Abstract
Retinal gene therapy with adeno-associated viral (AAV) vectors hold promises for treating inherited and non-inherited diseases of the eye. Although clinical data suggest that retinal gene therapy is safe and effective, delivery of large genes is hindered by the limited AAV cargo capacity. Protein trans-splicing mediated by split-inteins is used by single cell organisms to reconstitute proteins. Here we show that delivery of multiple AAV vectors each encoding one of the fragments of target proteins flanked by short split-inteins results in protein trans-splicing and full-length protein reconstitution in the retina of mice, pigs and in human retinal organoids. The reconstitution of large therapeutic proteins using this approach improved the phenotype of two mouse models of inherited retinal diseases. Our data support the use of split-inteins-mediated protein trans-splicing in combination with AAV subretinal delivery for gene therapy of inherited blindness due to mutations in large genes.
Introduction
The first adeno-associated viral (AAV) vector-based gene therapy product for an inherited form of blindness was approved in December 2017 (1). In addition, a number of other AAV-based products are currently under clinical development for gene therapy of rare and common forms of blindness (2). While it is now well established that AAV represents, to date, the most efficient gene therapy vehicle for the retina (2, 3) its limited cargo capacity (2) has hampered its use for conditions that require delivery of DNA sequences that exceed 5 kb in size including not only the transgene but also the cis regulatory elements that are necessary for its expression. We and others have shown that this limitation can be overcome by using either dual (up to 9 kb) (4–6) or triple (up to 14 kb) (7) AAV vectors, each containing fragments of the coding sequence (CDS) of the large transgene expression cassette. Dual and triple AAV vectors exploit concatemerization and recombination of AAV genomes to reconstitute the full-length genomes in cells co-infected by multiple AAV vectors. However, the efficiency of transgene expression achieved with either dual or triple AAV vectors in photoreceptors, which are the main therapeutic targets for most inherited retinal diseases, is lower than that achieved with single AAV vectors (4, 7, 8). This might be due to the various limiting steps required for efficient transduction including: proper DNA concatemer formation, stability of the heterogeneous mRNA and splicing efficiency across the junctions of the vectors.
Inteins are genetic elements, transcribed and translated within a host protein from which they self-excise similarly to a protein intron, without leaving amino acid modifications in the final protein product, in the absence of energy supply, exogenous host-specific proteases or co-factors (9, 10). Intein activity is context-dependent, with certain peptide sequences surrounding their ligation junction (called N- and C-exteins) that are required for efficient trans-splicing to occur, of which the most important is an amino acid containing a thiol or hydroxyl group (Cys, Ser or Thr) as first residue in the C-extein (11). Split-inteins are a subset of inteins that are expressed as two separate polypeptides at the ends of two host proteins, and catalyze their trans-splicing resulting in the generation of a single larger polypeptide (12). Inteins, including split-inteins, are widely used in biotechnological applications that include protein purification and labeling steps (12, 13), as well as the reconstitution of the widely used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 genome editing nuclease (14, 15).
In this study, we took advantage of the intrinsic ability of split-inteins to mediate protein trans-splicing to reconstitute large full-length proteins following their fragmentation into either two or three split-intein-flanked polypeptides whose sequences fit into single AAV vectors. We tested the efficiency of AAV intein in the retina by delivering the enhanced green fluorescent protein (EGFP) gene and the large ATP binding cassette subfamily A member 4 (ABCA4) and centrosomal protein 290 (CEP290) genes, which are defective in two common forms of severe inherited retinal diseases, Stargardt disease (STGD1) and Leber congenital amaurosis type 10 (LCA10), respectively. We compared the efficiency of transgene expression achieved with AAV intein to that of either single or dual AAV vectors in vitro and in vivo and used mouse, pig retina and human retinal organoids. We also investigated the efficacy of AAV intein in improving the retinal phenotypes in mouse models of STGD1 and LCA10.
Results
AAV-EGFP intein reconstitute full-length protein in vitro
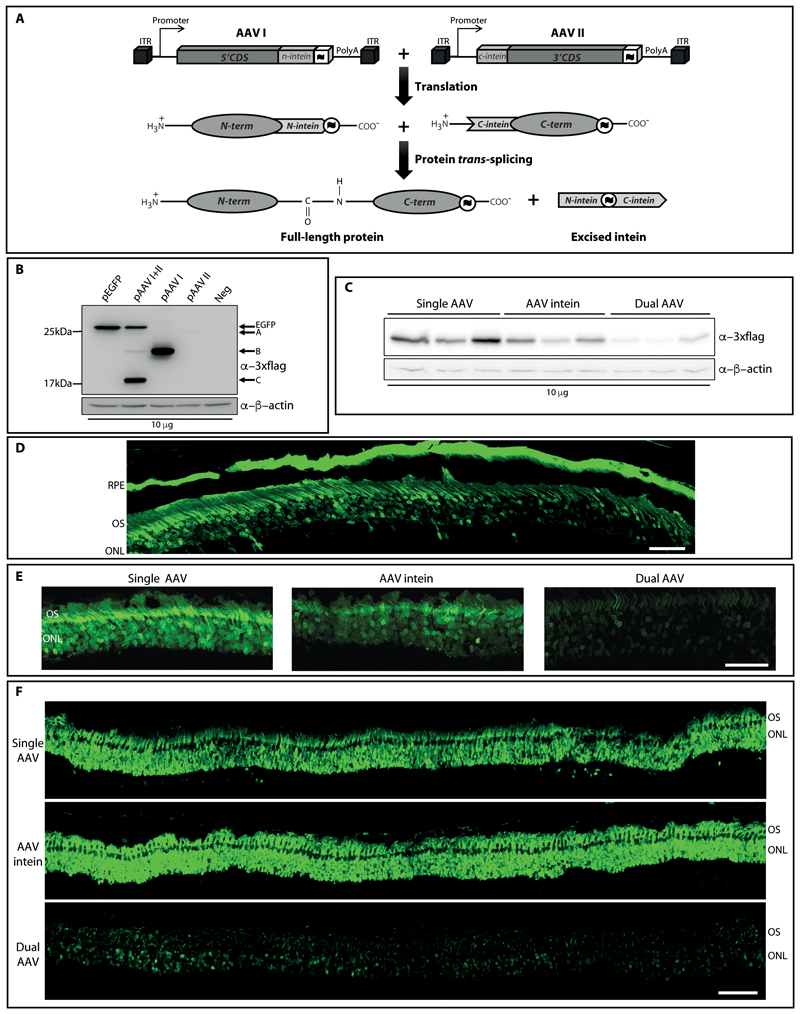
To test the efficiency of intein-mediated protein trans-splicing in the retina, we generated two AAV vectors each encoding either the N- or the C-terminal half of the reporter EGFP protein fused to the N- and C- terminal halves of the DnaE split-intein from Nostoc punctiforme [Npu (16, 17) Fig. 1A], respectively. Each AAV vector included appropriate regulatory elements (promoter and a polyadenylation signal) and a triple flag tag (3xflag) to allow detection of both halves as well as of the full-length reconstituted EGFP protein (Fig. 1A).
Figure 1. AAV intein reconstitute EGFP both in vitro and in mouse and pig retina at levels that are higher than dual AAV and up to those achieved with a single AAV.
(A) Schematic representation of AAV intein-mediated protein trans-splicing. ITR: AAV2 inverted terminal repeats; CDS: coding sequence; : 3xflag tag; PolyA: polyadenylation signal.
: 3xflag tag; PolyA: polyadenylation signal.
(B) Western blot (WB) analysis of lysates from HEK293 transfected with either full-length or AAV intein CMV-EGFP plasmids. pEGFP: full-length EGFP plasmid; pAAV I+II: AAV-EGFP I+II intein plasmids; pAAV I: single AAV-EGFP I intein plasmid; pAAV II: single AAV-EGFP II intein plasmid; Neg: untransfected cells. The arrows indicate both the full-length EGFP protein (EGFP), the N- and C-terminal halves of the EGFP protein (B and A, respectively), and the reconstituted intein excised from the full-length EGFP protein (C). The WB is representative of N=3 independent experiments.
(C) WB analysis of lysates from HEK293 infected with either single, intein or dual AAV2/2-CMV-EGFP vectors. The WB is representative of N=5 independent experiments.
(D) Retinal cryosection from C57BL/6J mice injected subretinally with AAV2/8-CMV-EGFP intein vectors. Scale bar: 50 μm. RPE: retinal pigment epithelium; OS: outer segments; ONL: outer nuclear layer. The image is representative of n=5 eyes.
(E-F) Retinal cryosections from either C57BL/6J mice (E) or Large White pigs (F) injected subretinally with either single, intein or dual AAV2/8-GRK1-EGFP vectors. Scale bar: 50 μm (E); 200 μm (F). OS: outer segment; ONL: outer nuclear layer.
AAV-EGFP intein plasmids were used to transfect human embryonic kidney 293 (HEK293) cells and evaluate the production of single N- and C-terminal halves as well as of the full-length EGFP protein. EGFP fluorescence was detected only in cells transfected either with a single AAV plasmid that encodes full-length EGFP or with the combination of AAV-EGFP intein plasmids but not with the single N- and C-terminal AAV-EGFP intein plasmids (Fig. S1). Trans-spliced EGFP protein of the expected size (~28 kDa) along with DnaE intein (~17 kDa) spliced out from the mature protein were detected by Western blot (WB) analysis of HEK293 cell lysates following co-transfection only of both AAV-EGFP intein plasmids (Fig. 1B). Quantification of EGFP bands’ intensity from AAV intein plasmids and from a single AAV plasmid is shown in Fig. S2. To define the accuracy of protein reconstitution, we immunopurified EGFP from HEK293 cells transfected with the AAV-EGFP intein plasmids and performed Liquid Chromatography-Mass Spectrometry (LC-MS) analysis to define its protein sequence. The 3532 peptides obtained from proteolytic digestion of this sample, 7 of which included the splitting point (Table 1), covered the whole protein and confirmed that the amino acidic sequence of EGFP reconstituted by AAV intein plasmids precisely corresponds to that of wild-type EGFP.
Table 1. Peptides which include the EGFP splitting point.
| Peptide sequence | Length |
|---|---|
| GVQCFSR | 7 |
| LPVPWPTLVTTLTYGVQCFSRY | 22 |
| PTLVTTLTYGVQCFSR | 16 |
| TYGVQCFSR | 9 |
| YGVQCFSR | 8 |
| VQCFSR | 6 |
| QCFSR | 5 |
N.B. C: Cysteine 71
AAV-EGFP intein are more efficient than dual AAV vectors in vitro
To confirm EGFP protein reconstitution from AAV intein vectors, we infected HEK293 cells with either AAV2/2-EGFP intein or with single and dual AAV vectors that included the same expression cassette under the control of the ubiquitous cytomegalovirus (CMV) promoter [multiplicity of infection (m.o.i): 5x10^4 genome copies (GC)/cell of each vector, which means a similar dose between the 3 systems assuming that dual vectors undergo complete DNA or protein recombination]. Seventy-two hours after infection, cell lysates were harvested and EGFP expression was evaluated by both WB (Fig. 1C) and enzyme-linked immunosorbent assay (ELISA) to quantify precisely EGFP amounts, which were found to be around half of those achieved with a single AAV and 8-times higher than those obtained with dual AAV vectors (Fig. S3). Additional quantification of the intensity of full-length EGFP relative to that of excised intein is shown in Fig. S4.
Subretinal administration of AAV-EGFP intein vectors results in efficient full-length protein reconstitution in both mouse and pig retina
To investigate whether AAV intein-mediated trans-splicing reconstitutes full-length protein expression in the retina, we injected subretinally 4-week-old C57BL/6J mice with AAV2/8-CMV-EGFP intein vectors (dose of each vector/eye: 5.8x10^9 GC). Eyes were harvested 1 month later and analyzed by microscopy analysis. EGFP fluorescence was detected in all eyes in the retinal pigment epithelium and, most importantly, in photoreceptors (Fig. 1D and S5). To compare transgene expression from AAV intein to that of single and dual AAV in photoreceptors, we injected subretinally in 4-week-old C57BL/6J mice AAV2/8 vectors (dose of each vector/eye: 5x10^9 GC) that encode EGFP under the control of the photoreceptor-specific human G protein-coupled receptor kinase 1 (GRK1) promoter. Eyes were harvested 1 month post-injection and EGFP fluorescence from photoreceptor cells (as the photoreceptor-specific GRK1 was used to drive EGFP expression) was detected in eyes injected with all set of vectors (Fig. 1E and S6). Precise quantification of EGFP protein amounts by ELISA confirmed that AAV intein reconstituted EGFP protein less efficiently than a single AAV and about 3-times more than dual AAV (Fig. S7). The relative amount of full-length EGFP to excised intein following quantification of WB band intensities is shown in Fig. S8.
We then evaluated the efficiency of AAV intein vectors at transducing photoreceptors in the pig retina, which is an excellent pre-clinical model to evaluate viral vector transduction due to its size and architecture (18). Thus, we injected subretinally Large White pigs with single, intein and dual AAV2/8-GRK1-EGFP vectors (dose of each vector/eye: 2x10^11 GC, delivered through two adjacent subretinal blebs). Eyes were harvested 1 month post-injection and EGFP protein reconstitution in the photoreceptor cell layer mediated by either single, dual or AAV intein vectors, as assessed by EGFP spontaneous fluorescence, is shown in Fig. 1F. Precise quantification of EGFP in retinal lysates confirmed that AAV intein reconstitute the protein to quantities that are similar to those achieved with a single AAV and about 3-times higher than those obtained with dual AAV vectors (Fig. S9). The relative amount of full-length EGFP to excised intein following quantification of WB band intensities is shown in Fig. S10.
Full-length EGFP is reconstituted by AAV-mediated protein trans-splicing in 3D human retinal organoids
As an additional pre-clinical model representative of the human retina, we generated 3D retinal organoids (19, 20) from human induced pluripotent stem cells (iPSCs). Six month-old organoids (Fig. 2A and S11A) contained cells stained by mature photoreceptor markers (Fig. 2B and S11B) and transduced by AAV2 vectors with a photoreceptor-specific promoter (Fig. 2C and S11C). Light (Fig. 2D and S11B) and electron (Fig. 2E-F and S11E-F) microscopies show the presence of buds of photoreceptor outer segments. Nine month-old 3D human retinal organoids incubated for 30 days with AAV-GRK1-EGFP intein vectors (dose of each vector/organoid: 1x10^12 GC) show EGFP fluorescence (Fig. 2G and S11G). The relative amount of full-length EGFP to excised intein following quantification of WB band intensities is shown in Fig. S12.
Figure 2. Characterization and AAV intein-mediated transduction of human iPSCs-derived 3D retinal organoids.
(A) Light microscopy analysis of retinal organoids at 183 days of culture.
(B) Immunofluorescence analysis with antibodies directed to mature photoreceptor markers. Scale bar: 100 μm.
(C) Fluorescence analysis of retinal organoids infected with both AAV2/2-CMV-EGFP and AAV2/2-IRBP-DsRed vectors. Scale bar: 100 μm.
(D) Outer segment-like structures were observed which protrude from the surface of retinal organoids at 230 days of culture. The inset shows the presence of outer segment (OS)-like structures with radial architecture. NR: neural retina; RPE: retinal pigment epithelium.
(E) Scanning electron microscopy analysis reveals the presence of inner segments (IS), connecting cilia (CC) and outer segment (OS)-like structures. Scale bar: 4 μm.
(F) Electron microscopy analysis reveals the presence of the outer limiting membrane (*), centriole (C), basal bodies (BB), connecting cilia (CC) and sketches of outer segments (OS). The inset shows the presence of disorganized membranous discs in the OS. Scale bar: 500 nm.
D: days of culture.
(G) Fluorescence analysis of retinal organoids infected with AAV2/2-GRK1-EGFP-intein vectors at 293 days of culture. Scale bar: 100 μm. The image is representative of n=4 organoids.
Identification of optimal ABCA4 and CEP90 splitting points is required for efficient AAV intein-mediated protein trans-splicing
To test whether protein trans-splicing can be developed as a mechanism to reconstitute large therapeutic proteins, we developed AAV-ABCA4 and -CEP290 intein vectors.
ABCA4 and CEP290 were split into either two (AAV I, AAV II) or three (AAV I, AAV II, AAV III) fragments whose coding sequences were separately cloned in single AAV vectors, fused to the coding sequences of the split-inteins N- and C-termini (Fig. S13). The AAV intein vectors included either the ubiquitous short CMV (shCMV) or the GRK1 promoter.
Splitting points for each protein were selected taking into account both amino acid residue requirements at the junction points for efficient protein trans-splicing (11, 21), as well as preservation of the integrity of critical protein domains, which should favor proper folding and stability of each independent polypeptide, and thus, of the final reconstituted protein. Additional split-inteins were also considered. CEP290 sets in which the protein was split in 3 polypeptides (sets 4 and 5, Fig. S13B) were generated to allow the inclusion of the Woodchuck hepatitis virus Post-transcriptional Regulatory Element [WPRE, (22)] to increase transgene expression. To prevent unwanted trans-splicing between AAV I and AAV III which could reduce the amount of full-length protein generated, sets 4 and 5 included two different split-inteins at the two splitting junctions, specifically DnaB intein from Rhodothermus marinus and either wild-type or a mutated DnaE intein which we show do not result in detectable EGFP expression, thus do not cross-react (Fig. S14).
We compared the ability of each set of AAV intein plasmids to reconstitute ABCA4 and CEP290 following transfection of HEK293 cells. WB analysis of cell lysates 72 hours post-transfection showed that full-length ABCA4 and CEP290 proteins of the expected size (~ 250 kDa and ~ 290 kDa, respectively) were reconstituted from each set of AAV intein plasmids, although with variable efficiency (Fig. 3A-B). Sets 1 and 5 tended to be the most efficient for ABCA4 and CEP290 protein reconstitution, respectively, and thus used in all the subsequent experiments.
Figure 3. Optimization of AAV intein allows proper reconstitution of the large ABCA4 and CEP290 proteins.
Western blot (WB) analysis of lysates from HEK293 transfected with different sets of either AAV-shCMV-ABCA4 or -CEP290 intein plasmids. A schematic representation of the various sets used is depicted in Fig. S13. The WB are representative of N=3 independent experiments. (A) Kruskal-Wallis test p value was not significant, thus no post hoc comparison was performed to evaluate statistical differences between groups. (B) Significant differences were assessed using One-way ANOVA followed by Tukey multiple pairwise-comparison. * p <0.05; ** p <0.01; *** p <0.001. Details on set 1 (A) and set 5 (B) variability can be found in the Statistical analysis paragraphs of the Materials and methods section.
To define the accuracy of protein reconstitution we immunopurified ABCA4 from HEK293 cells transfected with set 1 and performed LC-MS analysis to define its protein sequence. The 3108 peptides obtained from proteolytic digestion of this sample, 22 of which included the splitting point (Table 2), covered the whole protein and confirmed that the amino acidic sequence of ABCA4 reconstituted by AAV intein plasmids precisely corresponds to that of wild-type ABCA4 (Fig. S15).
Table 2. Peptides which include the ABCA4 splitting point.
| Peptide sequence | Length |
|---|---|
| KNCFGT | 6 |
| KNCFGTGL (x3) | 8 |
| KNCFGTGLY (x2) | 9 |
| FLKNCFGTGL | 10 |
| KNCFGTGLYLT | 11 |
| KNCFGTGLYLTL | 12 |
| LYCSGTPLFLKNC | 13 |
| YCSGTPLFLKNCF | 13 |
| KNCFGTGLYLTLVR (x7) | 14 |
| KNCFGTGLYLTLVRKM | 16 |
| IAIIAQGRLYCSGTPLFLKNCFGTGLYLT | 29 |
| QGRLYCSGTPLFLKNCFGTGLYLTLVRKMKNIQSQR | 36 |
| GTPLFLKNCFGTGLYLTLVRKMKNIQSQRKGSEGTCSCSS | 40 |
N.B. C: Cysteine 1150
We then assessed the intracellular localization of the protein products of the different intein containing plasmids comparing them to the localization of the full-length protein. Full-length ABCA4 is known to localize at the endoplasmic reticulum (ER) when expressed in cultured cell lines (23, 24). We found that the two ABCA4 polypeptides from set 1 show ER co-localization which is slightly lower for the polypeptide from AAV1. Low localization at the Trans-Golgi network was found for all products, in particular for the polypeptide from AAV1 (Fig. 4A). A similar localization was observed in cells co-transfected with both AAV intein plasmids, as well as in cells transfected with a plasmid encoding for the full-length ABCA4 protein, thus confirming the predominant localization in the ER of ABCA4 exogenously expressed in cell lines (23, 24).
Figure 4. ABCA4 and CEP290 proteins from AAV intein vectors have a distribution pattern similar to those from full-length plasmids.
Representative images of immunofluorescence analysis of HeLa cells transfected with either AAV-shCMV-ABCA4 (A) or -CEP290 (B) intein plasmids. pABCA4 (A) or pCEP290 (B): plasmid including the full-length expression cassette; pAAV intein: AAV-intein plasmids (either set 1 in A or set 5 in B); I+II+III: AAV I+II+III intein plasmids; I+II: AAV I+II intein plasmids; I+III: AAV I+III intein plasmids; II+III: AAV II+III intein plasmids; I: single AAV I intein plasmid; II: single AAV II intein plasmid; III: single AAV III intein plasmid; Neg: untransfected cells.
Cells were stained for 3xFLAG and either VAP-B (endoplasmic reticulum marker) and TGN46 (Trans-Golgi network marker) in A, or acetylated tubulin (marker of microtubules) in B. White arrows point at cells shown at higher magnification in Fig. S16.
Quantification of both ABCA4 co-localization with VAP-B and TGN46 markers (A) and cells showing the various CEP290 polypeptides patterns (B) are shown in the graphs. At least 150 cells for each condition were counted in N=3 independent experiments. (A) Significant differences between groups were assessed using Kruskal-Wallis test followed by pairwise comparisons using Wilcoxon rank sum test. ** p <0.01; *** p <0.001. Asterisks above pAAV intein I column in the upper graph indicate significant differences with both pABCA4, I+II and II. (B) Significant differences between patterns of each column were assessed using binomial distribution. # indicates the predominant pattern for each CEP290 polypeptide (p<< 0,00001).
As for CEP290, it has been reported that the full-length protein shows a mixed distribution pattern with a predominant punctate and a minor fibrillar pattern (25). The dissection of the domains responsible for the subcellular targeting of CEP290 (25) showed that N-terminal domain (a.a. 1-362) targets the protein to vesicular structures thanks to its ability to interact with membranes, while a region near the C-terminus of CEP290, encompassing much of the protein’s myosin-tail homology domain, mediates microtubule binding (a.a. 580-2479) and when expressed as truncated form has a prominent fibrillar distribution coincident with acetylated tubulin (Ac-Tub) (25). Consistent with this, products from AAV I and II have a predominant punctate pattern while products from AAV III (encompassing protein’s myosin-tail homology domain) show a predominant fibrillar pattern and is the one that mostly colocalizes with Ac-tub (Fig. 4B). Thus, the patterns shown by products from AAV I+II, AAV I+III and AAV II+III are a combination of those shown by the single AAV intein plasmids (Fig. 4B). Cells co-transfected with the three AAV-CEP290 intein plasmids or with the plasmid encoding for the full-length CEP290 protein showed a predominant punctate signal partially aligned along microtubules (Fig. 4B and Fig. S16).
We then compared the amount of protein obtained with the best set of AAV-ABCA4 and -CEP290 intein plasmids to those obtained from a single AAV plasmid encoding for the corresponding full-length protein. To this aim, HEK293 cells were transfected with same equimolar amounts of either the single or the AAV intein plasmids and 72 hours after transfection cell lysates were analyzed by WB (Fig. S17).
AAV intein vectors mediate expression of large therapeutic proteins in vitro and in the retina
We compared the efficiency of AAV intein-mediated large protein reconstitution to that of dual AAV vectors both in vitro and in the mouse and pig retina. HEK293 cells were infected with either AAV2/2 dual or intein vectors encoding for either ABCA4 or CEP290 (m.o.i: 5x 10^4 GC/cell of each vector) and cell lysates were analyzed 72 hours later by WB. As shown in Figure 5A, the intensity of the AAV intein ABCA4 band is higher than that of dual AAV vectors despite the amount of protein lysate loaded was 10-fold lower to avoid saturation. Similarly, as shown in Figure 5B, CEP290 expression is detected only in cells infected with AAV intein but not with dual AAV vectors. As expected, in addition to full-length proteins, shorter polypeptides derived from either the single AAV intein vectors (in the case of both ABCA4 and CEP290) or from trans-splicing occurring between AAV II and AAV III (in the case of CEP290) were observed (Fig. 5A and 5B).
Figure 5. ABCA4 and CEP290 large protein reconstitution by AAV intein and dual AAV vectors.
Western blot (WB) analysis of lysates from HEK293 cells infected with either dual or intein AAV2/2-shCMV-ABCA4 (A) or -CEP290 (B) vectors.
AAV intein: AAV-ABCA4 (set 1, A) or -CEP290 (set 5, B) intein vectors; I+II+III: AAV I+II +III intein vectors; I+II: AAV I+II intein vectors; I+III: AAV I+III intein vectors; II+III: AAV II+III intein vectors; I: single AAV I intein vector; II: single AAV II intein vector; III: single AAV III intein vector; dual AAV: dual AAV vectors; Neg: either AAV-EGFP vectors or PBS.
(A) The arrows indicate the full-length ABCA4 protein and A: protein product derived from AAV I; B: protein product derived from AAV II. * protein product with a potentially different post-translational modification.
(B) The arrows indicate the full-length CEP290 protein and A: protein product derived from AAV II+III; B: protein product derived from AAV I+II; C: protein product derived from AAV II; D: protein product derived from AAV III; E: protein product derived from AAV I.
The WB are representative of N=3 independent experiments.
We then injected subretinally 4-week-old wild-type mice with AAV-GRK1-ABCA4 or -CEP290 intein vs dual vectors (dose of each ABCA4 vector/eye: 3.3x10^9 GC, dose of each CEP290 vector/eye: 1.1x10^9 GC). Animals were sacrificed 4-7 weeks post-injection, and protein expression in retinal lysates was evaluated by WB. Full-length proteins were detected in 10/11 of AAV-ABCA4 intein-injected eyes (Fig. 6A and S18) and in 5/10 of AAV-CEP290 intein-injected eyes (Fig. 6B and S19). Conversely, full-length protein expression was evident in 5/9 and in 0/5 eyes injected with ABCA4 and CEP290 dual AAV vectors, respectively. In addition, we detected polypeptides derived from the single AAV intein vectors (in the case of both ABCA4 and CEP290) and from trans-splicing occurring between AAV II and AAV III (in the case of CEP290) (Fig. 6A and 6B), as previously observed in vitro.
Figure 6. AAV intein reconstitute large proteins in mouse, pig and human photoreceptors.
(A-C) Western blot (WB) analysis of retinal lysates from either wild-type mice (A, B) or Large White pigs (C) injected with either dual or intein AAV2/8-GRK1-ABCA4 (A, C) or -CEP290 (B) vectors. AAV intein: AAV intein vectors; Dual AAV: dual AAV vectors; Neg: either AAV-EGFP vectors or PBS. Quantification of ABCA4 expression in Large White pigs injected with either dual (n=3) or intein (n=2) AAV2/8-GRK1-ABCA4 is included in (C). Significant differences between groups were assessed using unpaired Student’s t-test. ** p <0.01.
(D) WB analysis of lysates from human iPSCs-derived 3D retinal organoids infected with AAV2/2-GRK1-ABCA4 intein vectors. AAV intein: AAV-ABCA4 intein vectors; Neg: not infected organoids; -/-: organoids derived from STGD1 patients.
(A, C, D) The arrows indicate the full-length ABCA4 protein (ABCA4) and A: protein product derived from AAV I; B: protein product derived from AAV II. * protein product with a potentially different post translational modification.
(B) The arrows indicate both the full-length CEP290 protein (CEP290); A: protein product derived from AAV II+III and D: protein product derived from AAV III.
The WB are representative of: n=10 AAV intein- and 9 dual AAV-injected eyes (A); n=10 AAV intein- and 5 dual AAV-injected eyes (B); n=7 AAV intein-infected organoids (D).
To investigate the efficiency of protein reconstitution mediated by AAV intein relative to endogenous, we injected subretinally 1-4 month-old Abca4-/- mice with AAV-GRK1-ABCA4 intein vectors (dose of each ABCA4 vector/eye: 5.5x10^9 GC). One month later, ABCA4 expression in retinal lysates from unaffected and AAV intein-injected Abca4-/- mice was analyzed by WB using an antibody which recognizes both murine and human ABCA4. The quantification of AAV intein ABCA4 expression versus endogenous is shown in Fig. S20.
To confirm efficient large protein reconstitution in the clinically-relevant pig retina, we injected subretinally Large White pigs with either AAV2/8-GRK1-ABCA4 intein or dual vectors (dose of each vector/eye: 2x10^11 GC, delivered through two adjacent subretinal blebs) and 1 month post-injection analyzed protein expression by WB. We found that AAV intein reconstitute full-length ABCA4 protein more efficiently than dual AAV vectors (Fig. 6C).
Lastly, we infected human retinal organoids from iPSCs of either healthy individuals or patients with STGD1 at 121 days of culture [when photoreceptor maturation starts (20)] with AAV2/2-GRK1-ABCA4 intein vectors (dose of each vector/organoid: 1x10^12 GC). Organoids were lysed between 20 and 40 days after infection and analyzed by WB. ABCA4 of the expected size was detected in all infected organoids (Fig. 6D and Fig. S21; n=3 and n=4 from normal control and STGD1 organoids, respectively).
Subretinal administration of AAV intein vectors improves the retinal phenotype of STGD1 and LCA10 mouse models
To determine whether the photoreceptors transduction obtained with AAV intein vectors could be therapeutically-relevant, we tested them in the retina of STGD1 and LCA10 mouse models (Abca4−/− and rd16 mice).
One month-old Abca4−/− mice were injected subretinally with AAV2/8-GRK1-ABCA4 intein vectors (dose of each vector/eye: 4.3-4.8x10^9 GC). Three months later we harvested the eyes and performed transmission electron microscopy analysis of retinal ultrathin sections to measure the amounts of lipofuscin, which accumulates in the retinal pigmented epithelium (RPE) of Abca4−/− mice (26, 27).
We found that RPE lipofuscin accumulation was significantly reduced in the Abca4−/− eyes injected with AAV intein vectors but not in negative control injected eyes (p value = 0.0163; Fig. 7A and Fig. S22). Naïve uninjected Abca4-/- mice were not included as we have shown that subretinal injections per se induce lipofuscin granules (28), therefore the appropriate controls for the effect of ABCA4 gene delivery on lipofuscin reduction are sham-injected Abca4-/- mice. In parallel, 4-6 day-old rd16 mice were injected subretinally with AAV2/8-GRK1-CEP290 intein vectors (dose of each vector/eye: 5.5x10^8 GC). Microscopy analysis of retinal sections 1 month after injection showed that the thickness of the outer nuclear layer (ONL), which includes photoreceptors nuclei, was significantly reduced in rd16 mice compared to wild-type mice (p value = 0.0057; Fig. 7B), as result of progressive retinal degeneration (25). We found that the ONL thickness in the rd16 retinas injected with AAV intein vectors was significantly higher (about 60%, p value = 0.0042) than that of negative control injected rd16 retinas (Fig. 7B). Accordingly, retinal function tests based on pupillary light responses (PLR) showed a significant higher pupil constriction (about 20%, p value = 0.0005939) in rd16 mice injected with AAV intein vectors than in negative control-injected rd16 eyes (Fig. 7C).
Figure 7. Subretinal administration of AAV intein improves the retinal phenotype of mouse models of inherited retinal degenerations.
(A) Quantification of the mean area occupied by lipofuscin in the RPE of Abca4-/- mice treated with AAV intein. Each dot represents the mean value measured for each eye. The mean value of the lipofuscin area for each group is indicated in the graph. +/+ or +/-: control injected Abca4+/+ or +/- eyes (PBS); -/-: negative control-injected Abca4-/- eyes (AAV I ABCA4 or AAV II ABCA4 or PBS); -/- AAV intein: Abca4-/- eyes injected with AAV intein vectors. Significant differences between groups were assessed using one-way ANOVA followed by Tukey multiple pairwise-comparison. * p <0.05; *** p <0.001.
(B) Representative images of retinal sections from wild-type uninjected and rd16 mice either not injected or injected subretinally with AAV2/8-GRK1-CEP290 intein vectors (AAV intein) or with negative controls (Neg: AAV I+II or AAV II+III or PBS). Scale bar: 25 μm. The thickness of the ONL measured in each image is indicated by the vertical black line. RPE: retinal pigment epithelium; ONL: outer nuclear layer; INL: inner nuclear layer; GCL: ganglion cell layer. Significant differences between groups were assessed using Kruskal-Wallis test followed by pairwise comparisons using Wilcoxon rank sum test. ** p <0.01.
(C) Representative images of eyes from wild-type uninjected and rd16 mice either not injected or injected subretinally with AAV2/8-GRK1-CEP290 intein vectors (AAV intein) or with negative controls (Neg: AAV I+II or AAV II+III or PBS). White circles define pupils. Significant differences between groups were assessed using one-way ANOVA followed by Tukey multiple pairwise-comparison. *** p <0.001.
The complete set of p values can be found in the Auxiliary Quantification File.
We additionally investigated the safety of AAV intein vectors in the retina. To this aim, we injected subretinally wild-type C57BL/6J mice with either AAV2/8-GRK1-ABCA4 or -CEP290 intein vectors (dose of each ABCA4 vector/eye: 4.3x10^9 GC; dose of each CEP290 vector/eye: 1.1x10^9 GC) and measured retinal electrical activity by Ganzfeld electroretinogram (ERG) at 6 and 4.5 months post-injection, respectively. In both studies a- and b-wave amplitudes were similar between mouse eyes that were injected with AAV intein vectors and eyes injected with negative controls (Fig. S23). In addition, the thickness of the ONL measured by optical coherence tomography was similar between AAV intein- and negative control- injected eyes (Fig. S24).
Discussion
Great efforts have been directed to overcome the challenge of delivering large genes with AAV vectors. One of the systems with potential for clinical translation relies on the use of multiple AAV vectors (dual or triple) each carrying one part of a large transgene expression cassette that gets reconstituted upon co-infection by the multiple AAV and their genome tail-to-head concatemerization/recombination (29). The retina, being small and enclosed, favors co-infection by multiple AAV vectors and indeed dual and triple AAV have been successfully used to expand AAV transfer capacity in the retina (4, 7, 30, 31). Yet, transduction efficiency of multiple AAV is lower than that of single AAV (4, 7, 8) and may not be sufficient for specific applications (7). Here, we have investigated the reconstitution of large proteins in the retina via intein-mediated protein trans-splicing, a method widely used to purify/modify recombinant proteins (9, 10, 12).
In the context of gene therapy, intein-mediated protein trans-splicing has been used to reconstitute large therapeutic proteins like Factor VIII in liver (32, 33), dystrophin in muscle (34) or the L-type calcium channel in cardiomyocytes (35), however with limited efficacy. Here we show that inteins reconstitute large proteins in the retina of small and large animal models and in human retinal organoids, and this improves the retinal phenotype of animal models of STGD1 and LCA10. This may be well favored by the small subretinal space, as it does with dual AAV vectors.
These amounts largely exceeds those achieved via AAV genome recombination by dual AAV vectors, the gold standard for large protein reconstitution, both in vitro and in pig photoreceptors. In the large pig retina, we found that the amounts of the reconstituted EGFP protein were comparable to those achieved by single AAV vectors. This is similar to what observed with dual AAV where the efficiency of multiple AAV transduction is higher in the pig than in the mouse retina (4, 7), and bodes well for future translation of this approach to the clinic. We show that AAV intein-mediated protein trans-splicing occurs in photoreceptors across species up to human from iPSCs-derived retinal organoids.
One of the limitations we noted when applying trans-splicing to large proteins was that construct design needs to take into account: i. preservation at the junction points of amino acid residues needed for efficient protein trans-splicing; ii. splitting of the proteins outside of structural domains to avoid incorrect polypeptide folding. Indeed, we generated several sets of AAV intein constructs both for ABCA4 and CEP290, and found that their efficiency varied in terms of amount of protein reconstitution. In some cases, one of the two protein halves was less stable than the other which presumably impacted the quantity of full-length protein produced. Also, in one case (when using AAV-ABCA4 intein set 3) we found a band higher than full-length, indicating a larger product, which could be the result of incomplete trans-splicing. According to our experience, construct design is more critical for AAV intein than for dual AAV where the use of exogenous recombinogenic sequences as well as splicing signals allows to split a large coding sequence independently of the native protein structure and organization.
Additionally, when using intein-mediated protein trans-splicing each polypeptide is expressed from an independent expression cassette that needs to carry the required transcriptional elements which will be included in each of the AAV intein vectors. This is different from dual AAV where the single large expression cassette is split into two halves with the promoter element present only in the 5’ vector and the polyA signal in the 3’ vector. Therefore, when using AAV intein vectors one limitation is that part of the cloning capacity will be taken up by regulatory elements that need to be replicated within each vector. This might be limiting in terms of size constraint, thus to accommodate in two AAV intein vectors the coding sequence of large proteins, as CEP290, short regulatory elements are required. However, these shorter elements are often weak in terms of transcriptional activity. Indeed, here we show that splitting CEP290 into three polypeptides rather than two allows to accommodate into each AAV vector larger or more potent regulatory elements (a full-length CMV promoter or WPRE) and this leads to more efficient full-length protein expression than that obtained with two AAV intein vectors with weaker regulatory elements, despite the need to achieve co-infection of the same cell by three vectors. Pivotal to the development of triple AAV intein vectors has been the use of different inteins, DnaE and DnaB, which do not cross-react thus preventing improper trans-splicing between the polypeptides produced by AAV I and AAV III.
Despite this extensive optimization, the amounts of therapeutic large proteins we achieved by intein-mediated trans-splicing tend to be lower than those achieved for EGFP compared to a full-length expression cassette, suggesting that large therapeutic proteins appear to be more challenging to reconstitute than the small and stable EGFP. Yet, the amount of AAV intein ABCA4 in whole retinal lysates was about 10% of the endogenous protein. This expression is the result of only 1/3 of the retina exposed to AAV.
For efficient trans-splicing to occur the different polypeptide fragments should reside in the same subcellular compartment. This might be particularly critical for trans-membrane proteins like ABCA4 in which the two halves might be targeted to different compartments. Our immunofluorescence analysis shows that both the N- and C-terminal halves of ABCA4 localize to the endoplasmic reticulum in vitro, suggesting that trans-splicing might occur early after translation, thus assuring proper further targeting of the full-length protein. Proper protein assembly is also confirmed by protein sequencing of trans-spliced ABCA4, as well as by the presence of the correctly excised split-intein after trans-splicing. Importantly, the improvement in the retinal phenotype of the two STGD1 and LCA10 animal models of inherited retinal diseases following subretinal injection of the AAV intein vectors strongly suggests that the reconstituted proteins have been properly processed and were functional.
An additional limitation of the AAV intein platform is the presence of both intein excised from the mature protein and truncated protein products that derive from either non trans-spliced polypeptides or trans-splicing occurring only between two of three polypeptides (for CEP290 protein). Although these additional protein products are in some instances more abundant than the full-length proteins, they do not appear to be toxic. Strategies aiming at maximizing the equimolar amount of single polypeptides in target cells (for instance by modulating their doses based on their different stability, for example by increasing the dose of the AAV intein vector expressing the less stable polypeptide) could result in optimal trans-splicing and could lower the quantity of the more stable single polypeptide.
Similarly, excised inteins are produced upon trans-splicing. A method to reduce them can be envisaged which is based on the inclusion between the two split inteins of a sequence which mediates their selective degradation after splicing, as we have previously successfully done with dual AAV vectors (28). Anyhow, our in vivo electrophysiological and morphological analysis show no signs of toxicity up to 4.5-6 months post-injection in eyes injected with either AAV-CEP290 or -ABCA4 intein vectors, respectively, although definitive assessment of the safety of AAV intein will require formal long-term toxicity studies.
In conclusion, we have shown that AAV intein-mediated protein trans-splicing reconstitutes large proteins both in vitro and in the retina of mouse, pig and in human retinal organoids. Whereas some proteins can be efficiently reconstituted with two AAV intein vectors, others that are larger and require the use of large robust regulatory elements require three AAV intein vectors. Thus, careful design and optimization of the AAV intein constructs are required to achieve efficient protein trans-splicing. Importantly, subretinal administration of AAV intein vectors resulted in improvement of the retinal phenotype of STGD1 and LCA10 mouse models, thus providing evidence of the therapeutic potential of AAV intein vectors for gene therapy of these and other blinding diseases due to mutations in large genes.
Materials and Methods
Study Design
This study was designed to define the efficiency of AAV intein-mediated protein trans-splicing in reconstituting large proteins in the retina. This was defined by comparing the efficiency of protein expression achieved via protein trans-splicing to that achieved using reference platforms (normal size and dual AAV vectors) and by evaluating the impact of subretinal delivery of AAV intein on the retinal phenotype of animal models of inherited retinal diseases. In all in vivo studies right and left eyes were randomly assigned to each treatment group. Additionally, in the studies in the disease models, female and male mice were considered equivalent and randomly assigned to treatment groups. Littermate controls were used when available. Protein expression in the in vitro and in vivo studies was quantified objectively using ImageJ software (for Western blot analysis) or spectrophotometer to measure the optical density at 450 nm (for ELISA analysis), without blinding. The experimenters in the efficacy and toxicity studies were blind to both genotype and treatment of the animals. Sample sizes were determined on the basis of previous experience and technical feasibility, at least three biological replicates in culture experiments or five animals per group were used in all the experiments, as indicated in the Results section and figure legends.
Statistical analysis
Data are presented as mean (±s.e.) which has been calculated using the number of independent in vitro experiments or eyes (not replicate measurements of the same sample). Statistical p values ≤ 0.05 were considered significant. The normality assumption was verified using Shapiro-Wilk test. Data were analysed by either Student’s t -test or one-way ANOVA. Kruskal-Wallis rank sum test (non-parametric test) was used when ANOVA assumptions were not met. Pairwise comparisons between group levels with corrections for multiple testing were performed to determine if the mean difference between specific pairs of groups are statistically significant.
In experiments where internal reference samples were used to normalize data across different replicates, their expression was set to either 100% or 1, as indicated in each graph. To show the internal variability of these internal reference samples, we calculated their expression as percentage relative to selected samples, as described below: Fig. S2 pEGFP vs pAAVI+II: 138,7 ± 17,4; Fig. S4 EGFP vs DnaE: 5,01 ± 0,14; Fig. S8 EGFP vs DnaE: 0,38 ± 0,13; Fig. S10 EGFP vs DnaE: 0,60 ± 0,19; Fig. S12 EGFP vs DnaE: 6,2 ± 0,35; Fig. 3A Set1 vs Set2: 274,2 ± 46,5; Fig. 3B Set5 vs Set1: 529,4 ± 69,4; Fig. S17A pABCA4 vs Set1: 213,9 ± 51,5; Fig. S17B pCEP290 vs Set5: 238,6 ± 38,2.
Original data including specific statistical values are provided in the Auxiliary Quantification File.
Supplementary Material
One Sentence Summary.
Adeno-associated viral vectors reconstitute large proteins in the retina via intein-mediated protein trans-splicing.
Acknowledgements
We thank the following people from TIGEM, Naples, Italy: Annamaria Carissimo (Bioinformatics Core) for the statistical analysis; the AAV Vector Core for AAV vector production; Edoardo Nusco for help with mice injections; the Microscopy Core for assistance in microscope imaging; Graciana Diez-Roux (Scientific Office) and Nicola Brunetti-Pierri for the critical reading of this manuscript.
Fundings
This work was supported by the European Research Council (ERC) (grant numbers, 694323 “EYEGET” to A.A.), the Telethon Foundation (grant TGM16MT1) and by the University of Naples Federico II under STAR Program (to I.T.). M.A.D acknowledges the support of Telethon grant TGM11CB1, Associazione Italiana per la Ricerca sul Cancro grant IG2013_14761, and European Research Council Advanced Investigator grant 670881 (SYSMET).
Footnotes
Author contribution: The study was conceived, designed, and written by A.A., P. Tornabene and I.T. All data were generated by P. Tornabene and I.T. with the technical help of R.M., M.C., M.L., S.dS.
P. Tornabene developed retinal organoids and performed the in vitro and in vivo experiments of EGFP and CEP290; I.T. performed the in vitro and in vivo experiments of ABCA4. F.D.A. and E.M. performed the morphological and functional analysis in vivo. P. Tiberi, C.I., C.G., S.R., E.M.S., F.S performed injections in mice and pigs. L.G and M.A.D.M performed studies on ABCA4 localization in cells. S.A, C.B.H., F.P.M.C provided iPSCs cell lines. E.P and R.P. supervised and performed electron microscopy analysis.
Competing interests: A.A., P. Tornabene and I.T. are co-inventors on patent application number EP 18200490.3 entitled “Intein proteins and uses thereof”. The other authors declare that they have no competing interests.
Data and materials availability
All data associated with this study are included in the paper and in the Supplementary Materials.
References
- 1.FDA approves hereditary blindness gene therapy. Nat Biotechnol. 2018;36:6. doi: 10.1038/nbt0118-6a. [DOI] [PubMed] [Google Scholar]
- 2.Trapani I, Auricchio A. Seeing the Light after 25 Years of Retinal Gene Therapy. Trends Mol Med. 2018 doi: 10.1016/j.molmed.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Auricchio A, Smith AJ, Ali RR. The Future Looks Brighter After 25 Years of Retinal Gene Therapy. Hum Gene Ther. 2017;28:982–987. doi: 10.1089/hum.2017.164. [DOI] [PubMed] [Google Scholar]
- 4.Trapani I, et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol Med. 2014;6:194–211. doi: 10.1002/emmm.201302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan D, Yue YJ, Engelhardt F. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 6.Yan Z, Zhang Y, Duan D, Engelhardt JF. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc Natl Acad Sci U S A. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maddalena A, et al. Triple Vectors Expand AAV Transfer Capacity in the Retina. Mol Ther. 2018;26:524–541. doi: 10.1016/j.ymthe.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colella P, et al. Efficient gene delivery to the cone-enriched pig retina by dual AAV vectors. Gene Ther. 2014;21:450–456. doi: 10.1038/gt.2014.8. [DOI] [PubMed] [Google Scholar]
- 9.Novikova O, Topilina N, Belfort M. Enigmatic distribution, evolution, and function of inteins. J Biol Chem. 2014;289:14490–14497. doi: 10.1074/jbc.R114.548255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills KV, Johnson MA, Perler FB. Protein splicing: how inteins escape from precursor proteins. J Biol Chem. 2014;289:14498–14505. doi: 10.1074/jbc.R113.540310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah NH, Eryilmaz E, Cowburn D, Muir TW. Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. J Am Chem Soc. 2013;135:5839–5847. doi: 10.1021/ja401015p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y. Split-inteins and their bioapplications. Biotechnol Lett. 2015;37:2121–2137. doi: 10.1007/s10529-015-1905-2. [DOI] [PubMed] [Google Scholar]
- 13.Shah NH, Muir TW. Inteins: Nature's Gift to Protein Chemists. Chem Sci. 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmelas C, Grimm D. Split Cas9, Not Hairs - Advancing the Therapeutic Index of CRISPR Technology. Biotechnol J. 2018;13:e1700432. doi: 10.1002/biot.201700432. [DOI] [PubMed] [Google Scholar]
- 15.Villiger L, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 16.Iwai H, Zuger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 17.Zettler J, Schutz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Mussolino C, et al. AAV-mediated photoreceptor transduction of the pig cone-enriched retina. Gene Ther. 2011;18:637–645. doi: 10.1038/gt.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Zhong X, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheriyan M, Chan SH, Perler F. Traceless splicing enabled by substrate-induced activation of the Nostoc punctiforme Npu DnaE intein after mutation of a catalytic cysteine to serine. J Mol Biol. 2014;426:4018–4029. doi: 10.1016/j.jmb.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 22.Donello JE, Loeb JE, Hope TJ. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N, et al. Protein misfolding and the pathogenesis of ABCA4-associated retinal degenerations. Hum Mol Genet. 2015;24:3220–3237. doi: 10.1093/hmg/ddv073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H, Smallwood PM, Nathans J. Biochemical defects in ABCR protein variants associated with human retinopathies. Nat Genet. 2000;26:242–246. doi: 10.1038/79994. [DOI] [PubMed] [Google Scholar]
- 25.Drivas TG, Holzbaur EL, Bennett J. Disruption of CEP290 microtubule/membrane-binding domains causes retinal degeneration. J Clin Invest. 2013;123:4525–4539. doi: 10.1172/JCI69448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mata NL, et al. Delayed dark-adaptation and lipofuscin accumulation in abcr+/- mice: implications for involvement of ABCR in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2001;42:1685–1690. [PubMed] [Google Scholar]
- 27.Weng J, et al. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt's disease from the phenotype in abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 28.Trapani I, et al. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum Mol Genet. 2015;24:6811–6825. doi: 10.1093/hmg/ddv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClements ME, MacLaren RE. Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J Biol Med. 2017;90:611–623. [PMC free article] [PubMed] [Google Scholar]
- 30.Dyka FM, Boye SL, Chiodo VA, Hauswirth WW, Boye SE. Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum Gene Ther Methods. 2014;25:166–177. doi: 10.1089/hgtb.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes VS, et al. Retinal gene therapy with a large MYO7A cDNA using adeno-associated virus. Gene Ther. 2013;20:824–833. doi: 10.1038/gt.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu F, Liu Z, Chi X, Qu H. Protein trans-splicing based dual-vector delivery of the coagulation factor VIII gene. Sci China Life Sci. 2010;53:683–689. doi: 10.1007/s11427-010-4011-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhu F, et al. Inter-chain disulfide bond improved protein trans-splicing increases plasma coagulation activity in C57BL/6 mice following portal vein FVIII gene delivery by dual vectors. Sci China Life Sci. 2013;56:262–267. doi: 10.1007/s11427-013-4455-7. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Sun W, Wang B, Xiao X, Liu XQ. Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Hum Gene Ther. 2008;19:958–964. doi: 10.1089/hum.2008.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanyam P, et al. Manipulating L-type calcium channels in cardiomyocytes using split-intein protein transsplicing. Proc Natl Acad Sci U S A. 2013;110:15461–15466. doi: 10.1073/pnas.1308161110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auricchio A, Hildinger M, O'Connor E, Gao GP, Wilson JM. Isolation of highly infectious and pure adeno-associated virus type 2 vectors with a single-step gravity-flow column. Hum Gene Ther. 2001;12:71–76. doi: 10.1089/104303401450988. [DOI] [PubMed] [Google Scholar]
- 37.Gao G, et al. Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum Gene Ther. 2000;11:2079–2091. doi: 10.1089/104303400750001390. [DOI] [PubMed] [Google Scholar]
- 38.Tsybovsky Y, Palczewski K. Expression, purification and structural properties of ABC transporter ABCA4 and its individual domains. Protein Expr Purif. 2014;97:50–60. doi: 10.1016/j.pep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bungert S, Molday LL, Molday RS. Membrane topology of the ATP binding cassette transporter ABCR and its relationship to ABC1 and related ABCA transporters: identification of N-linked glycosylation sites. J Biol Chem. 2001;276:23539–23546. doi: 10.1074/jbc.M101902200. [DOI] [PubMed] [Google Scholar]
- 40.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci U S A. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellissier LP, et al. Specific tools for targeting and expression in Muller glial cells. Mol Ther Methods Clin Dev. 2014;1:14009. doi: 10.1038/mtm.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khani SC, et al. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest Ophthalmol Vis Sci. 2007;48:3954–3961. doi: 10.1167/iovs.07-0257. [DOI] [PubMed] [Google Scholar]
- 43.Doria M, Ferrara A, Auricchio A. AAV2/8 vectors purified from culture medium with a simple and rapid protocol transduce murine liver, muscle, and retina efficiently. Hum Gene Ther Methods. 2013;24:392–398. doi: 10.1089/hgtb.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sangermano R, et al. Photoreceptor Progenitor mRNA Analysis Reveals Exon Skipping Resulting from the ABCA4 c.5461-10T-->C Mutation in Stargardt Disease. Ophthalmology. 2016;123:1375–1385. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
- 45.Albert S, et al. Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease. Am J Hum Genet. 2018;102:517–527. doi: 10.1016/j.ajhg.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang B, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang FQ, Anand V, Maguire AM, Bennett J. Intraocular delivery of recombinant virus. Methods Mol Med. 2001;47:125–139. doi: 10.1385/1-59259-085-3:125. [DOI] [PubMed] [Google Scholar]
- 48.Manfredi A, et al. Combined rod and cone transduction by adeno-associated virus 2/8. Hum Gene Ther. 2013;24:982–992. doi: 10.1089/hum.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen M, et al. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- 50.Li A, Zhu X, Brown B, Craft CM. Melatonin enhances retinoic acid induction of cone arrestin gene expression in retinoblastoma cells. Adv Exp Med Biol. 2003;533:361–368. doi: 10.1007/978-1-4615-0067-4_46. [DOI] [PubMed] [Google Scholar]
- 51.Li A, Zhu X, Brown B, Craft CM. Gene expression networks underlying retinoic acid-induced differentiation of human retinoblastoma cells. Invest Ophthalmol Vis Sci. 2003;44:996–1007. doi: 10.1167/iovs.02-0434. [DOI] [PubMed] [Google Scholar]
- 52.Polishchuk EV, Polishchuk RS. Analysis of Golgi complex function using correlative light-electron microscopy. Methods Cell Biol. 2013;118:243–258. doi: 10.1016/B978-0-12-417164-0.00015-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are included in the paper and in the Supplementary Materials.