Abstract
Swine are important in the ecology of influenza A virus (IAV) globally. Understanding the ecological role of wild pigs in IAV ecology has been limited because surveillance in wild pigs is often for antibodies (serosurveillance) rather than IAVs, as in humans and domestic swine. As IAV antibodies can persist long after an infection, serosurveillance data are not necessarily indicative of current infection risk. However, antibody responses to IAV infections cause a predictable antibody response, thus time of infection can be inferred from antibody levels in serological samples, enabling identification of risk factors of infection at estimated times of infection. Recent work demonstrates that these quantitative antibody methods (QAMs) can accurately recover infection dates, even when individual-level variation in antibody curves is moderately high. Also, the methodology can be implemented in a survival analysis (SA) framework to reduce bias from opportunistic sampling. Here we integrated QAMs and SA and applied this novel QAM–SA framework to understand the dynamics of IAV infection risk in wild pigs seasonally and spatially, and identify risk factors. We used national-scale IAV serosurveillance data from 15 US states. We found that infection risk was highest during January–March (54% of 61 estimated peaks), with 24% of estimated peaks occurring from May to July, and some low-level of infection risk occurring year-round. Time-varying IAV infection risk in wild pigs was positively correlated with humidity and IAV infection trends in domestic swine and humans, and did not show wave-like spatial spread of infection among states, nor more similar levels of infection risk among states with more similar meteorological conditions. Effects of host sex on IAV infection risk in wild pigs were generally not significant. Because most of the variation in infection risk was explained by state-level factors or infection risk at long-distances, our results suggested that predicting IAV infection risk in wild pigs is complicated by local ecological factors and potentially long-distance translocation of infection. In addition to revealing factors of IAV infection risk in wild pigs, our framework is broadly applicable for quantifying risk factors of disease transmission using opportunistic serosurveillance sampling, a common methodology in wildlife disease surveillance. Future research on the factors that determine individual-level antibody kinetics will facilitate the design of serosurveillance systems that can extract more accurate estimates of time-varying disease risk from quantitative antibody data.
Introduction
Respiratory disease due to influenza A virus (IAV) results in 290,000–650,000 human deaths worldwide annually (WHO 2019). Understanding seasonal changes in incidence of IAV in humans has helped to optimize vaccination programs (Cox 2014) and understand the mechanistic underpinnings of IAV transmission risk (Alonso et al. 2007). Temperature, precipitation, and humidity appear to be significant drivers of IAV dynamics in humans, and these meteorological factors affect transmission risk differently depending on regional meteorological conditions (Tamerius et al. 2013). For example, in temperate zones, IAV peaks occur during cold–dry conditions, whereas in tropical regions they occur during humid–rainy conditions (Tamerius et al. 2013). Also, in temperate regions, IAV incidence occurs in distinct seasonal peaks (Viboud et al. 2004), whereas in the tropics, cases occur more consistently throughout the year with elevated transmission during the rainy season (Moura 2010). In addition to local meteorological conditions, seasonal variation in host contact rate, virus survival outside the host, and host immunity are thought to contribute to seasonal dynamics of IAVs in humans (Tamerius et al. 2011). However, the mechanistic underpinnings of IAV seasonality in humans remain unresolved and differ by region (Tamerius et al. 2011).
The human pandemic of 2009–2010 that originated from a reassortment event in swine emphasized that swine can play a major role in the global dynamics of human IAVs (Dawood et al. 2009; Vijaykrishna et al. 2010). Transmission of IAVs between swine and humans occurs in both directions, and reassortment of swine strains with avian and human strains has been documented multiple times (Zhou et al. 1999; Brown 2000; Nelson et al. 2014; Martin et al. 2017). A high frequency of mutations that confer resistance to amantadine (Wan et al. 2013; Diaz et al. 2017), an antiviral used to treat human infections, was detected by longitudinal sampling of domestic swine in mid-western United States (Diaz et al. 2017). This suggests that there have been multiple introductions from humans to domestic swine (Feng et al. 2013; Sun et al. 2013; Nelson et al. 2014). Growing evidence for the important role of swine in the evolutionary ecology of IAV at the human–swine interface (Nelson et al. 2015), has underscored the need to understand patterns and drivers of IAV transmission in swine populations.
Building on knowledge of IAV transmission risk factors in humans, surveillance of IAV in commercial swine (Sus scrofa) has facilitated identification of drivers of transmission risk and evolutionary dynamics of IAVs in domestic swine—especially in the United States, one of the largest commercial swine producers in the world (Anderson et al. 2013; Corzo et al. 2013; Kaplan et al. 2015). Active surveillance of swine production systems demonstrated that IAV is present throughout the year with higher prevalence in winter through early summer (Corzo et al. 2013; Kaplan et al. 2015), similar to patterns observed from passive surveillance in Ontario, Canada (Poljak et al. 2014). Corzo et al. (2013) suggested that prevalence was higher in spring and summer because piglets of new gilts enter the population at higher frequency during this time period, and they are expected to have lower maternal antibodies. Arrival of new gilts also seems to be the most important driver of new genetic diversity of swine IAVs (Diaz et al. 2017). Similarly, Kaplan et al. (2015) found that development farms with piglets and commercial gilts had the highest risk of IAV, and that risk varied regionally. In contrast to the seasonal patterns identified during active surveillance, time series analysis of positive isolates from passive surveillance revealed bi-phasic seasonal dynamics with a major peak in October–November and a secondary peak in March–April (Anderson et al. 2013; Walia et al. 2019). In summary, there is ample evidence that transmission risk in domestic swine is elevated during January–June in North America, but that transmission occurs year-round and clinical signs are most apparent in October–November. Demographic dynamics and age–structure appear to be significant risk factors in domestic swine. The role of meteorology remains unknown, but meteorology as well as management practices have been suggested as explanations for the increased reporting of clinical disease in passive surveillance systems during times that active surveillance indicates lowered transmission risk (Anderson et al. 2013; Corzo et al. 2013).
One factor that has not been examined is the potential role of wild pigs (Sus scrofa; Keiter et al. 2016) in the overall transmission dynamics of IAVs. There are an estimated 4.4–11.3 million wild pigs in the United States (Mayer 2014), with the majority in the southern region of the country (Corn et al. 2009). Pandemic H1N1 (2009) has been isolated from wild pigs (Clavijo et al. 2013), indicating their potential importance in the transmission of IAV between wild pigs and domestic swine or humans. Swine strains of IAVs (mostly H1N1, H1N2, H3N2 strains) are most commonly detected in wild pigs, but they are also exposed to avian (Martin et al. 2017) and human IAVs (Feng et al. 2014). The overall seroprevalence rate of IAVs in wild pigs in the United States was ∼4.9% from 2010 to 2013 (Martin et al. 2017); however, the seroprevalence rate varied across regions (Martin et al. 2017). In another study, antibodies to IAVs were detected in 14.1% of wild pigs in abattoirs in Texas (Pedersen et al. 2017). Similar to domestic swine, seropositive wild pigs can be found throughout the year (Martin et al. 2017). A primary concern is that wild pigs exchange IAVs with domestic swine through fence-line contact with backyard domestic swine and even enter holding pens (Gipson et al. 1999). Wild pig populations also exist in close proximity with commercial operations (Martin et al. 2017), which may allow for indirect transmission through aerosols, other wildlife vector species, or fomites. Wild pigs are also popular to hunt and process for consumption in many states, presenting a potential contact mechanism between humans and infectious animals.
It has been challenging to understand the potential role of wild pigs in the dynamics of IAVs in domestic swine and humans because the virus has been rarely isolated from wild pigs such that comprehensive genetic analyses have not been possible. Thus, understanding risk patterns in wild pigs has been limited to serological data. However, although they are useful for understanding the spatial extent of IAV, serological data are not a measure of infection risk because antibodies can persist long after IAV exposure. Thus, serological data limit our understanding of IAV infection risk in wild pigs using conventional risk models (Sun et al. 2015). A novel and robust computational method to determine risk factors solely using antibody levels in wild pigs is lacking. In previous work, we and others have developed methods for inferring population-level incidence patterns using individual-level antibody status collected during serosurveillance programs (Borremans et al. 2016; Pepin et al. 2017; Wilber M, unpublished results). Here we apply these new methods to identify potential drivers of IAV dynamics in wild pigs in 15 states across the United States. Building on knowledge of risk factors of IAV infection in humans and domestic swine, we determined the relative role of meteorological factors (e.g., temperature, precipitation, and absolute humidity), host factors (e.g., sex), IAV activity in local populations of domestic swine and humans, and IAV infection risk in wild pigs in other states, on determining risk of IAV in wild pigs. Our analyses demonstrate how individual-level variation in immunity can be leveraged to understand the population-level risk factors of transmission in a reservoir host species for a disease of global importance.
Methods
Study system
Wild pigs are hosts to many zoonotic and animal diseases (Meng et al. 2009), and have the potential to contribute to IAV spillover events in livestock and humans (Miller et al. 2017). Wild pigs also threaten food security, natural resources, and endangered species through their foraging (especially rooting) and predation behaviors (Bevins et al. 2014). The US Department of Agriculture, Wildlife Services’ National Wildlife Disease Program collects serological samples from wild pigs that are removed during management activities, and screens the samples for a variety of important human and livestock pathogens, including IAV. We used IAV serosurveillance samples from wild pigs sampled across the USA from 2010 to 2017 (Supplementary Fig. S1). Two goals of the National Wildlife Disease Program are disease monitoring and surveillance across large temporal and spatial scales to aid in “managing wildlife disease threats to agriculture, human health, and safety.” The program collaborates with state agencies and necessarily uses opportunistic samples across space and time to achieve the monitoring and surveillance goals.
To infer time of infection for seropositive individuals and censor seronegative individuals, we used experimental, longitudinal data on IAV infections in wild pigs to estimate within-host antibody dynamics (Sun et al. 2015). In the experiment, 10 wild pigs were infected with an influenza A/swine/Texas/A01104013/2012(H3N2) strain of wild-pig origin and infections were monitored for 113 days (Sun et al. 2015). Enzyme-linked immunosorbent assay (ELISA) IAV antibody assays were performed on 21 serum samples per wild pig over the 113 days using the IDEXX Influenza A Ab Test (IDEXX, Westbrook, ME, USA). On Day 103, all experimental wild pigs were re-challenged with IAV. We excluded experimental data after re-challenge in parameterization of the within-host antibody curve (Sun et al. 2015). The primary infection experiment ended before the antibody curve completed its decay. We assumed that antibody quantities would continue to decline at approximately the same rate as observed in the empirical data and would level-off above the seroconversion threshold of (used by Sun et al. [2015])—slightly less stringent than the recommended seropositivity threshold (Tse et al. 2012). The within-host antibody dynamics (Supplementary Fig. S2) are qualitatively similar to those in waterfowl (Pepin et al. 2017). In previous work, we showed that true incidence dynamics can be captured well by the model using experimental measures of antibody dynamics in waterfowl (Pepin et al. 2017).
Framework to estimate IAV infection risk
In previous work, we developed a method for estimating incidence over time from cross-sectional serological samples collected opportunistically across a broad geographic area (Wilber M, unpublished results). Briefly, data from within-host antibody kinetics with a known time of infection were used to back-infer time of infection for antibody levels in serology samples (Simonsen et al. 2009; Borremans et al. 2016; Pepin et al. 2017). We also coupled time of infection inference for individuals with survival analysis to account for the uneven sampling design from the opportunistic surveillance system, as in previous work (Wilber M, unpublished results). Seronegative samples were right-censored based on the time of year the sample was collected and the age class of the host sampled. The rationale is that the cumulative probability of remaining seronegative as the year progresses (or as individuals’ age) declines over time, or conversely, the cumulative risk of becoming infected increases over time (or as individuals age). Age provided additional information because a seronegative individual that is younger than the width of the antibody kinetic curve (Fig. S2) is too young to have been infected and recovered. Previous work shows that maternally derived antibodies may affect IAV transmission dynamics, but transmission from individuals with maternal antibodies still occurs (i.e., reproduction number is substantially greater than 1 for individuals with maternally derived antibodies; (Cador et al. 2016). Thus, for simplicity we assumed that maternal antibodies did not affect immunity. Similarly, seropositive hosts were left-censored to account for extended periods of seropositivity following exposure and decline to baseline levels. We assumed extended periods of seropositivity because the infection experiment ended before the measured antibody curve completed its decay. In previous work, we used simulations to show that censoring the serosurveillance data to account for the sampling distribution produced unbiased inference of incidence (Wilber M, unpublished results). The full likelihood accounting for the censoring is described in the Supplementary material (Methods S1).
Each seropositive host in the dataset had a quantitative measure of antibody level based on an ELISA assay. By coupling the observed quantitative antibody level with the experimentally estimated antibody curve, we back-inferred the time of infection for each seropositive host, with some level of uncertainty (Pepin et al. 2017), using the experimental antibody curve described in ‘Study System’ and the methods developed in Pepin et al. (2017). We used the median estimated time of infections in the analyses described below. All seropositive hosts with time of infection greater than τend = 200 days were considered left-censored, where τend was derived from the experimental curve.
Host-level characteristics affecting IAV infection risk
Wild pigs were classified into three age classes: juveniles (2 months; 1288 samples), sub-adults (2 months–1 year; 3272 samples), adults (1 year; 11,944 samples), and sex was recorded (7902 males, 8618 females). We accounted for host age in the infection likelihood function (Supplementary Methods S1). This inherently accounts for the fact that we would expect increased cumulative risk for older pigs as they have been at risk for longer. We assumed that antibody dynamics were similar across age classes as we did not have data to suggest otherwise. We examined the effect of sex on infection risk.
To test the effect of sex on infection risk, we used a proportional hazard survival model with Equation (1) as our likelihood. Specifically, we modeled the log-hazard rate (log(h(t)), that is, the instantaneous IAV infection risk for a susceptible host at time t, hereafter referred to as ‘IAV infection risk’) as
| (1) |
where is a flexible baseline log-hazard function computed with B-splines (Hens et al. 2012; Rutherford et al. 2015), x is a vector of host-level characteristics, and β are coefficients describing how these host-level characteristics shift the baseline IAV infection risk for a host. We ran a separate proportional hazard analysis for each state to allow the baseline function to be state-specific (Fig. 1). The proportional hazard model included the effect of sex on log IAV infection risk.
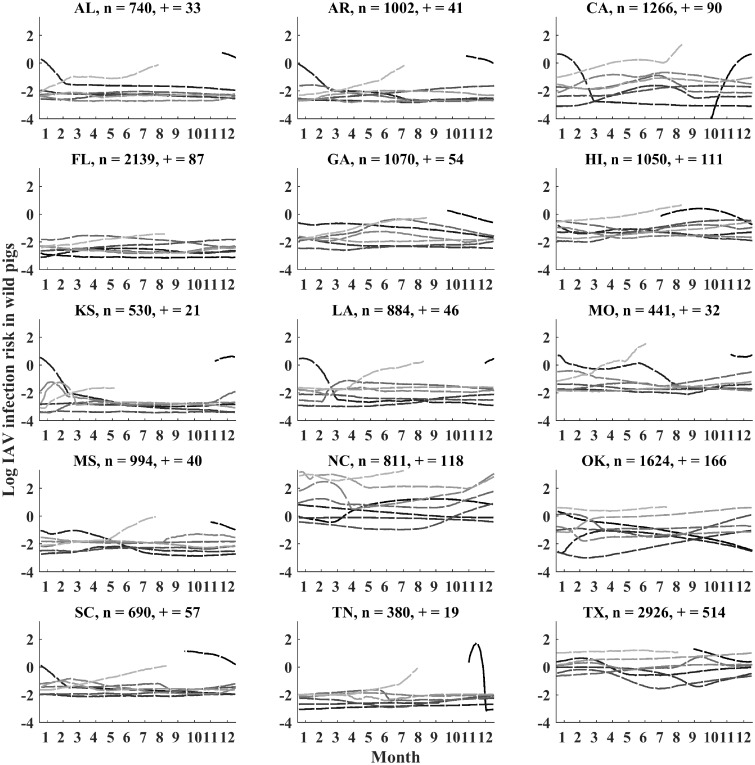
Fig. 1.
The estimated IAV infection risk for 15 states. Each line shows the seasonal pattern for a different year (2010–2017). Year 2010 is in black and subsequent years are indicated in progressively lighter shades of gray. Lines that are cut off indicated missing data (i.e., the start and endpoints of the time series). X-axes indicate months in the calendar year (January to December). The number of hosts sampled (n) and the number of seropositive samples (+) are shown for each state. The distribution of sampling and uncertainty are shown in Supplementary Fig. S3.
Meteorological and influenza related predictors of IAV infection risk
In a survival analysis framework with right-censored data, time-varying covariates can be easily incorporated into a standard Cox model (Cox 1975). While there has been substantial theoretical development for using parametric survival approaches with time-varying covariates (Petersen 1986; Sparling et al. 2006), linking flexible, spline-based parametric survival models with time-varying covariates can be computationally challenging. Instead, we took the following approach to understand how time-varying covariates affected IAV infection hazard in wild pigs.
We estimated log-hazard IAV infection risk in wild pigs through time for each of the 15 states included in the analysis using the likelihood function in the Supplementary Methods S1. Figure 1 shows the estimated IAV infection risk, specifically the log-hazard rate that gives the IAV infection risk through time. From these trajectories we calculated the average monthly estimates of log IAV infection risk for each state with associated uncertainty (namely, the width of the inter-quartile range about the monthly log-IAV infection risk estimate). Then we modeled the effects of meteorological conditions and IAV infection risk in other populations on log-IAV infection risk in wild pigs.
Meteorological covariates
Precipitation, maximum temperature, minimum temperature, and specific humidity were included as meteorological covariates in this study. We extracted all meteorological variables from gridMET (Abatzolglou 2013). Meteorological variables were available on the daily temporal scale and on the 4 km2 spatial scale. We averaged all variables to the monthly temporal scale across the counties in which pigs were sampled in a given state. We excluded Hawaii from this analysis as gridMET only provided meteorological covariates for the contiguous United States, and because of its distance from the spatial covariates.
Covariates describing proxies for IAV activity in domestic swine and humans
We used data from a national-scale passive surveillance system of domestic swine (USDA-APHIS 2018) as a proxy for IAV activity in domestic swine. Samples were submitted from anywhere in the country and classified into one of five regions as defined in (USDA-APHIS 2018). Sample submission is voluntary and based on animals that display influenza-like illness. Thus, the surveillance system represents IAV reporting trends rather than prevalence according to a random sample. We standardized the number of IAV-positive samples to the number of samples submitted per region and month (i.e., number of positive samples submitted/number of negative samples submitted) in order to account for variation in the total number of samples submitted in our analyses. IAV positivity is determined by screening matrix genes by real-time reverse transcriptase polymerase chain reaction. Similarly, the Centers for Disease Control and Prevention maintains a publicly available database of weekly IAV activity across the United States at the state level (https://www.cdc.gov/flu/weekly/fluviewinteractive.htm). The human IAV surveillance data (positive test result for IAV) is a qualitative measure with five increasing levels of geographic spread of IAV across a state (No activity, Sporadic, Local activity, Regional, Widespread). We used this latter covariate as a quantitative variable (with units 0, 1, 2, 3, 4) assuming that each incremental category was an equal amount of increased geographic spread over the last category. As IAV activity in domestic swine and humans are proxies for the presence of IAVs, they represent actual IAV activity at the time of sampling (rather than just seropositivity). Thus, we would expect that our estimate of IAV infection risk in wild pigs (a back-inference of infection time) should correlate with IAV activity in domestic swine and humans with minimal, if any, time lag, if these variables significantly predict IAV infection risk in wild pigs.
Spatial covariates
We also examined how infection risk in the previous month in other states affected infection risk in the current month for focal states to investigate whether there were patterns in spatial spread among states (methods and results described in Supplementary Results S2). This analysis was conducted on each state separately due to rank deficiencies from excluding state effects on themselves.
We fit linear mixed effects models to explore how the time-varying covariates correlated with IAV infection risk in wild pigs. The response variable was the monthly IAV infection risk that was weighted by its associated uncertainty by including the inverse of the 95% confidence interval for log-IAV infection risk as the ‘Weights’ variable in the fitlme function in Matlab’s Statistics Toolbox (Version R2018a, Mathworks). Details of the model selection procedure, model specification, and additional results are described in the Supplemental Information (Results S1).
Results
Seasonal dynamics
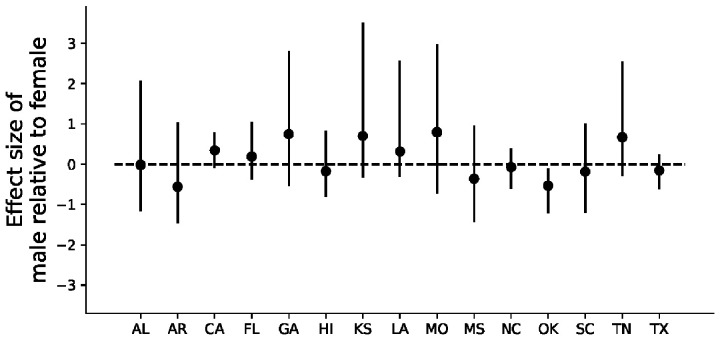
After accounting for the antibody dynamics of seropositive wild pigs, we detected clear patterns of seasonality in IAV infection risk in the 15 states included in our analysis (Fig. 1, Supplementary Fig. S3). Across all 15 states, peaks in IAV infection risk occurred most frequently between January and March (54% of 61 estimated peaks) with 24% of estimated peaks occurring from May to July. In all but one state (Oklahoma), we detected no significant effects of sex on infection risk (Fig. 2).
Fig. 2.
The effects of sex on log IAV infection risk in wild pigs. All coefficients were regularized about zero to avoid the detection of false positives over multiple comparisons.
Time-varying covariate effects
The time-varying covariates for human and domestic swine flu trends and weather only explained 5.5% of the variability in IAV infection, whereas including state-level random effects in the model described 77% of the variation in IAV infection risk (Supplementary Results S1). We also performed a county-level analysis in which we estimated IAV incidence for wild pigs in 30 counties in the dataset (with at least 10 positive samples; sampled over at least two consecutive years). The county-level analysis found nearly identical results as the state-level analysis, further suggesting that local conditions determine how covariate data affect estimated seasonal IAV infection risk in wild pigs (results not shown).
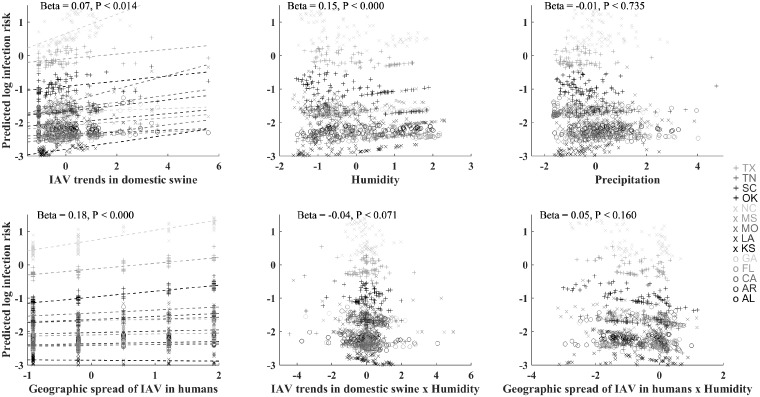
Although effects of covariates were weak, humidity and IAV trends in domestic swine and humans were significantly positively correlated with estimated IAV infection risk in wild pigs (Fig. 3), despite these covariates being uncorrelated themselves (Supplementary Table S1). The slope of the relationship between IAV infection risk and these covariates varied by state, being generally null or positive except in Alabama and Texas where the relationship between IAV infection risk in wild pigs and IAV infection trends in domestic swine was significantly negative (Fig. 3, Supplementary Fig. S5).
Fig. 3.
Predicted relationship of IAV infection risk in wild pigs to time-varying covariate data by state. States are distinguished by a combination of gray shades and symbols. Covariates are labeled on the X-axes. Dashed lines illustrate the linear trends by state for the variables that have random slope effects by state.
Spatial patterns
If state-to-state spread was primarily local, we would expect neighboring states to predict infection risk in focal states in the next month. However, there were no clear patterns for lagged infection risk in other states predicting current infection risk in focal states (Supplementary Figs. S5 and S6, Supplementary Table S5). For example, infection risk in Mississippi was significantly positively related to infection risk in California, Florida, and Kansas (distant states), but was unrelated to infection risk in Louisiana, Arkansas, and Tennesee (neighboring states of Mississippi), and negatively related to infection risk in Alabama (Supplementary Fig. S5). Along the same line of reasoning, infection risk in states with more similar meteorological conditions (e.g., Mississippi and Louisiana) was not more correlated than those with very different meteorological conditions (e.g., Mississippi and California). Similarly, the centroid of infection risk in neighboring states that had significant positive relationships with infection risk in focal states were not closer than those with weaker relationships (Supplementary Fig. S6), suggesting that spatial spread among states is not mainly from neighboring states. Interestingly, infection risk in most states, including California, was positively correlated with infection risk in Florida in the previous month, except Texas which was negatively correlated (Supplementary Fig. S5). States that had the most strongly correlated infection risks (positive relationship) were North Carolina and Texas and Oklahoma, and Tennessee and California.
Discussion
While there is ample evidence from genetic analyses that strains of IAVs are exchanged between domestic swine and humans (Nelson et al. 2014; 2015; 2016), less is known about the role of wild pigs in this multi-host system (Feng et al. 2014; Martin et al. 2017). A primary reason for this gap is that it is challenging to detect IAVs in wild pig populations. Because samples are serological and collected opportunistically, it has been difficult to interpret how wild pig serosurveillance data align with virus surveillance data that are collected in other host populations (e.g., humans, domestic swine). Our methodology provides a first attempt at linking IAV infection risk in wild pigs to that in domestic swine and humans. The positive correlation we found between IAV trends in domestic swine and humans, and IAV infection risk in wild pigs suggests that wild pigs could play a role in the national-scale circulation of IAVs in the United States. However, as these covariates did not explain much of the variation in wild pig infection risk without considering state-level effects, our results highlighted that within-state processes strongly influenced how covariates determined the patterns of IAV infection risk in wild pigs. These patterns were further corroborated by the finding that IAV infection risk among states with closer centroids of infection risk (and more similar meteorological conditions) did not predict IAV infection risk more strongly than those that were further apart (with more different meteorological conditions). This further suggests that IAV transmission in wild pigs occurs endemically within the sampled states with levels governed by unmeasured state-level processes or/and there is substantial long-distance movement of infectious individuals among states (similar to IAV in humans, e.g., Brownstein et al. 2006). We found no evidence that IAV transmission spreads through wild pig populations as a traveling wave as in some human populations (Alonso et al. 2007).
Wild pigs may interact directly or indirectly with domestic swine (Gipson et al. 1999; Wyckoff et al. 2009). Humans may be exposed to pathogens in wild pigs during hunting and trapping, or through hunting dogs (Pedersen et al. 2018). There is growing evidence that wild pigs are translocated frequently by humans (Spencer and Hampton 2005; Goedbloed et al. 2013; Tabak et al. 2017; Hernandez et al. 2018) to establish new hunting opportunities or backyard populations, and fencing is often inadequate to prevent them from escaping and becoming feral. In addition to promoting transmission between wild pigs and humans, this behavior may also partly explain why infection risk in wild pigs in neighboring states was not a primary driver of wild pig infection risk (i.e., due to long-distance translocation of infected individuals). Interactions that occur during trap-and-release typically involve close physical contact. This could serve as a mechanism of transmission to humans who could subsequently transmit IAV to domestic swine. Using a subset of the same IAV surveillance data from wild pigs (including years 2011–2013), Martin et al. (2017) found that of the 38.4% and 53.7% out of 294 seropositive samples cross-reacted with swine H1 and H3 subtypes respectively, 17.7% cross-reacted with both H1 and H3 swine subtypes, and 92.2% cross-reacted with both swine and human H1 and/or H3 subtypes. Thus, there is genetic evidence that the dominant IAVs circulating in wild pigs are closely related to human IAVs. These results combined with our results showing that infection risk in wild pigs is positively correlated with human flu and domestic swine activity suggests that spillover between wild pigs and humans could be occurring either directly or through domestic swine. Applying phylodynamic approaches (e.g., Dudas et al. 2018) to genetic surveillance of IAVs in all three host populations concurrently over time (domestic swine, humans, and wild pigs) are important for understanding the directionality of transmission pathways and the frequency at which IAVs are transmitted between these different host populations.
Similar to domestic swine (Corzo et al. 2013; Kaplan et al. 2015), our analyses revealed that IAV infections in wild pigs occurred year-round but that infection risk was highest from January to March, remaining moderately high until July. Wild pigs and domestic swine can have very different density and birth dynamics, despite similar seasonal trends in IAV infection risk. This suggests that demographic dynamics may not be a primary driver of seasonal IAV dynamics in swine. Although the analyses by us and others have not formally tested effects of demographic dynamics on IAV seasonality in swine, studies from humans have reached a similar conclusion—that human density is not the most prominent determinant of seasonal IAV infection risk (Tamerius et al. 2011). Although data on wild pig densities over time were unavailable for us to test demographic factors, a study on birth seasonality of wild pigs in South Carolina, United States, showed that births are highest in December–January, moderately high February–May, and lowest June–November, but they occurred all year-round (Mayer and Brisbin 2009). These trends in birth seasonality appear to correlate positively with the trends in IAV infection risk we identified. Studies to investigate the potential role of birth seasonality on infection risk could reveal whether birth seasonality explains some of the unexplained or among-state variation in infection risk. Other factors that can affect disease transmission risk for which we did not have data include wild pig social structure (Pepin et al. 2016; Pepin and VerCauteren 2016) and individual-level variation in movement patterns (McClure K, unpublished results).
Two meteorological conditions have been found to predict risk of IAV in humans: cold–dry conditions in climates with a wider range of temperatures across seasons, and humid–rainy conditions in climates with more constant temperature throughout the year (Tamerius et al. 2013). For wild pigs across southern United States, infection risk generally increased with increasing humidity, although the relationship trended negative in some states, and the interaction of humidity with IAV trends in domestic swine and humans was important for predicting their relationship to IAV infection risk in wild pigs. This further confirms that meteorological factors can modify IAV transmission levels, but that their effects can vary depending on local conditions or climate (Tamerius et al. 2013).
Wild pigs can also be exposed to avian IAVs (Martin et al. 2017). Avian IAVs have well-documented seasonality that is thought to be primarily determined by the migratory behavior and host demography of wild birds (van Dijk et al. 2014; Hill et al. 2016). It is possible that the seasonality of IAV infection risk in wild pigs is affected by the seasonal dynamics of avian IAVs in wild birds, in addition to those in domestic swine and the epidemics in humans (analyzed in this study). One possible mechanism could be environmental transmission as wild pigs seek out water for wallowing, and wild birds are known to shed IAVs that remain infectious in water. However, the subtype diversity for avian IAVs is much broader than for swine IAVs and many of these subtypes are not known to infect mammals. Thus, it would be important to analyze subtype-specific data to understand the potential transmission interface of avian IAVs and wild pigs. In a recent study of wild-pig serosurveillance data, only 1 of 294 serum samples selected for antigenic characterization cross-reacted with avian IAVs (but 16 cross-reacted with avian and swine IAVs), whereas 236 cross-reacted with swine IAVs (Martin et al. 2017). Thus, to date, the frequency of avian IAVs present in wild pigs appears low. Nevertheless, lack of consideration of seasonality of IAVs in wild birds in our analyses is a limitation of our study.
Our individual-level analysis did not detect a difference between the sexes in infection risk, similar to Feng et al. (2014) who used serological status. Generally, we expected to find similar results in this regard because sex is a lifetime characteristic and thus the timing of infection may have less relevance. However, if we only consider current seropositivity status, infections that occurred in the past may escape detection (because antibody levels can wane below the seropositivity threshold). Our survival analysis framework can be modified to account for the likelihood of prior infection in determining whether a seronegative sample was once seropositive (Wilber M, unpublished results), and thus could produce different results if variation in the seropositivity threshold is higher or different between the sexes. Similarly, both methodologies do not account for the frequency of IAV infections in the same host. If additional data that allowed us to identify prior infection history were available (e.g., levels of other types of antibodies such as IgM), our methodology could reveal different results than analysis of current seroprevalence (e.g., if there was a difference between the sexes in the likelihood of repeated infections). Disentangling the population-level distribution of antibody levels due to prior infections from individual-level sources of variation is an important challenge for improving inference of infection risk using quantitative antibody methods. A final difference of our approach compared with using seroprevalence is that we estimated a baseline, time-dependent hazard jointly while estimating the effect of sex. In systems where the effects of sex may be significant, our framework could allow quantification of the effect of sex over the course of a disease transmission season, an analysis that could not be performed using regression of seroprevalence data.
Our approach assumed that variation in antibody levels in nature is similar to those during experimental infections. This is unrealistic as individuals in natural settings likely experience a wider range of environmental conditions that can affect immunological responses (Hawley and Altizer 2011), relative to individuals in controlled experimental settings. It is even plausible that the overall magnitude of immune responses are different in experimental conditions relative to natural conditions due to phenotypic plasticity (Gervasi et al. 2015), especially when natural populations are experiencing resource-poor conditions. Thus, ideally, it would be better to use within-host data from individuals tracked over time in natural populations for a more appropriate perspective of individual-level variation in antibody levels, which could be accomplished in a small-scale research study. Separate studies that quantify factors affecting antibody kinetics within hosts in natural settings could reveal important covariate data (e.g., host physiological condition, genetics, or co-infection status) for improving inference of infection dates. Although our analysis does not currently provide precise quantitative relationships for IAV infection risk factors in wild pigs, it is suggestive of IAV infection risk factors that deserve further attention.
While we focused on translating individual-level variation in antibody levels to seasonal infection risk, other applications of our methodology are also possible. For example, by using serosurveillance date to infer time of infection for individuals, we could reconstruct transmission patterns in space (e.g., Jombart et al. 2011; Ypma et al. 2012). This could be done using the time of infection data alone, but could be more accurate if IAV genetic data were also collected, at least for some individuals (Ypma et al. 2012). Using serosurveillance data to unravel the spatial dynamics of transmission is important for spatial risk assessment, planning interventions, and improving our understanding of disease transmission mechanisms.
Conclusions
Our methodology provides a promising avenue for interpreting and harnessing immunological variation within and among hosts to improve inference of disease risk. Our methodology is important to many disease surveillance systems, particularly for wildlife species where surveillance for pathogens is frequently infeasible and sampling designs are necessarily opportunistic. Although our current inferences were coarse due to the resolution of our within-host antibody data (and low seropositivity of IAV), we identified significant correlations between IAV infection in wild pigs and those in humans and domestic swine that merit further investigation. For example, are these patterns merely correlative because human IAV and domestic swine IAV trends in these areas are driven by the same meteorological variables (e.g., Tamerius et al. 2019), or are these three host populations actively exchanging IAVs (e.g. Nelson et al. 2014), and if so, how often? Additional studies to understand factors that determine individual-level antibody kinetics are important for identifying appropriate surveillance data for extracting more accurate estimates of time-varying disease risk from serosurveillance data; and realizing the full potential of quantitative antibody methods.
Supplementary Material
Acknowledgments
We thank the Division of Ecoimmunology and Disease Ecology, Division of Comparative Endocrinology, Division of Animal Behavior, and Division of Ecology and Evolution of the Society for Integrative and Comparative Biology as well as the Macroecology of Infectious Disease Research Coordination Network funded by the National Science Foundation (NSF DEB 1316223) for supporting the symposium “The Scale of Sickness: How Immune Variation across Space and Species Affects Infectious Disease Dynamics” financially. We thank Thomas Deliberto and Brandon Schmit for sharing and help extracting the IAV surveillance data from wild pigs, and the many members of APHIS-Wildlife Services who have collected the wild pig samples and run ELISA assays over the years. We thank Ted Anderson for running the ELISA assays for testing the wild pig samples for exposure to IAV. We thank John Korslund for providing helpful insight about the domestic swine surveillance data.
Funding
K.M.P., C.T.W., F.L.C., and M.Q.W. were supported by USDA-APHIS National Feral Swine Damage Management Program. Funding for X-F.W. was provided by NIH NIAID R01AI116744.
Supplementary data
Supplementary data available at ICB online.
References
- Abatzolglou JT. 2013. Development of gridded surface meteorological data for ecological applications and modelling. Int J Climatol 33:121–31. [Google Scholar]
- Alonso WJ, Viboud C, Simonsen L, Hirano EW, Daufenbach LZ, Miller MA.. 2007. Seasonality of influenza in Brazil: a traveling wave from the Amazon to the subtropics. Am J Epidemiol 165:1434–42. [DOI] [PubMed] [Google Scholar]
- Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL.. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7 (Suppl 4):42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins SN, Pedersen K, Lutman MW, Gidlewski T, Deliberto TJ.. 2014. Consequences associated with the recent range expansion of nonnative feral swine. Bioscience 64:291–9. [Google Scholar]
- Borremans B, Hens N, Beutels P, Leirs H, Reijniers J.. 2016. Estimating time of infection using prior serological and individual information can greatly improve incidence estimation of human and wildlife infections. PLoS Comput Biol 12:e1004882.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IH. 2000. The epidemiology and evolution of influenza in pigs. Vet Microbiol 74:29–46. [DOI] [PubMed] [Google Scholar]
- Brownstein JS, Wolfe CJ, Mandl KD.. 2006. Empirical evidence for the effect of airline travel on inter-regional influenza spread in the United States. PLoS Med 3:e401.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cador C, Herve S, Andraud M, Gorin S, Paboeuf F, Barbier N, Queguiner S, Deblanc C, Simon G, Rose N.. 2016. Maternally-derived antibodies do not prevent transmission of swine influenza A virus between pigs. Vet Res 47:86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo A, Nikooienejad A, Esfahani MS, Metz RP, Schwartz S, Atashpaz-Gargari E, Deliberto TJ, Lutman MW, Pedersen K, Bazan LR, et al. 2013. Identification and analysis of the first 2009 pandemic H1N1 influenza virus from US feral swine. Zoonoses Public Health 60:327–35. [DOI] [PubMed] [Google Scholar]
- Corn JL, Cumbee JC, Barfoot R, Erickson GA.. 2009. Pathogen exposure in feral swine populations geographically associated with high densities of transitional swine premises and commercial swine production. J Wildl Dis 45:713–21. [DOI] [PubMed] [Google Scholar]
- Corzo CA, Culhane M, Juleen K, Stigger-Rosser E, Ducatez MF, Webby RJ, Lowe JF.. 2013. Active surveillance for influenza A virus among swine, midwestern United States, 2009-2011. Emerg Infect Dis 19:954–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. 1975. Partial likelihood. Biometrika 62:269–76. [Google Scholar]
- Cox N. 2014. Influenza seasonality: timing and formulation of vaccines. Bull World Health Organ 92:311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM.. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360:2605–15. [DOI] [PubMed] [Google Scholar]
- Diaz A, Marthaler D, Culhane M, Sreevatsan S, Alkhamis M, Torremorell M.. 2017. Complete genome sequencing of influenza A viruses within swine farrow-to-wean farms reveals the emergence, persistence, and subsidence of diverse viral genotypes. J Virol 91:e00745–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G, Carvalho LM, Rambaut A, Bedford T.. 2018. MERS-CoV spillover at the camel-human interface. Elife 7:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Baroch JA, Long LP, Xu Y, Cunningham FL, Pedersen K, Lutman MW, Schmit BS, Bowman AS, Deliberto TJ, Wan X. F. 2014. Influenza A subtype H3 viruses in feral swine, United States, 2011-2012. Emerg Infect Dis 20:843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng ZX, Gomez J, Bowman AS, Ye JQ, Long LP, Nelson SW, Yang JL, Martin B, Jia K, Nolting JM, et al. 2013. Antigenic characterization of H3N2 influenza A viruses from Ohio agricultural fairs. J Virol 87:7655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasi SS, Civitello DJ, Kilvitis HJ, Martin LB.. 2015. The context of host competence: a role for plasticity in host-parasite dynamics. Trends Parasitol 31:419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson PS, Veatch JK, Matlack RS, Jones DP.. 1999. Health status of a recently discovered population of feral swine in Kansas. J Wildl Dis 35:624–7. [DOI] [PubMed] [Google Scholar]
- Goedbloed DJ, van Hooft P, Megens HJ, Langenbeck K, Lutz W, Crooijmans RP, van Wieren SE, Ydenberg RC, Prins HH.. 2013. Reintroductions and genetic introgression from domestic pigs have shaped the genetic population structure of Northwest European wild boar. BMC Genet 14:43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DM, Altizer SM.. 2011. Disease ecology meets ecological immunology: understanding the links between organismal immunity and infection dynamics in natural populations. Funct Ecol 25:48–60. [Google Scholar]
- Hens N, Shkedy Z, Aerts M, Faes C, Van Damme P, Beutels P.. 2012. Modeling infectious disease parameters based on serological and social contact data. New York: Springer. [Google Scholar]
- Hernandez FA, Parker BM, Pylant CL, Smyser TJ, Piaggio AJ, Lance SL, Milleson MP, Austin JD, Wisely SM.. 2018. Invasion ecology of wild pigs (Sus scrofa) in Florida, USA: the role of humans in the expansion and colonization of an invasive wild ungulate. Biol Invasions 20:1865–80. [Google Scholar]
- Hill NJ, Ma EJ, Meixell BW, Lindberg MS, Boyce WM, Runstadler JA.. 2016. Transmission of influenza reflects seasonality of wild birds across the annual cycle. Ecol Lett 19:915–25. [DOI] [PubMed] [Google Scholar]
- Jombart T, Eggo RM, Dodd PJ, Balloux F.. 2011. Reconstructing disease outbreaks from genetic data: a graph approach. Heredity 106:383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BS, DeBeauchamp J, Stigger-Rosser E, Franks J, Crumpton JC, Turner J, Darnell D, Jeevan T, Kayali G, Harding A, et al. 2015. Influenza virus surveillance in coordinated swine production systems, United States. Emerg Infect Dis 21:1834–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiter D, Mayer JJ, Beasley JC.. 2016. What is in a “common” name? A call for consistent terminology for nonnative Sus scrofa: terminology for Sus scrofa. Wildl Soc Bull 40:384–7. [Google Scholar]
- Martin BE, Sun H, Carrel M, Cunningham FL, Baroch JA, Hanson-Dorr KC, Young SG, Schmit B, Nolting JM, Yoon KJ, Lutman MW, et al. 2017. Feral swine in the United States have been exposed to both avian and swine influenza A viruses. Appl Environ Microbiol 83: e01346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Brisbin IL, (eds.). 2009. Wild pigs: biology, damage, control techniques and management. Aiken (SC: ): Savannah River National Laboratory. [Google Scholar]
- Mayer JJ. 2014. Estimation of the number of wild pigs found in the United States. Washington, DC: SRNL.DOE.GOV, Department of Energy, Savannah River National Laboratory.
- Meng XJ, Lindsay DS, Sriranganathan N.. 2009. Wild boars as sources for infectious diseases in livestock and humans. Philos Trans R Soc Lond B Biol Sci 364:2697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RS, Sweeney SJ, Slootmaker C, Grear DA, Di Salvo PA, Kiser D, Shwiff SA.. 2017. Cross-species transmission potential between wild pigs, livestock, poultry, wildlife, and humans: implications for disease risk management in North America. Sci Rep 7:7821.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura FE. 2010. Influenza in the tropics. Curr Opin Infect Dis 23:415–20. [DOI] [PubMed] [Google Scholar]
- Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL.. 2015. Continual Reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol 89:6218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Stucker KM, Schobel SA, Trovao NS.. 2016. Introduction, evolution, and dissemination of influenza A viruses in exhibition swine in the United States during 2009 to 2013. 90:10963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC, Detmer SE.. 2014. Introductions and evolution of human-origin seasonal influenza a viruses in multinational swine populations. J Virol 88:10110–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Bauer NE, Rodgers S, Bazan LR, Mesenbrink BT, Gidlewski T.. 2017. Antibodies to various zoonotic pathogens detected in feral swine (Sus scrofa) at abattoirs in Texas, USA. J Food Prot 80:1239–42. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Turnage CT, Gaston WD, Arruda P, Alls SA, Gidlewski T.. 2018. Pseudorabies detected in hunting dogs in Alabama and Arkansas after close contact with feral swine (Sus scrofa). BMC Vet Res 14:388.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin KM, Davis AJ, Beasley J, Boughton R, Campbell T, Cooper SM, Gaston W, Hartley S, Kilgo JC, Wisely SM, et al. 2016. Contact heterogeneities in feral swine: implications for disease management and future research. Ecosphere 7:e01230. [Google Scholar]
- Pepin KM, Kay SL, Golas BD, Shriner SS, Gilbert AT, Miller RS, Graham AL, Riley S, Cross PC, Samuel MD, et al. 2017. Inferring infection hazard in wildlife populations by linking data across individual and population scales. Ecol Lett 20:275–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin KM, VerCauteren KC.. 2016. Disease-emergence dynamics and control in a socially-structured wildlife species. Sci Rep 6:25150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. 1986. Fitting parametric survival models with time-dependent covariates. J R Stat Soc Ser C 35:281–8. [Google Scholar]
- Poljak Z, Carman S, McEwen B.. 2014. Assessment of seasonality of influenza in swine using field submissions to a diagnostic laboratory in Ontario between 2007 and 2012. Influenza Other Respir Viruses 8:482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford MJ, Crowther MJ, Lambert PC.. 2015. The use of restricted cubic splines to approximate complex hazard functions in the analysis of time-to-event data: a simulation study. J Stat Comput Simul 85:777–93. [Google Scholar]
- Simonsen J, Molbak K, Falkenhorst G, Krogfelt KA, Linneberg A, Teunis PFM.. 2009. Estimation of incidences of infectious diseases based on antibody measurements. Stat Med 28:1882–95. [DOI] [PubMed] [Google Scholar]
- Sparling YH, Younes N, Lachin JM, Bautista OM.. 2006. Parametric survival models for interval-censored data with time-dependent covariates. Biostatistics 7:599–614. [DOI] [PubMed] [Google Scholar]
- Spencer PBS, Hampton JO.. 2005. Illegal translocation and genetic structure of feral pigs in Western Australia. J Wildl Manage 69:377–84. [Google Scholar]
- Sun H, Cunningham FL, Harris J, Xu Y, Long LP, Hanson-Dorr K, Baroch JA, Fioranelli P, Lutman MW, Li T, et al. 2015. Dynamics of virus shedding and antibody responses in influenza A virus-infected feral swine. J Gen Virol 96:2569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HL, Yang JL, Zhang T, Long LP, Jia K, Yang GH, Webby RJ, Wan XF.. 2013. Using sequence data to infer the antigenicity of influenza virus. MBio. 4:e00230–00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak MA, Piaggio AJ, Miller RS, Sweitzer RA, Ernest HB.. 2017. Anthropogenic factors predict movement of an invasive species. Ecosphere 8:e01844. [Google Scholar]
- Tamerius J, Nelson MI, Zhou SZ, Viboud C, Miller MA, Alonso WJ.. 2011. Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ Health Perspect 119:439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamerius J, Uejio C, Koss J.. 2019. Seasonal characteristics of influenza vary regionally across US. PLoS One 14:e0212511.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamerius JD, Shaman J, Alonso WJ, Bloom-Feshbach K, Uejio CK, Comrie A, Viboud C.. 2013. Environmental predictors of seasonal influenza epidemics across temperate and tropical climates. PLoS Pathog 9:e1003194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse M, Kim M, Chan C-H, Ho P-L, Ma S-K, Guan Y, Peiris JSM.. 2012. Evaluation of three commercially available influenza A type-specific blocking enzyme-linked immunosorbent assays for seroepidemiological studies of influenza A virus infection in pigs. Clin Vaccine Immunol 19:334–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA-APHIS. 2018. Influenza A virus in swine surveillance. Veterinary Services (https://www.aphis.usda.gov/animal_health/animal_dis_spec/swine/downloads/fy2018quarter1swinereport.pdf).
- van Dijk JGB, Hoye BJ, Verhagen JH, Nolet BA, Fouchier RAM, Klaassen M.. 2014. Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. J Anim Ecol 83:266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C, Boelle PY, Pakdaman K, Carrat F, Valleron AJ, Flahault A.. 2004. Influenza epidemics in the United States, France, and Australia, 1972-1997. Emerg Infect Dis 10:32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, Smith GJ, Peiris JS, Guan Y.. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia RR, Anderson TK, Vincent AL.. 2019. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 13:262–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XF, Carrel M, Long LP, Alker AP, Emch M.. 2013. Perspective on emergence and re-emergence of amantadine resistant influenza A viruses in domestic animals in China. Infect Genet Evol 20:298–303. [DOI] [PubMed] [Google Scholar]
- WHO. 2019. World Health Organization: Influenza updates. [Google Scholar]
- Wyckoff AC, Henke SE, Campbell TA, Hewitt DG, VerCauteren KC.. 2009. Feral swine contact with domestic swine: a serologic survey and assessment of potential for disease transmission. J Wildl Dis 45:422–9. [DOI] [PubMed] [Google Scholar]
- Ypma RJF, Bataille AMA, Stegeman A, Koch G, Wallinga J, van Ballegooijen WM.. 2012. Unravelling transmission trees of infectious diseases by combining genetic and epidemiological data. Proc R Soc Lond B Biol Sci 279:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG.. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.