Summary
Several lines of evidence implicate abnormalities in glutamatergic neural transmission in major depressive disorder (MDD) and treatment response. A high percentage of MDD patients do not respond adequately to antidepressants and are classified as having treatment-resistant depression (TRD). In this study we investigated five GRIK4 variants, previously associated with antidepressants response, in an Italian cohort of 247 MDD no-TRD and 380 TRD patients. We found an association between rs11218030 G allele and TRD. Moreover, significant associations between rs11218030 and rs1954787 and the presence of psychotic symptoms were observed. In conclusion, our data support the involvement of GRIK4 in TRD and in the risk of developing psychotic symptoms during depressive episodes.
Introduction
MDD is one of the most prevalent mental disorders worldwide with a profound impact on public health (Carvalho et al., 2014). Despite significant progress in antidepressant treatments, the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study has shown that only about one-third of patients experience full remission after the first treatment trial, and for many patients, achieving remission requires repeated trials of sufficiently dosed antidepressant medication. Moreover, approximately 30% or more MDD patients have refractory or TRD (Bentley et al., 2014).
Research from the past decade has accumulated a large amount of evidence involving the glutamate system in antidepressant response, suggesting it as a target for the development of novel antidepressants (Catena-Dell'Osso et al., 2013). Interestingly, traditional monoaminergic-based antidepressants have been repeatedly shown to interfere with glutamate system function, starting with modulation of N-methyl-D-aspartate (NMDA) receptors and it has been demonstrated that antidepressants selectively regulate glutamate receptors expression and function (Musazzi et al., 2013).
Several pharmacogenomic studies investigated Glutamate Receptor Ionotropic Kainate 4 (GRIK4) gene variants in relation to antidepressant outcome with interesting but contrasting findings (Paddock et al., 2007; Horstmann et al., 2010; Perlis et al., 2010; Serretti et al., 2012; Pu et al., 2013; Kawaguchi et al., 2014). GRIK4 encodes the kainic acid-type glutamate receptor 1 (KA1) subunit, which co-assembles with other glutamate receptor subunits to form cation-selective ion channels, but might also possess metabotropic functions (Horstmann & Binder, 2009). The role of ionotropic glutamate receptors, such as GRIK4, in antidepressant action has been supported by studies in animal models. Chronic treatment with antidepressants resulted in a region-specific reduction in binding activity of N-methyl-D-aspartate receptors (Nowak et al., 1996; Boyer et al., 1998), and an antidepressant phenotype associated with ablation of GRIK4 and a parallel disruption in hippocampal plasticity in mice was recently demonstrated (Catches et al., 2012).
Taking into account the involvement of GRIK4 in antidepressant actions and the dearth of studies focusing on GRIK4 SNPs, we performed an association study in Italian TRD and no-TRD MDD patients, investigating the role of the GRIK4 genetic variants that are the most significant in the STAR*D study (Paddock et al., 2007).
Method and materials
Study cohort
A total of 627 Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) MDD patients with at least moderately severe depression were voluntarily enrolled in the study; all of them had been referred to the ‘Villa S. Chiara’ Psychiatric Hospital in Verona. The study was approved by the Local Ethics Committees (CEIOC IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia and Ethics Committee of the province of Verona), and written informed consent was obtained.
Diagnosis of unipolar depression was confirmed using the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I) diagnostic scale. Exclusion criteria were: 1) mental retardation or cognitive disorder; 2) a lifetime history of schizophrenic, schizoaffective or bipolar disorder; 3) personality disorders, substance abuse, alcohol abuse or dependency, obsessive compulsive disorder, post-traumatic stress disorder, as primary diagnosis; and 4) comorbidity with eating disorders.
A total of 22·7% of the patients showed psychotic symptoms, 41·8% showed current comorbidity in Axis I (Generalized Anxiety Disorder, Panic Attacks or Anxiety Disorders not otherwise specified [NOS]), 21·9% showed symptoms of Axis II disorders (Paranoid, Dependent, Obsessive-Compulsive and Histrionic personality disorders) and 3·8% alcohol abuse, as a secondary diagnosis (the total number exceeds the number of subjects due to the presence of comorbidity).
A total of 380 subjects had been evaluated as TRD patients (Group 1: TRD). Treatment resistance to antidepressants was defined as the failure of the patient to respond to two or more adequate trials of two or more different classes of antidepressants and to an adequate trial of a tricyclic (TCA) drug as described in Stage III of Thase and Rush Staging Method (Thase & Rush, 1997).
The second group was composed of 247 MDD patients experiencing a depressive episode who responded to their current treatment (Group 2: no-TRD).
All patients were Caucasians of Italian descent for at least two generations, and unrelated to other participants. All socio-demographic and clinical features for the two groups are shown in Table 1.
Table 1.
Clinical and demographical features of no-TRD and TRD patients.
| Characteristics | no-TRD (n = 247) | TRD (n = 380) | p-value |
|---|---|---|---|
| Age (years), mean (SD) | 54·0 (14·6) | 56·7 (13·3) | 0·02 |
| Gender (% female) | 71·3 | 66·6 | 0·22 |
| Education (years), mean (SD) | 10·0 (4·0) | 8·9 (3·9) | 0·001 |
| % of recurrent MDD | 69·4 | 90·5 | <0·001 |
| % of severe vs. moderate MDD | 42·1 | 87·4 | <0·001 |
| % psychotic symptoms | 3·7 | 35·0 | <0·001 |
| % comorbidity with personality disorders | 14·0 | 26·7 | <0·001 |
| % comorbidity with anxiety disorders | 38·9 | 43·7 | 0·23 |
| % comorbidity with alcohol abuse | 4·8 | 3·2 | 0·34 |
SD, standard deviation.
Genotyping
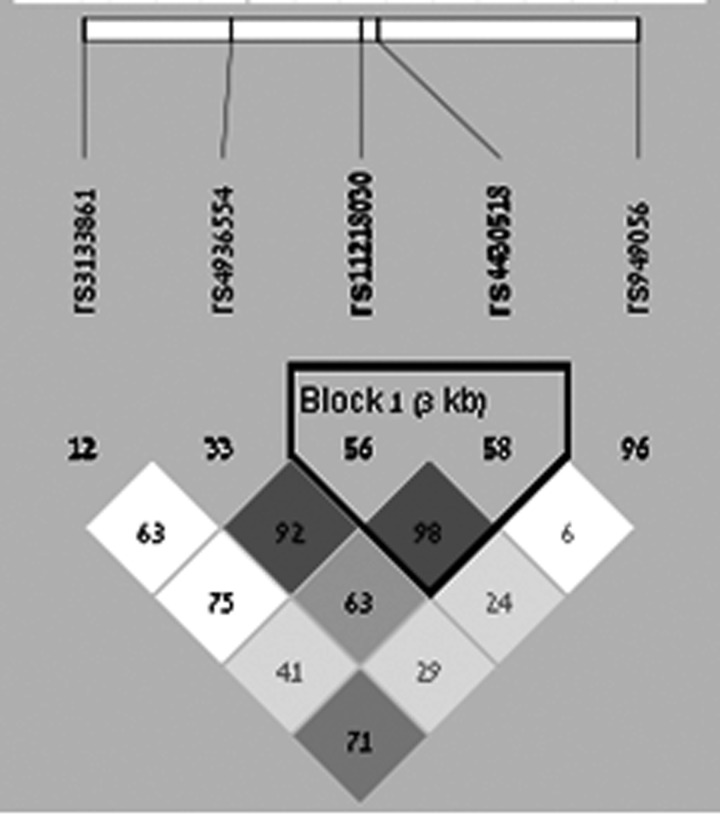
The five intronic GRIK4 polymorphisms (rs1954787, rs4936554, rs11218030, rs4430518 and rs949056) were genotyped using the BeadXpress System and the VeraCode Assay according to the manufacturer's protocols (http://www.illumina.com). The raw BeadXpress data were processed using the Illumina BeadStudio software suite (genotyping module 3·3·7), producing report files containing normalized intensity data and single nucleotide polymorphism (SNP) genotypes. Features of the investigated SNPs are reported in Table 2, whereas data regarding linkage disequilibrium (LD) pattern are described in the Supplementary Material (Table S1) and Fig. 1.
Table 2.
SNPs features and RegulomeDB results.
| Marker | Chr | Position | Location | Alleles | Regulome scorea | Regulome hits |
|---|---|---|---|---|---|---|
| rs1954787 | 11 | 120168573 | Intron | G/A | 5 | Minimal binding evidence |
| rs4936554 | 11 | 120196495 | Intron | C/T | 2b | Likely to affect binding |
| rs11218030 | 11 | 120225204 | Intron | A/G | 5 | Minimal binding evidence |
| rs4430518 | 11 | 120228643 | Intron | G/T | 5 | Minimal binding evidence |
| rs949056 | 11 | 120284569 | Intron | C/T | 4 | Minimal binding evidence |
Regulome score from 1 to 6: 1 strong evidence, 6 minimal evidence.
Fig. 1.
LD among the investigated SNPs. Dark gray and light gray colours represent a strong LD pattern (D’ >0·8) and white colour represents moderate to low LD (D’ <0·8 to >0·5).
Statistical analysis
Chi-square (χ2) tests and logistic regression analyses were conducted to evaluate the association between groups and categorical variables, while analysis of variance (ANOVA) was used to compute possible differences in age and education between groups. Odd ratios (OR) and corresponding 95% confidence intervals (CI) were used to quantify the association between genotype and response. As no-TRD and TRD patients differed in terms of socio-demographic variables such as age and education (see Table 1), for all the association analyses we used logistic regression adjusted by these variables.
All statistical analyses were conducted using SPSS version 17·0 (SPSS Inc., Chicago, IL).
Results
The analysis of single locus effect between no-TRD and TRD groups revealed differences in genotype and allele distributions between the two groups (p = 0·03 and p = 0·03, respectively) for rs1954787 (see Table 3A). AA homozygotes were more frequent in no-TRD patients (F = 3·73, p = 0·05). However, the risk effect of rs1954787 G allele for TRD subjects did not remain significant after adjustment for multiple comparisons in terms of the number of SNPs analysed.
Table 3.
Allele and genotype distributions in no-TRD and TRD patients for the significant variants GRIK4 rs1954787 (A) and GRIK4 rs11218030 (B).
| Table 3A | MAF (A) | Allele model | Genotypes n (frequency) | Genotype models | ||||
|---|---|---|---|---|---|---|---|---|
| n (frequency) | AA | AG | GG | Codominant | Dominant (GG vs AG + AA) | Recessive (GG + AG vs AA) | ||
| No-TRD (n = 247) | 241 (0·49) | 63 (0·25) | 115 (0·47) | 69 (0·28) | ||||
| χ2 = 5·01 p = 0·03 | F = 4·82 p = 0·03a | F = 2·94 p = 0·09a | F = 3·73 p = 0·05a | |||||
| TRD (n = 379) | 321 (0·42) | 75 (0·20) | 171 (0·45) | 133 (0·35) | ||||
| Table 3B | MAF (G) | Genotypes n (frequency) | Genotype models | |||||
| n (frequency) | Allele model | AA | AG | GG | Codominant | Dominant (AA vs AG + GG) | Recessive (AA + AG vs GG) | |
| No-TRD (n = 247) | 58 (0·12) | 195 (0·79) | 46 (0·19) | 6 (0·02) | F = 7·79 | |||
| χ2 = 5·41 p = 0·02 | F = 5·55 p = 0·02a | p = 0·005a OR = 1·67 | F = 0·14 p = 0·71a | |||||
| TRD (n = 379) | 125 (0·16) | 262 (0·69) | 109 (0·29) | 8 (0·02) | 95%C.I. = 1·15–2·44 | |||
p-values corrected for age and education.
MAF, minor allele frequency.
Stronger results were obtained for rs11218030. The analysis showed differences in genotype and allele distributions between the two populations (p = 0·02 and p = 0·02, respectively) (see Table 3B). In the dominant model, AA homozygotes were more frequently found among the no-TRD patients (F = 7·79, p = 0·005, OR = 1·67, 95% CI = 1·15–2·44). The risk effect of rs11218030 G allele for a non-response to treatment in MDD remained significant after adjustment for multiple comparisons, by Bonferroni correction, in terms of the number of SNPs analysed (p = 0·025).
No significant associations were observed for the other SNPs.
Because many clinical variables strongly affect the response to treatment (Table 1), we carried out secondary analyses on these two SNPs to investigate putative influences of clinical factors.
Significant effects were found for the presence of psychotic symptoms in association with both the SNPs. A different distribution of genotype and allele frequencies (p = 0·005 and p = 0·005, respectively, for rs1954787, and p = 0·03 and p = 0·03 for rs11218030) was observed between MDD patients with and without the presence of psychotic symptoms (see Table 4A and 4B). On the assumption of a co-dominant, recessive and dominant model, for rs1954787 the dominant model best fitted the data (p = 0·006), for which, homozygous GG patients presented an OR of 1·71 for rs1954787 of developing psychotic symptomatology during a depression episode.
Table 4.
Allele and genotype distributions in MDD patients with and without psychotic symptoms for the significant variants GRIK4 rs1954787 (A) and GRIK4 rs11218030 (B).
| Table 4A | MAF (A) | Allele model | Genotypes n (frequency) | Genotype models | ||||
|---|---|---|---|---|---|---|---|---|
| n (frequency) | AA | AG | GG | Codominant | Dominant (GG vs AG + AA) | Recessive (GG + AG vs AA) | ||
| MDD without psychotic symptoms (n = 483) | 455 (0·47) | χ2 = 7·88 p = 0·005 | 114 (0·24) | 227 (0·47) | 142 (0·29) | F = 8·07 p = 0·005a | F = 7·50 p = 0·006a OR = 1·71 95%C.I. = 1·16–2·51 | F = 3·69 p = 0·06a |
| MDD with psychotic symptoms (n = 142) | 107 (0·38) | 24 (0·17) | 59 (0·41) | 59 (0·42) | ||||
| Table 4B | MAF (G) | Allele model | Genotypes n (frequency) | Genotype models | ||||
| n (frequency) | AA | AG | GG | Codominant | Dominant (AA vs AG + GG) | Recessive (AA + AG vs GG) | ||
| MDD without psychotic symptoms (n = 483) | 129 (0·13) | χ2 = 4·97 p = 0·03 | 363 (0·75) | 111 (0·23) | 9 (0·02) | F = 4·60 p = 0·03a | F = 4·50 p = 0·03a OR = 1·55 95%C.I. = 1·03–2·31 | F = 0·81 p = 0·37a |
| MDD with psychotic symptoms (n = 142) | 53 (0·19) | 94 (0·66) | 43 (0·30) | 5 (0·04) | ||||
p-values corrected for gender and education.
Discussion
In the present study we investigated the role of five GRIK4 variants previously associated with response to antidepressants in a cohort of depressed patients, comparing TRD vs. no-TRD. We found a risk effect of rs11218030 G allele for the resistance to treatment in MDD. Moreover we showed that rs1954787 GG homozygotes and G allele carriers of rs11218030 have a higher risk of developing psychotic symptomatology during a depressive episode.
Clinical studies have shown that antidepressant treatments result in a correction of glutamate balance; however, the molecular mechanisms correlating with the lack of outcome have been investigated only in genetic association studies.
In the STAR*D cohort, the rs11218030 variant was found to be significantly associated with non-response when comparing non-responders vs. controls, but no data regarding this variant was reported for the comparisons of responders vs. non-responders in both the discovery and validation cohorts (Paddock et al., 2007). The most studied SNP in the GRIK4 gene in relation to treatment response is rs1954787. In the STAR*D cohort, associations between G allele with a better treatment response and remission were found (Paddock et al., 2007) as well as in the study conducted by Pu and collaborators (Pu et al., 2013) in a Chinese Han cohort. However, several other evidences revealed a lack of association in Caucasian MDD patients (Horstman et al., 2010; Perlis et al., 2010; Serretti et al., 2012) in line with our results. Recently, a meta-analysis was carried out on these five studies (Kawaguchi et al., 2014), which included more than 2000 MDD patients. The results showed that the T allele of GRIK4 rs1954787 represented a risk factor that reduced the likelihood of response to treatment in MDD.
The high heterogeneity in the results could be explained by the frequency of this GRIK4 variant that strongly shows significant ethnic and geographic variations (see http://hapmap.ncbi.nlm.nih.gov/). Indeed, the effect obtained in the meta-analysis (Kawaguchi et al., 2014) seems principally due to the Asian cohort (Pu et al., 2013), whereas the ORs for the other studies were poor.
Furthermore, the conflicting results could be due to different study designs, different definitions of responders or a different clinical MDD phenotype with more severe symptoms in our sample and in other studies such as those by Serretti and Hortsmann (Hortsmann et al., 2010; Serretti et al., 2012). This might be in line with our results concerning the positive associations between both rs1954787 and rs11218030, and the presence of psychotic symptoms in MDD. This is of particular importance since the presence of psychotic symptoms is one of the strongest negative predictors of resistance to treatment in MDD (Schlaepfer et al., 2012; Perlis et al., 2013). Considering the complexity of the TRD phenotype, literature suggests that pharmacogenetic study designs should take into account the manifold biological, clinical, socio-demographical and environmental variables that have an impact on treatment response. This would enable putative associations to be identified with clinical and environmental factors that influence the outcome in MDD. This would then suggest that GRIK4 variants may be implicated indirectly in the response through a direct involvement in negative or positive predictors of response.
In addition, our results are in agreement with evidence that GRIK4 was associated with psychosis and mood disorders and with haloperidol efficacy during acute treatment (Pickard et al., 2006; Blackwood et al., 2007; Drago et al., 2013).
However, all the SNPs investigated are in intronic regions and seem not to have a functional role or an effect on gene expression regulation (Table 2). Further studies should explore the association between GRIK4 variants with a putative functional role in the TRD phenotype, which may explain the associations reported in our present study.
The main limitation of this work is represented by the heterogeneity of the therapy followed by the patients enrolled in the study. This could represent a confounding factor in the understanding of the involvement of these polymorphisms in TRD as these gene variants may be associated with the resistance of specific classes of antidepressant drugs. This could be clarified by increasing the number of patients.
In conclusion, our data showed the involvement of the GRIK4 gene in treatment response mechanisms and psychotic symptoms. Independent replications with larger sample sizes are needed to better understand the potential role of these polymorphisms in TRD and in psychotic symptoms.
Acknowledgments
This research was supported by grants from the Italian Ministry of Health (RC and RF2007 Conv. 42). E.M. is supported by the Shabbetai Donnolo Fellowship between Italy and Israel.
Declaration of interest
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0016672315000142.
click here to view supplementary material
References
- Bentley S. M., Pagalilauan G. L. & Simpson S. (2014). Major depression. The Medical Clinics of North America 98, 981–1005. [DOI] [PubMed] [Google Scholar]
- Blackwood D. H., Pickard B. J., Thomson P. A., Evans K. L., Porteous D. J. & Muir W. J. (2007). Are some genetic risk factors common to schizophrenia, bipolar disorder and depression? Evidence from DISC1, GRIK4 and NRG1. Neurotoxicity Research 11, 73–83. [DOI] [PubMed] [Google Scholar]
- Boyer P., Skolnick P. & Fossom L. H. (1998). Chronic administration of imipramine and citalopram alters the expression of NMDA receptor subunit mRNAs in mouse brain. Journal of Molecular Neuroscience 10, 219–233. [DOI] [PubMed] [Google Scholar]
- Carvalho A. F., Berk M., Hyphantis T. N. & McIntyre R. S. (2014). The integrative management of treatment-resistant depression: a comprehensive review and perspectives. Psychotherapy and Psychosomatics 83, 70–88. [DOI] [PubMed] [Google Scholar]
- Catches J. S., Xu J. & Contractor A. (2012). Genetic ablation of the GluK4 kainate receptor subunit causes anxiolytic and antidepressant-like behavior in mice. Behavioural Brain Research 228, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena-Dell'Osso M., Fagiolini A., Rotella F., Baroni S. & Marazziti D. (2013). Glutamate system as target for development of novel antidepressants. CNS Spectrums 18, 188–198. [DOI] [PubMed] [Google Scholar]
- Drago A., Giegling I., Schäfer M., Hartmann A. M., Friedl M., Konte B., Möller H. J., De Ronchi D., Stassen H. H., Serretti A. & Rujescu D. (2013). AKAP13, CACNA1, GRIK4 and GRIA1 genetic variations may be associated with haloperidol efficacy during acute treatment. Neuropsychopharmacology 23, 887–894. [DOI] [PubMed] [Google Scholar]
- Horstmann S. & Binder E. B. (2009). Pharmacogenomics of antidepressant drugs. Pharmacology & Therapeutics 124, 57–73. [DOI] [PubMed] [Google Scholar]
- Horstmann S., Lucae S., Menke A., Hennings J. M., Ising M., Roeske D., Müller-Myhsok B., Holsboer F. & Binder E. B. (2010). Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology 35, 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi D. M. & Glatt S. J. (2014). GRIK4 polymorphism and its association with antidepressant response in depressed patients: a meta-analysis. Pharmacogenomics 15, 1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musazzi L., Treccani G., Mallei A. & Popoli M. (2013). The action of antidepressants on the glutamate system: regulation of glutamate release and glutamate receptors. Biological Psychiatry 73, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Nowak G., Li Y. & Paul I. A. (1996). Adaptation of cortical but not hippocampal NMDA receptors after chronic citalopram treatment. European Journal of Pharmacology 295, 75–85. [DOI] [PubMed] [Google Scholar]
- Paddock S., Laje G., Charney D., Rush A. J., Wilson A. F., Sorant A. J., Lipsky R., Wisniewski S. R., Manji H. & McMahon F. J. (2007). Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. The American Journal of Psychiatry 164, 1181–1188. [DOI] [PubMed] [Google Scholar]
- Perlis R. H. (2013). A clinical risk stratification tool for predicting treatment resistance in major depressive disorder. Biological Psychiatry 74, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis R. H., Fijal B., Dharia S., Heinloth A. N. & Houston J. P. (2010). Failure to replicate genetic associations with antidepressant treatment response in duloxetine-treated patients. Biological Psychiatry 67, 1110–1103. [DOI] [PubMed] [Google Scholar]
- Pickard B. S., Malloy M. P., Christoforou A., Thomson P. A., Evans K. L., Morris S. W., Hampson M., Porteous D. J., Blackwood D. H. & Muir W. J. (2006). Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, GRIK4, in schizophrenia and bipolar disorder. Molecular Psychiatry 11, 847–857. [DOI] [PubMed] [Google Scholar]
- Pu M., Zhang Z., Xu Z., Shi Y., Geng L., Yuan Y., Zhang X. & Reynolds G. P. (2013). Influence of genetic polymorphisms in the glutamatergic and GABAergic systems and their interactions with environmental stressors on antidepressant response. Pharmacogenomics 14, 277–288. [DOI] [PubMed] [Google Scholar]
- Schlaepfer T. E., Agren H., Monteleone P., Gasto C., Pitchot W., Rouillon F., Nutt D. J. & Kasper S. (2012). The hidden third: improving outcome in treatment-resistant depression. Journal of Psychopharmacology 26, 587–602. [DOI] [PubMed] [Google Scholar]
- Serretti A., Chiesa A., Crisafulli C., Massat I., Linotte S., Calati R., Kasper S., Bailer U., Lecrubier Y., Fink M., Antonijevic I., Forray C., Snyder L., Bollen J., Zohar J., De Ronchi D., Souery D. & Mendlewicz J. (2012). Failure to replicate influence of GRIK4 and GNB3 polymorphisms on treatment outcome in major depression. Neuropsychobiology 65, 70–75. [DOI] [PubMed] [Google Scholar]
- Thase M. E. & Rush A. J. (1997). When at first you don't succeed: sequential strategies for antidepressant nonresponders. Journal of Clinical Psychiatry 13, 23–29. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0016672315000142.
click here to view supplementary material