Abstract
Duck hepatitis A virus (DHAV) infection is characterized by severe hepatitis. In recent years, DHAV-A has become widespread in Asia and has led to economic losses. Conventional methods of DHAV-A detection must often be performed in the laboratory with inconvenience equipment. We have developed a rapid reverse transcription insulated isothermal (RT-iiPCR) technique for the on-site detection of DHAV-A based on the POCKITTM system in a convenient minitype device. We optimized the PCR primers and probes for the amplification of the DHAV-A 3C/3D genes, and successfully amplified a specific fragment of DHAV-A, but no fragment from 18 other duck pathogens. The limit of detection for viral RNA was 49 copies per reaction, and the sensitivity and specificity were each 100% in the analysis of 60 liver samples. By comparison, the sensitivities of RT-iiPCR was comparable in sensitivity to existing rRT-PCR. Furthermore, the RT-iiPCR results were 98.3% in agreement with those of the rRT-PCR, with a kappa value of 0.938. In conclusion, this new method not only offers a higher sensitivity and specificity than existing techniques, but also time-saving and better suited to field diagnoses because device is portable.
Keywords: detection, duck hepatitis A virus, insulated isothermal RT-PCR, real-time RT-PCR
Duck hepatitis A virus (DHAV) is the commonest lethal causative agent of viral hepatitis in young ducks, and is characterized by petechia and ecchymotic hemorrhage on the liver surface [16]. Duck hepatitis is on the list of notifiable diseases of the World Organization for Animal Health. In China, domestic ducks in nine provinces were infected with the virus nearly five years, and evidence of its economic impact is growing [22]. The infected ducklings often displayed increased numbers of liver vacuoles, liver necrosis and hemorrhage, and a high mortality rate [26]. Severe cytokine storms caused by DHAV-induced hemorrhagic liver lesions, result in the rapid death of infected ducklings [24]. The levels of IL-2 and IL-6 are shown up-regulated during the first 24 hr of DHAV infection, and this is believed to be linked to the severe hepatic injury observed [27]. DHAV, which belongs to the family Piconaviridae, is genetically divided into three serotypes: DHAV-A is the original serotype, and is widely epidemic on duck-breeding farms worldwide; DHAV-B was isolated in Taiwan [21]; DHAV-C was isolated in China and South Korea [12]. No cross-neutralization between the genotypes and no antigenic relationships among these serotypes have been observed [12, 20]. DHAV-A has become widely distributed throughout Asia in recent years, with mortality rates of up to 95% in young ducklings <1 week of age [22]. An outbreak occurred in June 2015 on a duckling farm in Japan, with a 76% (251/330) mortality rate, and this was the first DHAV-A to be isolated in Japan since its outbreak in Japan in 1963 [11]. In Korea, although ducks have been vaccinated against DHAV-A since the 2000s, DHAV infections are still one of the most devastating diseases to the Korean duck industry. Epidemiological investigations revealed that 14.6% of DHAV infections in South Korea between 2013 and 2015 were caused by DHAV-A [13, 18]. Sixteen DHAV-A strains were identified between 2009 and 2013 in Vietnam [7]. In China, the highest number of outbreaks caused by DHAV-1 occurred in 2010–2015, and 23 DHAV-A field strains were isolated from different commercial duck farms in nine provinces (Jangsu, Shandong, Guangxi, Liaoning, Zhejiang, Fujian, Sichuan, Hube and Guangxi Provinces) [22]. Several duck pathogens present similar clinical symptoms, causing hepatitis, and which makes DHAV-A infections difficult to identify [8]. Therefore, immunological and molecular methods have been used for their rapid diagnosis. An enzyme-linked immunosorbent assay (ELISA) based on the recombinant VP3 protein of DHAV-A was developed to efficiently monitor antibody levels in the sera of duck flocks with suspected DHAV infections [17]. However, this assay does not distinguish the different DHAV genotypes because of cross-reactivity. PCR-based assays are another mainstream method for laboratory test in etiologic diagnose, which possess the advantages of high accuracy, sensitiveness and cost effectiveness. Therefore, many techniques for DHAV-A detection have been developed including RT-PCR [5], real-time Taqman RT-PCR assay for the detection of DHAV-A [9, 25], and a real-time RT-PCR with SYBR green I for DHAV-A [10]. Although these methods are rapid and efficient and have low limits of detection, they must often be performed in the laboratory, with complex equipment.
Insulated isothermal PCR (iiPCR) is a kind of Rayleigh–Bénard convection technique to amplify gene in vitro. It involves the amplification of the target cDNA by cycling the reaction components through different temperature ranges to achieve the denaturation, annealing, and extension steps of PCR [4, 19]. Since it was first reported in 2002 [15], iiPCR assays have been used for the detection of many animal pathogens, including the H3N8 subtype of equine influenza virus [2] and foot and mouth disease virus [1]. The iiPCR assay has also been used as a qualitative method for the rapid on-site detection of malaria, achieving 96.9% efficiency and a lower detection limit of ≥100 copies of plasmid DNA [6]. All iiPCRs are performed with the POCKITTM system (GeneReach, Lexington, MA, U.S.A.), a small device that is extremely convenient for rapid on-site detection.
To develop a rapid method of detecting DHAV-A, we targeted the 3C/3D genes of DHAV-A a rapid on-site RT-iiPCR assay. A one-step duplex rRT-PCR assay for DHAV [9] was evaluated in parallel with RT-iiPCR to compare their analytical sensitivities and specificities, and the accuracy of the two assays in detecting DHAV-A in liver samples. Two RNA extraction methods for use in liver samples were also compared.
MATERIALS AND METHODS
Viruses, bacteria, and clinical samples
Four field strains of DHAV-A and 18 other pathogenic microorganisms that potentially infect ducklings were used to evaluate the specificity of the RT-iiPCR assay (Table 1). Sixty archived sequential DHAV-A-positive liver samples collected from sick ducklings (less than 3 weeks old) in May 2017 in China were sequenced at the Laboratory for Preventive Veterinary Science of Southwest University for Nationalities. All the animal procedures used in the present study were conducted in accordance with good animal practices, defined by the Laboratory Animal Use License (certificate no. SYXK [CHUAN] 2014-187).
Table 1. Reference strains and isolates used in this study for RT-iiPCR specificity testing.
| Pathogeny species | No. | Source | RT-iiPCR detection |
|---|---|---|---|
| DHAV-A strains SWUN3518 to SWUN3521 | 4 | a | + |
| DHAV-C strain SWUN3501 to SWUN3507 | 7 | a | − |
| NDV SWUN 2690 | 1 | a | − |
| GPV SWUN 5301 | 1 | a | − |
| MPV SCAU 251 | 1 | b | − |
| AIV CAHEC 343 | 1 | c | − |
| FAV-1 SWUN 7002 | 1 | a | − |
| DAstV-1 CAU121 | 1 | d | − |
| DAstV-2 CAU122 | 1 | d | − |
| Pasteurella multocida SWUN 0300 | 1 | a | − |
| Escherichia coli (O46) SWUN 0314 | 1 | a | − |
| Salmonella Enteritidis SWUN 5223 | 1 | a | − |
| Riemrella anatipestifer SWUN 0233 | 1 | a | − |
a, Southwest University for Nationalities, China; b, Sichuan Agricultural University, China; c, China Animal Health and Epidemiology Center, China; d, China Agricultural University, China. +, positive; −, negative. DHAV, duck hepatitis A virus; NDV, Newcastle disease virus; GPV, gosling parvovirus; MPV, muscovy parvovirus; AIV, avian influenza virus; FAV-1, fowl adenovirus-1; DAstV: duck astrovirus.
Viral nucleic acid extraction and cDNA synthesis
Two different nucleic acid extraction methods, performed according to the manufacturer’s instructions, were evaluated in this study. Initially, DHAV-A nucleic acid was extracted from 20 duck liver samples (12 of which were previously identified as positive) with TRIzol Reagent (Applied Biosystems Inc., Carlsbad, CA, U.S.A.). The total RNA was then reverse-transcribed to cDNA with the PrimeScriptTM reverse transcription kit (TaKaRa Biotechnology, Dalian, China). These samples were then compared with those obtained with the PetNADTM nucleic acid rapid extraction kit (GENERADAR Biotechnology Corp., Xiamen, China), which were prepared as follows. The clinical samples were centrifuged at 13,000 × g for 2 min before nucleic acid extraction. An aliquot (200 µl) of the supernatant was mixed with 600 µl of buffer PB1 and shaken for 1 min. Buffer PB2 (600 µl) was added and the mixture was transferred to a spin column. The washing steps were performed as described in the manufacturer’s manual. The total RNA was eluted in 50 µl of PB5 elution buffer. All the nucleic acid preparations were stored at −80°C before testing.
Bacterial nucleic acid extraction
The DNA extraction kit (TaKaRa Biotechnology) was used to extract nucleic acid from plate cultures of bacteria. The nucleic acid samples were then used to test the specificity of the RT-iiPCR assays.
Development of RT-iiPCR for DHAV-A
The primers and probe were designed according to the complete genome sequence (GenBank database) of 22 different DHAV-A strains (accession nos. KU923754, GU066825, JF828997, JQ804521, EU264072, FJ496340, FJ496341, FJ496344, FJ496339, FJ496342, FJ157179, FJ157173, FJ157178, FJ436047, GU944671, JF828999, JF829000, EF585200, NC008250, FJ971623, JF914945 and FJ496343), 10 different DHAV-C strains (accession nos. DQ256133, KC993890, GQ485311, KU860090, JF835025, GQ122332, EU755009, KP995438, JF914944 and JX312194), and two DHAV-2 strains (accession nos. EF067923.1 and EF067924.1). A 179 bp segment of the 3C/3D genes of DHAV-A (accession no. KU923754) was set as the taget by using Primer Express software, version 1.0, for Taqman technology. The primer sequences were: 3C3DF, 5′-AGATCAGGAYCAGTATATG-3′ and 3C3DR, 5′-GTATCCCAAGGATCTTCA-3′; and the Taqman probe sequence was VP3P, FAM-CACCACCACAGGARCCAG-BHQ1.
DHAV-A strain SWUN3518 was used to optimize the RT-iiPCR assay. Based on Uni-ii PCR Starter Kit instructions (GENERADAR Biotechnology Corp.), various concentrations of components were tested and screened in a total volume of 50 µl, which included the forward and reverse primers at 10 µmol/µl (0.5–4 µl), the 10 µmol/µl probe (0.05–0.4 µl), 5 U/µl Taq DNA polymerase (1–5 µl), and 20 µmol/µl reverse transcriptase (0.5–2 µl). Optimum conditions were determined by the absorption ratios at A520/B520.
The RT-iiPCR was carried out with the POCKITTM device, with RT performed at 42°C for 10 min and iiPCR then performed at 95°C for 30 min. The reaction signals were processed with an optical detection module and shown automatically on the display screen. The results were converted automatically to “+” (positive), “–” (negative), or “?” (inconclusive), according to the default S/N thresholds of POCKITTM [19].
Sensitivity of RT-iiPCR
Total RNA was extracted from DHAV-A strain SWUN3521 with PetNADTM nucleic acid rapid extraction. The Copy number was determined by rRT-PCR, as previously described [9]. Ten-fold serially diluted standard RNA templates with RNase-free water were simultaneously assayed by RT-iiPCR and rRT-PCR to compare the sensitivity. All samples were amplified under the optimum conditions.
Specificity of RT-iiPCR
Twelve major duck pathogens were used to determine the specificity of the RT-iiPCR (Table 1). Nucleic acid templates were extracted from DHAV-C (strains SWUN3501–SWUN3507), duck astrovirus 1 (DAstV-1), duck astrovirus 2 (DAstV-2), gosling parvovirus (GPV), muscovy parvovirus (MPV), avian influenza virus (AIV [H5N1]), fowl adenovirus-1 (FAV-1), Pasteurella. multocida, Escherichiacoli (O46), Salmonella Enteritidis, and Riemrella anatipestifer.
Repeatability and reproducibility of RT-iiPCR
Six different RNA dilutions of positive samples (1 × 10−2~1 × 10−7) were used to evaluate the reproducibility of the RT-iiPCR. Each sample was amplified in triplicate.
Analysis of RT-iiPCR and comparison with rRT-PCR
A one-step duplex rRT-PCR assay, which was established as previously described, was compared with the RT-iiPCR. The primer sequences used were: F3, 5′-CCATCTGTGTCATTGTGTTAGGCA-3′ and R3, 5′-CAAATCAGTTTCAAGGAGTTCTCCA-3′; Taqman probe was P-VP0, HEX-ACCGACATGGCAATGGAACCTCCA-BHQ. The concentrations of relevant reagents and the thermocycling conditions for the detection of DHAV-A were as described previously [9]. A total of 60 clinical liver samples collected from sick ducklings were analyzed simultaneously by rRT-PCR and RT-iiPCR. The sensitivity and specificity of the assays were determined with 2 × 2 contingency tables. The degree of agreement between the two assays was assessed by calculating Cohen’s kappa (κ) values.
RESULTS
Protocol optimization
Optimization showed that the combination of 3 µl of each (10 µmol/µl) primers, 0.3 µl (10 µmol/µl) probes, 2 µl (5 U/µl) Taq DNA polymerase of 25 µl Premix Ex Taq and 1.25 µl (20 U/µl) reverse transcriptase in a total volume of 50 µl achieved the maximum A520/B520 value of 4.4. These reagent volumes were used for all subsequent experiments.
RT-iiPCR sensitivity: The rRT-PCR assay detected a DHAV-A RNA concentration of 4.91 × 1010 copies/µl, which was serially diluted and tested in duplicate. A regression analysis of rRT-PCR indicated that good linearity was achieved (slope= −3.3157, R2=0.993; Fig. 1). The rRT-PCR amplification efficiency (%), calculated with the formula E=[10−1/slope −1] ×100%, was 100%. The rRT-PCR assay detection limit of detection was a dilution of 10−7–10−8 (Table 2 ). However, the RT-iiPCR limit of detection was a dilution of 10−8–10−9, indicating that RT-iiPCR was more sensitive than rRT-PCR.
Fig. 1.

The linear relationship of every dilution detected by rRT-PCR.
Table 2. Evaluation of rRT-PCR and RT-iiPCR sensitivity for duck hepatitis A virus (DHAV) detection.
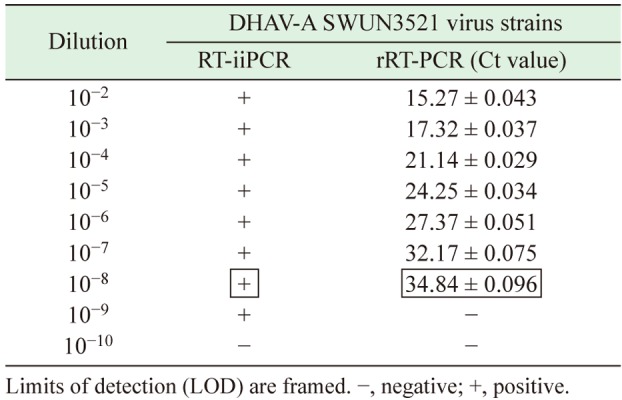
RT-iiPCR specificity
DHAV-A RNA could be detected from all four DHAV-A strains by RT-iiPCR, but nucleic acids extracted from 18 other duck pathogens could not be amplified (Table 1). These data indicate the high specificity of RT-iiPCR for DHAV-A.
Comparison of two nucleic acid extraction methods
Samples extracted with TRIzol Reagent or PetNADTM nucleic acid rapid extraction were compared simultaneously in the rRT-PCR and RT-iiPCR assays. Both methods of RNA extraction achieved the same positive detection rate for 20 DHAV-A samples of 60% (12/20) (Table 3), which agreed with the previous sequencing results. Therefore, the PetNADTM nucleic acid rapid extraction achieved the same results as TRIzol Reagent, but is faster and does not require a high-speed centrifuge, which can be inconvenient for the on-site detection of pathogens.
Table 3. Performance evaluation of PetNADTM and TRIzol RNA extraction methods for duck hepatitis A virus detection by rRT-PCR and RT-iiPCR.
| Sample ID | Sequencing result |
PetNADTM | TRIzol | ||
|---|---|---|---|---|---|
| rRT-PCR (Ct value) |
RT-iiPCR | rRT-PCR (Ct value) |
RT-iiPCR | ||
| SCGH04 | + | 27.09 | + | 26.51 | + |
| SCGH 05 | + | 30.93 | + | 28.14 | + |
| SCGH08 | + | 32.11 | + | 30.07 | + |
| SCGH09 | + | 29.73 | + | 27.46 | + |
| SCGH10 | + | 29.16 | + | 29.05 | + |
| SCGH11 | + | 25.31 | + | 23.17 | + |
| SCGH12 | + | 26.44 | + | 24.32 | + |
| SCGH13 | + | 30.24 | + | 28.76 | + |
| SWUN18 | + | 31.57 | + | 25.06 | + |
| SWUN19 | + | 30.18 | + | 23.19 | + |
| SWUN20 | + | 29.36 | + | 24.18 | + |
| SWUN21 | + | 27.19 | + | 22.34 | + |
| SCMY14 | − | Neg | − | Neg | − |
| SCMY 27 | − | Neg | − | Neg | − |
| SCMY 31 | − | Neg | − | Neg | − |
| SCMY 36 | − | Neg | − | Neg | − |
| SCMY 49 | − | Neg | − | Neg | − |
| DA 14 | − | Neg | − | Neg | − |
| DY 17 | − | Neg | − | Neg | − |
| DY 32 | − | Neg | − | Neg | − |
−, negative; +, positive.
Comparison of rRT-PCR and RT-iiPCR in the analysis of liver samples
In order to compare the accordance rate of rRT-PCR with RT-iiPCR by referencing the sequencing results of amplification products to evaluate their sensitivity in detecting DHAV-A in liver samples, RNA was extracted from 60 archived liver samples with the PetNADTM nucleic acid rapid extraction kit to be used for detection. Of these, 51 samples had been shown to be positive by sequencing analysis (Table 4). The rRT-PCR assay successfully detected DHAV-A nucleic acid in 50/51 established positive liver samples, while the 10 established negative liver samples were also confirmed by rRT-PCR (Table 4). By comparison, the RT-iiPCR assay successfully detected all 51 DHAV-A RNA-positive liver samples, identified previously with sequenceing (51/51). This demonstrates that the detection rate of RT-iiPCR is comparable in sensitivity to rRT-PCR. The sensitivity and specificity of the DHAV-A rRT-PCR and RT-iiPCR were determined from 2 × 2 contingency tables as compared to sequencing analysis. The sensitivity and specificity of the DHAV-A RT-iiPCR were both 100%, whereas they were 98.04 and 100% for the rRT-PCR, respectively. When compared with the sequencing results, the accuracy of the rRT-PCR was 98.3% (95% CI: 91.1–100%) (κ=0.98), whereas it was 100% (95% CI: 94–100%) (κ=1) for the RT-iiPCR. Therefore, rRT-PCR and RT-iiPCR were in good agreement with the real consequences in liver samples. Finally, the agreement between the two assays was 98.3% with a kappa value of 0.938 (Table 4), confirming the substantial level of agreement between the RT-rPCR and RT-iiPCR assays in detecting DHAV-A in liver samples.
Table 4. Contingency table for the level of agreement between rRT-PCR and RT-iiPCR assays for the detection of duck hepatitis A virus in clinical sample.
| Sequencing results | ||||
| Positive | Negative | Total | ||
| rRT-PCR | Positive | 50 | 0 | 50 |
| Negative | 1 | 9 | 10 | |
| Total | 51 | 9 | 60 | |
| Sequencing results | ||||
| Positive | Negative | Total | ||
| RT-iiPCR | Positive | 51 | 0 | 51 |
| Negative | 0 | 9 | 9 | |
| Total | 51 | 9 | 60 | |
| rRT-PCR | ||||
| Positive | Negative | Total | ||
| RT-iiPCR | Positive | 50 | 1 | 51 |
| Negative | 0 | 9 | 9 | |
| Total | 50 | 10 | 60 | |
Evaluation of RT-iiPCR reproducibility
RT-iiPCR was used to analyze six samples in triplicate, which were obtained from the serial 10-fold dilution (from 10−2 to 10−7) of DHAV-A strain SWUN3521. The results for all samples were positive. The consistency of this analysis is indicative of the high reproducibility of the RT-iiPCR.
DISCUSSION
DHAV-A has recently decimated Asian poultry farms, especially in China, and has caused enormous economic losses for commercial duck farms. Liver injury is a characteristic of DHAV-A infection, but this can also be caused by many other duck pathogens including DHAV-B, DHAV-C, Newcastle disease virus, DAstv-1, and bacteria, so diagnosis can be difficult. Therefore, a rapid, effective, and reliable detection method for DHAV-A is required. Although viral isolation and purification are currently the recognized gold standard method of viral detection, their efficacy depends on the type of culture medium used, the susceptibility of the host cell, coinfection of multiple pathogens, and many other uncertain conditions [14].
Molecular methods are commonly used in laboratory diagnoses. For example, PCR is a rapid and efficient method with a low detection limit. However, it requires complex equipment, involves multiple steps, and is a time-consuming process. In recent years, RT-iiPCR has been widely used in the detection of pathogens [1, 3]. The reagents and reaction systems are contained in a small device (POCKITTM), and results are obtained after the data are processed automatically with the default algorithm, and no manual data analysis or interpretation is required. Therefore, this method facilitates the rapid diagnosis of disease and can be applied to many situations.
In the present study, we developed a new RT-iiPCR assay for the detection of DHAV-A based on the POCKITTM system. Similar to quantitative RT–PCR, this assay is based on TaqMan® probe hydrolysis, which generates a fluorescent signal. All the components for a single reaction, including 30 µmol of each primers, 3 µmol of probes, 10 U of Taq DNA polymerase, and 25 U of reverse transcriptase, are contained in one tube and processed with vacuum freeze-drying technology. The premixed reagents simply require dissolution in sterile deionized water before use. The technique also reduces the operating time, which minimizes sample contamination [23]. Four DHAV-A strains collected from different areas were correctly diagnosed by our RT-iiPCR assay, which displayed high specificity for DHAV-A (Table 1). The low limit of detection of the RT-iiPCR assay was 49.1 copies per reaction, which is better than that reported for duplex real-time RT-PCR, and its detection range was similar to that of rRT-PCR (10° −10−8 dilutions) (Table 2). The results of rRT-PCR and RT-iiPCR were in excellent agreement with the actual findings in 60 liver samples, and RT-iiPCR was in 98.3% agreement with rRT-PCR, with a κ value of 0.938 (Table 4). Moreover, the established RT-iiPCR assays had higher sensitivity and accuracy compared with rRT-PCR, and its advantages of rapid nucleic acid extraction, short processing time, and portability should make it popular for field diagnoses.
In this study, we developed an RT-iiPCR method based on the POCKITTM system that can be applied with a minitype device that is convenient to transport. The entire detection process from RNA extraction to the results, takes only about 1 hr. The technique is both sensitive and specific. We propose that the RT-iiPCR/POCKITTM system can be used as a reliable and effective tool for the diagnosis and detection of DHAV-A, and that it will facilitate the molecular epidemiological investigations of DHAV-A and prevent and control DHAV breeding reservoirs.
Acknowledgments
We would like to thank Prof. Hua Yue for insightful comments regarding study design and the manuscript, and Dr. Bin Zhang for providing the nucleic acids from various duck bacterial pathogens used in this study. This present study was supported simultaneously by The National Key Research and Development Program of China (2017YFD0501101) and the Fundamental Research Funds for the Central Universities (2017NZYQN01).
REFERENCES
- 1.Ambagala A., Fisher M., Goolia M., Nfon C., Furukawa-Stoffer T., Ortega Polo R., Lung O.2017. Field-Deployable Reverse Transcription-Insulated Isothermal PCR (RT-iiPCR) Assay for Rapid and Sensitive Detection of Foot-and-Mouth Disease Virus. Transbound. Emerg. Dis. 64: 1610–1623. doi: 10.1111/tbed.12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasuriya U. B., Lee P. Y., Tiwari A., Skillman A., Nam B., Chambers T. M., Tsai Y. L., Ma L. J., Yang P. C., Chang H. F., Wang H. T.2014. Rapid detection of equine influenza virus H3N8 subtype by insulated isothermal RT-PCR (iiRT-PCR) assay using the POCKITTM Nucleic Acid Analyzer. J. Virol. Methods 207: 66–72. doi: 10.1016/j.jviromet.2014.06.016 [DOI] [PubMed] [Google Scholar]
- 3.Carossino M., Lee P. Y., Nam B., Skillman A., Shuck K. M., Timoney P. J., Tsai Y. L., Ma L. J., Chang H. F., Wang H. T., Balasuriya U. B.2016. Development and evaluation of a reverse transcription-insulated isothermal polymerase chain reaction (RT-iiPCR) assay for detection of equine arteritis virus in equine semen and tissue samples using the POCKITTM system. J. Virol. Methods 234: 7–15. doi: 10.1016/j.jviromet.2016.02.015 [DOI] [PubMed] [Google Scholar]
- 4.Chang H. F., Tsai Y. L., Tsai C. F., Lin C. K., Lee P. Y., Teng P. H., Su C., Jeng C. C.2012. A thermally baffled device for highly stabilized convective PCR. Biotechnol. J. 7: 662–666. doi: 10.1002/biot.201100453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L. L., Xu Q., Zhang R. H., Yang L., Li J. X., Xie Z. J., Zhu Y. L., Jiang S. J., Si X. K.2013. Improved duplex RT-PCR assay for differential diagnosis of mixed infection of duck hepatitis A virus type 1 and type 3 in ducklings. J. Virol. Methods 192: 12–17. doi: 10.1016/j.jviromet.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 6.Chua K. H., Lee P. C., Chai H. C.2016. Development of insulated isothermal PCR for rapid on-site malaria detection. Malar. J. 15: 134. doi: 10.1186/s12936-016-1183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doan H. T., Le X. T., Do R. T., Hoang C. T., Nguyen K. T., Le T. H.2016. Molecular genotyping of duck hepatitis A viruses (DHAV) in Vietnam. J. Infect. Dev. Ctries. 10: 988–995. doi: 10.3855/jidc.7239 [DOI] [PubMed] [Google Scholar]
- 8.Gan Y., Huang C., Wang X., Chen S., Li J.2014. Epidemiological investigation of duck hepatitis a virus (DHAV) isolated from sichuan basin by RT-PCR disclose the existence of mixed infection and the feasibility of DHAV-A evolved from C80 Strain. Pakistan. J. Vet. 34: 356–360. [Google Scholar]
- 9.Hu Q., Zhu D., Ma G., Cheng A., Wang M., Chen S., Jia R., Liu M., Sun K., Yang Q., Wu Y., Chen X.2016. A one-step duplex rRT-PCR assay for the simultaneous detection of duck hepatitis A virus genotypes 1 and 3. J. Virol. Methods 236: 207–214. doi: 10.1016/j.jviromet.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 10.Huang Q., Yue H., Zhang B., Nie P., Tang C.2012. Development of a real-time quantitative PCR for detecting duck hepatitis a virus genotype C. J. Clin. Microbiol. 50: 3318–3323. doi: 10.1128/JCM.01080-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamomae M., Kameyama M., Ishii J., Nabe M., Ogura Y., Iseki H., Yamamoto Y., Mase M.2017. An outbreak of duck hepatitis A virus type 1 infection in Japan. J. Vet. Med. Sci. 79: 917–920. doi: 10.1292/jvms.16-0646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M. C., Kwon Y. K., Joh S. J., Kim S. J., Tolf C., Kim J. H., Sung H. W., Lindberg A. M., Kwon J. H.2007. Recent Korean isolates of duck hepatitis virus reveal the presence of a new geno- and serotype when compared to duck hepatitis virus type 1 type strains. Arch. Virol. 152: 2059–2072. doi: 10.1007/s00705-007-1023-0 [DOI] [PubMed] [Google Scholar]
- 13.Kim M. C., Kwon Y. K., Joh S. J., Lindberg A. M., Kwon J. H., Kim J. H., Kim S. J.2006. Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae. J. Gen. Virol. 87: 3307–3316. doi: 10.1099/vir.0.81804-0 [DOI] [PubMed] [Google Scholar]
- 14.Kumar N., Barua S., Riyesh T., Chaubey K. K., Rawat K. D., Khandelwal N., Mishra A. K., Sharma N., Chandel S. S., Sharma S., Singh M. K., Sharma D. K., Singh S. V., Tripathi B. N.2016. Complexities in Isolation and Purification of Multiple Viruses from Mixed Viral Infections: Viral Interference, Persistence and Exclusion. PLoS One 11: e0156110. doi: 10.1371/journal.pone.0156110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan M., Ugaz V. M., Burns M. A.2002. PCR in a Rayleigh-Bénard convection cell. Science 298: 793. doi: 10.1126/science.298.5594.793 [DOI] [PubMed] [Google Scholar]
- 16.Lin S. L., Cong R. C., Zhang R. H., Chen J. H., Xia L. L., Xie Z. J., Wang Y., Zhu Y. L., Jiang S. J.2016. Circulation and in vivo distribution of duck hepatitis A virus types 1 and 3 in infected ducklings. Arch. Virol. 161: 405–416. doi: 10.1007/s00705-015-2648-z [DOI] [PubMed] [Google Scholar]
- 17.Shen Y., Cheng A., Wang M., Chen S., Jia R., Zhu D., Liu M., Sun K., Yang Q., Chen X.2015. Development of an indirect ELISA method based on the VP3 protein of duck hepatitis A virus type 1 (DHAV-1) for dual detection of DHAV-1 and DHAV-3 antibodies. J. Virol. Methods 225: 30–34. doi: 10.1016/j.jviromet.2015.08.016 [DOI] [PubMed] [Google Scholar]
- 18.Soliman M., Alfajaro M. M., Lee M. H., Jeong Y. J., Kim D. S., Son K. Y., Kwon J., Choi J. S., Lim J. S., Choi J. S., Lee T. U., Cho K. O., Kang M. I.2015. The prevalence of duck hepatitis A virus types 1 and 3 on Korean duck farms. Arch. Virol. 160: 493–498. doi: 10.1007/s00705-014-2264-3 [DOI] [PubMed] [Google Scholar]
- 19.Tsai Y. L., Wang H. T., Chang H. F., Tsai C. F., Lin C. K., Teng P. H., Su C., Jeng C. C., Lee P. Y.2012. Development of TaqMan probe-based insulated isothermal PCR (iiPCR) for sensitive and specific on-site pathogen detection. PLoS One 7: e45278. doi: 10.1371/journal.pone.0045278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng C. H., Tsai H. J.2007. Molecular characterization of a new serotype of duck hepatitis virus. Virus Res. 126: 19–31. doi: 10.1016/j.virusres.2007.01.012 [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Pan M., Fu Y., Zhang D.2008. Classification of duck hepatitis virus into three genotypes based on molecular evolutionary analysis. Virus Genes 37: 52–59. doi: 10.1007/s11262-008-0233-1 [DOI] [PubMed] [Google Scholar]
- 22.Wen X., Zhu D., Cheng A., Wang M., Chen S., Jia R., Liu M., Sun K., Zhao X., Yang Q., Wu Y., Chen X.2018. Molecular epidemiology of duck hepatitis a virus types 1 and 3 in China, 2010-2015. Transbound. Emerg. Dis. 65: 10–15. doi: 10.1111/tbed.12741 [DOI] [PubMed] [Google Scholar]
- 23.Wilkes R. P., Lee P. Y., Tsai Y. L., Tsai C. F., Chang H. H., Chang H. F., Wang H. T.2015. An insulated isothermal PCR method on a field-deployable device for rapid and sensitive detection of canine parvovirus type 2 at points of need. J. Virol. Methods 220: 35–38. doi: 10.1016/j.jviromet.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J., Wang M., Cheng A., Zhao X. X., Liu M., Zhu D., Chen S., Jia R., Yang Q., Wu Y., Zhang S., Liu Y., Yu Y., Zhang L., Sun K., Chen X.2018. Cytokine storms are primarily responsible for the rapid death of ducklings infected with duck hepatitis A virus type 1. Sci. Rep. 8: 6596. doi: 10.1038/s41598-018-24729-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M., Cheng A., Wang M., Xing H.2008. Development and application of a one-step real-time Taqman RT-PCR assay for detection of Duck hepatitis virus type1. J. Virol. Methods 153: 55–60. doi: 10.1016/j.jviromet.2008.06.012 [DOI] [PubMed] [Google Scholar]
- 26.Yugo D. M., Hauck R., Shivaprasad H. L., Meng X. J.2016. Hepatitis Virus Infections in Poultry. Avian Dis. 60: 576–588. doi: 10.1637/11229-070515-Review.1 [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y., Wang M., Cheng A., Zhu D., Jia R., Liu M., Chen S., Zhao X., Yang Q., Wu Y., Chen X.2018. Distribution of Duck Hepatitis A Virus Type 3 in Artificial Infected Ducklings and Analysis of the Relationship between Histopathological Changes and Expression of Cytokines. Acta. Veterinaria. et. Zootechnica. Sinica. 49: 164–172. [Google Scholar]


