Abstract
Rubinstein-Taybi syndrome (RSTS) is a rare genetic disorder in humans characterized by growth and psychomotor delay, abnormal gross anatomy, and mild to severe mental retardation (Rubinstein and Taybi, Am J Dis Child 105:588–608, 1963, Hennekam et al., Am J Med Genet Suppl 6:56–64, 1990). RSTS is caused by de novo mutations in epigenetics-associated genes, including the cAMP response element-binding protein (CREBBP), the gene-encoding protein referred to as CBP, and the EP300 gene, which encodes the p300 protein, a CBP homologue. Recent studies of the epigenetic mechanisms underlying cognitive functions in mice provide direct evidence for the involvement of nuclear factors (e.g., CBP) in the control of higher cognitive functions. In fact, a role for CBP in higher cognitive function is suggested by the finding that RSTS is caused by heterozygous mutations at the CBP locus (Petrij et al., Nature 376:348–351, 1995). CBP was demonstrated to possess an intrinsic histone acetyltransferase activity (Ogryzko et al., Cell 87:953–959, 1996) that is required for CREB-mediated gene expression (Korzus et al., Science 279:703–707, 1998). The intrinsic protein acetyltransferase activity in CBP might directly destabilize promoter-bound nucleosomes, facilitating the activation of transcription. Due to the complexity of developmental abnormalities and the possible genetic compensation associated with this congenital disorder, however, it is difficult to establish a direct role for CBP in cognitive function in the adult brain. Although aspects of the clinical presentation in RSTS cases have been extensively studied, a spectrum of symptoms found in RSTS patients can be accessed only after birth, and, thus, prenatal genetic tests for this extremely rare genetic disorder are seldom considered. Even though there has been intensive research on the genetic and epigenetic function of the CREBBP gene in rodents, the etiology of this devastating congenital human disorder is largely unknown.
Keywords: Rubinstein-Taybi syndrome, RSTS, CREBBP, CBP, EP300, p300, Memory, Epigenetic, Histone acetylation, HDAC
3.1. Introduction
The regulation of gene expression requires not only activation of transcription factors but also the recruitment of multifunctional coactivators that are independently regulated and utilized in a cell- and promoter-specific fashion to stimulate or repress transcription [1]. Dynamic changes in the organization of chromatin control gene expression and histone acetylation are one mechanism for the local and global control of chromatin structure [2, 3]. Studies have shown that chromatin acetylation at a region of ongoing transcription is essential for high-level gene expression [2, 3]. The cAMP response element-binding protein (CBP, which is encoded by the CREBBP gene) and its homologue p300 protein (encoded by the EP300 gene) are transcriptional coactivators [4, 5] that interact with multiple transcriptional regulators and facilitate the assembly of the basic transcriptional machinery [6] (Fig. 3.1). In addition to serving as molecular scaffolds, CBP and p300 each possess intrinsic histone acetyltransferase (HAT) activities [10] that can be specifically and directly inhibited by phosphorylation or by association with viral proteins [6, 11]. Owing to the ability of CBP and p300 to control the function of chromatin via mechanisms that involve the covalent modification of chromatin, resulting in long-lasting marks on histones, these proteins are defined as epigenetic “writers” (Fig. 3.2). More recently, the HAT activity of both CBP and p300 has been referred to as lysine acetyltransferase (KAT11) activity because it targets lysine not only on histones but also on nonhistone proteins [7]. CBP and p300 proteins share more than 70% sequence homology, but they retain some distinct cellular functions and behavioral functions and cannot always replace each other [8, 15, 16].
Fig. 3.1.
Structure of CBP/p300 protein family. CBP and p300 proteins belong to the same family of coactivator of transcription and share 63% identity and 73% similarity with the highest homology as mapped to functional domains. A central part of CBP/p300 protein encompasses chromatin association and the modification region. The N-terminal and C-terminal regions, which include a variety of motives that provide a platform for specific protein-protein interactions, enable the formation of multiprotein complexes critical for a cellular signal- and promoter-specific gene expression regulation. CBP was discovered as a CREB-binding protein and a phosphorylation-dependent interaction between CREB’s KID domain and CBP’s KIX domain. Further research demonstrated that more than 400 proteins could interact with CBP/p300 proteins. The schematic shows examples of these interaction partners’ interacting predominantly with regions containing zinc finger motives. The central region of CBP/p300 contains domains supporting chromatin-modifying functions. While bromodomain (Bromo) provides ability for chromatin recognition, the KIT11 domain has lysine acetyltransferase enzymatic activity targeting primary histone N-terminals and nonhistone nuclear proteins [7, 8]. For more detailed information, see the text. The diagram does not show proper proportions and is based on data from UniGene (CREBBP, NP_004371.2; EP300, NP_001420.2; and [8, 9]). CBP’s known domains from the N-terminal are NRID nuclear receptor interaction domain, TAZ1 transcriptional adapter zinc-binding domain 1, KIX CREB binding, Bromo bromodomain, PHD plant homeodomain, KIT11 lysine acetyltransferase, ZZ zinc finger domain, TAZ2, Q polyglutamine stretch, NRs nuclear hormone receptors, SRC steroid receptor coactivator
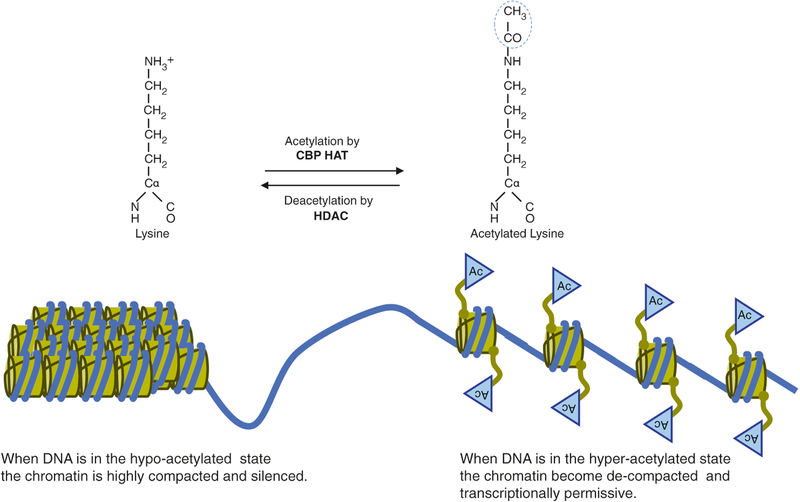
Fig. 3.2.
Histone acetylation controls chromatin structure and function. Although CBP’s function as a platform to recruit other required coactivators appears to be indispensable, the requirement for HAT activity is transcription unit specific and may depend on the structure of chromatin at a specific locus [12, 13]. Histone deacetylase (HDAC)-mediated hypo-acetylation of histones promotes a compact chromatin structure state, subsequently silencing transcription. Promoter-specific recruitment of chromatin remodeling factors, such as CBP HAT, facilitates de-compaction of the chromatin structure, where genes are accessible for large multiprotein complexes that mediate gene expression (i.e., RNA polymerase II holoenzyme). CBP HAT has been implicated in epigenetic mechanisms that control higher cognitive functions [14]
A role for the CBP gene in higher cognitive function is suggested by the finding that Rubinstein-Taybi syndrome (RSTS), a disorder in humans characterized by growth and psychomotor delay, abnormal gross anatomy, and mild to severe mental retardation [17, 18], is caused by heterozygous mutations at the CBP locus [19]. CBP is one of the major regulatory nuclear proteins that control gene expression associated with multiple critical cell functions during development and in the adult. Due to the complexity of developmental abnormalities and possible genetic compensation associated with this congenital disorder, however, it is difficult to establish a direct role for CBP in cognitive function in the adult brain.
3.2. Genotype
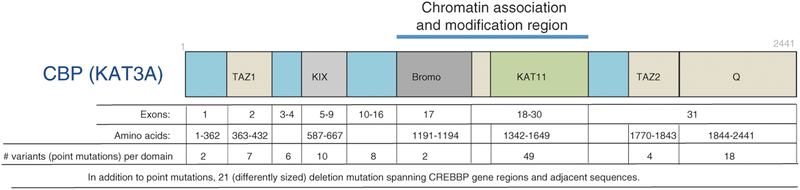
Mutations in the human CREBBP gene were reported to be associated with RSTS (OMIM #180849, #613684), a haploinsufficiency disorder characterized by multiple developmental defects and severe mental retardation (Rubinstein and Taybi, 1963). RSTS is a rare genetic syndrome caused by de novo heterozygous mutations in epigenetic genes and was first described in 1963 [17]. RSTS is found in one case per 100,000 to 125,000 live births. In 1991, Petrij et al. demonstrated for the first time the genetic origin of RSTS by reporting de novo reciprocal (balanced) translocations with breakpoints in chromosomal regions 16p13.3 in RSTS patients [19]. The CREBBP gene (CREBBP; OMIM #600140) is located on chromosome 16p13.3. A mutation in the gene that encodes the CREBBP gene was reported in approximately 55% of RSTS cases (Fig. 3.3), defining RSTS type 1 (RSTS1; OMIM #180849) [19, 22, 23]. The CREBBP gene spans ~155 kb, and there are 31 exons in coding region. Transcription of the CREBBP gene proceeds from centromere to telomere with start codon located in exon 1 and stop codon in 31. ~10 kb CREBBP mRNA contains 7.3 kb of coding sequence. The CREBBP gene encodes a large protein (CBP) with a molecular weight of 26,531 Da that consists of 2441 amino acids.
Fig. 3.3.
CREBBP germline mutations in RSTS1 patients. CREBBP germline mutations account for 50–60% of RSTS cases. About 50% of mutations associated with RSTS1 have been mapped to chromatin association and the modification region in the CREBBP gene (Fig. 3.1). CREBBP germline mutations in RSTS patients include 106 point mutations and 21 deletions, such as exonic and whole-gene deletions, with some encompassing flanking genes. Only a few mosaic mutations in the CREBBP gene have been reported (not shown). Data used for analysis were reported in Leiden Open Variation Database [20], Gervasini et al. (2010) [21], and UniGene database (CREBBP: NP_004371.2)
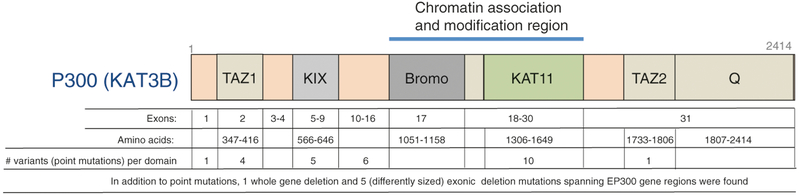
In addition, 10% of cases of RSTS have been associated with mutations present in a CBP homologue, the E1A binding protein p300 [24–26] (Fig. 3.4). Protein p300 is encoded by the human EP300 gene (OMIM #602700) located at 22q13.2. Cases of mutated EP300 are described as RSTS type 2 (RSTS2; OMIM #613684). Mutations found in CREBBP and EP300 (i.e., frameshift, nonsense, splice site, and missense mutations) are heterozygous, rare, and de novo [27]. In addition, less frequently occurring large deletions (exonic or whole gene) or balanced inversions and translocations also have been characterized.
Fig. 3.4.
EP300 germline mutations in RSTS patients. To date, 34 EP300 mutations in known RSTS2 cases have been reported worldwide [25]. Mutations in the EP300 gene account for about 10% of all RSTS cases. EP300 germline mutations in RSTS patients include 27 point mutations, six exonic deletions, and one whole-gene deletion. About 40% of mutations associated with RSTS2 have been mapped to chromatin association and the modification region in the EP300 gene (Fig. 3.1). Data used for analysis were reported in the Leiden Open Variation Database [20], Negri et al. (2016) [25], and the UniGene database (EP300: NP_001420.2)
3.3. Clinical Phenotypes
There are no precise diagnostic criteria for RSTS, but there is a spectrum of clinical presentations with unique clinical hallmarks (Table 3.1). A number of studies have found characteristic developmental anomalies in patients with mutations in the CREBBP and EP300 genes, including growth and psychomotor delay, abnormal gross anatomy, and intellectual disabilities [17, 18, 28]. In general, phenotypes associated with mutations in EP300 are less marked than those found with CREBBP mutations. A high number of individuals with mutations in CREBBP and EP300 show broad thumbs and halluces (96% and 69%, respectively); facial abnormalities, such as a grimacing smile (94% and 47%, respectively); and intellectual disabilities (99% and 94%, respectively) (Table 3.1). Retarded motor and mental development is visible during the first year of life. The average IQ of individuals with RSTS varies between 35 and 50, but some patients show better performance [28].
Table 3.1.
Comparison of symptoms found in RSTS individuals who carry mutations in CREBBP and EP300
| Symptom | CREBBPa (n = 308) | EP300b (n = 52) | p-valuec | ||
|---|---|---|---|---|---|
| Intellectual disability | 99% | 250/253 | 94% | 48/51 | NS |
| Severe | 36% | 33/92 | 7% | 2/29 | <0.005 |
| Moderate | 48% | 44/92 | 31% | 9/29 | NS |
| Mild | 14% | 13/92 | 62% | 18/29 | <0.00005 |
| Broad thumbs | 96% | 277/290 | 69% | 36/52 | <0.00005 |
| Broad halluces | 95% | 221/233 | 81% | 42/52 | <0.005 |
| Grimacing smile | 94% | 99/105 | 47% | 21/45 | <0.00005 |
| Long eyelashes | 89% | 75/84 | 90% | 44/49 | NS |
| Columella below alae nasi | 88% | 195/222 | 92% | 48/52 | NS |
| Arched eyebrows | 85% | 71/84 | 65% | 34/52 | <0.05 |
| Convex nasal ridge | 81% | 225/278 | 44% | 23/52 | <0.00005 |
| Downslanted palpebral fissures | 79% | 208/263 | 56% | 29/52 | <0.001 |
| Highly arched palate | 77% | 160/208 | 67% | 30/45 | NS |
| Hypertrichosis | 76% | 93/122 | 51% | 23/45 | <0.005 |
| Postnatal growth retardation | 75% | 160/214 | 66% | 33/50 | NS |
| Micrognathia | 61% | 131/214 | 42% | 22/52 | <0.05 |
| Microcephaly (OFC < 3rd centile) | 54% | 77/143 | 87% | 45/52 | <0.00005 |
| Autism/autistiform behavior | 49% | 51/105 | 25% | 12/49 | <0.005 |
| Angulated thumbs | 49% | 135/273 | 2% | 1/51 | <0.00005 |
3.3.1. Epigenetic Mechanisms Underlying RSTS
Extensive studies of CBP protein structure and functions, reported in more than 2000 manuscripts published since its discovery by Dr. Goodman’s laboratory [4], have revealed a high level of complexity of structural features and that not all aspects of CBP molecular functions are understood (Fig. 3.1). CBP was discovered as the protein binding to CREB transcriptional factor, and the interaction between these two molecules depends on CREB phosphorylation at serine 133 (Ser 133) [4] located within the kinase-inducible domain (KIK). It turned out that CBP is a required coactivator for CREB-dependent transcriptional activation [29]. It is well documented that phosphorylation of CREB on serine 133 is the sufficient requirement for CREB-dependent transcriptional activation and involves direct interaction between the kinase-inducible domain interacting (KIX) domain (CBP) and phosphorylated KIK domain (CREB, Ser 133) [30].
CBP is a large, multifunctional transcriptional coactivator protein with molecular weight 26,531 Da that consists of 2441 amino acids. The CBP has been recognized as interacting with more than 400 nuclear proteins to mediate transcriptional activation from multiple promoters [31]. Through direct interactions with DNA-bound transcriptional factors and components of basal transcriptional machinery, including TBP, TFIIB, TFIIE, and TFIIF, CBP provides a platform for the generation of multiprotein complexes and serves as a bridge between transcriptional factors and the RNA polymerase II holoenzyme during transcriptional activation [32, 33]. CBP is recruited to the transcription sites via mechanisms that involve direct protein-protein interactions between the activation domain of transcription factors and CBP’s multiple protein-protein interaction domains, including the transcriptional adapter zinc-binding (TAZ) domains (reviewed in [34]). The CBP structure includes four zinc-binding domains that are localized in the three cysteine-/histidine-rich regions (i.e., CH1, CH2, and CH3). The TAZ1 domain is mapped to CH1. CH2 is the plant homeodomain (PHD)-type zinc finger motive. The CH3 region comprises two independent zinc finger motives, ZZ and TAZ2. Both TAZ1 and TAZ2 possess four zinc finger motives. Three of these motives are identical, but the fourth zinc finger motive is substantially different between the two domains and is believed to provide recognition specificity, as both TAZ1 and TAZ2 bind to two different groups of nuclear factors (or different regions on the same molecule, e.g., p53). In fact, the CH1 and CH3 regions mediate the majority of CBP’s protein- protein interactions. Although interaction with CBP’s nuclear receptor interaction domain (NRID) provides a mechanism for nuclear receptor (i.e., steroid hormones) transcriptional activation, the KIX domain binds to phosphorylated CREB. In addition, the glutamine-rich (Q) coactivator binding domain (CABD) is another domain that is critical for the assembly of multiprotein complexes [35]. In addition to its function as a platform, CBP belongs to a family chromatin-modifying enzymes and enhances transcription by altering chromatin structure via histone acetylation (Fig. 3.2) [7, 10, 12, 36]. While bromodomain (Bromo) may be involved in chromatin recognition, the KIT11 domain is a lysine acetyltransferase able to transfer acetyl groups primary on histone N-terminals and also nonhistone nuclear proteins [7, 8].
In the eukaryotic cell nucleus, DNA is packaged by histones into nucleosomes, which are repeated subunits of chromatin. One of the central questions in the regulation of gene expression is how the transcriptional machinery gains access to DNA that is tightly packed in chromatin. Over the last two decades, our understanding of the causal relationship between histone acetylation and gene expression has been enhanced dramatically by the identification of intrinsic HAT domains in several newly discovered coactivators of transcription, including CBP, p300, GCN5, and PCAF [10, 37] (Fig. 3.2). The finding that transcriptional coactivators are HATs recruited to specific gene promoters by activated transcription factors are consistent with an idea of the targeted chromatin acetylation as a critical step in transcriptional activation [37]. Moreover, there is selectivity in the specific HAT activity required for the function of distinct classes of transcription factors [12]. The HAT activity of CBP, recruited directly by phosphorylated CREB, is selectively required for the transcriptional function of CREB, whereas the HAT activity of PCAF is indispensable only for nuclear receptor activity [12]. It is now known that CBP interacts with a variety of nuclear factors (Fig. 3.1), highlighting CBP’s critical role in the regulation of a variety of cell programs during development and in the adult.
Histone acetylation is one of the major epigenetic mechanisms that controls chromatin structure and function in postmitotic mammalian neurons. As noted, CBP and p300 are critical for human and rodent development due to their ubiquitous expression patterns and their ability to control the functions of chromatin as epigenetic writers. In addition, they were found to bind to specific loci in daughter cells immediately following cell division and act as epigenetic chromatin “bookmarks” [38]. Owing to the high complexity of RSTS’s etiology, genetically engineered mice have been employed to gain insight into the mechanisms underlying this devastating disorder.
Both acetyltransferases, CBP and p300, can enhance transcription by relaxing the structure of chromatin nucleosomes via a mechanism that involves the direct acetylation of lysines (K) located at the N-terminals of histones H2A, H2B, H3, and H4, including the acetylation of H4-K5, H3-K14, H3-K18, H3-K27, and H3-K56 [39, 40]. CBP and p300 can be involved in two major epigenetic mechanisms, including the control of global acetylation of chromatin and mediating local chromatin modification in a promoter-specific manner via specific interactions with DNA regulatory element-bound and activated transcriptional factors [41].
The following describes a study of two CBP mutant mice. In a CBP-deficient mutant mouse model, amino acids 29–265 of CBP were replaced with a targeting vector [42]. In second study, a CBP truncated mutant mouse model was generated by insertional mutagenesis, resulting in the expression of a truncated version of CBP (amino acids 1–1084) that contained the CREB-binding domain amino acids (462–661) [43, 44]. In both cases, the homozygous mutants died in utero between 8 and 10.5 days post coitum (d.p.c.) [44, 45]. The CBP+/−-deficient mice exhibited various developmental abnormalities, partially resembling RSTS in humans [42]. The CBP+/−-truncated mutant leads to classical RSTS anomalies, showing a much more severe phenotype, including deficiencies in learning and memory [43].
CBP and EP300 show very similar patterns of molecular functions, but their expression during development and in the adult does not fully overlap. Although it is believed that there are subtle differences between CBP and EP300, patients with mutations in CBP or EP300 do not present any marked phenotypic differences (Table 3.1). In addition to certain skeletal abnormalities found specifically in heterozygous CBP null mice, there are no marked phenotypic distinctions between CBP heterozygous null mice and EP300 heterozygous null mice [42, 43, 46]. Each of three mutants, homozygous CBP null mice, homozygous EP300 null mice, and double heterozygous mice for CBP and p300, presents strikingly similar phenotypes and dies in utero [46]. These findings lead to hypotheses that combined levels of CBP and EP300 direct developmental processes rather than that CBP and p300 each have very distinct developmental roles. Although this might be true in general, there is also evidence that CBP and p300 functions do not always compensate for each other (e.g., [16]).
3.3.2. Testing the Biological Function of CBP in Rodents: Epigenetics and Memory
Life depends on the fidelity of DNA replication and the decoding of DNA into RNA and protein. Although our ability to accumulate knowledge involves genetic mechanisms, the nature of cognitive processes, how information is encoded, and what controls functional brain anatomy are not obvious. We thus ask: To what extent does the blueprint of life affect who we are? What is the relationship between neural information processing and chromatin, the functional form of DNA?
The stabilization of learned information into long-term memories requires both new gene expression and alterations in synaptic strength. Whether nondeclarative or declarative memory systems [47] are examined in invertebrates or vertebrates, information is stored first in a transient short-term memory that can eventually be stabilized into long-term memory [48, 49]. A variety of inhibitors of protein and RNA synthesis have been shown to effectively block long-term memory without altering short-term memory [50, 51]. Environmental stimuli or high levels of neuronal activity are known to induce a variety of immediate-early genes, such as Fos, Jun, and Zif268, in many brain areas [52, 53]. In addition, genetic studies in mice suggest that Zif268, CREB, and c-Fos may be involved in memory formation and consolidation [54–59]. Thus, regulatory mechanisms that direct transcription subsequent to the molecular changes in neurons during transient memory formation play a pivotal role in the conversion of short- to long-term memory.
CRE-binding factors, such as CREB, seem to be conserved from mollusks to mammals, and their activity is regulated by both cAMP and calcium influx (reviewed in [60]). These CREB/ATF or CREM families of activators and repressors belong to the bZip transcription factor class. A proposed mechanism by which CREB activates its target promoters is based on the observation that PKA phosphorylates CREB at Ser-133 in response to elevated levels of cAMP. CREB mediates transcriptional induction upon its phosphorylation by PKA [61, 62] or a calcium-dependent nuclear kinase [63], followed by direct interaction with a coactivator of transcription, CBP [4], which facilitates the assembly of the basic transcriptional machinery. CREB has a bipartite transactivation domain that consists of a constitutive domain, Q2, and an inducible domain, KID. It has been shown that the Q2 domain can potentially interact with TFIID via an hTAF135 bridging protein. In contrast, phosphorylation of the KID domain (Ser-133) induces interaction with the KIX domain (CREB interaction domain in CBP). Although it is unclear how the transactivation occurs, it is believed that CBP/CREB-P complex formation enables direct association with the RNA polymerase II complex.
Studies in Aplysia demonstrate that the cAMP-signaling pathway appears to play a central role in memory encoding [64–66]. A single electric shock to the tail of the mollusk produces a transient enhancement of the gill-withdrawal reflex. This short-term memory could be transformed into long-term memory by applying multiple stimuli. Reconstitution of the neurons that mediate the gill-withdrawal reflex by co-culturing a single Aplysia sensory neuron with the motor neuron that mediates the reflex allows the study of the molecular and cellular mechanisms involved in this simple form of learning. In response to one pulse of serotonin, this synapse underwent short-term facilitation, while five repeated pulses of serotonin resulted in long-term facilitation. The physiological changes that accompanied presynaptic facilitation were observed after intracellular injection of the catalytic subunit of cAMP-dependent protein kinase A (PKA) into Aplysia sensory neurons [65], while inhibitors of PKA blocked both forms of facilitation. Taken together, the conversion of short- to long-term memory requires the removal of certain inhibitory constraints on the storage of long-term memory followed by an activation of CREB-controlled gene expression; both mechanisms are required for the stabilization of transient memory [64–70].
These observations in Aplysia are in agreement with reported genetic studies in fruit flies (Drosophila) on memory, which demonstrate that fruit flies that overexpress a CREB repressor transgene under the control of the inducible heat shock promoter and were tested for memory retention after Pavlovian olfactory learning showed drastically impaired long-term memory formation but without an effect on transient short-term memory [71]. Moreover, overexpression of a CREB activator decreased the number of training trials needed to establish long-term memory but did not affect short-term memory formation [71]. Thus, the level of active CREB was associated with the number of training trials required for long-term memory in Drosophila olfactory associative learning.
In 1973, Bliss and Lomo discovered that the synaptic connections within the rabbit hippocampus undergo long-term potentiation (LTP) [72], which encodes memories [73–75]. There are two classes of mechanisms for long-lasting modification of synaptic strength that follow different patterns of tetanic electrical stimulation: LTP (for mechanisms that increase synaptic strength) and long-term depression (LTD) for mechanisms that decrease synaptic strength [75]. Pharmacological studies have demonstrated that blockers of N-methyl-D-aspartate receptor (NMDAR) eliminated both hippocampal LTP and LTD [76, 77] and hippocampus-dependent spatial memory formation in rodents treated with an NMDAR antagonist [78].
Advances in the study of learning and memory, using the genetic approach, provide an opportunity to study the correlation between LTP and memory storage in mutant mice. Mutant mice that carry a targeted deletion of the gene encoding CaMKII, the fyn tyrosine kinase, PKCg, NMDAR, or CREB exhibited deficits in spatial memory when tested in the Morris water maze paradigm (which is known to depend on the integrity of the hippocampus) [54, 76–84]. These mutants also showed modified LTP between CA3 and CA1 neurons (Schaffer collateral LTP) tested in hippocampal slices [54, 76–84].
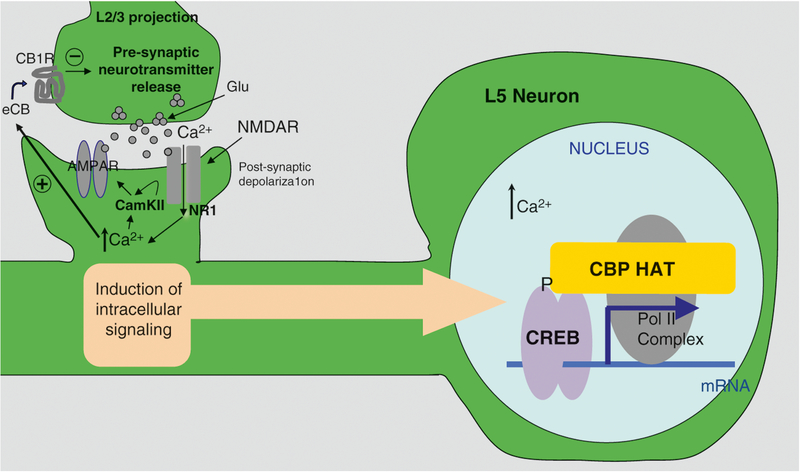
A number of studies in mammalian systems have shown that synaptic activity in neurons controls CBP’s ability to function as a transcriptional coactivator [85–88]. Voltage-gated calcium channels, such as the N-methyl-D-aspartate receptor (NMDAR), or local calcium transients can induce CBP-dependent transactivation in cultured hippocampal and cortical neurons [85–87]. In addition, CBP itself is a target for calcium influx-induced CaM kinase IV via a mechanism that involves CBP phosphorylation at Ser301, a requirement for NMDA-induced transcription [87]. Although the question of whether NMDA-dependent transcription requires CBP HAT activity has not been addressed, these findings suggest that, in addition to the well-characterized function of transcription factors, such as CREB in NMDA-induced gene expression, coactivators, such as CBP, are regulated by a separate pathway that plays a critical role in the activation of gene expression. To investigate these functions of CBP, Korzus et al. generated transgenic mice that express a reversible CBP-lacking HAT activity in a subset of excitatory neurons in the adult [14]. These mutant mice exhibited long-term memory deficits, while the encoding of new information and short-term memory were both normal. The behavioral phenotype was due to an acute requirement for CBP HAT activity in the adult, as it is rescued by both suppression of the transgene expression and by administration of a histone deacetylase (HDAC) inhibitor. This mouse model eliminates data interpretation problems that result from developmental changes and mimics a heterozygous CBP mutation that causes severe mental retardation [18]. These data indicate that, independent of its developmental role, CBP HAT activity is essential for higher cognitive functions in adults. Furthermore, this research provides the first demonstration that CBP’s HAT activity is required for brain information processing and demonstrates that histone modulation by deacetylase/acetylase activities affects specific processes of cognitive function. Thus, CBP-dependent transcriptional activation in response to synaptic signals appears to be regulated by multiple pathways through mechanisms that may underlie experience-induced neuronal gene expression during information encoding and memory formation (Fig. 3.5).
Fig. 3.5.
Putative molecular mechanism underlying alterations of synaptic strength potentially associated with cognitive performance, including memory formation. Studies have revealed that a number of synaptic (e.g., NMDAR, CaMKII) and nuclear (e.g., CREB and CBP) molecules could be critical for long-term memory consolidation. Long-term potentiation, or LTP, is an induced increase in synaptic efficacy. Many believe that LTP is a laboratory model for learning and memory [89]. Involvement of glutamate receptors, such as NMDAR, in LTP was demonstrated by Susumu Tonagawa at a molecular level [90] and by Richard Morris in behavioral studies [91]. Neuronal activity induces glutamate release into the synaptic cleft. Glutamate acts on the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) and NMDARs. However, initially, Na+ flows only through AMPAR because NMDAR is blocked by Mg. Postsynaptic depolarization removes the Mg2+ block, and then Ca2+ (and Na+) can flow through NMDAR. The resultant rise in Ca2+ levels within the dendritic spine is the critical trigger for LTP. Ca2+ influx activates CaMKII through autophosphorylation, and activated CaMKII induces molecular changes in postsynaptic neurons, yielding a change in synaptic strength called LTP. Ca2+ influx in dendritic spines activates intracellular signaling pathways, directing CREB phosphorylation, which is required but not sufficient for NMDA-induced gene expression. Studies of Drosophila, Aplysia, and mice clearly demonstrated the requirement for CREB in long-term memory. Further studies implicated CREB’s partner, CBP, as an obligatory component of the molecular mechanism underlying learning and memory [14, 92, 93]. AMPAR is α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor, NR1 is an obligatory subunit of postsynaptic NMDAR, CaMKII is Ca2+/Calmodulin-dependent protein kinase, CREB is a transcription factor, and CBP is coactivator of transcription and histone acetyltransferase (HAT)
In independent studies, Kandel and collaborators [92] tested a developmental model for RSTS that carried only a single crebbp allele, referred to as the CBP+/−-deficient mice [42]. These CBP mutant mice exhibited certain typical RSTS developmental abnormalities [42], but brain gross anatomy, emotional responses, and working memory were spared [92]. These studies revealed that chromatin acetylation, memory, and synaptic plasticity were abnormal in CBP+/−-deficient mutant mice, providing further evidence that implicates CBP as an important component of the molecular mechanism underlying learning and memory. Remarkably, inhibiting histone deacetyltransferase activity, the molecular counterpart of the histone acetylation function of CBP, ameliorated the memory impairment and the deficit in synaptic plasticity found in CBP+/−-deficient mutant mice.
Further studies of CBP function that involve different mouse models of RSTS that employs various CBP mutations and covalent histone modifications add substantially to the understanding of CBP function in memory consolidation [93–97]. Table 3.2 provides examples of genetic studies in mouse models. As seen in the table, disruption of the interaction between the transcriptional coactivator CBP and a gene promoter binding transcription factor, such as CREB, led to impaired synaptic plasticity, fear memory, and poor performance on tasks that tested the ability to remember familiar objects [94]. The ability of HDAC inhibitors to rescue some these phenotypes demonstrated that recruitment of CBP’s HAT to specific gene promoters is critical for long-term memory consolidation [102].
Table 3.2.
Examples of mutations targeted to the CREBBP gene in a mouse model that revealed high complexity of this gene function in cell regulation, development, adult neurogenesis, and cognition
| Mouse model | Genotype | Phenotype |
|---|---|---|
| Mutations that specifically target CBP HAT activity in adults: | ||
| CBP{HAT-}Exc. Neur [93] | Transgenic/hemizygote (germline mutation) | Deficit in LTM |
| Tetracycline-inducible dominant negative CBP transgene that lacks HAT activity expressed in CaMKIIα positive excitatory neurons (exc. Neur.) in the forebrain | Normal STM | |
| No effects on prenatal development | ||
| No effects on postnatal development | ||
| CBP {HAT-}mPFC [98, 99] | Viral-mediated gene transfer (somatic mutation) dominant negative CBP transgene that lacks HAT activity expressed in the medial prefrontal cortex (mPFC) | Deficit in LTM (limited to mPFC-dependent behavior) |
| Normal STM | ||
| Deficit in discrimination between safety and danger | ||
| No effects on prenatal development | ||
| No effects on postnatal development | ||
| Mutations that target CBP function as a “platform”: | ||
| CBP −/− [45] | Null mutant (germline mutation) | Death in utero |
| CBP Δ/Δ [44] | Null mutant (germline mutation) | Death in utero |
| CBP +/Δ [43] | Heterozygote (germline mutation) | Deficit in LTM |
| Truncated allele that expresses dominant negative CBP truncated protein (aa1–1084) | Normal STM | |
| Skeletal abnormalities/growth retardation | ||
| CBP +/− [42, 92, 100] | Heterozygote (germline mutation) | Deficit in LTM |
| Normal STM | Deficit in neurogenesis | |
| Skeletal abnormalities/growth retardation | ||
| CBP Δ1 [93] | Transgenic/hemizygote (germline mutation) | Normal STM |
| Transgenic mice that express dominant negative truncated CBP protein (aa1–1083) under control of CaMKIIα promoter transgene activated during postnatal brain development | Deficit in LTM | |
| No effects on prenatal development | ||
| CBPKIX/KIX [95, 101] | Homozygote (germline mutation) triple point mutation in the KIX domain | Deficit in LTM |
| Focal homozygous k.o. No effects on prenatal development in the dorsal hippocampus | Normal STM | |
| No effects on postnatal development | ||
The system-level effects of CBP hypofunction depend not only on whether the mutation was present during development or only in the adult but also on whether the locus of the mutation is critical. A number of studies have addressed this specific problem with great success. Studies have shown that CBP hypofunction targeted specifically to adult excitatory neurons [14], adult hippocampal neurons [95], adult excitatory neurons in the medial prefrontal cortex (mPFC) [98, 99], or intra-lateral amygdala infusion of c646 (a selective pharmacological inhibitor of p300/CBP HAT activity) in adult mice shortly following fear conditioning [103] resulted in selective impairment of synaptic plasticity in the targeted brain region [95, 103] and deficits in long-term memory consolidation [14, 95, 98, 99, 103]. Further, changes in histone acetylation in the mPFC were connected to the extinction of conditioned fear [104, 105], whereas intrahippocampal delivery of histone deacetylase inhibitors facilitated fear extinction [106].
CBP also has been implicated in adult neurogenesis, which has been linked to higher cognitive function and depression. The reduction of CBP targeted to the adult brain leads to abnormalities in environmental enrichment-induced neurogenesis [100], which provides strong evidence for the role of CBP in adult neurogenesis-dependent enhancement of adaptability toward novel experiences [107, 108].
A CBP deficiency that results from elimination of its multiple functional domains, including HAT, is associated with abnormal synaptic plasticity [92, 93], and altered histone acetylation in the hippocampus correlates with new learning experiences [92, 109]. Thus, chromatin dynamics appear to control memory consolidation via CBP-mediated acetylation during gene activation, altering chromatin structure and mediating gene-specific removal of epigenetically controlled repression.
Synaptic stimulation of excitatory neurons leads to increases in intracellular Ca2+ and modulation of gene expression [110, 111]. Ca2+ signaling to the nucleus has been studied extensively in neurons in culture, where it is clear that a primary Ca2+ signal can act both directly through CaM in the nucleus [112–114] and indirectly through the activation of other second-messenger pathways, such as cAMP [115] and MAP kinase [114, 116]. Mayford laboratory discovered that nuclear blocking of Ca2+/calmodulin-mediated signaling in the nuclei of hippocampal neurons blocks stabilization of new memories but spares learning and short-term memory [117]. Thus, both impaired nuclear Ca2+ signaling in excitatory neurons [117] and a deficiency in CBP acetyltransferase activity in excitatory neurons [14] produce exactly the same biological effects, preventing memory consolidation processes (Fig. 3.5).
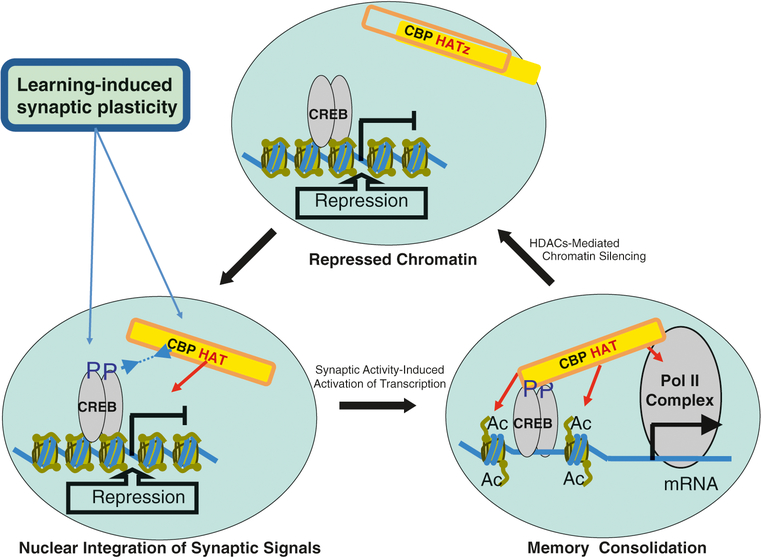
Understanding how chromatin is involved in neural information processing also is of interest. It has been established that CREB phosphorylation (a critical step for CBP recruitment) is insufficient to induce neuronal gene expression [87, 118]. A separate, still unknown, CBP-signaling pathway was shown to be required for synaptic plasticity-dependent neuronal gene expression. Conversely, Frey and Morris proposed that heterosynaptic coactivation of both glutamate and modulatory receptors may be necessary to trigger the upregulation of the relevant protein synthesis [119]. These findings suggest that multiple signaling pathways must be activated to allow neuronal activity-dependent gene expression and that such pathways are critical for memory consolidation by recruiting functional CBP HAT activity and possibly by altering chromatin structure (Fig. 3.6). Chromatin-based information processing provides an advanced threshold control, whereby the nuclear signal generated as a sum of individual synaptic signals determines the outcome. If transcription is specifically induced by combining synaptic signals in the presence (or absence) of other irrelevant signals, chromatin may function to filter neural information. Thus, the integration of brief synaptic signals by CBP in the nucleus leads to (a) reversible changes in chromatin structure that result in removal of gene-specific epigenetically controlled repression, (b) transient alterations in the gene expression that last a few hours after the initial learning experience, and, subsequently, (c) permanent learning-dependent changes in neuronal networks (Fig. 3.6).
Fig. 3.6.
Proposed model for CBP HAT involvement in long-term memory consolidation. Before activation, target genes are repressed by chromatin structure. Signal-inducing CREB phosphorylation must occur but is not sufficient. It is well known that CBP is independently regulated in response to NMDA. A second signal is required to remove chromatin repression. This can be accomplished by CBP-dependent histone acetylation. The covalent modification by HAT activity leaves long-lasting marks on chromatin at the target genes. This represents a very attractive mechanism for the regulation of long-lasting transcriptional changes associated with long-term synaptic and behavioral plasticity. The proposed model postulates that two signals—the first signal induces CREB and the second signal removes repression of target genes by chromatin acetylation—are required to occur during initial learning. This sort of the acetylation-mediated covalent modification by CBP could change requirements for subsequent transcriptional activation of genes in response to future signals. This would open a temporal window in which cellular signals, which would not recruit acetyltransferase, would nevertheless stimulate transcription required for memory consolidation. Chromatin opening at the target genes by acetylation would allow for prolonged transcription, even in the absence of an initial stimulus. After execution of “memory program,” these transiently activated gene expression would shut down via default mechanisms controlled by ubiquitous HDACs, allowing for a homeostatic update of integrated circuits. Model initially presented in Korzus et al. (2004) [14], modified by author
The model presented in Fig. 3.6 has several implications. The gating and filtering of neural signals by chromatin suggests an additional layer of complexity for computational aspects of neural networks. The chromatin gating/filtering hypothesis provides a novel way to discriminate stabilized changes in neural networks. Another implication of this model has more practical use in biomedical research. This implication is that epigenetic mechanisms permit biological systems to respond and adapt to environmental stimuli by altering gene expression. Alternatively, environmental influences can provoke undesired and persistent epigenetic effects that may increase susceptibility to mental retardation, long-latency ailments of the nervous system, and unsuccessful aging. Recently, strong evidence has implicated epigenetic mechanisms based on chromatin remodeling as the cause of neuronal dysfunction [120, 121]. Future research should broaden the study of molecular mechanisms that are involved in the stabilization of learning-initiated changes in neuronal networks underlying memory consolidation and provide new avenues to investigate mental illnesses, including memory disorders and psychostimulant-elicited plasticity in the brain-reward system that underlies drug addiction.
The model presented in Fig. 3.6 is consistent with the prevailing hypothesis of learning and memory in the mammalian brain, which postulates that the ability to learn involves a selective and bidirectional modification of individual synapses, such that the same stimulus elicits different responses before and after learning [73]. Each learning experience initiates changes in specific synapses that, subsequently, translate into alterations of neuronal network properties and alter behavioral responses. During initial learning, protein synthesis inhibitors allow animals to learn and remember a given task for a few hours but severely impair memory after 24 h [51]. The link between gene expression and synapse alteration suggests that protein synthesis is required to permanently alter synapses modified by a learning experience [122]. Thus, the molecular neuronal changes during information acquisition have pivotal effects on gene expression, facilitating the conversion of short- to long-term memory. Recent research has revealed that the rate of transcription is a function of the physical state of chromatin and that chromatin plasticity mediated by chemical modification allows dynamic changes in gene expression without changing the DNA sequence [123, 124].
Chromatin functions are not limited to controlling the transmission of genetic and epigenetic information. Based on current data, it has been proposed that chromatin dynamics in individual neurons may shape neural information processing for what is or is not permanently stored. This hypothesis is consistent with the view that knowledge is not encoded in a simple molecular form but reflects the dynamics of neural interconnectivity throughout the brain. This hypothesis postulates that epigenetic regulation controls cognitive performance and may explain some of the cognitive phenotypes associated with epigenetic disorders, such as RSTS.
3.3.3. Potential Therapeutic Applications of Histone Deacetylase Inhibitors for RSTS
It is feasible to infer that epigenetic aberrations can be “healed” because the major feature of epigenetic changes is their reversibility. Whereas the specificity of these changes creates a major challenge, the global patterning of epigenetic marks can be controlled by generating an appropriate balance between histone acetylation and deacetylation (e.g., [14]). In addition, there is growing evidence that shows that a variety of available drugs are capable of reactivating epigenetically silenced genes, including a variety of HDAC inhibitors, including suberoylanilide hydroxamic acid (SAHA), valproic acid (VPA), and trichostatin A (TSA). It also has been demonstrated that DNA demethylation drugs, such as zebularine and 5-aza-20- deoxycytidine (5-ADC), are capable of unlocking previously inactive genes. Although some of these drugs are being used in cancer treatment therapies, epigenetic drugs have recently been considered for neurological and mental disorders [125]. Preclinical studies of mouse models for RSTS with reduced HAT activity demonstrated that some of the cognitive impairments could be ameliorated with HDAC inhibitor treatment [14, 92]. Recently, Lopez-Atalaya et al. reported that deficits in the levels of histone H2A and H2B acetylation in lymphoblastoid cell lines derived from nine patients with RSTS2 were rescued by treatment with the HDAC inhibitor TSA [126]. Although some HDAC inhibitors show positive effects on memory and synaptic plasticity [102, 106, 109, 127, 128], the HDAC3 gene was found to negatively regulate long-term memory [129]. In fact, HDAC inhibitors have been used in the past by psychiatrists as mood stabilizers and antiepileptics (e.g., VPA). Epigenetic regulators are being intensively investigated as a possible treatment for cancers [130], parasitics [131], inflammatory diseases [124] and, more recently, mental and neurological disorders, including RSTS [132–134].
References
- 1.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276(40):36865–8. [DOI] [PubMed] [Google Scholar]
- 2.Neely KE, Workman JL. Histone acetylation and chromatin remodeling: which comes first? Mol Genet Metab. 2002;76(1):1–5. [DOI] [PubMed] [Google Scholar]
- 3.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15(2):172–83. [DOI] [PubMed] [Google Scholar]
- 4.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–9. [DOI] [PubMed] [Google Scholar]
- 5.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8(8):869–84. [DOI] [PubMed] [Google Scholar]
- 6.Janknecht R. The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol Histopathol. 2002;17(2):657–68. [DOI] [PubMed] [Google Scholar]
- 7.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–6. [DOI] [PubMed] [Google Scholar]
- 8.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114(Pt 13):2363–73. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. 2013;70(21):3989–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87(5):953–9. [DOI] [PubMed] [Google Scholar]
- 11.Yuan LW, Soh JW, Weinstein IB. Inhibition of histone acetyltransferase function of p300 by PKCdelta. Biochim Biophys Acta. 2002;1592(2):205–11. [DOI] [PubMed] [Google Scholar]
- 12.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279(5351):703–7. [DOI] [PubMed] [Google Scholar]
- 13.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16(2):369–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Atalaya JP, Valor LM, Barco A. Epigenetic factors in intellectual disability: the Rubinstein-Taybi syndrome as a paradigm of neurodevelopmental disorder with epigenetic origin. Prog Mol Biol Transl Sci. 2014;128:139–76. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira AM, Abel T, Brindle PK, Wood MA. Differential role for CBP and p300 CREB-binding domain in motor skill learning. Behav Neurosci. 2006;120(3):724–9. [DOI] [PubMed] [Google Scholar]
- 17.Rubinstein JH, Taybi H. Broad thumbs and toes and facial abnormalities. Am J Dis Child. 1963;105:588–608. [DOI] [PubMed] [Google Scholar]
- 18.Hennekam RC, Stevens CA, Van de Kamp JJ. Etiology and recurrence risk in Rubinstein-Taybi syndrome. Am J Med Genet Suppl. 1990;6:56–64. [DOI] [PubMed] [Google Scholar]
- 19.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–51. [DOI] [PubMed] [Google Scholar]
- 20.LOVD v.2.0: the next generation in gene variant databases (Updated February 12, 2016) [Internet]. Wiley-Liss, Inc; 2011. http://www.LOVD.nl. [DOI] [PubMed] [Google Scholar]
- 21.CREBBP [Internet]. Huret Jean-Loup (Editor-in-Chief); INIST-CNRS; (Publisher). 2010. http://atlasgeneticsoncology.org//Genes/CBPID42.html. [Google Scholar]
- 22.Coupry I, Roudaut C, Stef M, Delrue MA, Marche M, Burgelin I, Taine L, Cruaud C, Lacombe D, Arveiler B. Molecular analysis of the CBP gene in 60 patients with Rubinstein-Taybi syndrome. J Med Genet. 2002;39(6):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentivegna A, Milani D, Gervasini C, Castronovo P, Mottadelli F, Manzini S, Colapietro P, Giordano L, Atzeri F, Divizia MT, Uzielli ML, Neri G, Bedeschi MF, Faravelli F, Selicorni A, Larizza L. Rubinstein-Taybi syndrome: spectrum of CREBBP mutations in Italian patients. BMC Med Genet. 2006;7:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, Hennekam RC, Peters DJ. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76(4):572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negri G, Magini P, Milani D, Colapietro P, Rusconi D, Scarano E, Bonati MT, Priolo M, Crippa M, Mazzanti L, Wischmeijer A, Tamburrino F, Pippucci T, Finelli P, Larizza L, Gervasini C. From whole gene deletion to point mutations of EP300-positive Rubinstein-Taybi patients: new insights into the mutational spectrum and peculiar clinical hallmarks. Hum Mutat. 2016;37(2):175–83. [DOI] [PubMed] [Google Scholar]
- 26.Spena S, Milani D, Rusconi D, Negri G, Colapietro P, Elcioglu N, Bedeschi F, Pilotta A, Spaccini L, Ficcadenti A, Magnani C, Scarano G, Selicorni A, Larizza L, Gervasini C. Insights into genotype-phenotype correlations from CREBBP point mutation screening in a cohort of 46 Rubinstein-Taybi syndrome patients. Clin Genet. 2015;88(5):431–40. [DOI] [PubMed] [Google Scholar]
- 27.van Belzen M, Bartsch O, Lacombe D, Peters DJ, Hennekam RC. Rubinstein-Taybi syndrome (CREBBP, EP300). Eur J Hum Genet. 2011;19(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fergelot P, Van Belzen M, Van Gils J, Afenjar A, Armour CM, Arveiler B, Beets L, Burglen L, Busa T, Collet M, Deforges J, de Vries BB, Dominguez Garrido E, Dorison N, Dupont J, Francannet C, Garcia-Minaur S, Gabau Vila E, Gebre-Medhin S, Gener Querol B, Genevieve D, Gerard M, Gervasini CG, Goldenberg A, Josifova D, Lachlan K, Maas S, Maranda B, Moilanen JS, Nordgren A, Parent P, Rankin J, Reardon W, Rio M, Roume J, Shaw A, Smigiel R, Sojo A, Solomon B, Stembalska A, Stumpel C, Suarez F, Terhal P, Thomas S, Touraine R, Verloes A, Vincent-Delorme C, Wincent J, Peters DJ, Bartsch O, Larizza L, Lacombe D, Hennekam RC. Phenotype and genotype in 52 patients with Rubinstein-Taybi syndrome caused by EP300 mutations. Am J Med Genet A. 2016;170(12): 3069–82. [DOI] [PubMed] [Google Scholar]
- 29.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370(6486):223–6. [DOI] [PubMed] [Google Scholar]
- 30.Cardinaux JR, Notis JC, Zhang Q, Vo N, Craig JC, Fass DM, Brennan RG, Goodman RH. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20(5):1546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiama N. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7(6):230–6. [DOI] [PubMed] [Google Scholar]
- 33.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14(13):1553–77. [PubMed] [Google Scholar]
- 34.De Guzman RN, Martinez-Yamout MA, Dyson HJ, Wright PE. Structure and function of the CBP/p300 TAZ domains In: Iuchi S, Kuldell N, editors. Zinc finger proteins: from atomic contact to cellular function. Molecular biology intelligence unit. Georgetown, TX, New York: Landes Bioscience; Kluwer Academic/Plenum; 2005. [Google Scholar]
- 35.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415(6871):549–53. [DOI] [PubMed] [Google Scholar]
- 36.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384(6610):641–3. [DOI] [PubMed] [Google Scholar]
- 37.Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6(2):176–84. [DOI] [PubMed] [Google Scholar]
- 38.Wong MM, Byun JS, Sacta M, Jin Q, Baek S, Gardner K. Promoter-bound p300 complexes facilitate post-mitotic transmission of transcriptional memory. PLoS One. 2014;9(6):e99989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McManus KJ, Hendzel MJ. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol Cell Biol. 2003;23(21):7611–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci U S A. 1997;94(19):10215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oike Y, Hata A, Mamiya T, Kaname T, Noda Y, Suzuki M, Yasue H, Nabeshima T, Araki K, Yamamura K. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum Mol Genet. 1999;8(3):387–96. [DOI] [PubMed] [Google Scholar]
- 44.Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood. 1999;93(9):2771–9. [PubMed] [Google Scholar]
- 45.Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95(1–2):133–45. [DOI] [PubMed] [Google Scholar]
- 46.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–72. [DOI] [PubMed] [Google Scholar]
- 47.Squire LR. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans [published erratum appears in Psychol Rev 1992 Jul;99(3):582]. Psychol Rev. 1992;99(2):195–231. [DOI] [PubMed] [Google Scholar]
- 48.McGaugh JL, Hertz MJ. Memory consolidation. San Francisco: Albion; 1972. [Google Scholar]
- 49.Squire LR. Memory and brain. New York: Oxford; 1987. [Google Scholar]
- 50.Andrew RJ. The functional organization of phases of memory consolidation In: Hinde RA, Beer C, Bunsel M, editors. Advances in the study of behaviour, vol. 11 New York: Academic; 1980. p. 337–67. [Google Scholar]
- 51.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96(3): 518–59. [PubMed] [Google Scholar]
- 52.Worley PF, Cole AJ, Murphy TH, Christy BA, Nakabeppu Y, Baraban JM. Synaptic regulation of immediate-early genes in brain. Cold Spring Harb Symp Quant Biol. 1990;55:213–23. [DOI] [PubMed] [Google Scholar]
- 53.Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cell Mol Life Sci. 1999;55(4):564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP- responsive element-binding protein. Cell. 1994;79(1):59–68. [DOI] [PubMed] [Google Scholar]
- 55.Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34(3):447–62. [DOI] [PubMed] [Google Scholar]
- 56.Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4(3):289–96. [DOI] [PubMed] [Google Scholar]
- 57.Josselyn SA, Shi C, Carlezon WA Jr, Neve RL, Nestler EJ, Davis M. Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J Neurosci. 2001;21(7):2404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, Wagner EF, Gass P. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23(27):9116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kida S, Josselyn SA, de Ortiz SP, Kogan JH, Chevere I, Masushige S, Silva AJ. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 2002;5(4):348–55. [DOI] [PubMed] [Google Scholar]
- 60.Brindle PK, Montminy MR. The CREB family of transcription activators. Curr Opin Genet Dev. 1992;2(2):199–204. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59(4):675–80. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalez GA, Yamamoto KK, Fischer WH, Karr D, Menzel P, Biggs 3rd W, Vale WW, Montminy MR. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337(6209):749–52. [DOI] [PubMed] [Google Scholar]
- 63.Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16(1): 89–101. [DOI] [PubMed] [Google Scholar]
- 64.Byrne JH, Kandel ER. Presynaptic facilitation revisited: state and time dependence. J Neurosci. 1996;16(2):425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castellucci VF, Kandel ER, Schwartz JH, Wilson FD, Nairn AC, Greengard P. Intracellular injection of t he catalytic subunit of cyclic AMP- dependent protein kinase simulates facilitation of transmitter release underlying behavioral sensitization in Aplysia. Proc Natl Acad Sci U S A. 1980;77(12):7492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castellucci VF, Nairn A, Greengard P, Schwartz JH, Kandel ER. Inhibitor of adenosine 3′:5′-monophosphate-dependent protein kinase blocks presynaptic facilitation in Aplysia. J Neurosci. 1982;2(12):1673–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abel T, Martin KC, Bartsch D, Kandel ER. Memory suppressor genes: inhibitory constraints on the storage of long- term memory. Science. 1998;279(5349):338–41. [DOI] [PubMed] [Google Scholar]
- 68.Alberini CM, Ghirardi M, Metz R, Kandel ER. C/EBP is an immediate-early gene required for the consolidation of long- term facilitation in Aplysia. Cell. 1994;76(6):1099–114. [DOI] [PubMed] [Google Scholar]
- 69.Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95(2):211–23. [DOI] [PubMed] [Google Scholar]
- 70.Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83(6):979–92. [DOI] [PubMed] [Google Scholar]
- 71.Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79(1):49–58. [DOI] [PubMed] [Google Scholar]
- 72.Bliss TV, Gardner-Medwin AR. Long-lasting potentiation of synaptic transmission in the dentate area of the unanaestetized rabbit following stimulation of the perforant path. J Physiol (Lond). 1973;232(2):357–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361(6407):31–9. [DOI] [PubMed] [Google Scholar]
- 74.Bliss TV, Richter-Levin G. Spatial learning and the saturation of long-term potentiation [comment]. Hippocampus. 1993;3(2):123–5. [DOI] [PubMed] [Google Scholar]
- 75.Malenka RC. Synaptic plasticity in the hippocampus: LTP and LTD. Cell. 1994;78(4):535–8. [DOI] [PubMed] [Google Scholar]
- 76.Collingridge GL, Kehl SJ, McLennan H. The antagonism of amino acid-induced excitations of rat hippocampal CA1 neurones in vitro. J Physiol (Lond). 1983;334:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudek SM, Bear MF. Homosynaptic long-term depression in area CA1 of hippocampus and effects of N-methyl-D-aspartate receptor blockade. Proc Natl Acad Sci U S A. 1992;89(10):4363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis S, Butcher SP, Morris RG. The NMDA receptor antagonist D-2-amino-5-phosphonopentanoate (D-AP5) impairs spatial learning and LTP in vivo at intracerebral concentrations comparable to those that block LTP in vitro. J Neurosci. 1992;12(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abeliovich A, Chen C, Goda Y, Silva AJ, Stevens CF, Tonegawa S. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell. 1993;75(7):1253–62. [DOI] [PubMed] [Google Scholar]
- 80.Abeliovich A, Paylor R, Chen C, Kim JJ, Wehner JM, Tonegawa S. PKC gamma mutant mice exhibit mild deficits in spatial and contextual learning. Cell. 1993;75(7):1263–71. [DOI] [PubMed] [Google Scholar]
- 81.Grant SG, O’Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice [see comments]. Science. 1992;258(5090):1903–10. [DOI] [PubMed] [Google Scholar]
- 82.Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, et al. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature. 1995;373(6510):151–5. [DOI] [PubMed] [Google Scholar]
- 83.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium- calmodulin kinase II mutant mice. Science. 1992;257(5067):201–6. [DOI] [PubMed] [Google Scholar]
- 84.Silva AJ, Wang Y, Paylor R, Wehner JM, Stevens CF, Tonegawa S. Alpha calcium/calmodulin kinase II mutant mice: deficient long-term potentiation and impaired spatial learning. Cold Spring Harb Symp Quant Biol. 1992;57:527–39. [DOI] [PubMed] [Google Scholar]
- 85.Hardingham GE, Chawla S, Cruzalegui FH, Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22(4):789–98. [DOI] [PubMed] [Google Scholar]
- 86.Hu SC, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22(4):799–808. [DOI] [PubMed] [Google Scholar]
- 87.Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34(2):235–44. [DOI] [PubMed] [Google Scholar]
- 88.Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111(4):483–93. [DOI] [PubMed] [Google Scholar]
- 89.Malenka RC, Nicoll RA. Long-term potentiation—a decade of progress? Science. 1999;285(5435):1870–4. [DOI] [PubMed] [Google Scholar]
- 90.Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87(7):1327–38. [DOI] [PubMed] [Google Scholar]
- 91.Morris RG, Anderson E, Lynch GS, Baudry M. Selective impairment of learning and blockade of long-term potentiation by an N-methyl-D-aspartate receptor antagonist, AP5. Nature. 1986;319(6056):774–6. [DOI] [PubMed] [Google Scholar]
- 92.Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–59. [DOI] [PubMed] [Google Scholar]
- 93.Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12(2):111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13(5):609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36(8):1545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci. 2011;31(5):1652–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levenson JM, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol Life Sci. 2006;63(9):1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vieira PA, Korzus E. CBP-dependent memory consolidation in the prefrontal cortex supports object-location learning. Hippocampus. 2015;25(12):1532–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vieira PA, Lovelace JW, Corches A, Rashid AJ, Josselyn SA, Korzus E. Prefrontal consolidation supports the attainment of fear memory accuracy. Learn Mem. 2014;21(8):394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lopez-Atalaya JP, Ciccarelli A, Viosca J, Valor LM, Jimenez-Minchan M, Canals S, Giustetto M, Barco A. CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J. 2011;30(20):4287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, Brindle PK. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419(6908):738–43. [DOI] [PubMed] [Google Scholar]
- 102.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27(23):6128–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maddox SA, Watts CS, Schafe GE. p300/CBP histone acetyltransferase activity is required for newly acquired and reactivated fear memories in the lateral amygdala. Learn Mem. 2013;20(2):109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol Psychiatry. 2012;72(1): 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14(4):268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121(5):1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aimone JB, Deng W, Gage FH. Resolving new memories: a critical look at the dentate gyrus, adult neurogenesis, and pattern separation. Neuron. 2011;70(4):589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sahay A, Wilson DA, Hen R. Pattern separation: a common function for new neurons in hippocampus and olfactory bulb. Neuron. 2011;70(4):582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279(39):40545–59. [DOI] [PubMed] [Google Scholar]
- 110.Bading H. Transcription-dependent neuronal plasticity the nuclear calcium hypothesis. Eur J Biochem. 2000;267(17):5280–3. [DOI] [PubMed] [Google Scholar]
- 111.West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98(20):11024–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385(6613):260–5. [DOI] [PubMed] [Google Scholar]
- 113.Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392(6672):198–202. [DOI] [PubMed] [Google Scholar]
- 114.Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4(3):261–7. [DOI] [PubMed] [Google Scholar]
- 115.Chetkovich DM, Gray R, Johnston D, Sweatt JD. N-methyl-D-aspartate receptor activation increases cAMP levels and voltage-gated Ca2+ channel activity in area CA1 of hippocampus. Proc Natl Acad Sci U S A. 1991;88(15):6467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294(5541):333–9. [DOI] [PubMed] [Google Scholar]
- 117.Limback-Stokin K, Korzus E, Nagaoka-Yasuda R, Mayford M. Nuclear calcium/calmodulin regulates memory consolidation. J Neurosci. 2004;24(48):10858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281(5382):1505–9. [DOI] [PubMed] [Google Scholar]
- 119.O’Carroll CM, Morris RG. Heterosynaptic co-activation of glutamatergic and dopaminergic afferents is required to induce persistent long-term potentiation. Neuropharmacology. 2004;47(3):324–32. [DOI] [PubMed] [Google Scholar]
- 120.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–63. [DOI] [PubMed] [Google Scholar]
- 121.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–25. [DOI] [PubMed] [Google Scholar]
- 122.Frey U, Morris RG. Synaptic tagging and long-term potentiation [see comments]. Nature. 1997;385(6616):533–6. [DOI] [PubMed] [Google Scholar]
- 123.Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83(4):468–76. [DOI] [PubMed] [Google Scholar]
- 124.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14(1):5–16. [DOI] [PubMed] [Google Scholar]
- 125.Day JJ, Sweatt JD. Epigenetic treatments for cognitive impairments. Neuropsychopharmacology. 2012;37(1):247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lopez-Atalaya JP, Gervasini C, Mottadelli F, Spena S, Piccione M, Scarano G, Selicorni A, Barco A, Larizza L. Histone acetylation deficits in lymphoblastoid cell lines from patients with Rubinstein-Taybi syndrome. J Med Genet. 2012;49(1):66–74. [DOI] [PubMed] [Google Scholar]
- 127.Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Roozendaal B, Hernandez A, Cabrera SM, Hagewoud R, Malvaez M, Stefanko DP, Haettig J, Wood MA. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci. 2010;30(14):5037–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31(2):764–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Berdasco M, Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum Genet. 2013;132(4):359–83. [DOI] [PubMed] [Google Scholar]
- 131.Duraisingh MT, Horn D. Epigenetic regulation of virulence gene expression in parasitic protozoa. Cell Host Microbe. 2016;19(5):629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8(1):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Graff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur J Neurosci. 2009; 30(1):1–8. [DOI] [PubMed] [Google Scholar]
- 134.Rudenko A, Tsai LH. Epigenetic modifications in the nervous system and their impact upon cognitive impairments. Neuropharmacology. 2014;80:70–82. [DOI] [PubMed] [Google Scholar]
- 135.Schorry EK, Keddache M, Lanphear N, Rubinstein JH, Srodulski S, Fletcher D, Blough-Pfau RI, Grabowski GA. Genotype-phenotype correlations in Rubinstein-Taybi syndrome. Am J Med Genet A. 2008;146A(19):2512–9. [DOI] [PubMed] [Google Scholar]
- 136.Rusconi D, Negri G, Colapietro P, Picinelli C, Milani D, Spena S, Magnani C, Silengo MC, Sorasio L, Curtisova V, Cavaliere ML, Prontera P, Stangoni G, Ferrero GB, Biamino E, Fischetto R, Piccione M, Gasparini P, Salviati L, Selicorni A, Finelli P, Larizza L, Gervasini C. Characterization of 14 novel deletions underlying Rubinstein-Taybi syndrome: an update of the CREBBP deletion repertoire. Hum Genet. 2015;134(6):613–26. [DOI] [PubMed] [Google Scholar]
- 137.Wincent J, Luthman A, van Belzen M, van der Lans C, Albert J, Nordgren A, Anderlid BM. CREBBP and EP300 mutational spectrum and clinical presentations in a cohort of Swedish patients with Rubinstein-Taybi syndrome. Mol Genet Genomic Med. 2016;4(1): 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shanmugam MK, Sethi G. Role of epigenetics in inflammation-associated diseases. Subcell Biochem. 2013;61:627–57. [DOI] [PubMed] [Google Scholar]