Abstract
Increasingly sophisticated instrumentation for chemical separations and identification has facilitated rapid advancements in our understanding of the metabolome. Since many analyses are performed using either mass spectroscopy (MS) or nuclear magnetic resonance (NMR) spectroscopy, the spin ½ stable 13C isotope is now widely used as a metabolic tracer. There is strong interest in quantitative analysis of metabolic flux through pathways in vivo, particularly in human patients. Although instrumentation advances and scientific interests in metabolism are increasing in parallel, a practical and rational design of a 13C tracer study can be challenging. Prior to planning the details of a tracer experiment, is it important to consider whether the analytical results will be sensitive to flux through the pathways of interest. Here, we briefly summarize the various approaches that have been used to design carbon tracer experiments, outline the sources of complexity, and illustrate the use of a software tool, tcaSIM, to aid in the experimental design of both MS and NMR data in complex systems including patients.
1. Introduction
Intermediary metabolism is a complex intertwined network of pathways and nodes. The activity of specific pathways within the larger network can be experimentally assessed in a living system by providing one or more substrates that have been labeled with a carbon isotopic tracer (11C, 13C or 14C) and analyzing metabolic products for the position and amount of the isotopic tracer. In such studies, metabolic models of varying degrees of sophistication can be used to quantitatively evaluate the activity of specific underlying pathways. 11C and 14C radiotracers offer exquisite sensitivity but limited chemical information about specific metabolites derived from that tracer. In addition, the practical constraints of using radioactive materials are significant barriers associated with use of 11C and 14C radiotracers. For this reason, many investigators, particularly those working with human subjects, have chosen to study metabolic networks using non-radioactive 13C-enriched substrates with analysis of products done by either mass spectroscopy (MS) or 13C nuclear magnetic resonance (NMR) spectroscopy. This 13C approach has been particularly powerful for examining intermediary metabolism in a number of specific contexts such as human cancers [1–6] or human liver [7–11]. To evaluate tumor metabolism in a human subject, a 13C-enriched substrate can be infused intravenously for a period of 1 to 3 hours prior to tumor resection. The resected tumor tissue is then quickly frozen and subjected to acid extraction to isolate 13C enriched metabolic products, which are then analyzed by MS or 13C NMR or both. The prolonged infusion of substrate ensures steady-state isotopic flow throughout the metabolic network and this simplifies the metabolic analysis considerably. NMR and MS are already standard analytical tools used in metabolomics experiments (i.e., systems biochemistry) so adding 13C detection to a metabolomics analysis is relatively straightforward [12].
An important challenge is to design a 13C metabolic tracing experiment that addresses the clinical or biochemical question of interest. Some questions involving linear pathways such as glycolysis are relatively simple to address with a 13C tracer but due to the complexity of carbon flow in pathways intersecting in the tricarboxylic acid (TCA) cycle, selection of the optimal substrate and its labeling pattern may not be so obvious. Given that 13C metabolic tracing experiments are expensive, metabolic simulations can be helpful for designing informative studies. The purpose of this article is to illustrate that the distribution of 13C in product metabolites after passage of a 13C-enriched tracer through the TCA cycle and intersecting metabolic networks can be predicted using the tcaSIM software package. tcaSIM was originally written in the C programming language and has been in use for a number of years [13,14]. However, the display components eventually became incompatible with later operating systems. This issue led to the decision to recode the software in MATLAB, a widely used scientific computing language that runs on multiple operating systems. In the process, the user interface was improved and the options for displaying and archiving the predicted results were expanded. This article describes the key features of this new version of tcaSIM and demonstrates the utility of using tcaSIM to design a typical 13C metabolic tracing experiment to examine, as a simple example, liver and tumor metabolism using either MS or NMR as analysis tools.
2. Terminology used in 13C Metabolic Tracing Experiments
Isotopomer, a contraction of isotope and isomer, refers to all possible combinations of 13C enrichment in a particular molecule. For example, lactate with three carbons has 23 = 8 possible isotopomers and glutamate, which has five carbons, has 25 = 32 possible isotopomers [15,16]. Various isotopomer notations have been used. All notations identify particular carbon atoms in metabolites with numbers according to conventions developed for organic chemistry and biochemistry. For example, the carboxylic carbon in lactate is identified as C1 and the methyl carbon of lactate is identified as C3. The International Union for Pure and Applied Chemistry (IUPAC) notation uses [1,2,3-13C3]lactate for uniformly-labeled lactate, [3-13C1] for methyl-labeled lactate. Alternate shorthand notations as shown below have also come into use. The 8 lactate isotopomers can be written as
| IUPAC Notation [12C]lactate |
Alternate Notation Lac 12C |
|
| [1-13C1]lactate | Lac 13C1 | |
| [2-13C1]lactate | Lac 13C2 | |
| [1,2-13C2]lactate | Lac 13C1,2 | |
| [3-13C1]lactate | Lac 13C3 | |
| [1,3-13C2]lactate | Lac 13C1,3 | |
| [2,3-13C2]lactate | Lac 13C2,3 | |
| [1,2,3-13C3]lactate | Lac 13C1,2,3 |
where Lac 12C is defined as lactate having no 13C enrichment and the subscripts identify which carbon number is labeled with 13C. In computer software, the following notation is convenient for the purposes of identifying specific isotopomers:
Lac_ooo
Lac_xoo
Lac_oxo
Lac_xxo
Lac_oox
Lac_xox
Lac_oxx
Lac_xxx
In this notation ‘o’ denotes 12C and ‘x’ denotes 13C; carbon numbers are read from left to right. Ordering the isotopomers in this way is convenient because the isotopomers can be identified numerically with lactate isotopomer number 1 being Lac_ooo (uniformly 12C) and lactate isotopomer number 8 being Lac_xxx (uniformly 13C). Each variable refers to the relative fraction of that isotopomer in the total lactate pool. The sum of the concentrations of all 8 isotopomers is normalized to 1. This numbering system is identical to earlier usage [14,17].
The term isotopologue refers to all possible combinations of 13C and 12C that will result in unique mass numbers that can be readily distinguished by MS. For example, for a molecule with n carbons, there are n + 1 isotopologues. Lactate has four isotopologues [15] that are related to the lactate isotopomers in the following groups:
For more complex molecules such as glutamate or glucose, more detailed information about groups of isotopomers can be obtained by tandem MS methods [18]. In this example we see that MS quantifies some isotopomers uniquely (such as [12C]lactate or [1,2,3-13C3]lactate) and also quantifies some groups of lactate isotopomers as a fraction of the total lactate pool.
Multiplets refer to components of a 13C NMR resonance that arise from nearest neighbor spin-spin (J) couplings. In a 13C NMR spectrum of glutamate for example, each of the five carbons of glutamate resonates at a different frequency and each carbon resonance can appear as a multiplet if one or two of adjacent carbons atoms is labeled with 13C. A single signal (a singlet) identified as GluC4S at the glutamate C4 frequency is produced by all glutamate isotopomers labeled in position 4 but not 3 or 5. In other words, the GluC4S multiplet is produced by a group of isotopomers: Glu_oooxo, Glu_xooxo, Glu_oxoxo, Glu_xxoxo.
Two signals symmetrically positioned around the C4 frequency (a doublet identified as GluC4D34) reflects spin-spin (J3,4) coupling between carbon 4 and carbon 3 (arising from all isotopomers labeled in position 4 and 3 but not 5). Similarly, a second doublet (GluC4D45) can also be present due to coupling between carbon 4 and carbon 5 (arising from all isotopomers labeled in position 4 and 5 but not 3). A doublet of doublet, or quartet (GluC4Q) arising from all isotopomers labeled in positions 3 and 4 and 5 can also be present. The terms GluC4S, GluC4D34, GluC4D45 and GluC4Q denote the normalized multiplet integrated signal intensities relative to the total signal from the C4 resonance. Thus, GluC4S + GluC4D34 + GluC4D45 + GluC4Q = 1, by definition. The relation between isotopomers and the observed 13C NMR spectrum, variable names, and resonance assignments for other carbons were described previously [14,17,19]. As with MS, in general NMR quantifies groups of isotopomers.
Since 13C is present at about 1.1% natural abundance, most often an investigator may choose to ignore the effects of natural abundant levels of 13C. However, when the concentration or utilization of 13C-enriched substrates is very low, then natural abundance levels of 13C can contribute significantly to a metabolite spectrum. If this is the case the above described notation may be readily used to account for natural abundance. For example, during the oxidation of lactate (input parameters are described below), natural abundance Lac_xoo, Lac_oxo and Lac_oox can each be set to 0.011. The program will automatically provide for natural abundance if chosen by the operator.
As can be seen from the above, an enormously useful feature of both 13C NMR and MS is the capacity to quantify groups of isotopomers. With NMR, this property arises from the 13C-13C spin-spin coupling that create multiplets in the 13C NMR spectrum plus the information from the chemical shift which identifies a particular carbon in the molecular framework. With MS this information arises from isotopologues, and the information yield can be further delineated using tandem MS methods [18]. A major advantage of 13C NMR over MS, arising from the combined information of chemical shift and J coupling, is the simplicity of studying mixtures of 13C-labeled materials. For example, studies of oxidative metabolism [22–24] used complex mixtures of labeled fatty acids, ketones and carbohydrates. By judicious selection of 13C labeling patterns it is possible to resolve substrate oxidation profiles using up to 3 substrates, and by difference from 1, the unlabeled contribution).
Steady-state indicates an experimental condition in which the concentration of all intermediates and the concentration of all 13C isotopomers does not change over time. By definition, the rate of removal of carbon from the metabolic network equals the rate of addition. For relatively simple experimental preparations such as isolated cells or perfused tissues, 13C spectra can be acquired at multiple time points to demonstrate that steady-state has been achieved. Steady-state is typically reached in 30 to 45 min in metabolically active tissues. However, in vivo and especially with human studies, it can be more difficult to assure steady-state. Under those conditions, alternative NMR methods may be used [20,21].
Anaplerosis refers to the group of metabolic pathways that serve biosynthetic needs and allow entry of carbon into the TCA cycle via pathways other than citrate synthase [25–27]. Investigators at the University of California, Berkeley, first developed 14C tracer methods for analysis of anaplerosis via pyruvate carboxylase [28, 29]. The ratio of the pyruvate carboxylase flux relative to the citrate synthase flux was defined as y and this term has subsequently been used by others to express the rate of an anaplerotic pathway. Metabolic pathways that yield succinyl-CoA, a TCA intermediate, are also anaplerotic pathways. For example, metabolism of propionate via propionyl-CoA carboxylase generates succinyl-CoA. Other substrates entering the TCA cycle via α -ketoglutarate can be accommodated by considering succinyl-CoA as the entry to the cycle. For example, if one is planning an infusion study in a cancer patient, possible entry of a labeled glutamine tracer into the TCA cycle via glutaminase can be accommodated in the simulation program by placing the label into the appropriate carbon positions in succinyl-CoA, the downstream product of decarboxylation of α-ketoglutarate. Anaplerosis via pyruvate carboxylation is given the symbol, ypc (or YPC in the interface, see Table 1) while anaplerosis through succinyl-CoA is given the symbol, yS (or YS in the interface). Thus, yS refers to any group of anaplerotic pathways converging on succinyl-CoA which could include glutamine → glutamate → α-ketoglutarate → succinyl-CoA or propionate carboxylation.
Table 1:
Metabolic variables.
| Abbreviation | Reaction | Range |
|---|---|---|
| CS | Citrate synthase flux | 1 |
| PDH | Pyruvate Dehydrogenase flux | 0 to 1 |
| yPC | Pyruvate Carboxylase flux | 0 to ∞ |
| yS | Anaplerotic Flux at Succinyl-CoA | 0 to ∞ |
| PK | Pyruvate Kinase + Malic Enzyme flux | see text |
| ROF | Oxaloacetate-Fumarate Exchange | 0 to 1 |
| RSM | Conserved Orientation Transfer | 0 to 1 |
| GK | Glycerol Kinase flux | 0 to 1 |
| TPI | Triose Phosphate Isomerase | 0 to 1 |
| Eoaa | 4-Carbon Intermediate Pool Sizes | 0 to∞ |
| EaKG | 5-Carbon Intermediate Pool Sizes | 0 to ∞ |
| ECit | 6-Carbon Intermediate Pool Sizes | 0 to ∞ |
Exchanging pools refers to the concept that some TCA intermediates may be in exchange with a much larger metabolite pool that may take time to become enriched. Good examples are the exchange of oxaloacetate with aspartate or α-ketoglutarate with glutamate. The presence of a large exchanging pool does not influence the steady-state distribution of 13C but the rate of approach to steady-state is delayed. The simulation allows the user to select any pool size (from 0 to ∞) for an exchanging 4 carbon pool, 5 carbon pool or 6 carbon pool; the variables are EOAA, EAKG and ECIT, respectively. Although this capability is not emphasized in the examples, it is very useful for visualizing the evolution of 13C- labeling as a function of time under various metabolic conditions.
3. Motivation for Modeling Using Computational Iterations
In the classical studies of tracer distribution in the TCA cycle [25–27], each turn of the cycle was envisioned as a step in a series of multiplications. Under some substrate labeling conditions the concentrations of isotopomers can be expressed using a relatively simple Taylor series. An alternative approach, termed “input-output analysis” focused on conservation of mass. These resulting analytical expressions enabled more sophisticated analysis [30–33]. In both methods, each carbon position in an intermediate of the TCA cycle is fed by an upstream pool. For example, selective 14C labeling of the C1 of acetyl-CoA will always appear in the C5 of α-ketoglutarate. This approach yields expressions that are suitable for fitting 14C tracer data obtained from carbon-by-carbon degradation of molecules such as glucose or amino acids. Thus, five possible 14C specific activities can in principle be measured by step-by-step chemical degradation of α-ketoglutarate.
The results from 13C tracer studies require a different approach because the patterns of labeling in product molecules contain more information than simple site-specific enrichment. This requires a more comprehensive approach to evaluate the impact of exchanges occurring in the TCA cycle on the relative concentration of all isotopomers in a specific metabolite pool. For example, since α-ketoglutarate has 5 non-equivalent carbons, there are 25 = 32 possible isotopomers. Managing the combinations of 32 possible isotopomers quickly becomes unwieldy for analytical expressions but fortunately matrix mathematics is ideally suited to evaluate the distribution of carbon in the TCA cycle. Using this approach, changes in isotopomer concentrations may be specified as:
where A is a matrix describing the activity of various pathways that feed substrate into the TCA cycle and the 13C enrichment in each substrate of those pathways. The column vector, x, consists of 32 rows and each element represents the relative concentration of one of the isotopomers of α-ketoglutarate at a particular time, t [13,17,19]. This approach offers two advantages. First, at steady-state xt = xt+1 and it is a simple matter to calculate the relative concentration of each isotopomer, given the values of each element of A. Second, if the relative concentrations of all isotopomers can be calculated, then the resulting mass spectrum, 1H NMR spectrum, 13C NMR spectrum, etc., are easily calculated. The above approach is computationally simple and applicable to complex situations such as the possible entry of 13C via propionyl-CoA, acetyl-CoA or pyruvate carboxylase.
The mathematics becomes more challenging under certain more complex metabolic conditions. For example, the carbon used for glucose synthesis in the liver is provided, for the most part, by oxaloacetate derived from carboxylation of pyruvate (an anaplerotic pathway). In 1971, Friedman and colleagues reported that the futile cycle,
is active in liver [34,35]. This combined reaction pathway includes three well-known enzymes, pyruvate carboxylase (PC), phosphoenolpyruvate carboxykinase (PEPCK) and pyruvate kinase (PK). One consequence of flux in this pathway is that a 13C label in oxaloacetate (OAA) may be transferred to pyruvate which can then re-enter the TCA cycle as either OAA after carboxylation, or as acetyl-CoA after decarboxylation in the pyruvate dehydrogenase (PDH) reaction. Flux through the malic enzyme (malate → pyruvate) would have the same effect. The terms “pyruvate cycling” or “pyruvate recycling” have been used to describe the conversion of a 4-carbon intermediate of the TCA cycle to pyruvate, followed by re-entry of pyruvate back into cycle intermediates via PC or PDH. In tcaSIM, we use “PK” to refer to generation of pyruvate by decarboxylation of either malate or OAA because the dominant pathway in gluconeogenic tissues is via PEPCK and PK, keeping in mind that activity of the malic enzyme and perhaps other enzymes in certain tissue are also possible. Most investigators focused on liver metabolism have included the activity of this combined pathway, although it is often described only as pyruvate kinase (PK) [30,32,36–42]. If PK or the malic enzyme are active, then the 13C labeling in pyruvate, which feeds both PDH and PC, is sensitive to labeling in the medium plus pyruvate cycling. This generates a more complex situation that is generally less suitable for matrix algebra because the elements in A become sensitive to the concentrations of isotopomers of TCA cycle intermediates [43].
4. tcaSIM Simulation Processes
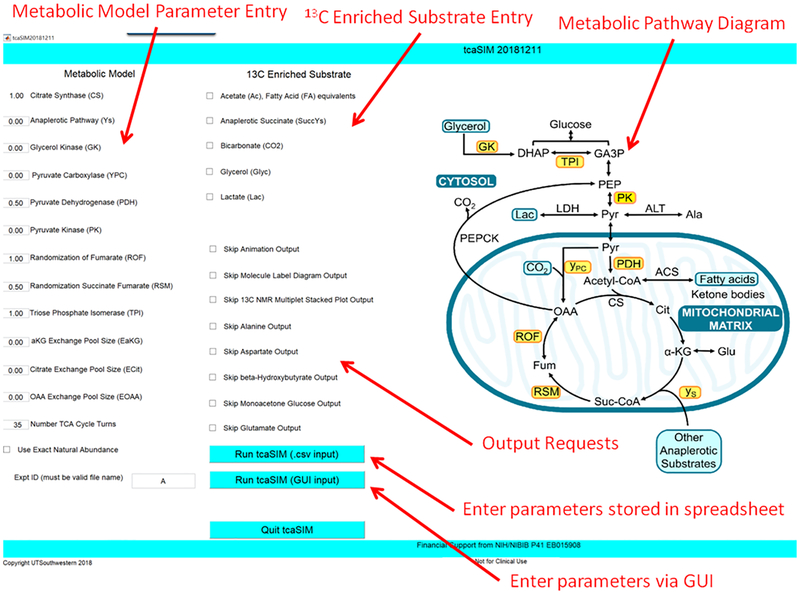
The primary user interface for the updated tcaSIM is provided in Figure 1. The interface includes user input fields for 1) Metabolic model parameters, 2) 13C-enriched substrate and its labeling pattern and 3) selection of which types of output are required.
Figure 1:
tcaSIM User Interface
tcaSIM simulates flow of carbon isotopic tracer atoms through the TCA cycle and connected networks. It uses a finite element approach in which the elements model the behavior of individual enzymes or small groups of enzymes at a single time epoch. Isotopic transformations resulting from all of the biochemical reactions in the TCA cycle and its connected metabolic networks are modeled based on the known reaction chemistry. For instance, citrate synthase (CS), which catalyzes condensation of oxaloacetate (OAA) and acetyl-CoA (AcA) to form citrate, produces the following carbon atom transformations:
| OAA C1 | → | citrate C6 |
| OAA C2 | → | citrate C3 |
| OAA C3 | → | citrate C2 |
| OAA C4 | → | citrate C1 |
| AcA C1 | → | citrate C5 |
| AcA C2 | → | citrate C4 |
This knowledge enables one to write 64 equations that calculate the relative concentrations of each 64 possible citrate isotopomers from the relative concentrations of the 16 possible OAA isotopomers and the 4 possible acetyl-CoA isotopomers. The relative concentration of each citrate isotopomer results from multiplying the relative concentration of the relevant acetyl-CoA isotopomer and oxaloacetate isotopomer. For example, the concentration of [1,3,5-13C3]citrate relative to total citrate is simply the product of [1-13C]acetyl-CoA (normalized to total acetyl-CoA) times [2,4-13C2]oxaloacetate (normalized to total OAA). Analogous expressions that define isotopic interconversion can be written for all isotopomers of all substrates, intermediates and products. Each of these expressions is encapsulated within the tcaSIM software.
At certain points in the metabolic network there is a convergence of reaction products. For instance succinyl-CoA is formed from α-ketoglutarate via α-ketoglutarate dehydrogenase and also from various anaplerotic substrates via the yS reaction. At such points of convergence the software models the formation of two product pools using the above described methodology and then mixes these product pools according to the relative rates of the two contributing input flows.
One purpose of tcaSIM is to allow the investigator to set up a series of metabolic conditions that plausibly describes the system of interest. For example, one could simulate cardiac metabolism by setting all anaplerotic fluxes to zero and predict the NMR and MS of any metabolite as the heart switches from fatty acid to carbohydrate oxidation. For liver studies, the additional complexity of carbons flowing in and out of the TCA cycle due to gluconeogenesis can be added. Obscure or controversial details such as activity of pyruvate kinase, “backwards” scrambling of oxaloacetate into fumarate, or a variable extent of free rotation of succinate and fumarate (termed orientation-conserved transfer, 44–47] are easily simulated. All pathway fluxes have values relative to TCA cycle turnover (i.e., citrate synthase flux), which is defined as 1. The range of each variable depends on the specific pathway. For example, the range of PDH is 0 to 1 because flux through PDH by definition cannot be greater than flux through citrate synthase. On the other hand, anaplerotic fluxes, yS and yPC, have a range of 0 - ∞ because input of carbons into the cycle is always balanced by equal export of carbons. Hence, gluconeogenesis can be significantly higher than TCA cycle flux without altering the steady-state levels of TCA cycle intermediates. The definitions of the variables describing metabolic pathways or reactions and their ranges are summarized in Table 1.
The model assumes that the concentrations of all substrates, intermediates and products shown in the figure are in steady state. The steady state assumption serves to make some of the rates dependent on other rates. For example it is assumed that the pyruvate concentration (i.e. the total of all 8 possible pyruvate isotopomers) is in steady state, and therefore the inward flows into the pyruvate pool equal the outward flows from the pool.
In this expression, LDH refers to pyruvate generated from lactate. This assumption makes one of the above four rates dependent completely on the other three, LDH = yPC + PDH - PK, and therefore the user enters the values of yPC, PDH and PK to be used in the simulation but not LDH because it can be calculated from the available input. If the operator input values for yPC, PDH and PK forces LDH to be < 0, the software reports the error.
5. Experimental Variables: Available Substrates and Labeling Patterns
The user can designate 13C enrichment patterns at five different input molecules: lactate, fatty acids, glycerol, CO2, and molecules equivalent to succinyl-CoA. It then performs a step-wise finite element simulation of isotopic transformations of the type described above for each of the reactions in the TCA cycle and its connected metabolic network to determine the isotopomer distributions for products β-hydroxybutyrate, glutamate, aspartate, alanine and glucose for a single time epoch, defined as the time needed to complete one turn of the TCA cycle. In order to accomplish this, the software keeps records of the relative concentrations of all possible isotopomers for all the substrates, intermediates and products. It then repeats the simulation for additional time epochs (TCA cycle turns) to allow the simulated isotopomer distributions in the products to reach steady state. Typical simulations use 35 TCA cycle turns to ensure isotopic steady-state.
In tcaSIM, lactate refers to any molecule provided to a system that can be metabolized to pyruvate. Consequently, if the investigator wants to study metabolism of alanine, lactate, glucose or pyruvate, the input is lactate. This convention was chosen because pyruvate may be an input to the system as well as a product of PEPCK. tcaSIM assumes that lactate is the exclusive exogenous input to the metabolically active pool of intracellular pyruvate. Up to 8 different isotopomers of lactate may be entered, although for practical purposes most investigators will select a simple molecule such as [U-13C3]lactate or [3-13C1]lactate or [2-13C1]lactate. Mixtures of inputs may be examined for substrate competition experiments. As is stated above, by definition, the sum of all possible lactate input isotopomers is 1.
Fatty acids and ketones are considered equivalent inputs to tcaSIM because both yield only acetyl-CoA. There are only 4 possible combinations of labeling in acetyl-CoA. Since there are many possible 13C enriched fats and ketones available from commercial sources, including [U-13C] fatty acids, alternately-labeled fatty acids such as [2,4-13C2]butyrate or [1,3-13C2]butyrate, or asymmetrically-labeled fatty acids such as [1,2,3,4-13C4]octanoate, the user must determine how the fat or ketone will be metabolized to acetyl-CoA. For example, [U-13C16]]palmitate will generate exclusively [1,2-13C2]acetyl-CoA. [2,4,6,8-13C4]octanoate will generate exclusively [2-13C]acetyl-CoA, and [1-13C]acetoacetate will generate a 1:1 mixture of [1-13C]acetyl-CoA and unlabeled acetyl-CoA.
13C-enriched glycerol tracers are also available for study. Like lactate, glycerol has 3 carbons but, because of symmetry, glycerol has only 6 unique isotopomers: unlabeled glycerol, [1-13C]glycerol, [2-13C]glycerol, [1,2-13C2]glycerol, [1,3-13C2]glycerol and [U-13C3]glycerol.
The user may also elect to use 13CO2 as a substrate in a simulation. The fraction of 13CO2 must be in the range of 0 to 1.
Finally, there are 16 possible labeling patterns entering the TCA cycle via succinyl-CoA. This pathway, YS in tcaSIM, is used to simulate multiple possible anaplerotic reactions. Propionate is avidly metabolized in mammalian systems after carboxylation to succinyl-CoA. Amino acids such as glutamate and glutamine enter the TCA cycle at the level of α-ketoglutarate. Since the product of decarboxylation is succinyl-CoA, anaplerotic entry of [3,4,5-13C3]]glutamate would be modeled as [2,3,4-13C3]]succinyl-CoA. In astroglia, succinate semialdehyde is metabolized via succinate and again would be modeled as entry via succinyl-CoA.
6. Results Provided by tcaSIM
The core of tcaSIM is the calculation of all isotopomers of all TCA intermediates and exchanging pools at every turn of the cycle. This generates an enormous amount of information but only selected aspects are useful for most purposes. Therefore tcaSIM produces various forms of output that characterize the simulation results for each of the five output molecules (β-hydroxybutyrate, glutamate, aspartate, glucose and alanine). Those outputs include: 1) plots and tables of the isotopomer distribution in each product, 2) plots of simulated 13C NMR spectra, 3) tables of 13C NMR multiplet intensities and 4) plots and tables of the isotopologue distributions. All plots are stored as electronic ‘picture’ or ‘video’ files (.png and .mp4 formatted files). All tables are stored as electronic comma separated value (.csv) files that are readable by common spreadsheet software such as Microsoft excel. All possible isotopomers for the above output metabolites are made available by tcaSIM. This information is useful for teaching purposes and gaining an understanding of the program. For example, interested users should be able to take the listing of all glutamate isotopomers and calculate the predicted mass spectrum or NMR spectrum. In general, however, this is unnecessary since tcaSIM provides such information. Site specific 13C fractional enrichments is also provided. This information is useful for simulating traditional 14C experiments because those studies, if carbon-by-carbon degradation was performed, provide specific enrichment at each site.
7. Examples
tcaSIM is designed to answer specific questions related to the design of metabolic tracing experiments. For instance, one could ask ‘Does a change in flux in one pathway cause a detectable change in tracer distribution in a product?’ Moreover an investigator may be interested in the effects of a drug on, for example, PDH flux in liver, or the effect of a nutritional supplement on flux through propionyl-CoA carboxylase or the effects of variable expression of an enzyme on activity of the network. The investigator must also explicitly decide whether certain pathways are active or inactive. For example, should flux through PDH and PC allowed or should one be completely inactive? Some metabolic processes are controversial such as “backwards” scrambling of oxaloacetate after pyruvate carboxylation (pyruvate + CO2 → oxaloacetate → fumarate and succinate → oxaloacetate) or orientation conserved transfer, meaning the failure of free rotation of the symmetric intermediates, succinate and fumarate, as they pass through the TCA cycle. tcaSIM assumes full backwards scrambling and free reorientation of succinate and fumarate by default, but the investigator has the option to alter these defaults as a means of exploring these possible effects.
Simulation of an experiment in which PDH activity might vary
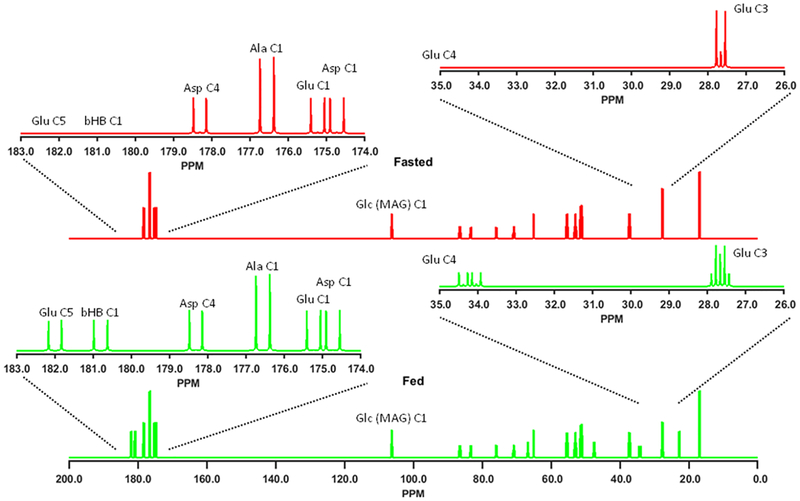
This example illustrates how the 13C NMR spectrum of the acid extract of a liver perfused with [U-13C3]lactate is predicted to vary depending upon whether the liver is obtained from a fasted or fed animal. In general, these simulations assume that liver is poised to undergo gluconeogenesis in both fed and fasted conditions (ie yPC is non-zero), but that fasted liver does not oxidize pyruvate (PDH = 0.0) while fed liver does oxidize pyruvate (PDH = 0.4 means that 40% of the oxidized substrate comes from lactate and the remaining 60% comes from fats or ketone bodies). The simulated 13C NMR spectra shown in Figure 2 illustrate many differences that would be quite evident; the Glu C3 multiplet patterns would differ as would the ratio of the Glu C4 to Glu C3. Furthermore, both bHB C1 and Glu C5 would become labeled in the fed condition due to label flux into the TCA cycle via PDH but not get labeled under the assumed fasting conditions. Note that tcaSIM makes no predictions about relative metabolite concentrations, i.e., the amount of bHB would be expected to be higher under fasting conditions, it simply predicts the isotopomer distribution in those metabolites. These differences could be predicted given a sophisticated understanding of flow of 13C isotopic tracers within the TCA cycle but the point of this example is that tcaSIM enables an investigator who does not have this sophisticated understanding to form a quantitative hypothesis about how the 13C NMR spectra from two metabolic conditions will differ.
Figure 2:
13C NMR spectra simulated by tcaSIM for liver perfused with uniformly enriched lactate under fed (green) and fasted conditions (red). The fed simulation used Ys = 0.0, GK = 0.0, yPC = 3.0, PDH = 0.4, PK = 0.0, ROF = 1.0, RSM = 0.5 with 35 TCA cycle turns. The fasted simulation used the same metabolic model parameters except PDH = 0.0. The inset spectra display the Glu C4 and Glu C3 region of the spectra (26.0 to 35.0 ppm) and the 174.0 to 183.0 ppm regions for both simulations. The simulated spectra assumed equimolar concentrations of Alanine (Ala), Aspartate (Asp),β-hydroxybutyrate (bHB), Glucose (Glc) and Glutamate (Glu). Glucose is typically processed to form monoacetone-glucose (MAG).
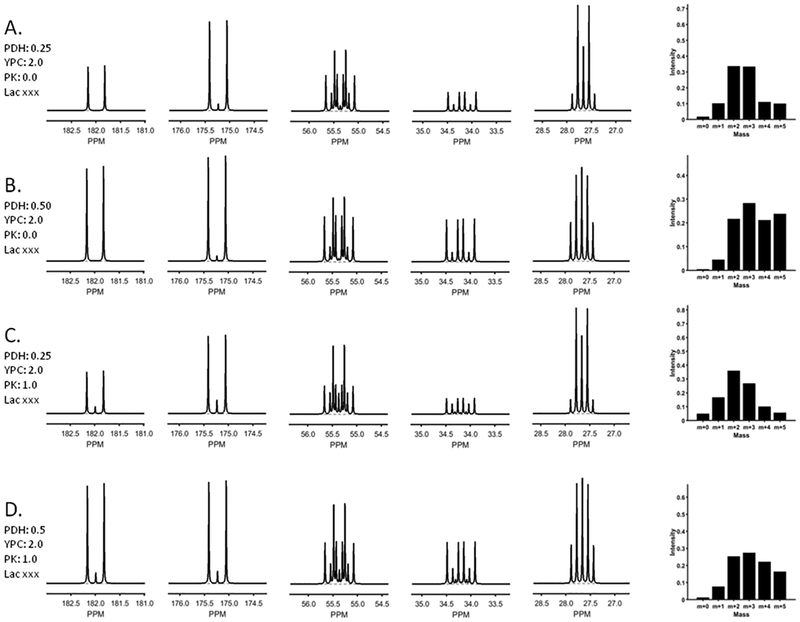
Figure 3 illustrates how the glutamate 13C NMR spectrum and MS spectra would change with increasing PDH. A comparison of Figures 3A and 3B illustrate the easily identified differences in both the 13C NMR spectrum and MS spectrum when PDH is increased from 0.25 to 0.50 in the presence of gluconeogenesis (yPC = 2.0). There are readily identifiable changes in the multiplets in Glu C5, Glu C4 and Glu C3 but not in Glu C2. The MS spectra are also sensitive to changes in PDH. A comparison of Figures 3A with 3C or 3B with 4D show that the addition of pyruvate cycling (PK > 0) yields only subtle changes in the 13C NMR spectra while the MS spectra are more sensitive to the presence of PK.
Figure 3:
13C NMR and MS simulated by tcaSIM for perfused liver having variable PDH and PK fluxes. Other key conditions are shown in the insets.
Figure 4:
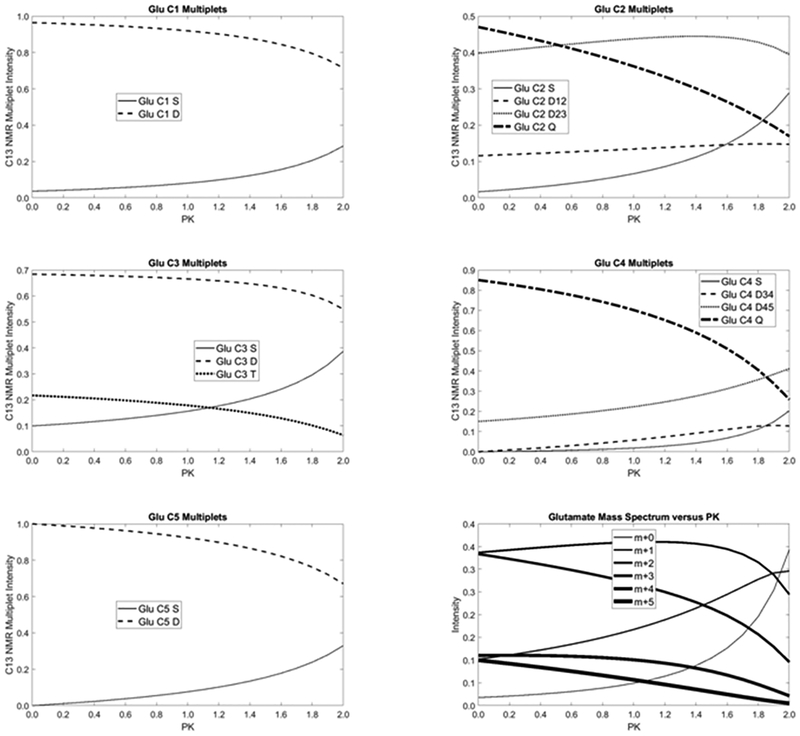
tcaSIM simulations of pyruvate kinase dependence of glutamate 13C NMR and mass spectra for perfused liver. The plots illustrate how the measured glutamate signals depend on PK. Note that tcaSIM produced the results for each of the 21 simulation used to generate these plots, but it does not produce these plots. This was done by writing a small MATLAB program that read the results of the multiple tcaSIM simulations, which were stored as .csv spreadsheet files and assembling/plotting the information.
Simulation of an experiment in which both PDH and PK activity might vary
The influence of small changes in pyruvate cycling on the glutamate 13C NMR multiplets and on the MS spectrum of glutamate are further illustrated by the plots shown in Figure 4. Such plots are easily generated using tcaSIM by changing one variable, in this case PK, by incremental amounts while keeping all other fluxes fixed. This example shows that most 13C NMR multiplets are rather insensitive to changes in PK < ~1 except for Glu C2Q and Glu C4Q while several of the MS peaks appear to be more sensitive to this flux. Such predictions are useful for experimentalists because if the goal is to determine PK quantitatively, then the user would know which analytical measurements that one should focus on measuring.
tcaSIM is designed to perform batched simulations in which metabolic conditions or substrates are altered. This is most easily accomplished by entering the metabolic conditions and substrates via an electronic spreadsheet rather than manually entering the information for each of the conditions through the user interface. To generate the results shown in Figure 4 a batch of 21 tcaSIM simulations was performed. For this batch PK was varied from 0.0 to 2.0 in steps of 0.1 for a perfused liver in a fed state (PDH = 0.25) with all other model parameters held constant. The figure illustrates how all 15 glutamate 13C NMR multiplets and the 5 glutamate isotopologues quantitatively depend on PK flux. This example illustrates that none of the dependences are simple linear functions. Some of the multiplets increase with increasing PK while others decrease. The same is true for the isotopologues. The figure also illustrates that it is possible to design an experiment from which a quantitative estimate of the PK flux can be derived.
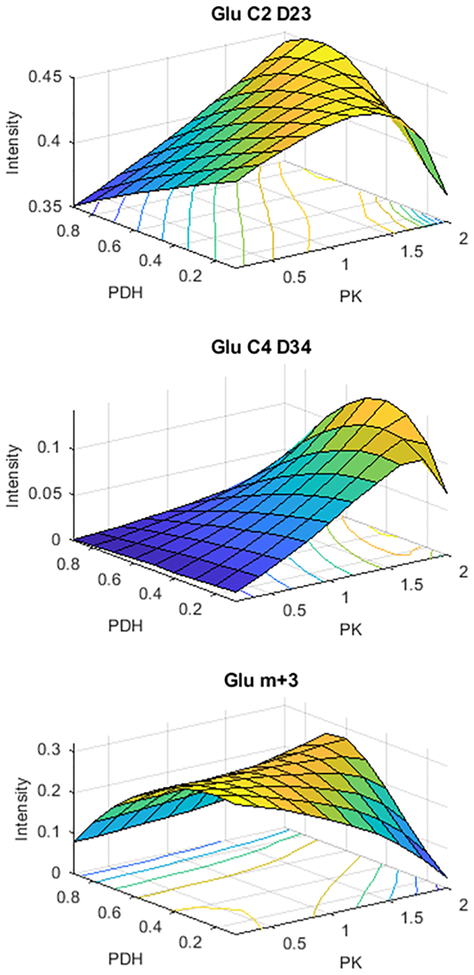
Quantitative estimatation of the PK flux would not be trivial because it would be necessary to jointly estimate PK flux and PDH flux with such an experiment. A batch of tcaSIM simulations was performed to illustrate this issue. In this batch of simulations, PK and PDH were both varied. PDH was varied from 0.0 to 1.0 in regular steps of 0.1. PK was varied from 0.0 to 2.0 in regular steps of 0.2. Figure 5 shows 3-dimensional surface plots that illustrate how two Glu NMR multiplets (GluC2D23 and GluC4D34) and the Glu m+3 isotopologue depend jointly on PK and PDH fluxes. These particular multiplets and isotopologues were chosen for presentation because they show the strongest PK and PDH dependence. However, it should be understood that much more data is produced by tcaSIM than is shown in this figure. In this batched simulation, tcaSIM calculated the PK and PDH dependencies for all multiplets and all isotopologues of glutamate, aspartate, alanine, hydroxybutyrate and glucose. These plots show that the there is no simple relationship between the measured multiplets or isotopologues and the PK and PDH fluxes. In fact some combinations of PDH and PK produce multiplets or isotopologues that are indistinguishable from those produced by other combinations of PDH and PK.
Figure 5:
Three-dimensional plots illustrating the dependence of selected glutamate 13C NMR multiplets and mass spectral intensities on PHD and PK flux. The data shown was derived from a series of tcaSIM simulations in which the PDH and PK flux was varied for perfused liver. Note that tcaSIM produced the results for each of the > 100 simulations used to generate these plots, but it does not produce these plots. This was done by writing a small MATLAB program that reads the results of the multiple tcaSIM simulations, which were stored as .csv spreadsheet files, and assembling/plotting the information.
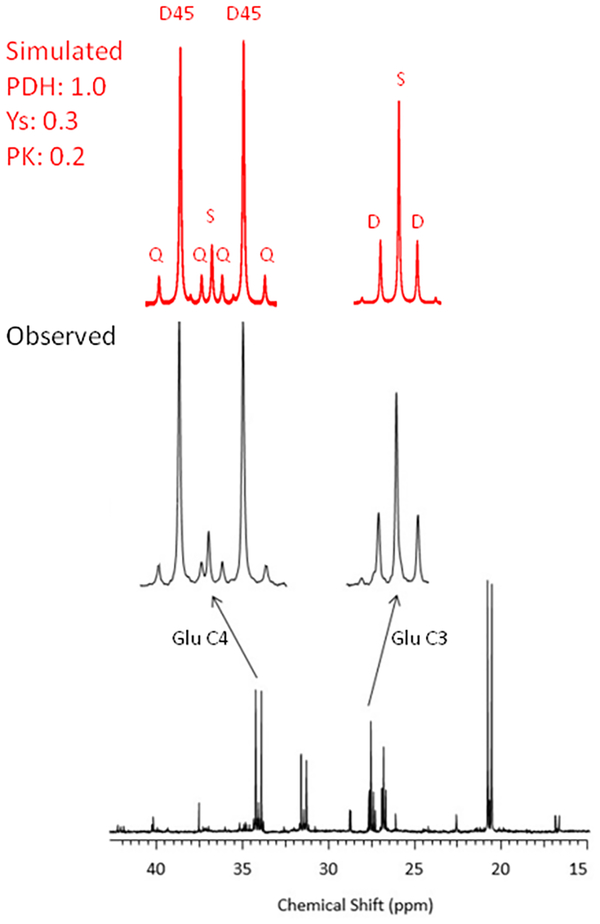
Simulation of an experiment involving in vivo human glioblastoma metabolism
Figure 6 provides an example illustrating that tcaSIM can be used to accurately model metabolism taking place in vivo in a human glioblastoma multiforme (GBM). The concepts underlying the example are provided in Maher et al [5]. In this experiment a human GBM patient received an intravenous infusion of uniformly enriched 13C-glucose prior to tumor resection. 13C- metabolites resulting from glucose metabolism within the tumor were analyzed by 13C NMR as described above. 13C enrichment in glutamate directly shows that PDH is active and the appearance of a D45 in glutamate C4 shows that the acetyl-CoA entering the TCA cycle was derived largely from [U13C6]glucose. The remaining 13C multiplet patterns in C4 and C3 are only consistent with active anaplerosis and pyruvate cycling in GBM. A batch of tcaSIM simulations was performed in which the PK (pyruvate cycling) and Ys (anaplerotic flux) were varied. The simulation that best matched the observed Glu C3 and C4 13C NMR multiplet intensities was selected for presentation in the figure. The close match between observed and simulated 13C NMR multiplet patterns illustrates that tcaSIM can be used to estimate anaplerosis and pyruvate cycling rates relative to TCA cycle flux in vivo in human GBM.
Figure 6:
In black is shown a 13C NMR spectrum of an extract of a human glioblastoma multiforme (GBM) that was resected following intravenous infusion of uniformly enriched 13C glucose as described in Maher et al [5]. In red is shown the corresponding 13C NMR simulation that matched the observed spectrum. This example illustrates that tcaSIM can be used as tool for quantitative estimation of pyruvate cycling and anaplerosis rates in human GBM.
9. Summary
Over many years, our center has developed and sustained a software tool named tcaSIM that can be used to predict results of complex metabolic tracing experiments that use 13C isotopic tracers. tcaSIM has two general uses. 1) It can serve as an educational tool that helps one learn about intermediary metabolism and the methodology associated with metabolic studies that use 13C tracers and 2) It can be used to predict outcome of complex metabolic experiments that use 13C tracers.
The use of tcaSIM as a teaching tool has not been emphasized in the present work but from the figures presented, it can be appreciated that tcaSIM helps to elucidate details related to the complexity of metabolic pathways and that the 13C isotopomer distribution is easily obtained from 13C NMR multiplets and MS data.
The second use of tcaSIM, which has been emphasized in this presentation, is the simulation of metabolic transformations of any 13C-enriched substrate through complex networks of metabolic pathways containing multiple transformations and variable rates. This use permits the modeling of experiments that are too complex for even those individuals who have detailed knowledge of metabolism to predict an outcome. One component of the present study, the illustration of PK as a variable, suggests that it should be possible experimentally to resolve a controversy related to whether PK is active in liver undergoing active gluconeogenesis [48, 49]. Furthermore, the ability to predict outcome of metabolic tracing experiments allows one to design experiments that are more efficient, precise and potentially less costly. The cost aspect is particularly important because of the expense of 13C enriched materials and the time and effort needed to set up and analyze a metabolic tracer experiment.
Much of the discussion in this paper suggests that only glutamate is subjected to isotopic analysis with 13C NMR and/or MS. The focus on glutamate arises largely because this amino acid is present at high levels in most biological tissues and it produces high quality NMR and MS data. However, in certain metabolic tracing experiments, particularly those done with living human subjects where blood rather than tissue is only available for analysis, an evaluation of other circulating molecules such as glucose or β-hydroxybutyrate is more appropriate. tcaSIM recognizes this need by providing output for each of these metabolites. tcaSIM also provides simulation of aspartate in case it is desired to evaluate 13C labeling of a 4-carbon TCA cycle derived metabolite. This is especially helpful for labeled pyruvate entering through PC and if one is interested in asking a question about backward flux from OAA to malate to fumarate which, if fast, should equalize labeling of carbons 1 and 4 and carbons 2 and 3.
The current version was written in MatLab and therefore can be run on multiple operating systems including Windows 10 and Mac OS. The software is available free-of-charge to interested members of the scientific community at http://invivometabolism.org/tca.html. The authors are evaluating approaches for providing tcaSIM functional capabilities as an online cloud service.
Acknowledgements
This study was supported by NIH grants DK058398, HL034557 and EB015908.
Footnotes
Conflict of Interest
Jeffry R. Alger has no conflicts of interest related to this paper.
A. Dean Sherry has no conflicts of interest related to this paper.
Craig R. Malloy has no conflicts of interest related to this paper.
References
- [1].Sellers K; Fox MP; Bousamra M 2nd; Slone SP; Higashi RM; Miller DM; Wang Y; Yan J; Yuneva MO; Deshpande R; Lane AN; Fan TW. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest 2015, 125(2), 687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Faubert B; Li KY; Cai L; Hensley CT; Kim J; Zacharias LG; Yang C; Do QN; Doucette S; Burguete D; Li H; Huet G; Yuan Q; Wigal T; Butt Y; Ni M; Torrealba J; Oliver D; Lenkinski RE; Malloy CR; Wachsmann JW; Young JD; Kernstine K; DeBerardinis RJ. Lactate Metabolism in Human Lung Tumors. Cell 2017, 171(2), 358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hensley CT; Faubert B; Yuan Q; Lev-Cohain N; Jin E; Kim J; Jiang L; Ko B; Skelton R; Loudat L; Wodzak M; Klimko C; McMillan E; Butt Y; Ni M; Oliver D; Torrealba J; Malloy CR; Kernstine K; Lenkinski RE; DeBerardinis RJ Metabolic Heterogeneity in Human Lung Tumors. Cell 2016, 164(4), 681–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mashimo T; Pichumani K; Vemireddy V; Hatanpaa KJ; Singh DK; Sirasanagandla S; Nannepaga S; Piccirillo SG; Kovacs Z; Foong C; Huang Z; Barnett S; Mickey BE; DeBerardinis RJ; Tu BP; Maher EA; Bachoo RM Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 2014, 159(7), 1603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Maher EA; Marin-Valencia I; Bachoo RM; Mashimo T; Raisanen J; Hatanpaa KJ; Jindal A; Jeffrey FM; Choi C; Madden C; Mathews D; Pascual JM; Mickey BE; Malloy CR; DeBerardinis RJ. Metabolism of [U-13C]glucose in human brain tumors in vivo. NMR Biomed 2012, 25(11), 1234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Courtney KD; Bezwada D; Mashimo T; Pichumani K; Vemireddy V; Funk AM; Wimberly J; McNeil SS; Kapur P; Lotan Y; Margulis V; Cadeddu JA; Pedrosa I; DeBerardinis RJ; Malloy CR; Bachoo RM; Maher EA Isotope Tracing of Human Clear Cell Renal Cell Carcinomas Demonstrates Suppressed Glucose Oxidation In Vivo. Cell Metab 2018, pii: S1550–4131(18), 30463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin ES; Browning JD; Murphy RE; Malloy CR Fatty Liver Disrupts Glycerol Metabolism in Gluconeogenic and Lipogenic Pathways in Humans. J Lipid Res 2018, 59(9), 1685–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Neeland IJ; Hughes C; Ayers CR; Malloy CR; Jin ES Effects of visceral adiposity on glycerol pathways in gluconeogenesis. Metabolism 2017, 67, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jin ES; Sherry AD; Malloy CR An oral load of [13C3]glycerol and blood NMR analysis detect fatty acid esterification, pentose phosphate pathway and glycerol metabolism through the tricarboxylic acid cycle in human liver. J Biol Chem 2016, 291(36), 19031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sunny NE; Parks EJ; Browning JD; Burgess SC Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab 2011, 14(6), 804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jones JG; Solomon MA; Cole SM; Sherry AD; Malloy CR An integrated 2H and 13C NMR study of gluconeogenesis and TCA cycle flux in humans. Am J Physiol Endocrinol Metab 2001. 281(4), E848–56. [DOI] [PubMed] [Google Scholar]
- [12].Jeffrey FM; Rajagopal A; Malloy CR; Sherry AD 13C-NMR: a simple yet comprehensive method for analysis of intermediary metabolism. Trends Biochem Sci 1991, 16(1), 5–10. [DOI] [PubMed] [Google Scholar]
- [13].Malloy CR; Sherry AD; Jeffrey FM Analysis of tricarboxylic acid cycle of the heart using 13C isotope isomers. Am J Physiol 1990, 259(3 Pt 2), H987–95. [DOI] [PubMed] [Google Scholar]
- [14].Fan TW; Lane AN Applications of NMR spectroscopy to systems biochemistry. Prog Nucl Magn Reson Spectrosc 2016, 92–93, 18–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].IUPAC. Compendium of Chemical Terminology, 2nd ed. (the Gold Book). Compiled by McNaught AD and Wilkinsn A. Blackwell Scientific Publications, Oxford: (1997). [Google Scholar]
- [16].London RE 13C labeling in studies of metabolic regulation. Prog NMR Spectrosc 1988, 20, 317–383. [Google Scholar]
- [17].Malloy CR; Sherry AD; Jeffrey FM Evaluation of carbon flux and substrate selection through alternate pathways involving the citric acid cycle of the heart by 13C NMR spectroscopy. J Biol Chem 1988, 263(15), 6964–71. [PubMed] [Google Scholar]
- [18].Jeffrey FM; Roach JS; Storey CJ; Sherry AD; Malloy CR. 13C isotopomer analysis of glutamate by tandem mass spectrometry. Anal Biochem 2002, 300(2), 192–205. [DOI] [PubMed] [Google Scholar]
- [19].Malloy CR; Sherry AD; Jeffrey FM Carbon flux through citric acid cycle pathways in perfused heart by 13C NMR spectroscopy. FEBS Lett 1987, 212(1), 58–62. [DOI] [PubMed] [Google Scholar]
- [20].Sherry AD; Malloy CR; Zhao P; Thompson JR Alterations in substrate utilization in the reperfused myocardium: a direct analysis by 13C NMR. Biochemistry 1992, 31(20), 4833–7. [DOI] [PubMed] [Google Scholar]
- [21].Malloy CR; Thompson JR; Jeffrey FM; Sherry AD Contribution of exogenous substrates to acetyl coenzyme A: measurement by 13C NMR under non-steady-state conditions. Biochemistry 1990, 29(29), 6756–61. [DOI] [PubMed] [Google Scholar]
- [22].Moreno KX; Sabelhaus SM; Merritt ME; Sherry AD; Malloy CR. Competition of Pyruvate with Physiological Substrates for Oxidation by the Heart: Implications for Studies with Hyperpolarized [1-13C]Pyruvate. Am J Physiol Heart Circ Physiol 2010, 298(5), H1556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jeffrey FM; Diczku V; Sherry AD; Malloy CR Substrate selection in the isolated working rat heart: effects of reperfusion, afterload, and concentration. Basic Res Cardiol 1995, 90(5), 388–96. [DOI] [PubMed] [Google Scholar]
- [24].Jeffrey FM; Alvarez L; Diczku V; Sherry AD; Malloy CR Direct evidence that perhexiline modifies myocardial substrate utilization from fatty acids to lactate. J Cardiovasc Pharmacol 1995, 25(3): 469–72. [DOI] [PubMed] [Google Scholar]
- [25].Kornberg HL Anaplerotic sequences and their role in metabolism. Essays Biochem 1966, 2, 31. [Google Scholar]
- [26].Owen OE; Kalhan SC; Hanson RW The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 2002, 277(34), 30409–12. [DOI] [PubMed] [Google Scholar]
- [27].Sherry AD; Malloy CR; Roby RE; Rajagopal A; Jeffrey FM Propionate metabolism in the rat heart by 13 C n.m.r. spectroscopy. Biochem J 1988, 254(2): 593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Weinman EO; Strisower EH; Chaikoff IL Conversion of fatty acids to carbohydrate; application of isotopes to this problem and role of the Krebs cycle as a synthetic pathway. Physiol Rev 1957, 37(2), 252–72. [DOI] [PubMed] [Google Scholar]
- [29].Strisower EH; Kohler GD; Chaikoff IL Incorporation of acetate carbon into glucose by liver slices from normal and alloxan-diabetic rats. J Biol Chem 1952, 198(1), 115–26. [PubMed] [Google Scholar]
- [30].Schumann WC; Magnusson I; Chandramouli V; Kumaran K; Wahren J; Landau BR Metabolism of [2-14C]acetate and its use in assessing hepatic Krebs cycle activity and gluconeogenesis. J Biol Chem 1991, 266(11), 6985–90. [PubMed] [Google Scholar]
- [31].Magnusson I; Schumann WC; Bartsch GE; Chandramouli V; Kumaran K; Wahren J; Landau BR Noninvasive tracing of Krebs cycle metabolism in liver. J Biol Chem 1991, 266(11), 6975–84. [PubMed] [Google Scholar]
- [32].Katz J Determination of gluconeogenesis in vivo with 14C-labeled substrates. Am J Physiol 1985, 248(4 Pt 2), R391–9. [DOI] [PubMed] [Google Scholar]
- [33].Sherry AD; Jeffrey FM; Malloy CR Analytical solutions for 13C isotopomer analysis of complex metabolic conditions: substrate oxidation, multiple pyruvate cycles, and gluconeogenesis. Metab Eng 2004, 6(1), 12–24. [DOI] [PubMed] [Google Scholar]
- [34].Friedman B; Goodman EH Jr.; Saunders HL; Kostos V; Weinhouse S Estimation of pyruvate recycling during gluconeogenesis in perfused rat liver. Metabolism 1971, 20(1), 2–12. [DOI] [PubMed] [Google Scholar]
- [35].Freidmann B; Goodman EH Jr.; Saunders HL; Kostos V; Weinhouse S An estimation of pyruvate recycling during gluconeogenesis in the perfused rat liver. Arch Biochem Biophys 1971, 143(2), 566–78. [DOI] [PubMed] [Google Scholar]
- [36].Fernandez CA; Des Rosiers C Modeling of liver citric acid cycle and gluconeogenesis based on 13C mass isotopomer distribution analysis of intermediates. J Biol Chem 1995, 270(17), 10037–42. [DOI] [PubMed] [Google Scholar]
- [37].Des Rosiers C; Fernandez CA; David F; Brunengraber H Reversibility of the mitochondrial isocitrate dehydrogenase reaction in the perfused rat liver. Evidence from isotopomer analysis of citric acid cycle intermediates. J Biol Chem 1994, 269(44): 27179–82. [PubMed] [Google Scholar]
- [38].Di Donato L; Des Rosiers C; Montgomery JA; David F; Garneau M; Brunengraber H Rates of gluconeogenesis and citric acid cycle in perfused livers, assessed from the mass spectrometric assay of the 13C labeling pattern of glutamate. J Biol Chem 1993, 268(6), 4170–80. [PubMed] [Google Scholar]
- [39].Grunnet N; Katz J Effects of ammonia and norvaline on lactate metabolism by hepatocytes from starved rats. The use of 14C-labelled lactate in studies of hepatic gluconeogenesis. Biochem J 1978, 172(3), 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cohen SM; Glynn P; Shulman RG 13C NMR study of gluconeogenesis from labeled alanine in hepatocytes from euthyroid and hyperthyroid rats. Proc Natl Acad Sci USA 1981, 78(1), 60–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Petersen KF; Blair JB; Shulman GI Triiodothyronine treatment increases substrate cycling between pyruvate carboxylase and malic enzyme in perfused rat liver. Metabolism 1995, 44(11), 1380–3. [DOI] [PubMed] [Google Scholar]
- [42].Petersen KF; Cline GW; Blair JB; Shulman GI Substrate cycling between pyruvate and oxaloacetate in awake normal and 3,3’−5-triiodo-L-thyronine-treated rats. Am J Physiol 1994, 267(2 Pt 1), E273–7. [DOI] [PubMed] [Google Scholar]
- [43].Jeffrey FM; Storey CJ; Sherry AD; Malloy CR. 13C isotopomer model for estimation of anaplerotic substrate oxidation via acetyl-CoA. Am J Physiol 1996, 271(4 Pt 1), E788–99. [DOI] [PubMed] [Google Scholar]
- [44].Malaisse WJ; Ladrière L; Zhang TM; Verbruggen I; Willem R Enzyme-to-enzyme channelling of symmetric Krebs cycle intermediates in pancreatic islet cells. Diabetologia 1996, 39(8), 990–2. [DOI] [PubMed] [Google Scholar]
- [45].Malaisse WJ; Zhang TM; Verbruggen I; Willem R Enzyme-to-enzyme channelling of Krebs cycle metabolic intermediates in Caco-2 cells exposed to [2-13C]propionate. Biochem J 1996, 317(Pt 3), 861–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sherry AD; Sumegi B; Miller B; Cottam GL; Gavva S; Jones JG; Malloy CR Orientation-conserved transfer of symmetric Krebs cycle intermediates in mammalian tissue. Biochemistry 1994, 33(20), 6268–75. [DOI] [PubMed] [Google Scholar]
- [47].Sumegi B; Sherry AD; Malloy CR; Srere PA Evidence for orientation-conserved transfer in the TCA cycle in Saccharomyces cerevisiae: 13C NMR studies. Biochemistry 1993, 32(47), 12725–9. [DOI] [PubMed] [Google Scholar]
- [48].Befroy DE; Perry RJ; Jain N; Dufour S; Cline GW; Trimmer JK; Brosnan J; Rothman DL; Petersen KF; Shulman GI Direct assessment of hepatic mitochondrial oxidative and anaplerotic fluxes in humans using dynamic 13C magnetic resonance spectroscopy. Nat Med 2014, 20(1), 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Burgess SC; Merritt ME; Jones JG; Browning JD; Sherry AD; Malloy CR Limitations of detection of anaplerosis and pyruvate cycling from metabolism of [1-13C] acetate. Nat Med 2015, 21, 108–9. [DOI] [PMC free article] [PubMed] [Google Scholar]