenk M.R., De Camilli P., Reinisch K.M. Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science. 2017;355 doi: 10.1126/science.aah6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Young B.P., Tavassoli S., Levine T.P. Inheritance of cortical ER in yeast is required for normal septin organization. J. Cell Biol. 2007;179:467–483. doi: 10.1083/jcb.200708205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorensen W.E., Cline H.E. Marching cubes: A high resolution 3D surface construction algorithm. SIGGRAPH Comput. Graph. Proceedings of the 14th SIGGRAPH 1987 (ACM) 1987;21:163–169. [Google Scholar]
- Lučić V., Fernández-Busnadiego R., Laugks U., Baumeister W. Hierarchical detection and analysis of macromolecular complexes in cryo-electron tomograms using Pyto software. J. Struct. Biol. 2016;196:503–514. doi: 10.1016/j.jsb.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Maass K., Fischer M.A., Seiler M., Temmerman K., Nickel W., Seedorf M. A signal comprising a basic cluster and an amphipathic alpha-helix interacts with lipids and is required for the transport of Ist2 to the yeast cortical ER. J. Cell Sci. 2009;122:625–635. doi: 10.1242/jcs.036012. [DOI] [PubMed] [Google Scholar]
- Machida K., Ohnishi S.I. Effect of bilayer membrane curvature on activity of phosphatidylcholine exchange protein. Biochim. Biophys. Acta. 1980;596:201–209. doi: 10.1016/0005-2736(80)90355-7. [DOI] [PubMed] [Google Scholar]
- Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A., Emr S.D. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Martens S., Kozlov M.M., McMahon H.T. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez A., Garcia I., Asano S., Lucic V., Fernandez J.J. Robust membrane detection based on tensor voting for electron tomography. J. Struct. Biol. 2014;186:49–61. doi: 10.1016/j.jsb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Miller S.B., Mogk A., Bukau B. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J. Mol. Biol. 2015;427:1564–1574. doi: 10.1016/j.jmb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Min S.W., Chang W.P., Südhof T.C. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc. Natl. Acad. Sci. USA. 2007;104:3823–3828. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser von Filseck J., Vanni S., Mesmin B., Antonny B., Drin G. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat. Commun. 2015;6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- Murphy S.E., Levine T.P. VAP, a Versatile Access Point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Nickell S., Förster F., Linaroudis A., Net W.D., Beck F., Hegerl R., Baumeister W., Plitzko J.M. TOM software toolbox: acquisition and analysis for electron tomography. J. Struct. Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Niggemann G., Kummrow M., Helfrich W. The bending rigidity of phosphatidylcholine bilayers - dependences on experimental-method, sample cell sealing and temperature. J. Phys. II France. 1995;5:413–425. [Google Scholar]
- Omnus D.J., Manford A.G., Bader J.M., Emr S.D., Stefan C.J. Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol. Biol. Cell. 2016;27:1170–1180. doi: 10.1091/mbc.E16-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto T.P. 2017. The graph-tool python library. figshare. 106084/m9figshare1164194. [Google Scholar]
- Pérez-Sancho J., Tilsner J., Samuels A.L., Botella M.A., Bayer E.M., Rosado A. Stitching organelles: organization and function of specialized membrane contact sites in plants. Trends Cell Biol. 2016;26:705–717. doi: 10.1016/j.tcb.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Pérez-Sancho J., Vanneste S., Lee E., McFarlane H.E., Esteban Del Valle A., Valpuesta V., Friml J., Botella M.A., Rosado A. The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol. 2015;168:132–143. doi: 10.1104/pp.15.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H., Gaigg B., Hrastnik C., Achleitner G., Kohlwein S.D., Zellnig G., Perktold A., Daum G. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 2001;268:2351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- Quon E., Sere Y.Y., Chauhan N., Johansen J., Sullivan D.P., Dittman J.S., Rice W.J., Chan R.B., Di Paolo G., Beh C.T. Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLOS Biol. 2018;16:e2003864. doi: 10.1371/journal.pbio.2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch K.M., De Camilli P. SMP-domain proteins at membrane contact sites: structure and function. Biochim. Biophys. Acta. 2016;1861:924–927. doi: 10.1016/j.bbalip.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigort A., Bäuerlein F.J., Villa E., Eibauer M., Laugks T., Baumeister W., Plitzko J.M. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos E., Figueroa L., Manno C., Kraeva N., Riazi S. The couplonopathies: A comparative approach to a class of diseases of skeletal and cardiac muscle. J. Gen. Physiol. 2015;145:459–474. doi: 10.1085/jgp.201411321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Südhof T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Robinson J.S., Klionsky D.J., Banta L.M., Emr S.D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y., Bian X., Schauder C.M., Sawaki Y., Surma M.A., Klose C., Pincet F., Reinisch K.M., De Camilli P. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 2016;18:504–515. doi: 10.1038/ncb3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y., De Camilli P. Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 2017;86:659–684. doi: 10.1146/annurev-biochem-061516-044932. [DOI] [PubMed] [Google Scholar]
- Saheki Y., De Camilli P. The extended-synaptotagmins. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1490–1493. doi: 10.1016/j.bbamcr.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire A.L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., Salinas J., Mancuso S., Valpuesta V., Baluska F. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell. 2008;20:3374–3388. doi: 10.1105/tpc.108.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder C.M., Wu X., Saheki Y., Narayanaswamy P., Torta F., Wenk M.R., De Camilli P., Reinisch K.M. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, W., Martin, K.M., and Lorensen, W.E. (2006). The visualization toolkit: an object-oriented approach to 3d graphics (Kitware).
- Schulz T.A., Creutz C.E. The tricalbin C2 domains: lipid-binding properties of a novel, synaptotagmin-like yeast protein family. Biochemistry. 2004;43:3987–3995. doi: 10.1021/bi036082w. [DOI] [PubMed] [Google Scholar]
- Sclip A., Bacaj T., Giam L.R., Südhof T.C. Extended synaptotagmin (ESyt) triple knock-out mice are viable and fertile without obvious endoplasmic reticulum dysfunction. PLoS One. 2016;11:e0158295. doi: 10.1371/journal.pone.0158295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai N., Yifrach E., van Roermund C.W.T., Cohen N., Bibi C., IJlst L., Cavellini L., Meurisse J., Schuster R., Zada L. Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 2018;9:1761. doi: 10.1038/s41467-018-03957-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siao W., Wang P., Voigt B., Hussey P.J., Baluska F. Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum-plasma membrane contact sites. J. Exp. Bot. 2016;67:6161–6171. doi: 10.1093/jxb/erw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B., Callan-Jones A., Manneville J.B., Nassoy P., Joanny J.F., Prost J., Goud B., Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C.J. Building ER-PM contacts: keeping calm and ready on alarm. Curr. Opin. Cell Biol. 2018;53:1–8. doi: 10.1016/j.ceb.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., Emr S.D. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Szule J.A., Fuller N.L., Rand R.P. The effects of acyl chain length and saturation of diacylglycerols and phosphatidylcholines on membrane monolayer curvature. Biophys. J. 2002;83:977–984. doi: 10.1016/s0006-3495(02)75223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli S., Chao J.T., Young B.P., Cox R.C., Prinz W.A., de Kroon A.I., Loewen C.J. Plasma membrane–endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 2013;14:434–440. doi: 10.1038/embor.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., Prinz W.A. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M.G., Moss T. Loss of all 3 Extended Synaptotagmins does not affect normal mouse development, viability or fertility. Cell Cycle. 2016;15:2360–2366. doi: 10.1080/15384101.2016.1203494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm A.M., Cohen S., Legant W.R., Melunis J., Hershberg U., Wait E., Cohen A.R., Davidson M.W., Betzig E., Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan E.M., You J.S., Elsie Yu H.Y., Lasek A., Vitale N., Hornberger T.A., Bement W.M. Lipid domain-dependent regulation of single-cell wound repair. Mol. Biol. Cell. 2014;25:1867–1876. doi: 10.1091/mbc.E14-03-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J., Abrams J., Wang Y., Morano K.A. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss C., Lahiri S., Young B.P., Loewen C.J., Prinz W.A. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J. Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J., Schaffer M., Fernández-Busnadiego R. Cryo-electron tomography-the cell biology that came in from the cold. FEBS Lett. 2017;591:2520–2533. doi: 10.1002/1873-3468.12757. [DOI] [PubMed] [Google Scholar]
- West M., Zurek N., Hoenger A., Voeltz G.K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J.M., Hartzell H.C. Anoctamins/TMEM16 proteins: chloride channels flirting with lipids and extracellular vesicles. Annu. Rev. Physiol. 2017;79:119–143. doi: 10.1146/annurev-physiol-022516-034031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf W., Kilic A., Schrul B., Lorenz H., Schwappach B., Seedorf M. Yeast Ist2 recruits the endoplasmic reticulum to the plasma membrane and creates a ribosome-free membrane microcompartment. PLoS One. 2012;7:e39703. doi: 10.1371/journal.pone.0039703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.H., Čopič A., Levine T.P. Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 2017;42:516–530. doi: 10.1016/j.tibs.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Carvalho P., Voeltz G.K. Here, there, and everywhere: the importance of ER membrane contact sites. Science. 2018;361 doi: 10.1126/science.aan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Kawamura Y., Minami A., Uemura M. Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell. 2008;20:3389–3404. doi: 10.1105/tpc.108.062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Liu Y., Gulbranson D.R., Paine A., Rathore S.S., Shen J. Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc. Natl. Acad. Sci. USA. 2016;113:4362–4367. doi: 10.1073/pnas.1517259113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M.N., Bello O.D., Wang J., Coleman J., Cai Y., Sindelar C.V., Rothman J.E., Krishnakumar S.S. Ring-like oligomers of Synaptotagmins and related C2 domain proteins. ELife. 2016;5:e17262. doi: 10.7554/eLife.17262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Macgurn J.A., Liu M., Emr S. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. ELife. 2013;2:e00459. doi: 10.7554/eLife.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The tomogram displayed in Figure 5A showing cER-PM MCS in a heat-shocked WT cell has been deposited at the Electron Microscopy Data Base (EMDB) with accession numbers EMD-10378 (raw tomogram) and EMD-10379 (tomogram filtered by non-linear anisotropic diffusion, as in Figure 5A).
The Python software used to estimate membrane curvature and calculate distances between membranes is available at https://github.com/kalemaria/pycurv. The software depends on the following external packages: Pyto (Lučić et al., 2016), Graph-tool (Peixoto, 2017) and VTK (Schroeder et al., 2006). Graph-tool and VTK are respectively available at https://graph-tool.skewed.de/ and https://vtk.org/.