Abstract
Setting appropriate conservation measures to halt the loss of biodiversity requires a good understanding of species' habitat requirements and potential distribution. Recent (past few decades) ecological data are typically used to estimate and understand species’ ecological niches. However, historical local extinctions may have truncated species–environment relationships, resulting in a biased perception of species' habitat preferences. This may result in incorrect assessments of the area potentially available for their conservation. Incorporating long-term (centuries-old) occurrence records with recent records may provide better information on species–environment relationships and improve the modelling and understanding of habitat suitability. We test whether neglecting long-term occurrence records leads to an underestimation of species’ historical niche and potential distribution and identify which species are more vulnerable to this effect. We compare outputs of species distribution models and niche hypervolumes built using recent records only with those built using both recent and long-term (post-1500) records, for a set of 34 large mammal species in South Africa. We find that, while using recent records only is adequate for some species, adding historical records in the analyses impacts estimates of the niche and habitat suitability for 12 species (34%) in our dataset, and that this effect is significantly higher for carnivores. These results show that neglecting long-term biodiversity records in spatial analyses risks misunderstanding, and generally underestimating, species' niches, which in turn may lead to ill-informed management decisions, with significant implications for the effectiveness of conservation efforts.
This article is part of a discussion meeting issue ‘The past is a foreign country: how much can the fossil record actually inform conservation?’
Keywords: habitat suitability models, historical ecology, megafauna, natural range, niche, reintroduction
1. Introduction
To avert the ongoing human-induced biodiversity decline, scientists have recently called for conservation efforts to be intensified, including through increased habitat protection and restoration [1]. Data on species' distribution patterns and species assemblages are key to identify candidate areas for conservation [2]. However, distribution patterns have been drastically modified by humans, notably through global extinctions and regional to local extirpations [3,4], and thus contemporary patterns do not necessarily reflect species’ natural distribution and habitat preferences. Analyses of species distributions that tend to ignore these modifications will likely result in a biased understanding of species' biogeography and ecological requirements and generate misleading perceptions of the options available for conservation [5,6]. This phenomenon of spatially shifted baselines poses clear challenges for conservation and management. By providing information on species’ historical rather than current-day relictual distributions, long-term biodiversity data have the potential to improve our understanding of the biogeography of species and participate in setting appropriate spatial and ecological baselines for environmental conservation and restoration [7].
Mammals are one of the most studied taxa, and their current distribution patterns are well known [8]. Historical and prehistoric human-driven global and local extinctions have, however, caused a strong deviation between current and pre-anthropogenic impact diversity patterns, in particular, for large terrestrial mammals (greater than 10 kg) [4]. In South Africa, while relatively few megafaunal extinctions occurred at the end of the Pleistocene compared to other continents [9], habitat loss, competition with livestock and direct exploitation, in particular following European colonization, have resulted in the global extinction of one mammal species in the recent past (the blue antelope Hippotragus leucophaeus, around AD 1800) [10], and the collapse of large mammal diversity in large parts of the country [11,12]. To halt this decline and restore populations, conservation efforts have focused on establishing protected areas and actively managing large mammal populations through reinforcement to increase population viability and reintroductions to re-establish populations within species' historical ranges [13]. Defining species’ historical distributions and suitable habitat is thus a critical aspect for conservation planning in South Africa [14], as it is for most restoration attempts elsewhere [15]. Setting appropriate baselines is even more critical for species that are a high priority for conservation, such as those identified as threatened by the IUCN Red List (25% of mammal species) [16] and those with an important ecological role that can be the focus of restoration efforts. Species at higher trophic levels, and large carnivores in particular, are functionally important to ecosystems and have undergone considerable historical range contractions [17], making them a major focus of conservation and trophic rewilding efforts [18]. It is thus critical to understand the extent to which historical data are needed to inform wildlife conservation and management, for threatened species and large carnivores in particular.
Habitat suitability models (HSMs) [19,20] and n-dimensional hypervolumes [21] are two widely used tools that relate species' occurrences to environmental variables in order to, respectively, map species’ potential distributions in geographical space and characterize species' niches in environmental space. They have notably been used in conservation and management contexts to improve our knowledge of species ranges, support management plans for species’ recovery, prioritize areas for biodiversity protection and predict changes in suitable habitat in response to human impacts [22–24]. HSMs and hypervolume approaches rely on the assumption that the observed geographical distribution of a species reflects its ecological requirements, making them highly contingent on the quality of occurrence records [20,24]. Range contractions that have affected the array of conditions that the species occupy risk truncating species–habitat relationships [25], thus hindering our ability to estimate species niches and predict the distribution of suitable habitat [6]. Failing to consider past local extinction events may thus misguide conservation efforts by overlooking potentially suitable sites for reintroduction or restrict protection to suboptimal habitats [5]. Despite providing useful information on the historical distribution of species, historical written records and museum specimens have long been overlooked in habitat suitability modelling approaches, being perceived as untrustworthy for their intrinsic biases and limitations [26] (but see [27–29]). The development of methods to address sampling biases in HSMs [30–33], however, provides an avenue for more confident incorporation of these records in spatial modelling analyses, and hence in conservation interventions.
Here we investigate how long-term biodiversity records can contribute to setting appropriate baselines for species' distributions. We test the hypothesis that neglecting historical data in niche quantification and HSM approaches leads to biased perceptions of species’ historical niches and suitable habitat distribution, and the patterns of potential species richness at the regional level. We focus on large terrestrial mammals in South Africa, for which we have access to a unique dataset of long-term occurrence records spanning the last five centuries, as well as recent (post-1950) occurrence records, for a community of 34 mammal species.
2. Material and methods
(a). Overview of the approach
We considered two datasets of occurrence: recent records (post-1950, RECENT) and recent + long-term records (post-1500, TOTAL) to quantify the effect of neglecting long-term occurrence data. We compared results obtained from these two datasets in three different approaches, two that are species based: estimation of the climatic niche in environmental space using n-dimensional hypervolumes [24] and prediction of suitable habitat in geographical space using HSMs; and one at the community level, namely prediction of the distribution of potential species richness using stacked HSMs. For the species-level approaches, we used two indices that summarize the cost of neglecting long-term biodiversity data and tested how the combination of these indices relates to species' conservation status and diet. For the community-level approach, we investigated spatial differences in predicted potential species richness, notably by comparing predictions between different South African bioregions. A 500-year time period being relatively short over an evolutionary timescale, we assume niche conservatism over the period considered [34]. Because we aim to test how using recent records only might lead to a shifting baseline in our understanding of species niche and potential habitat suitability, we did not perform this set of analyses on a ‘PAST’ (pre-1950) dataset, as we believe it to be less relevant for the objective of the study. Indeed, it is uncommon to model habitat suitability with long-term records only when recent data are available (notably due to the reduced availability and increased gaps and errors in long-term datasets).
(b). Species data
The general study area, hereafter referred to as South Africa, covers the countries of South Africa, Lesotho and eSwatini (former Swaziland), covering a total area of ca 1 270 000 km² for these three nations. We considered all extant South African large (greater than 20 kg) terrestrial mammals, except for species with fewer than 25 long-term observations in the dataset. In total, we analysed 15 315 recent (post-spatial thinning, range 55–1274) and 5446 long-term (range 25–501) records for the 34 species of large terrestrial mammals: 23 from the order Artiodactyla, 6 Carnivora, 4 Perissodactyla and 1 Proboscidea. In this analysis, Burchell's zebra and quagga are considered to be the same species (Equus quagga).
(i). Theoretically accessible areas
Barve et al. [35] outline the concept of the theoretically accessible area (the area that is climatically suitable and has been accessible to the species via dispersal over relevant periods of time) and show that restricting a model's training and validation areas to this theoretically accessible area greatly improves HSM performance and provides more accurate predictions of species richness and community composition [35,36]. As an approach to estimating the theoretically accessible area for each species, we identified the bioregions in which the species are known to have occurred historically, based on information on their ecology and interpretation of historical occurrences, and built a polygon using the boundaries of these bioregions. We defined the accessible area for each species as a buffer of 20 km around this polygon, to include ecotone regions where the species could disperse. This option is suggested by Barve et al. [35] to be the most operational compared to more intricate alternatives. We acquired spatial information on bioregions from the 2012 Vegetation Map of South Africa, Lesotho and Swaziland [37].
(ii). Modern and historical occurrence records
The long-term occurrence dataset used in this study covers the period 1500–1950 and includes records extracted from the historical literature, museum specimens and subfossil records. For historical records and museum specimens, we used the database presented in Boshoff et al. [11], completed with records from the KwaZulu-Natal, eSwatini and the rest of South Africa, and using the same approach and criteria defined in Boshoff et al. [11], so that the dataset covers all of South Africa. The reliability of these records in terms of identification and locality is discussed in Boshoff & Kerley [38] and their spatial, environmental and taxonomic biases in Monsarrat et al. [30] and Monsarrat & Kerley [39].
Subfossil records were obtained from Avery [40], the most comprehensive compilation of available taxonomical and distributional information on terrestrial mammalian species recorded from palaeontological and archaeological sites in mainland South Africa. We focused on records from the Holocene period (approx. 11 700 BP–present), for the set of mammal species present in the historical dataset. We recovered radiocarbon dates from primary sources and kept only those records with an upper date range after AD 1500, to match the period covered by the historical dataset. Only taxa identified to species in the archaeological samples were included. In total, the dataset used in the analyses represents 801 subfossil records from 67 archaeological sites throughout the study area. Additional information on the fossil record, type of sites and dating methods can be found in Avery [40] and references therein. We hereafter refer to this material as fossils.
The modern (post-1950) occurrence records dataset was consolidated as part of the national mammal Red List project conducted by the South African National Biodiversity Institute and the Endangered Wildlife Trust, for which over 460 000 geo-referenced unique occurrence records for South African mammals were centralized (see [41] for a list of data providers). As part of this process, data were vetted and underwent several rounds of data cleaning to check accuracy. These data are spatially biased, with the highest densities of records typically found in protected areas [41], artificially increasing spatial auto-correlation of occurrences. This in turn may affect the performance of HSMs built with these data [42]. To reduce the effect of sampling bias and spatial clustering on model performance, we subsampled the modern occurrence dataset using spatial thinning of the data (no occurrence records closer than 0.1°), as recommended by Boria et al. [43].
We considered all occurrence records located outside of a species' theoretically accessible area to be extralimital and we excluded them from the analyses. Modern extralimital records often correspond to introductions of individuals or populations outside of their historical range, often in suboptimal habitat, and are not informative of the habitat preferences of the species [44]. We, however, acknowledge that, by using bioregions as the filter for modern records, we may include some records that are outside the historical range, this being due to the relatively unique situation in South Africa of game translocations for commercial purposes [44].
(c). Environmental data
We considered six bioclimatic variables derived from BioClim [45]: mean annual temperature (BIO1) and annual precipitation (BIO12), describing the average climatic conditions; temperature seasonality (BIO4) and precipitation seasonality (BIO15), describing climatic seasonality; and maximum temperature of the warmer month (BIO5) and precipitation of the warmest quarter (BIO18), describing extreme climatic conditions. We also considered topography (TOPO), using altitude data from the ASTER Global Digital Elevation Model (ASTGTM) on https://lpdaac.usgs.gov [46]. These variables were chosen because they were biologically meaningful to predict large mammal species richness in South Africa [47] and because they potentially represent environmental characteristics that limit species’ distributions. All environmental variables were estimated at a 0.1° × 0.1° resolution, using the raster package [48] in R 3.5.1 [49].
(d). Hypervolume analysis
The n-dimensional hypervolume was originally proposed by Hutchinson [50] to describe the fundamental niche of a species, i.e. the environmental space where the species can exist indefinitely. In the modern understanding of the hypervolume function, a set of n variables that represent biologically important and independent axes are identified and the hypervolume is defined by a set of points within this n-dimensional space that reflects suitable values of the variables for the species' persistence [24]. Here, we consider five environmental axes: BIO1, BIO4, BIO12, BIO15 and TOPO, rescaled to a common and comparable scale before analysis. We used the Gaussian kernel density estimation with the Silverman bandwidth estimator method in the hypervolume package [21] in R 3.5.1 [49]. The bandwidth was estimated from the RECENT dataset with the Silverman estimator and the same value was used for the TOTAL dataset, to allow direct comparison.
The volume of the hypervolume is approximately linearly proportional to the number of observations in the dataset [21]. To ensure results are insensitive to sample size, we randomly subsampled the TOTAL dataset to have the same number of records as the RECENT dataset. We repeated the process 10 times and used averaged hypervolume measures of these 10 repetitions in the statistical analyses.
(e). Habitat suitability modelling
(i). Background data
Because the species occurrence records are highly biased spatially [30,41], we addressed the potential effect of sampling bias in the models. To do so, we produced background data with similar geographical bias as the RECENT and TOTAL occurrence datasets, following Phillips et al. [31]. We first created a sampling effort raster using a two-dimensional kernel density estimation applied on the occurrence dataset. Background data were then created by sampling without replacement within this raster grid, where the probability of a cell being sampled was proportional to the sampling density values (weighted target group approach, following Sanín & Anderson [51]). We selected the same number of background points as the number of occurrence records, so as to achieve a prevalence of 50%, as advised by Liu et al. [52].
(ii). Ensemble modelling
We created ensemble HSM [53] for each species by assembling five statistical methods (GAM, MAXENT, MARS, RF and GBM) to account for inter-model variability, using the ssdm package [54]. We ran 10 repetitions for each of the algorithms and produced an average of the models’ outputs, weighting each model according to its predictive ability. We measured predictive ability with a cross-validation approach, by using a random 70% of the data for calibration of the models (keeping the prevalence constant) and testing their predictive ability on the remainder of the dataset using the True Skill Statistic (TSS) [55]. We repeated this approach 10 times for each model and used an average of the predictive accuracy measure. In total, for each species and each dataset, we ran 500 models using five different statistical models, 10 repetitions of each algorithm and 10 repetitions of the random-splitting strategy. The outputs of these models are maps of predicted habitat suitability over the study area that provides hypotheses for the potential distribution of species for both datasets. We identified areas where the predicted habitat suitability differs between the RECENT and TOTAL datasets by subtracting the predicted values obtained from the RECENT model from those obtained with the TOTAL model in each cell within the study area. Areas with positive (negative) values are where we underestimate (overestimate) habitat suitability when considering only recent records.
(f). Species richness
Stacked species distribution models (SSDMs) combine multiple individual HSMs to produce a community-level model and predictive maps of potential species richness [56]. We used the ssdm package [54] to compute maps of local species richness by summing the probabilities from continuous habitat suitability maps provided by the ensemble HSMs, a method that performs better than stacking methods based on thresholding site-level occurrence probabilities [57]. To highlight areas where the potential species diversity is under- or overestimated because of neglecting long-term occurrence records in the models, we subtracted the map of species richness produced with the RECENT dataset from the one produced with the TOTAL dataset. We also compared the mean difference in predicted species richness for each bioregion of South Africa, both in absolute (i.e. the difference in species richness predicted with the TOTAL versus the RECENT dataset) and relative numbers (i.e. the percentage change relative to the species richness predicted with the RECENT dataset for each bioregion).
(g). Statistical analyses
For each species, we considered two indices to summarize the effects of neglecting long-term records on the estimation of climatic niche and habitat suitability: (1) the niche dissimilarity in environmental space (Ndis) [21] and (2) the dissimilarity in predicted habitat suitability in geographical space (PREDdis) [58] (see table 1 for a definition of these indices). For each index, higher values indicate a higher disparity between the results obtained with the RECENT and the TOTAL dataset.
Table 1.
Description of the two indices used in principal component analysis to quantify the effect of neglecting historical records on the estimation of climatic niche and suitable habitat.
| index | name | estimated from | formula | description |
|---|---|---|---|---|
| Ndis | dissimilarity of climatic niche | five-dimensional hypervolume | 1 − Jaccard similarity index | the Jaccard similarity index measures the overlap between niche hypervolumes [21] Ndis is comprised between 0 and 1, with higher values meaning higher dissimilarity |
| PREDdis | dissimilarity of predicted suitable environments | HSM | 1 − ESP | the expected fraction of shared presences (ESP) is a derivation from the Sørensen index of similarity of species' distributions that measure the overlap in predicted habitat suitability [58,59] PREDdis is comprised between 0 and 1, with higher values meaning higher dissimilarity |
We rescaled all indices by subtracting the mean and dividing by the standard deviation so that they are comparable and conducted a Principal Component Analysis (PCA) to convert these indices into a one-dimension variable (the first principal component PC1), quantifying the effect of neglecting historical records. We ran a two-way ANOVA with Type II errors to test for differences in PC1 between conservation status (threatened versus non-threatened) and broad diet guilds (herbivores versus carnivores). Conservation status was defined from the IUCN Red List categories [16], where species listed as Vulnerable, Endangered or Critically Endangered were considered ‘threatened’, and ‘non-threatened’ otherwise. We used a linear model to test how the change in mean predicted habitat suitability (ΔPRED, calculated as the proportional difference in mean predicted habitat suitability over the study area when it is estimated from the TOTAL dataset, compared to the RECENT dataset) varies with PC1 values. We also estimated the difference in the ability of HSMs to predict all the known occurrences for the species (ΔB) by measuring the proportional increase (or decrease) in the continuous Boyce index, a threshold-independent evaluator of the ability of HSMs to predict species presences [54], when it is estimated from the TOTAL dataset compared to the RECENT dataset.
3. Results
The ensemble modelling approach yielded a very good agreement between the different modelling methods, as indicated by low standard deviation around the predicted habitat suitability values (electronic supplementary material, S3). For 19 out of 34 species, the inclusion of historical records improved the ability of the model to predict all known occurrences of the species (ΔB > 0). The highest improvement in predictive ability was for the roan Hippotragus equinus, blesbok Damaliscus pygargus phillipsi and African wild dog Lycaon pictus (ΔB equal to 24%, 20% and 11%, respectively). By contrast, 12 species showed a decrease in predictive ability when historical data are included in the model, with the bushpig Potamochoerus larvatus and bontebok Damaliscus pygargus pygargus showing the strongest decrease (ΔB equal to −8% and −5%, respectively). The dissimilarity in climatic niche Ndis calculated from the five-dimensional hypervolume ranged from 0.11 to 0.70 (mean = 0.29 ± 0.17 s.d.), with Ndis > 0.60 for the African elephant Loxodonta africana, lion Panthera leo and African wild dog Lycaon pictus. The dissimilarity in predicted habitat suitability PREDdis ranged from 0.41 to 0.71 (mean = 0.57 ± 0.07 s.d.), with PREDdis > 0.70 for the African wild dog, lion and spotted hyaena Crocuta crocuta (electronic supplementary material, table S1).
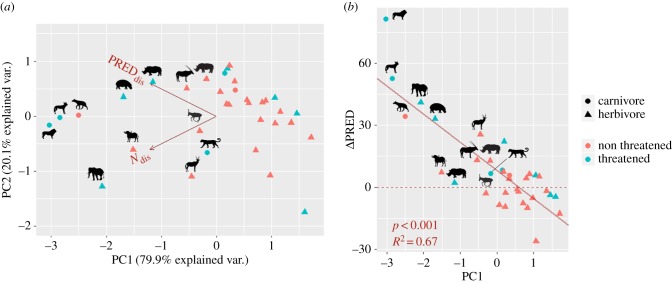
Overall, by combining Ndis and PREDdis in a PCA, 12 species (34% of our dataset) come out as impacted by neglecting historical records (PC1 < 0, with PC1 explaining 80% of the variance), with five species identified as the topmost impacted: the lion, African wild dog, spotted hyaena, African elephant and hippopotamus Hippopotamus amphibius (figure 1a). Of these five species, four are listed as threatened on the IUCN Red List. Three are carnivores and the other two are megaherbivores (body mass greater than 1000 kg). PC1 values were significantly lower for carnivores compared to herbivores (two-way ANOVA Type II, F1,34 = 7.30, m.s.e. = 8.96, p = 0.011) and marginally lower for threatened compared to non-threatened species (two-way ANOVA Type II, F F1,34 = 1.23, m.s.e. = 1.50, p = 0.28).
Figure 1.
Effects of incorporating historical records on the estimation of species climatic niche and predictions of habitat suitability. (a) Principal component analysis (PCA) of the two indices used to measure discrepancy between estimations of climatic niche (Ndis) and habitat suitability (PREDdis) with the RECENT and TOTAL datasets, for the 34 species of large mammals considered. Higher values of indices (lower values of PC1) indicate a higher discrepancy. We highlighted (silhouettes) 12 species with negative PC1 values most affected by neglecting historical records. We differentiate carnivores versus herbivores and threatened versus non-threatened species. The differences between these groups along the first principal component (PC1) are significant for the former (p = 0.011) and marginal for the latter (p = 0.28). (b) Plot showing the negative relationship between values of PC1 and the proportional difference in mean predicted habitat suitability over the study area when it is estimated from the TOTAL dataset compared to the RECENT dataset (ΔPRED). Positive values of ΔPRED mean that habitat suitability is underestimated without using historical records. Species that are most affected by neglecting historical data have increased mean predicted habitat suitability when historical records are included in HSMs. See electronic supplementary material, table S1 for a key of silhouettes.
We found a significant inverse linear relationship between PC1 and the change in mean predicted habitat suitability over the study area ΔPRED (p < 0.001, R2 = 0.68; figure 1b), i.e. species that are most affected by neglecting historical data have higher mean predicted habitat suitability over their study area when historical records are included in HSMs. The lion, African wild dog, African elephant, spotted hyaena and hippopotamus show the largest increase in mean predicted habitat suitability when historical records are included (ΔPRED equal to 81%, 53%, 41%, 34% and 33%, respectively).
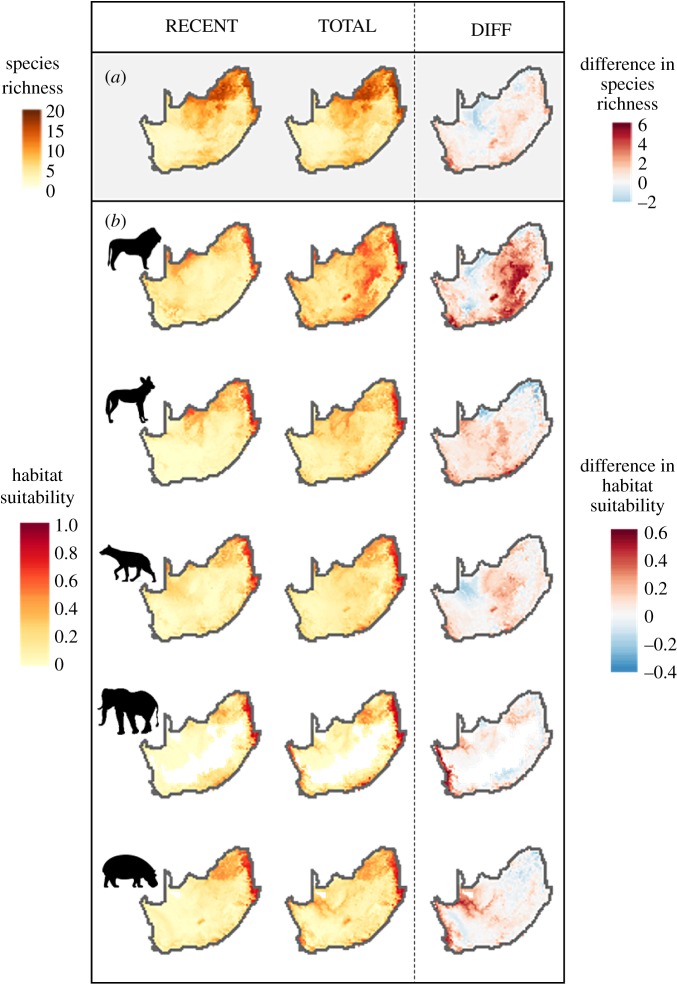
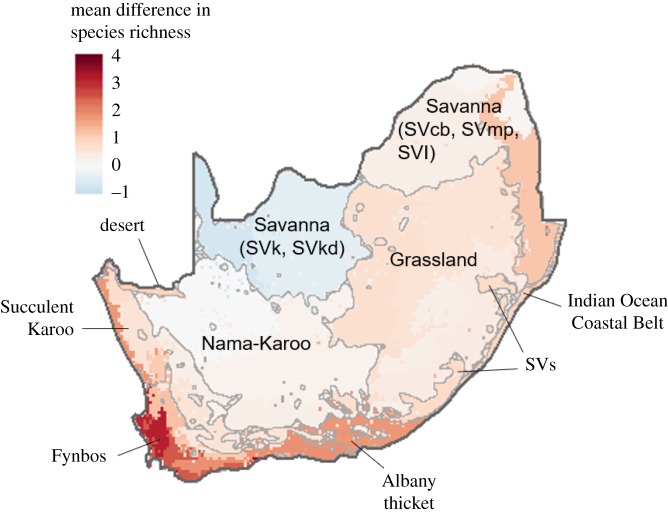
This results in differences in predicted potential species richness at the community level (figure 2a) and in the geographical distribution of predicted habitat suitability at the species level (see maps in figure 2b for the five most impacted species and electronic supplementary material, S3 for maps of all 34 species). Differences in predicted potential species richness are higher for the Albany Thicket, Fynbos and Savanna Lowveld biomes (figure 3). The figure of the relative difference in species richness shows that these shifts are particularly important for the Fynbos and Namaqualand Sandveld biomes, which currently have a low richness of large mammal species (electronic supplementary material, figure S1). The Nama-Karoo has on average very similar predicted species richness with the RECENT or TOTAL dataset, whereas the potential species richness tends to be overestimated in arid savannah.
Figure 2.
Effect of incorporating historical records in the spatial prediction of (a) species richness and (b) habitat suitability for the five species most impacted by neglecting historical records. The first column is the prediction of species richness/habitat suitability obtained from the RECENT dataset (post-1950 records) and the second column is obtained from the TOTAL dataset (historical + recent records). The last column is the difference between column 2 and column 1. We highlight the five species most impacted through neglecting historical records according to their PC1 score: lion Panthera leo, African wild dog Lycaon pictus, African elephant Loxodonta africana, spotted hyaena Crocuta crocuta and hippopotamus Hippopotamus amphibius.
Figure 3.
Mean difference in species richness estimated from the TOTAL versus RECENT dataset, calculated for each of the bioregions of South Africa. Darker shades of red indicate that potential species richness is higher when predicted with the TOTAL dataset than with the RECENT dataset. The main biomes are identified with grey contouring, obtained from simplifying polygons of the 2012 ‘Vegetation Map of South Africa, Lesotho and Swaziland’ [60]. A distinction is made between arid savannahs (SVk: Eastern Kalahari Bushveld Bioregion; and SVkd: Kalahari Duneveld Bioregion) and mesic savannahs (SVcb: Central Bushveld Bioregion; SVmp: Mopane bioregion; SVl: Lowveld Bioregion; and SVs: Sub-Escarpment Savanna Bioregion) [60].
4. Discussion
We show that neglecting long-term records can bias estimates of species climatic niche and suitable habitat and underestimate potential regional species richness. The implications are more severe for carnivore species, and marginally more so for threatened species, for which appropriate conservation actions and management decisions are the most critical. These results have implications for conservation planning and distribution modelling in general, given that globally most mapping of species' distributions and habitat use exclude long-term occurrence records, and for the conservation and management of South African mammalian fauna. These findings highlight the importance of considering long-term data in modern ecological analyses and may also provide explanatory insights into the limits of conservation approaches when they fail to consider appropriate species distribution baselines. We expand on these points below.
(a). Species implications
For several species, we observe only a limited effect of including historical records in the analyses. This indicates that modern occurrence records provide a reasonably good coverage of the climatic conditions found in their historical distribution. This possibly reflects that they have been less impacted by past range contractions, that range contraction did not affect the range of environmental conditions occupied by the species or that they have successfully recovered throughout their historical range, whether by natural recolonization or through active reintroductions. This result generally highlights the success of conservation efforts in South Africa, where many species have been successfully reintroduced throughout their historical range [61]. The most striking example is probably that of the white rhinoceros Ceratotherium simum. On the brink of extinction by the end of the nineteenth century, surviving in just a small population of approximately 20–50 animals in the area now known as Hluhluwe-iMfolozi in KwaZulu-Natal, the species has recovered throughout its historical range in South Africa and is now the most numerous of the rhino taxa, thanks to ambitious conservation and translocation efforts [62]. Other examples of striking recovery from near-extinction thanks to early twentieth-century conservation programmes are the bontebok, the black wildebeest Connochaetes gnou and the Cape mountain zebra Equus zebra zebra, all endemic mammals from South Africa [61]. We can only assume that the picture would be much grimmer for the South African large mammal fauna if not for these conservation success stories.
For other species that did not have such successful recovery, considering historical data hugely affects estimates of their climatic niche and potential distribution. For these, the geographical distribution of predicted habitat suitability is wider than expected from recent data only and this effect is higher for species of high conservation value. Three of the five most impacted species are carnivores (lion, African wild dog and spotted hyaena) and the two others are megaherbivores (elephant and hippopotamus). These species are highly charismatic [63], very sensitive to humans [64] and play important roles in ecosystems [65], thus acting as focal species for management efforts and trophic rewilding initiatives [66]. They are all listed as threatened by the IUCN Red List, except the spotted hyaena. This latter species is an interesting example of a shifting baseline where a species that is considered common has actually undergone a marked change in distribution over time, which is not captured by the IUCN Red List criteria [67]. This species has, however, recently been uplisted to Near Threatened on the Red List of Mammals of South Africa, Swaziland and Lesotho [68], pertaining to recent population declines in some protected areas. For these species of high conservation importance, analyses based on recent data only will lead to truncated estimates of bioclimatic relationships and underestimations of the extent of suitable areas for their protection. Important suitable areas might be overlooked when selecting appropriate sites for reintroductions and trophic rewilding, and protection efforts might focus on marginal habitat [5,6]. Forecasts of climate change impacts on biodiversity are also unlikely to be reliable without acknowledging past anthropogenic range contraction [69]. The implications of such missed opportunities on management outcomes and the conservation status of species need to be better understood.
(b). Community implications
At the community level, neglecting historical records underestimates potential regional species richness, with some areas being more impacted than others. In South Africa, the southwestern and western parts of the coastline, as well as the central Free State and Eastern Cape provinces, have higher potential richness than expected from recent records only. These areas were highly impacted historically, with the establishment of the Cape Colony by the Dutch in the mid-seventeenth century and the subsequent colonization of the interior, leading to increased pressures from land-use change and direct hunting [12,70]. In most bioregions, overlooking historical records underestimates the potential species richness, with particularly strong effects in the Fynbos and Albany thicket biomes. These shifted distribution baselines clearly have implications for our understanding of broader biogeographic patterns and processes. As an example, the underestimation of large mammal species richness in the Fynbos biome illustrated here demonstrates that the role of mammals in this biome, traditionally considered to support a low diversity of large mammals [14,71], needs to be reassessed. In addition to having suffered major biodiversity declines in the past [14,72], these areas are also where conservation efforts are thus most likely to be misguided (but see [14] for conservation planning in the Fynbos), which carries major implications for wildlife management and conservation in South Africa.
(c). Setting baselines
Shifting baselines [73] emphasize the need for setting appropriate references when exploring ecological patterns and how these may change, especially for detecting long-term processes. We have demonstrated here the occurrence of shifted baselines for the distribution of South African mammals, against which one can assess recent and future shifts in the geographical patterns of this fauna. Such phenomena can be expected elsewhere, and there is thus a need to study this on a global scale. The appropriate baseline should be adapted to each individual study, based on the objectives of the study, the system considered and a knowledge of the timeframe of past human activities that may have impacted the distribution of the focal species.
This study uses a 1500 cut-off date, which is often held up as a relevant baseline from which to define restoration objectives and quantify success [14,74]. This period is the approximate start of European expansion and pre-dates industrialization and massive human population growth. In South Africa, this baseline corresponds to pre-European settlement conditions, with the Cape colony only officially established by the Dutch East India Company in 1652. In the following centuries, the establishment of European settlements in the interior of the country and the concomitant introduction of horses and firearms will have induced a collapse of populations of large mammals from overhunting and loss of habitat, with the most documented episode occurring in the Highveld in the nineteenth century [12]. This baseline is thus relevant to identify the human-related impact on extant large mammal species in the recent past. It does not, however, consider earlier human impacts, which may have occurred before European colonization. Direct exploitation starting in the Middle Stone Age and the introduction of livestock in the Iron Age (approx. AD 200–1820s), likely associated with altered fire regimes, may have influenced the distribution of wildlife during prehistory (e.g. [75,76]). The extinction of the blue antelope around AD 1800 is an interesting case where direct competition with livestock herded by Khoen-khoen pastoralists in the Later Stone Age probably caused a drop in the population about 2000 years ago [77], before hunting by European colonists provided the coup de grace that brought the species to extinction [10]. While quantifying these ancient impacts is beyond the scope of this study, we encourage the use of palaeoecological approaches to bring additional insights into prehistorical baselines for the large mammal fauna and ecosystems of Southern Africa.
(d). Limitations
The time lag between the historical occurrence records (1500–1950) and the bioclimatic variables (1970–2000) used in the models could affect the habitat suitability model estimations if the connection between the occurrence location and the local climatic values has changed during this period. This issue is difficult to resolve because continuous series of high-resolution late-Holocene palaeoclimatic data are still rare in the Southern Hemisphere, and to our knowledge, no climatic reconstructions for the period 1500–1950 in South Africa are available to date. South African climate has been relatively stable during the past millennia. For the period considered, a cool, dry 500-year manifestation of the ‘Little Ice Age’ from AD 1300 to about 1800 can be noted [78], with a warmer episode occurring between about 1500 and 1675 [79]. The bulk of the historical data used in the analysis are for the nineteenth century, a period with a relatively similar climate to today, though with variations in precipitation patterns [80]. By using a long-term climatology over the period 1970–2000, we aimed to mitigate the impact of these inter-annual variations in temperature and precipitation. We acknowledge the limitation of this approach but believe this is an acceptable compromise at the resolution of this analysis.
The order of magnitude difference in the time-bin between the RECENT and TOTAL dataset could be a confounding factor explaining the higher species richness estimated when considering long-term records. This scale-dependence phenomenon is also known as the species–time relationship (STR), in which species number is a function of the time span of sampling [81]. STR could notably drive observed changes in species richness if species that were present in a given area in recent times were not described in the RECENT dataset but were detected in the long-term dataset due to a longer sampling period. However, while false absences may occur in the RECENT dataset (e.g. failure to report sensitive information such as the occurrence of rhinoceros), the current distribution of conspicuous and charismatic large mammals such as those considered in this analysis is generally well known. On the other hand, historical occurrence records are the result of opportunistic sampling and are more prone to false negatives, making the long-term dataset relatively less effective at detecting species. The bulk of occurrence records in the historical dataset is also for the nineteenth century, with a decrease in the number of records going back in time. This reduces concerns over the difference in temporal coverage between the two datasets and how this affects estimates of species richness. The slope of the STR also decreases as area sampled increases, i.e. species accumulation rates in time decrease with area sampled [81]. Here, the sampled area is very large and sampling over a longer period of time should not significantly change the rate of species identification. Sampling the whole of South Africa also limits the possibility of species turnover due to dispersion from neighbouring countries. For these reasons, the increase in predicted species richness as we consider older data is more likely to result from past anthropogenic impacts than from a sampling artefact driven by STR.
The set of bioclimatic variables used in this analysis does not necessarily capture the full range of environmental characteristics that limit species' distributions. They are, however, the most common set of variables used for modelling species distributions and thus fit our objective of showing how neglecting historical records in HSMs affects our perception of species’ climatic niche and potential distribution. As spatially explicit reconstructions of past environmental conditions are released in the future, including changes in human population and land cover, these could be incorporated into the analyses to improve predictions of species' distribution.
The study area follows the political boundaries of South Africa due to the unique amount of occurrence records available for the country. This is, however, an artificial cut-off that could affect our perception of niches, in particular, for those species with ranges that extend further north in sub-Saharan Africa. While this limits the transferability of predictions in space or time, our results remain valid at the regional level because we do not extrapolate outside the environmental space sampled in the occurrence dataset. South Africa is an exceptional ecoregion, with unique climatic regimes, high species richness and endemism [82,83]. Being at the southern margin of some species’ global distribution, it is a particularly important area for conservation since it may harbour populations with unique local adaptations that will be critical for species' ability to persist in the face of future climate change [84]. Range contractions that truncate species–climate relationships in this area are thus even more critical for our understanding of species’ niches than those occurring at the centre of the range.
5. Conclusion
Our study provides evidence that using recent distribution records only can underestimate species' bioclimatic niches, which in turn can misguide conservation efforts and is likely to provide biased forecasting of species’ responses to climate change [69]. The recognition that neglecting long-term biodiversity might lead to setting inappropriate spatial baselines is the first step towards a better integration of these data in decision-making for biodiversity conservation and management. A better understanding of the potential distribution of species can notably form the basis of the identification of areas that would be good candidates for protection and restoration efforts, in particular, if it is combined with knowledge on the distribution of anthropogenic pressures [85]. With the recent recognition of the value of these datasets for conservation, there is an encouraging development towards assembling long-term biodiversity datasets (e.g. 86–88]), including for underrepresented taxa (e.g. [89,90]). The release of global databases of historical distributions [91] is a promising avenue to integrate long-term perspectives into future ecological studies. We join previous calls for international, multidisciplinary effort to compile historical data [92], and urge that, whenever possible, these should be included in conservation and biogeographic studies. Unless efforts are made to integrate this historical perspective into biodiversity conservation, shifted distribution baselines risk undermining our efforts to define appropriate protected areas and halt the ongoing biodiversity crisis, as well as appropriately manage biodiversity under global change.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Margaret Avery and Shaw Badenhorst for assistance regarding the archaeological records, Matthew Child and the Endangered Wildlife Trust for providing access to the recent occurrence dataset, and Ana Rodrigues for providing comments on an earlier version of the manuscript.
Data accessibility
The species occurrence data that form the basis of this study were provided by a large number (greater than 60) of individual institutions in South Africa. These institutions are the sole owners of these data, which they provided under the condition that they would not be shared or released. Unfortunately, the authors of the study are thus not in a position to make these data publicly available. The R code for the analyses presented in the manuscript is available in FigShare (https://figshare.com/s/547b8606d672e39d2bec).
Authors' contributions
S.M. and G.K. developed the ideas; G.K., P.N. and I.R. contributed data; S.M. performed the analyses and wrote the paper. All authors commented on the paper.
Competing interests
We declare we have no competing interests.
References
- 1.Dinerstein E, et al. 2017. An ecoregion-based approach to protecting half the terrestrial realm. BioScience 67, 534–545. ( 10.1093/biosci/bix014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asaad I, Lundquist CJ, Erdmann MV, Costello MJ. 2017. Ecological criteria to identify areas for biodiversity conservation. Biol. Conserv. 213, 309–316. ( 10.1016/j.biocon.2016.10.007) [DOI] [Google Scholar]
- 3.Dirzo R, Young HS, Galetti M, Ceballos G, Isaac NJB, Collen B. 2014. Defaunation in the Anthropocene. Science 345, 401–406. ( 10.1126/science.1251817) [DOI] [PubMed] [Google Scholar]
- 4.Faurby S, Svenning J-C. 2015. Historic and prehistoric human-driven extinctions have reshaped global mammal diversity patterns. Divers. Distrib. 21, 1155–1166. ( 10.1111/ddi.12369) [DOI] [Google Scholar]
- 5.Kerley GIH, Kowalczyk R, Cromsigt JPGM. 2012. Conservation implications of the refugee species concept and the European bison: king of the forest or refugee in a marginal habitat? Ecography 35, 519–529. ( 10.1111/j.1600-0587.2011.07146.x) [DOI] [Google Scholar]
- 6.Cromsigt JPGM, Kerley GIH, Kowalczyk R. 2012. The difficulty of using species distribution modelling for the conservation of refugee species—the example of European bison. Divers. Distrib. 18, 1253–1257. ( 10.1111/j.1472-4642.2012.00927.x) [DOI] [Google Scholar]
- 7.Willis KJ, Araujo MB, Bennett KD, Figueroa-Rangel B, Froyd CA, Myers N. 2007. How can a knowledge of the past help to conserve the future? Biodiversity conservation and the relevance of long-term ecological studies. Phil. Trans. R. Soc. B 362, 175–187. ( 10.1098/rstb.2006.1977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schipper J, et al. 2008. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230. ( 10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 9.Stuart AJ. 2015. Late Quaternary megafaunal extinctions on the continents: a short review. Geol. J. 50, 338–363. ( 10.1002/gj.2633) [DOI] [Google Scholar]
- 10.Kerley GIH, Sims-Castley R, Boshoff AF, Cowling RM. 2009. Extinction of the blue antelope Hippotragus leucophaeus: modeling predicts non-viable global population size as the primary driver. Biodivers. Conserv. 18, 3235–3242. ( 10.1007/s10531-009-9639-x) [DOI] [Google Scholar]
- 11.Boshoff A, Landman M, Kerley GIH. 2016. Filling the gaps on the maps: historical distribution patterns of some larger mammals in part of southern Africa. Trans. R. Soc. South Afr. 71, 1–65. ( 10.1080/0035919X.2015.1084066) [DOI] [Google Scholar]
- 12.Boshoff AF, Kerley GIH. 2015. Lost herds of the Highveld: evidence from the written, historical record. Afr. J. Wildl. Res. 45, 287–300. ( 10.3957/056.045.0287) [DOI] [Google Scholar]
- 13.Spear D, Chown SL. 2009. The extent and impacts of ungulate translocations: South Africa in a global context. Biol. Conserv. 142, 353–363. ( 10.1016/j.biocon.2008.10.031) [DOI] [Google Scholar]
- 14.Kerley GIH, Pressey RL, Cowling RM, Boshoff AF, Sims-Castley R. 2003. Options for the conservation of large and medium-sized mammals in the Cape Floristic Region hotspot, South Africa. Biol. Conserv. 112, 169–190. ( 10.1016/S0006-3207(02)00426-3) [DOI] [Google Scholar]
- 15.IUCN/SSC. 2013. Guidelines for reintroductions and other conservation translocations, version 1.0. Gland, Switzerland: IUCN Species Survival Commission. [Google Scholar]
- 16.IUCN. 2018. The IUCN Red List of threatened species. Version 2018-2. http://www.iucnredlist.org.
- 17.Wolf C, Ripple WJ. 2017. Range contractions of the world's large carnivores. R. Soc. Open Sci. 4, 170052 ( 10.1098/rsos.170052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf C, Ripple WJ. 2018. Rewilding the world's large carnivores. R. Soc. Open Sci. 5, 172235 ( 10.1098/rsos.172235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guisan A, Zimmermann NE. 2000. Predictive habitat distribution models in ecology. Ecol. Model. 135, 147–186. ( 10.1016/S0304-3800(00)00354-9) [DOI] [Google Scholar]
- 20.Guisan A, Thuiller W. 2005. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 8, 993–1009. ( 10.1111/j.1461-0248.2005.00792.x) [DOI] [PubMed] [Google Scholar]
- 21.Blonder B, Lamanna C, Violle C, Enquist BJ. 2014. The n-dimensional hypervolume. Glob. Ecol. Biogeogr. 23, 595–609. ( 10.1111/geb.12146) [DOI] [Google Scholar]
- 22.Rodríguez JP, Brotons L, Bustamante J, Seoane J. 2007. The application of predictive modelling of species distribution to biodiversity conservation. Divers. Distrib. 13, 243–251. ( 10.1111/j.1472-4642.2007.00356.x) [DOI] [Google Scholar]
- 23.Franklin J. 2013. Species distribution models in conservation biogeography: developments and challenges. Divers. Distrib. 19, 1217–1223. ( 10.1111/ddi.12125) [DOI] [Google Scholar]
- 24.Blonder B. 2017. Hypervolume concepts in niche- and trait-based ecology. Ecography 41, 1441–1455. ( 10.1111/ecog.03187) [DOI] [Google Scholar]
- 25.Thuiller W, Brotons L, Araújo MB, Lavorel S. 2004. Effects of restricting environmental range of data to project current and future species distributions. Ecography 27, 165–172. ( 10.1111/j.0906-7590.2004.03673.x) [DOI] [Google Scholar]
- 26.Newbold T. 2010. Applications and limitations of museum data for conservation and ecology, with particular attention to species distribution models. Prog. Phys. Geogr. 34, 3–22. ( 10.1177/0309133309355630) [DOI] [Google Scholar]
- 27.Hendricks SA, Sesink Clee PR, Harrigan RJ, Pollinger JP, Freedman AH, Callas R, Figura PJ, Wayne RK. 2016. Re-defining historical geographic range in species with sparse records: implications for the Mexican wolf reintroduction program. Biol. Conserv. 194, 48–57. ( 10.1016/j.biocon.2015.11.027) [DOI] [Google Scholar]
- 28.Lentini PE, Stirnemann IA, Stojanovic D, Worthy TH, Stein JA. 2018. Using fossil records to inform reintroduction of the kakapo as a refugee species. Biol. Conserv. 217, 157–165. ( 10.1016/j.biocon.2017.10.027) [DOI] [Google Scholar]
- 29.Monsarrat S, Pennino MG, Smith TD, Reeves RR, Meynard CN, Kaplan DM, Rodrigues ASL. 2015. Historical summer distribution of the endangered North Atlantic right whale (Eubalaena glacialis): a hypothesis based on the environmental niche of a congeneric species. Divers. Distrib. 21, 925–937. ( 10.1111/ddi.12314) [DOI] [Google Scholar]
- 30.Monsarrat S, Boshoff AF, Kerley GIH. 2018. Accessibility maps as a tool to predict sampling bias in historical biodiversity occurrence records. Ecography 42, 125–136. ( 10.1111/ecog.03944) [DOI] [Google Scholar]
- 31.Phillips SJ, Dudík M, Elith J, Graham CH, Lehmann A, Leathwick J, Ferrier S. 2009. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl. 19, 181–197. ( 10.1890/07-2153.1) [DOI] [PubMed] [Google Scholar]
- 32.Fourcade Y, Engler JO, Rödder D, Secondi J. 2014. Mapping species distributions with MAXENT using a geographically biased sample of presence data: a performance assessment of methods for correcting sampling bias. PLoS ONE 9, e97122 ( 10.1371/journal.pone.0097122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruete A. 2015. Displaying bias in sampling effort of data accessed from biodiversity databases using ignorance maps. Biodivers. Data J. 3, e5361 ( 10.3897/BDJ.3.e5361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiens JJ, Graham CH. 2005. Niche conservationism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 36, 519–539. ( 10.1146/annurev.ecolsys.36.102803.095431) [DOI] [Google Scholar]
- 35.Barve N, Barve V, Jiménez-Valverde A, Lira-Noriega A, Maher SP, Peterson AT, Soberón J, Villalobos F. 2011. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol. Model. 222, 1810–1819. ( 10.1016/j.ecolmodel.2011.02.011) [DOI] [Google Scholar]
- 36.Cooper JC, Soberón J. 2018. Creating individual accessible area hypotheses improves stacked species distribution model performance. Glob. Ecol. Biogeogr. 27, 156–165. ( 10.1111/geb.12678) [DOI] [Google Scholar]
- 37.South African National Biodiversity Institute. 2012. 2012 Vegetation map of South Africa, Lesotho and Swaziland [vector geospatial dataset]. http://bgis.sanbi.org/SpatialDataset/Detail/18.
- 38.Boshoff AF, Kerley GI. 2010. Historical mammal distribution data: how reliable are written records? South Afr. J. Sci. 106, 26–33. ( 10.4102/sajs.v106i1/2.116) [DOI] [Google Scholar]
- 39.Monsarrat S, Kerley GIH. 2018. Charismatic species of the past: biases in reporting of large mammals in historical written sources. Biol. Conserv. 223, 68–75. ( 10.1016/j.biocon.2018.04.036) [DOI] [Google Scholar]
- 40.Avery DM. 2019. A fossil history of Southern African land mammals. Cambridge, UK: Cambridge University Press; https://www.cambridge.org/core/books/fossil-history-of-southern-african-land-mammals/41969EC1E7739F4775954E6ADA8EA036. [Google Scholar]
- 41.Child MF, Roxburgh L, Do Linh San E, Raimondo D, Davies-Mostert HT. 2017. Mammal Red list 2016: introduction and methodology. Lethabong, South Africa: South African National Biodiversity Institute and Endangered Wildlife Trust. [Google Scholar]
- 42.Veloz SD. 2009. Spatially autocorrelated sampling falsely inflates measures of accuracy for presence-only niche models. J. Biogeogr. 36, 2290–2299. ( 10.1111/j.1365-2699.2009.02174.x) [DOI] [Google Scholar]
- 43.Boria RA, Olson LE, Goodman SM, Anderson RP. 2014. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Model. 275, 73–77. ( 10.1016/j.ecolmodel.2013.12.012) [DOI] [Google Scholar]
- 44.Castley JG, Boshoff AF, Kerley GIH. 2001. Compromising South Africa's natural biodiversity—inappropriate herbivore introductions. South Afr. J. Sci. 97, 344–348. [Google Scholar]
- 45.Fick SE, Hijmans RJ. 2017. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315. ( 10.1002/joc.5086) [DOI] [Google Scholar]
- 46.NASA/METI/AIST/Japan Spacesystems, U.S./Japan ASTER Science Team. 2009. ASTER global digital elevation model [dataset]. NASA EOSDIS Land Processes DAAC. ( 10.5067/ASTER/ASTGTM.002) [DOI]
- 47.Andrews P, O'Brien EM. 2000. Climate, vegetation, and predictable gradients in mammal species richness in southern Africa. J. Zool. 251, 205–231. ( 10.1111/j.1469-7998.2000.tb00605.x) [DOI] [Google Scholar]
- 48.Hijmans RJ. 2014. raster: geographic data analysis and modeling. R package version 2.2-12. http://CRAN.R-project.org/package=raster.
- 49.R Core Team. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- 50.Hutchinson GE. 1957. Concluding remarks. Cold Spring Harb. Symp. Quant. Biol. 22, 415–427. ( 10.1101/SQB.1957.022.01.039) [DOI] [Google Scholar]
- 51.Sanín C, Anderson RP. 2018. A framework for simultaneous tests of abiotic, biotic, and historical drivers of species distributions: empirical tests for North American wood warblers based on climate and pollen. Am. Nat. 192, E000 ( 10.1086/697537) [DOI] [PubMed] [Google Scholar]
- 52.Liu C, Berry PM, Dawson TP, Pearson RG. 2005. Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28, 385–393. ( 10.1111/j.0906-7590.2005.03957.x) [DOI] [Google Scholar]
- 53.Araujo M, New M. 2007. Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42–47. ( 10.1016/j.tree.2006.09.010) [DOI] [PubMed] [Google Scholar]
- 54.Schmitt S, Pouteau R, Justeau D, de Boissieu F, Birnbaum P. 2017. ssdm: an R package to predict distribution of species richness and composition based on stacked species distribution models. Methods Ecol. Evol. 8, 1795–1803. ( 10.1111/2041-210X.12841) [DOI] [Google Scholar]
- 55.Allouche O, Tsoar A, Kadmon R. 2006. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232. ( 10.1111/j.1365-2664.2006.01214.x) [DOI] [Google Scholar]
- 56.Ferrier S, Guisan A. 2006. Spatial modelling of biodiversity at the community level. J. Appl. Ecol. 43, 393–404. ( 10.1111/j.1365-2664.2006.01149.x) [DOI] [Google Scholar]
- 57.Calabrese JM, Certain G, Kraan C, Dormann CF. 2014. Stacking species distribution models and adjusting bias by linking them to macroecological models. Glob. Ecol. Biogeogr. 23, 99–112. ( 10.1111/geb.12102) [DOI] [Google Scholar]
- 58.Godsoe W, Case BS. 2015. Accounting for shifts in the frequency of suitable environments when testing for niche overlap. Methods Ecol. Evol. 6, 59–66. ( 10.1111/2041-210X.12307) [DOI] [Google Scholar]
- 59.Godsoe W. 2014. Inferring the similarity of species distributions using species' distribution models. Ecography 37, 130–136. ( 10.1111/j.1600-0587.2013.00403.x) [DOI] [Google Scholar]
- 60.Mucina L, Rutherford MC (eds). 2006. The vegetation of South Africa, Lesotho and Swaziland. Pretoria, South Africa: SANBI. [Google Scholar]
- 61.Penzhorn BL. 1971. A summary of the re-introduction of ungulates into South African National Parks (to 31 December 1970). Koedoe 14, 145–159. ( 10.4102/koedoe.v14i1.725) [DOI] [Google Scholar]
- 62.Emslie R.2012. Ceratotherium simum . In IUCN Red List of threatened species , e .T4185A16980466. . [DOI]
- 63.Albert C, Luque GM, Courchamp F. 2018. The twenty most charismatic species. PLoS ONE 13, e0199149 ( 10.1371/journal.pone.0199149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riggio J, Kija H, Masenga E, Mbwilo F, Van de Perre F, Caro T.. 2018. Sensitivity of Africa's larger mammals to humans. J. Nat. Conserv. 43, 136–145. ( 10.1016/j.jnc.2018.04.001) [DOI] [Google Scholar]
- 65.Malhi Y, Doughty CE, Galetti M, Smith FA, Svenning J-C, Terborgh JW. 2016. Megafauna and ecosystem function from the Pleistocene to the Anthropocene. Proc. Natl Acad. Sci. USA 113, 838–846. ( 10.1073/pnas.1502540113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarvie S, Svenning J-C. 2018. Using species distribution modelling to determine opportunities for trophic rewilding under future scenarios of climate change. Phil. Trans. R. Soc. B 373, 10 ( 10.1098/rstb.2017.0446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodrigues ASL, Monsarrat S, Charpentier A, Brooks TM, Hoffmann M, Reeves R, Palomares MLD, Turvey ST. 2019. Unshifting the baseline: a framework for documenting historical population changes and assessing long-term anthropogenic impacts. Phil. Trans. R. Soc. B 374, 20190220 ( 10.1098/rstb.2019.0220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hunnicutt A, Power RJ, Lerm L, Page-Nicholson S, Mills MGL, Camacho G, Dalerum F, Child MF. 2016. A conservation assessment of Crocuta crocuta. In The Red List of mammals of South Africa, Swaziland and Lesotho (eds Child MF, Roxburgh L, Do Linh San E, Raimondo D, Davies-Mostert HT). Lethabong, South Africa: South African National Biodiversity Institute and Endangered Wildlife Trust. [Google Scholar]
- 69.Faurby S, Araújo MB. 2018. Anthropogenic range contractions bias species climate change forecasts. Nat. Clim. Change 8, 252–256. ( 10.1038/s41558-018-0089-x) [DOI] [Google Scholar]
- 70.Pringle JA, Bond C, Clark J. 1982. The conservationists and the killers: the story of game protection and the wildlife society of Southern Africa. Cape Town, South Africa: Bulpin; https://books.google.co.za/books?id=LmnPAAAAMAAJ. [Google Scholar]
- 71.Skinner JD, Chimimba CT. 2005. The mammals of the Southern African sub-region. Cambridge University Press; https://books.google.co.za/books?id=I6RhVKyFfjkC. [Google Scholar]
- 72.Biggs R, Reyers B, Scholes RJ. 2006. A biodiversity intactness score for South Africa. South Afr. J. Sci. 102, 277–283. [Google Scholar]
- 73.Soga M, Gaston KJ. 2018. Shifting baseline syndrome: causes, consequences, and implications. Front. Ecol. Environ. 16, 263 ( 10.1002/fee.1794) [DOI] [Google Scholar]
- 74.Akçakaya HR, et al. 2018. Quantifying species recovery and conservation success to develop an IUCN Green List of Species. Conserv. Biol. 32, 1128–1138. ( 10.1111/cobi.13112) [DOI] [PubMed] [Google Scholar]
- 75.Badenhorst S. 2015. Intensive hunting during the Iron Age of Southern Africa. Environ. Archaeol. 20, 41–51. ( 10.1179/1749631414Y.0000000039) [DOI] [Google Scholar]
- 76.Faith JT. 2008. Eland, buffalo, and wild pigs: were Middle Stone Age humans ineffective hunters? J. Hum. Evol. 55, 24–36. ( 10.1016/j.jhevol.2007.11.005) [DOI] [PubMed] [Google Scholar]
- 77.Klein RG. 1987. The extinct blue antelope. Sagittarius 2, 20–23. [Google Scholar]
- 78.Holmgren K, et al. 1999. A 3000-year high-resolution stalagmite based record of palaeoclimate for northeastern South Africa. The Holocene 9, 295–309. ( 10.1191/095968399672625464) [DOI] [Google Scholar]
- 79.Tyson PD, Lindesay JA. 1992. The climate of the last 2000 years in southern Africa. The Holocene 2, 271–278. ( 10.1177/095968369200200310) [DOI] [Google Scholar]
- 80.Nash DJ, Endfield GH. 2002. A 19th century climate chronology for the Kalahari region of central southern Africa derived from missionary correspondence. Int. J. Climatol. 22, 821–841. ( 10.1002/joc.753) [DOI] [Google Scholar]
- 81.Adler PB, White EP, Lauenroth WK, Kaufman DM, Rassweiler A, Rusak JA. 2005. Evidence for a general species–time–area relationship. Ecology 86, 2032–2039. ( 10.1890/05-0067) [DOI] [Google Scholar]
- 82.Olson DM, Dinerstein E. 2002. The Global 200: priority ecoregions for global conservation. Ann. MO Bot. Gard. 89, 199 ( 10.2307/3298564) [DOI] [Google Scholar]
- 83.Cowling RM, Rundel PW, Desmet PG, Esler KJ. 1998. Extraordinary high regional-scale plant diversity in southern African arid lands: subcontinental and global comparisons. Divers. Distrib. 4, 27–36. [Google Scholar]
- 84.Rehm EM, Olivas P, Stroud J, Feeley KJ. 2015. Losing your edge: climate change and the conservation value of range-edge populations. Ecol. Evol. 5, 4315–4326. ( 10.1002/ece3.1645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Monsarrat S, Jarvie S, Svenning J-C. 2019. Anthropocene refugia: integrating history and predictive modelling to assess the space available for biodiversity in a human-dominated world. Phil. Trans. R. Soc. B 374, 20190219 ( 10.1098/rstb.2019.0219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clavero M, Delibes M. 2013. Using historical accounts to set conservation baselines: the case of Lynx species in Spain. Biodivers. Conserv. 22, 1691–1702. ( 10.1007/s10531-013-0506-4) [DOI] [Google Scholar]
- 87.Turvey ST, Crees JJ, Di Fonzo MMI. 2015. Historical data as a baseline for conservation: reconstructing long-term faunal extinction dynamics in Late Imperial–modern China. Proc. R. Soc. B 282, 20151299 ( 10.1098/rspb.2015.1299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burgio K, Carlson C, Bond A. 2018. Georeferenced sighting and specimen occurrence data of the extinct Carolina parakeet (Conuropsis carolinensis) from 1564–1944. Biodivers. Data J. 6, e25280 ( 10.3897/BDJ.6.e25280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McClenachan L, O'Connor G, Neal BP, Pandolfi JM, Jackson JB. 2017. Ghost reefs: nautical charts document large spatial scale of coral reef loss over 240 years. Sci. Adv. 3, e1603155 ( 10.1126/sciadv.1603155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Popejoy T, Randklev CR, Neeson TM, Vaughn CC. 2018. Prioritizing sites for conservation based on similarity to historical baselines and feasibility of protection: zooarchaeological baseline. Conserv. Biol. 32, 1118–1127. ( 10.1111/cobi.13128) [DOI] [PubMed] [Google Scholar]
- 91.Faurby S, Davis M, Pedersen RØ, Schowanek SD, Antonelli A, Svenning J-C. 2018. PHYLACINE 1.2: the phylogenetic atlas of mammal macroecology. Ecology 99, 2626 ( 10.1002/ecy.2443) [DOI] [PubMed] [Google Scholar]
- 92.Clavero M, Revilla E. 2014. Biodiversity data: mine centuries-old citizen science. Nature 510, 35 ( 10.1038/510035c) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The species occurrence data that form the basis of this study were provided by a large number (greater than 60) of individual institutions in South Africa. These institutions are the sole owners of these data, which they provided under the condition that they would not be shared or released. Unfortunately, the authors of the study are thus not in a position to make these data publicly available. The R code for the analyses presented in the manuscript is available in FigShare (https://figshare.com/s/547b8606d672e39d2bec).