Abstract
Chronic kidney disease (CKD) affects more than 10% of the population worldwide and burdens citizens with heavy medical expenses in many countries. Because a vital erythroid growth factor, erythropoietin (EPO), is secreted from renal interstitial fibroblasts [renal EPO-producing (REP) cells], anemia arises as a major complication of CKD. We determined that hypoxia-inducible factor 2α (HIF2α), which is inactivated by HIF-prolyl hydroxylase domain-containing proteins (PHDs) in an oxygen-dependent manner, tightly regulates EPO production in REP cells at the gene transcription level to maintain oxygen homeostasis. HIF2α-mediated disassembly of the nucleosome in the EPO gene is also involved in hypoxia-inducible EPO production. In renal anemia patients, anemic and pathological hypoxia is ineffective toward EPO induction due to the inappropriate over-activation of PHDs in REP cells transformed into myofibroblasts (MF-REP cells) due to kidney damage. Accordingly, PHD inhibitory compounds are being developed for the treatment of renal anemia. However, our studies have demonstrated that the promoter regions of the genes encoding EPO and HIF2α are highly methylated in MF-REP cells, and the expression of these genes is epigenetically silenced with CKD progression. This finding notably indicates that the efficacy of PHD inhibitors depends on the CKD stage of each patient. In addition, a strategy for harvesting renal cells, including REP cells from the urine of patients, is proposed to identify plausible biomarkers for CKD and to develop personalized precision medicine against CKD by a non-invasive strategy.
Keywords: chronic kidney disease, DNA methylation, fibrosis, hypoxia, renal anemia, urine exfoliated cells
Renal Anemia
Currently, over 10% of the population worldwide suffers from chronic kidney disease (CKD), which is characterized by kidney dysfunction and/or proteinuria that persists for more than 3 months (Levery et al., 2005). A gradual decline in kidney function results in sclerotic lesions, cardiovascular disease, and mortality (Imai et al., 2009; Hill et al., 2016). While the etiologies of CKD are diverse, ranging from lifestyle-related diseases to autoimmune disorders, CKD progression is commonly accompanied by kidney fibrosis, in which myofibroblasts emerge and proliferate in the renal tubular interstitium (Quaggin and Kapus, 2011). Because kidneys are the major organs producing erythroid growth factor erythropoietin (EPO) in adult mammals (Suzuki, 2015; Hirano and Suzuki, 2019), erythropoiesis is often impaired in CKD patients (Nangaku and Eckardt, 2006). The liver supportively produces EPO under anemic conditions, but hepatic EPO production cannot adequately compensate for renal EPO production in renal anemia patients. In fact, mice lacking renal EPO gene expression exhibit severe anemia, although EPO-gene expression is induced in their hepatocytes (Yamazaki et al., 2013; Hirano et al., 2017).
Because EPO is required for erythropoiesis, gene-modified mouse lines lacking EPO production exhibit embryonic lethality due to severe anemia (Wu et al., 1995; Yamazaki et al., 2013). Since red blood cells are essential for oxygen delivery to every organ, renal anemia severely decreases the quality of life (QOL) of CKD patients. To maintain oxygen homeostasis, EPO production in the kidney is dramatically enhanced under hypoxic/anemic conditions (Suzuki and Yamamoto, 2016). As CKD progresses, renal EPO production becomes impaired, and renal anemia then develops (Nangaku and Eckardt, 2006; Souma et al., 2015). Intriguingly, recent studies have shown that proper treatment of renal anemia is associated with the prognosis of CKD patients and that the plasma EPO concentration tightly correlates with kidney function and fibrosis (Inomata et al., 1997; Singh et al., 2006; Pfeffer et al., 2009). Thus, plasma EPO is expected to be a plausible biomarker to estimate the CKD grade (Tsubakihara et al., 2015).
For treatment of renal anemia, recombinant human EPO reagents have been used as erythropoiesis-stimulating agents (ESAs) for more than 30 years, and these reagents have dramatically improved the QOL of CKD patients (Jones et al., 2004). However, the invasiveness of subcutaneous ESA injections and the formulation costs of ESAs are problems that need to be solved (Schiller et al., 2008). Additionally, ESAs are frequently ineffective for patients suffering from chronic inflammation because EPO-dependent erythropoiesis is strongly suppressed by high serum concentrations of inflammatory cytokines and hepcidin, which negatively regulates iron usage for hemoglobin (Ganz, 2003; Smrzova et al., 2005; Suzuki et al., 2016; Petrulienė et al., 2017).
Renal Erythropoietin-Producing Cells
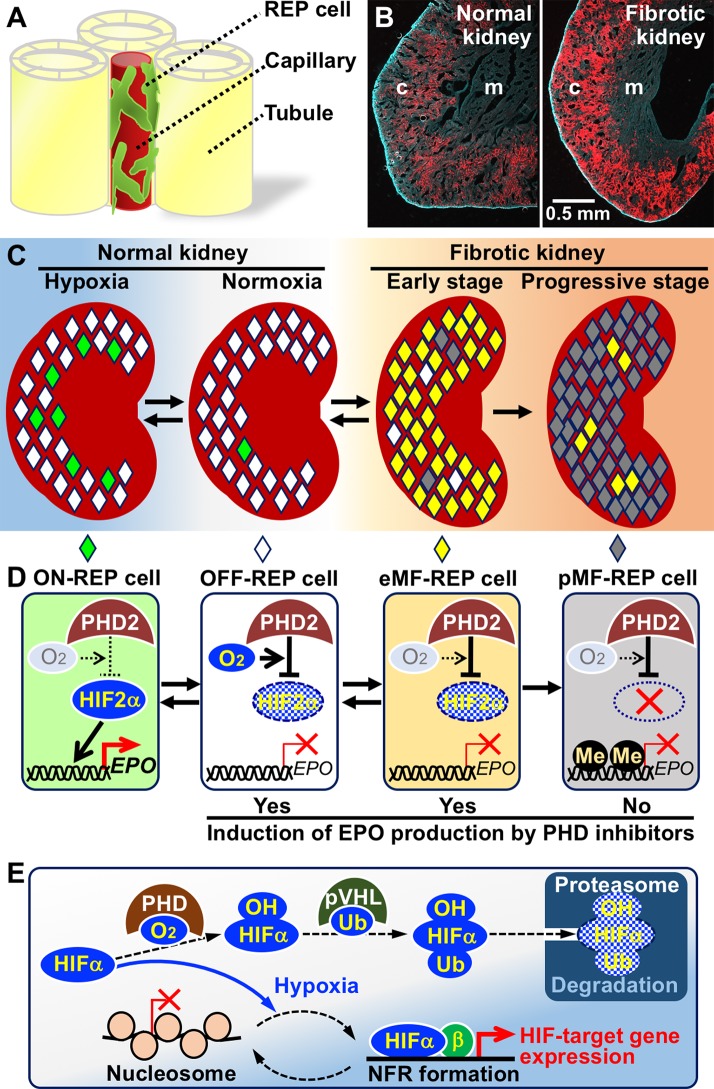
Using genetically modified mouse lines, we and others demonstrated that the ability to produce EPO is present in most fibroblasts that are positive for CD73 and platelet-derived growth factor receptor β (PDGFRβ) in the interstitium spreading from the cortico-medullary boundary to the renal cortex ( Figures 1A, B ; Maxwell et al., 1993; Pan et al., 2011; Yamazaki et al., 2013). The cells that produce EPO in response to a hypoxic microenvironment are known as REP (renal EPO-producing) cells (Suzuki et al., 2007; Obara et al., 2008). REP cells are fundamentally quiescent in terms of the cell cycle, and EPO production in the majority of REP cells is absent in healthy mice (Souma et al., 2013; Yamazaki et al., 2013). Under hypoxic/anemic conditions, the percentage of “ON-REP cells,” in which EPO production is ongoing, in the total REP cell population is increased. However, only up to 10% of REP cells are ON-REP cells, even under very severe chronic anemia conditions, suggesting that most REP cells are reservoirs (referred to as OFF-REP cells) in preparation for much more severe conditions that require high amounts of EPO ( Figures 1C, D ; Yamazaki et al., 2013; Souma et al., 2015). Thus, the total amount of EPO secretion from a kidney is correlated with the ratio of ON-REP cells to total REP cells, rather than the extent of EPO-production levels in each cell (Eckardt et al., 1993; Obara et al., 2008; Suzuki, 2015). Additionally, these data indicate that small numbers of ON-REP cells are sufficient for recovery from anemia because EPO-production levels in each ON-REP cell are very high.
Figure 1.
Mechanisms of hypoxia-inducible erythropoietin (EPO) production in renal EPO-producing (REP) cells and failure of EPO production in fibrotic kidney. (A) A schema of REP cell localization in the interstitia between renal tubules. REP cells directly associate with capillaries (Souma et al., 2016). (B) REP cells (red) distributed to the outer medulla (m) and cortex (c) of a normal healthy kidney (left) are expanded in a fibrotic kidney (right) of a genetically modified mouse line specifically expressing tdTomato fluorescence in REP cells (Yamazaki et al., 2013). (C) Distributions of ON-REP (green), OFF-REP (white), early myofibroblast (eMF)-REP (yellow), and progressive MF (pMF)-REP (gray) in normal kidneys and fibrotic kidneys. Note that a small fraction of REP cells produce EPO even under hypoxic conditions (left). (D) EPO-gene regulation by the PHD2-HIF2α pathway in REP cells and MF-REP cells. In eMF-REP cells (reversibly transformed REP cells), PHD2 over-activation results in inactivation of EPO-gene transcription. Therefore, PHD inhibitors may induce EPO production. Because the genes for EPO and HIF2α are epigenetically inactivated due to DNA methylation (Me) in pMF-REP cells (irreversibly transformed REP cells), PHD inhibitors are ineffective. (E) Molecular mechanism of hypoxia-inducible transcriptional regulation. HIFα proteins are always synthesized and degraded by the ubiquitin (Ub)-proteasome pathway via PHD-mediated hydroxylation (OH) in oxygen-replete cells. In hypoxic cells, PHD is inactivated, and HIFα proteins are stabilized. In some HIF-target gene promoters, HIFα/β complexes mediate the disassembly of nucleosome structures to form nucleosome-free regions under hypoxic conditions.
Whereas the origins of myofibroblasts coming into existence in the fibrotic kidneys of CKD patients are controversial and considered various (LeBleu et al., 2013), we and others have demonstrated that resident interstitial fibroblasts, including REP cells in healthy kidneys, are transformed into myofibroblasts under pathological conditions ( Figures 1C, D ; Humphreys et al., 2010; Asada et al., 2011; Souma et al., 2013). Importantly, REP cells gain proliferative activity and lose EPO-production ability after transformation (Souma et al., 2013). Thus, REP cells are closely related to the two major pathologies of CKD: renal anemia and fibrosis. Therefore, investigations of REP cells and myofibroblast-transformed REP (MF-REP) cells hold the key to elucidating the molecular pathology of CKD.
Various studies have proposed that the transformation of REP cells into MF-REP cells is promoted by the SMAD and NFκB transcription factors, which are activated by transforming growth factor beta (TGFβ) and tumor necrosis factor alpha, respectively (Wynn and Ramalingam, 2012; Souma et al., 2015). Additionally, DNA methylation in the EPO-gene promoter is thought to be involved in the loss of EPO-production ability in MF-REP cells (Chang et al., 2016). To further elucidate the molecular pathology of CKD by characterizing MF-REP cells, we recently established a myofibroblast cell line derived from mouse REP cells, and the cell line was referred to as Replic (REP cell-lineage immortalized and cultivable) cells (Sato et al., 2019). The genomic region of the EPO-gene promoter is highly methylated in Replic cells, and cell-autonomous TGFβ signaling supports their myofibroblast properties, which include the expression of genes for α smooth muscle actin, fibronectin, and collagens, among others.
EPO-Gene Regulation in REP Cells
EPO production in REP cells is strictly regulated at the gene transcription level, and transcription is likely regulated by an ON/OFF mechanism in each cell (Obara et al., 2008). Gene expression data of separately isolated ON- and OFF-REP cells indicated that hypoxia-inducible genes are highly expressed in ON-REP cells compared to OFF-REP cells, suggesting that there is a hypoxic threshold to activate EPO-gene expression in REP cells and that the oxygen levels of the microenvironments around ON-REP cells are below the threshold (Yamazaki et al., 2013). The expression levels of almost all hypoxia-inducible genes, including genes related to angiogenesis, glycolysis, and cell survival, are commonly regulated by hypoxia-inducible transcription factors (HIFs) ( Figure 1E ; Wang and Semenza, 1993; Lendahl et al., 2009; Suzuki et al., 2017).
HIFs consist of two subunits, namely, HIFα and HIFβ (also known as ARNT), and they bind to specific DNA sequences (A/GCGTG) in the regulatory regions of their target genes (Semenza et al., 1991; Lendahl et al., 2009; Haase, 2013). Under normal air conditions (normoxia), specific prolyl residues of HIFα are hydroxylated with HIF-specific prolyl hydroxylase domain proteins (PHDs) by means of intracellular oxygen, and hydroxylated HIFα proteins are degraded by the ubiquitin-proteasome system ( Figure 1E ; Lendahl et al., 2009). In cells with insufficient oxygen for PHD-mediated HIFα hydroxylation, HIFα proteins avoid degradation and activate transcription of their target genes. There are three isoforms encoded by the different genes for the PHD and HIFα proteins, respectively. Among the isoforms, PHD2 and HIF2α primarily control EPO-gene expression in a hypoxia-inducible manner in REP cells ( Figure 1D ; Castrop and Kurtz, 2010; Souma et al., 2016). Therefore, dysfunction of the PHD2-HIF2α-EPO axis in REP cells is considered the molecular cause of renal anemia. Notably, polycythaemia-related polymorphisms are found in the genes for PHD2 and HIF2α but not in those for the other isoforms, and these polymorphisms are predicted to lead to HIF2α stabilization followed by EPO-gene induction without hypoxic stimuli (Bento et al., 2014).
Due to the difficulty of isolating sufficient levels of REP cells for molecular biology analyses, hepatocytes and genetically modified mice have been used for studies on EPO-gene regulation. With transgenic mouse strategies, the murine Epo-gene regulatory region for REP-cell-specific and hypoxia-inducible expression was determined to be approximately 10 kb upstream from the transcription start site of the Epo gene (Hirano et al., 2017). We also discovered that histones located in the EPO-gene promoter are always acetylated regardless of hypoxic EPO induction and that histones are dissociated from the nucleosome structure in the EPO-gene promoter of hepatocytes under hypoxic conditions through HIF2α activation (Suzuki et al., 2011; Tojo et al., 2015). Nucleosome disassembly results in the formation of a nucleosome-free region (NFR) that has an open chromatin structure for the direct association between transcription factors and promoters and allows the induction of EPO-gene transcription ( Figure 1E ; Suzuki et al., 2017; Suzuki et al., 2018a).
Stepwise Mechanisms of EPO-Gene Silencing in MF-REP Cells
Since mice lacking PHD2 expression in REP cells are resistant to renal EPO deficiency caused by kidney injury, inappropriate over-activation of PHD2 is considered responsible for EPO-gene inactivation in MF-REP cells ( Figure 1D ; Souma et al., 2016). Although the oxygen affinities of PHDs are ordinarily very low compared to those of other oxygen-dependent enzymes, including collagen hydroxylases and epigenetic regulators (see below; Hancock et al., 2017; Chakraborty et al., 2019), unknown mechanisms are speculated to allow PHDs to use oxygen in MF-REP cells even under pathological hypoxic conditions. Indeed, PHD inhibitory compounds are being developed as medicines for renal anemia treatment, and clinical trials of these compounds are showing anticipated effects ( Figure 1D ; Akizawa et al., 2019).
In addition to PHD over-activation, DNA methylation in the EPO promoter is involved in EPO-gene silencing in MF-REP cells (Chang et al., 2016; Sato et al., 2019). Because hyper-methylation of gene promoter regions blunts gene transcription by tightly compacting the chromatin structure and blocking associations with transcription factor complexes (Jones, 2012; Schübeler, 2015), PHD inhibitors are predicted to be ineffective in cells in which the EPO promoter is highly methylated ( Figure 1D ). Consistent with this hypothesis, in Replic cells, neither PHD inhibitors nor HIF2α overexpression activated the Epo gene, which is highly methylated (Sato et al., 2019). Thus, the transformation of REP cells into myofibroblasts is divided into at least two consecutive stages: the early MF-REP (eMF-REP) cell stage with over-activation of PHD and the progressive MF-REP (pMF-REP) cell stage with hyper-methylation of the EPO promoter. PHD inhibitors are theoretically effective at inducing EPO production in the former cell type but ineffective in the latter cell type, which likely corresponds to Replic cells. Intriguingly, transformation of REP cells is reversible in the early stages of kidney injury ( Figure 1D ; Souma et al., 2013).
We recently discovered that the gene promoter for HIF2α is also highly methylated and that both the mRNA and protein of HIF2α are undetectable in pMF-REP cells, even under hypoxic conditions (Sato et al., 2019). This finding indicates that DNA methylation in specific gene promoters is one of the causes of EPO deficiency in CKD. DNA methylation is mediated by 3 DNA methyltransferases (DNMTs): DNMT1, DNMT3A, and DNMT3B. DNMT1 is essential for the maintenance of DNA methylation patterns beyond mitosis to inherit epigenetic memory (Jeltsch, 2006), while de novo DNA methylation is mediated by DNMT3A and DNMT3B (Hsieh, 1999). This transformation enhances the expression of mRNAs for DNMT1 and DNMT3B by TGFβ signaling (Souma et al., 2013), suggesting that these DNMTs are involved in the loss of EPO-production ability in MF-REP cells. In fact, 5-aza-2’-deoxycytidine (5-aza), an inhibitor of DNMT1, restores EPO production in primary-cultured mouse MF-REP cells by reducing DNA methylation in the Epo-gene promoter (Chang et al., 2016). In contrast, DNA methylation in the gene promoters for EPO and HIF2α in Replic cells was resistant to 5-aza treatment, whereas the other genomic regions tested were sensitive (Sato et al., 2019). This discrepancy in 5-aza efficacy between the primary-cultured MF-REP cells and Replic cells, which may represent eMF-REP and pMF-REP cells, respectively, is explained by differences in the activity of de novo DNA methylation because expression of de novo DNMTs (DNMT3A and DNMT3B) is induced by TGFβ signaling (Cardenas et al., 2014), which is autonomously promoted in Replic cells.
Efficacy of PHD Inhibitors in EPO-Induction Is Related to the Transformation Stage of Myofibroblast-Transformed REP Cells
As an alternative to ESAs, PHD inhibitors are a promising group of next-generation medicines for renal anemia treatment because they are orally administrable small compounds (Martin et al., 2017). The first PHD inhibitor, roxadustat, was launched in 2018 in China, where there are more than 100 million CKD patients (Zhang et al., 2012). However, it is concerned that PHD inhibitors cause unexpected side effects through their widespread activation of HIF-target genes in addition to EPO, and these genes include genes that are related to energy metabolism, angiogenesis, and cell survival (Wang and Semenza, 1993). Although obvious adverse events, such as tumor malignancy, have not been observed in clinical trials thus far (Akizawa et al., 2019), further long-term observation is necessary to confirm both the beneficial and unfavorable side effects of PHD inhibitors.
PHDs catalyze oxygenation reactions of the specific prolyl residues of HIFαs to produce hydroxylated HIFαs using oxygen, iron, ascorbate, and α-ketoglutarate. These substrates are also used by a variety of α-ketoglutarate-dependent dioxygenases, including important epigenetic regulators, TET (ten-eleven translocation) family DNA demethylases and KDM (histone lysine demethylase) family histone demethylases (Itoh et al., 2013; Kohli and Zhang, 2013). Since PHDs show the lowest affinity for oxygen among these dioxygenases, PHDs are the first dioxygenases inactivated by hypoxia and can thus sensitively detect hypoxia in cells. On the other hand, the other dioxygenases are less susceptible to hypoxia than PHDs. Notably, very recent studies have shown that some KDMs are as sensitive to hypoxia as PHDs, and further studies are expected to unveil mechanisms involving direct sensing of hypoxia by epigenetic regulators in addition to PHDs (Batie et al., 2019; Chakraborty et al., 2019). Since the current PHD inhibitors commonly block the specific association of α-ketoglutarate with PHDs, other α-ketoglutarate-dependent dioxygenases are unresponsive to these compounds.
In summary, PHD inhibitors are considered to be effective in eMF-REP cells but not in pMF-REP cells with methylation-based silencing of the genes for HIF2α and EPO ( Figure 1D ). Our preliminary experiments using mouse models have shown that EPO production is induced by PHD inhibitors in undamaged or slightly damaged REP Cells of fibrotic kidneys through HIF2α accumulation but not in severely damaged areas. In contrast, PHD inhibitors activate EPO production in almost all the REP cells of healthy kidneys within 6 H after peritoneal injection of the drug (Suzuki et al., 2018b). Clinical trials have demonstrated that PHD inhibitors induce erythropoiesis in nephric patients suffering from any CKD stage and end-stage renal disease, but anephric patients barely respond to PHD inhibitors with regard to EPO induction (Bernhardt et al., 2010). Taken together, these results suggest that EPO produced by a small number of REP cells in the kidney is sufficient to induce erythropoiesis in renal anemia patients. Indeed, as mentioned above, ON-REP cells constitute less than 10% of the REP cells in mouse models of severe chronic anemia ( Figure 1C ; Yamazaki et al., 2013). These observations also suggest that the efficacy of PHD inhibitors differs among renal anemia patients and that the population of eMF-REP and healthy REP Cells in each patient defines their responsiveness to PHD inhibitors.
Perspectives: Non-Invasive Strategies for Personalized Precision Medicine for Chronic Kidney Disease
Here, we summarize the epigenetic and molecular mechanisms of EPO-gene silencing in CKD patients and propose the stepwise transformation of REP cells into eMF-REP and pMF-REP cells in injured kidneys ( Figures 1C, D ). We also suggest that PHD-inhibitor responsiveness varies in patients and is dependent on the degree of REP cell transformation, which fundamentally correlates with the degree of kidney fibrosis in CKD. Thus, diagnosing the degree of kidney fibrosis is expected to inform us not only about CKD conditions/prognoses but also about PHD-inhibitor responsiveness of CKD patients. Currently, an invasive biopsy is widely adopted for the diagnosis of the complicated pathology of CKD (Mise et al., 2014). However, non-invasive biomarkers for the progression of CKD are being explored. For example, urine concentrations of N-acetyl-β-D-glucosaminidase (Bazzi et al., 2002) can be used. However, the quantitative relationship of the biomarkers to the degree of kidney fibrosis should be investigated in detail.
We propose that urine exfoliated cells can be used for the diagnosis and prediction of CKD. Urine contains several types of kidney cells, including tubular epithelial cells and podocytes, which are living and proliferative in ex vivo culture (Dörrenhaus et al., 2000; Kumagai et al., 2000; Vogelmann et al., 2003; Oliveira Arcolino et al., 2015). Therefore, these cells have been utilized as the experimental source of human renal epithelial cells and investigated as biomarkers for the early detection of bladder cancer (Rahmoune et al., 2005; Shimizu et al., 2013). Additionally, urine from CKD patients contains more cultivable exfoliated cells than urine from healthy individuals, which is advantageous for diagnosis (Detrisac et al., 1983). Importantly, our preliminary RT-PCR experiments detected the expression of mRNAs for EPO, HIF2α, and CD73 in cultured cells from the urine of patients with kidney disease, indicating that the exfoliated cell cultures contain REP cells and/or MF-REP cells. In addition, REP cells and MF-REP cells can be purified from the mixtures of exfoliated cells with cell surface expression of CD73 or PDGFRβ using cell sorters (Armulik et al., 2011; Pan et al., 2011).
With small numbers of urine exfoliated cells, high-sensitivity PCR-based techniques are expected to detect HIF2α mRNA expression and EPO-gene methylation. NFRs are also detectable with PCR, as we have identified NFRs in hypoxia-inducible gene promoters (Tojo et al., 2015; Suzuki et al., 2018a). Taking advantage of living cells, drug sensitivity may be directly investigated in urine exfoliated cells. Although further studies are needed, exfoliated cells in urine would provide novel diagnostic strategies to distinguish pMF-REP and eMF-REP for the prediction of PHD-inhibitor responsiveness, as well as plausible biomarkers for kidney fibrosis and CKD prognosis.
Author Contributions
NS conceived the idea. KS, NK, and NS wrote the manuscript and created the figures.
Funding
This work was supported in part by Takeda Life Science Foundation and SENSHIN Medical Research Foundation (for NS). The funders have no role in this study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Atsuko Konuma, Riona Asai, Taku Nakai, Rio Sasaki, Koichiro Kato and Masayuki Yamamoto (Tohoku University) for technical help and scientific comments. It has been a pleasure conducting this study with Tohoku University Advanced Research Center for Innovations in Next-Generation Medicine (INGEM).
References
- Akizawa T., Nangaku M., Yamaguchi T., Arai M., Koretomo R., Matsui A. (2019). A placebo-controlled, randomized trial of enarodustat in patients with chronic kidney disease followed by long-term trial. Am. J. Nephrol. 49, 165–174. 10.1159/000496929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A., Genové G., Betsholtz C. (2011). Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 21, 193–215. 10.1016/j.devcel.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Asada N., Takase M., Nakamura J., Oguchi A., Asada M., Suzuki N. (2011). Dysfunction of fibroblasts of external origin underlies renal fibrosis and renal anemia in mice. J. Clin. Invest. 121, 3981–3990. 10.1172/JCI57301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batie M., Frost J., Frost M., Wilson J. W., Schofield P., Rocha S. (2019). Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363, 1222–1226. 10.1126/science.aau5870 [DOI] [PubMed] [Google Scholar]
- Bazzi C., Petrini C., Rizza V., Arrigo G., Napodano P., Paparella M. (2002). Urinary N-acetyl-beta-glucosaminidase excretion is a marker of tubular cell dysfunction and a predictor of outcome in primary glomerulonephritis. Nephrol. Dial Transplant 17, 1890–1896. 10.1093/ndt/17.11.1890 [DOI] [PubMed] [Google Scholar]
- Bento C., Percy M. J., Gardie B., Maia T. M., van Wijk R., Perrotta S. (2014). Genetic basis of congenital erythrocytosis: mutation update and online databases. Hum. Mutat. 35, 15–26. 10.1002/humu.22448 [DOI] [PubMed] [Google Scholar]
- Bernhardt W. M., Wiesener M. S., Scigalla P., Chou J., Schmieder R. E., Günzler V. (2010). Inhibition of prolyl hydroxylases increases erythropoietin production in ESRD. J. Am. Soc. Nephrol. 21, 2151–2156. 10.1681/ASN.2010010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas H., Vieth E., Lee J., Segar M., Liu Y., Nephew K. P. (2014). TGF-β induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics 9, 1461–1472. 10.4161/15592294.2014.971608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrop H., Kurtz A. (2010). Functional evidence confirmed by histological localization: overlapping expression of erythropoietin and HIF-2alpha in interstitial fibroblasts of the renal cortex. Kidney Int. 77, 269–271. 10.1038/ki.2009.470 [DOI] [PubMed] [Google Scholar]
- Chakraborty A. A., Laukka T., Myllykoski M., Ringel A. E., Booker M. A., Tolstorukov M. Y. (2019). Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222. 10.1126/science.aaw1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. T., Yang C. C., Pan S. Y., Chou Y. H., Chang F. C., Lai C. F. (2016). DNA methyltransferase inhibition restores erythropoietin production in fibrotic murine kidneys. J. Clin. Invest. 126, 721–731. 10.1172/JCI82819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrisac C. J., Mayfield R. K., Colwell J. A., Garvin A. J., Sens D. A. (1983). In vitro culture of cells exfoliated in the urine by patients with diabetes mellitus. J. Clin. Invest. 71, 170–173. 10.1172/JCI110747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörrenhaus A., Müller J. I., Golka K., Jedrusik P., Schulze H., Föllmann W. (2000). Cultures of exfoliated epithelial cells from different locations of the human urinary tract and the renal tubular system. Arch. Toxicol. 74, 618–626. 10.1007/s002040000173 [DOI] [PubMed] [Google Scholar]
- Eckardt K. U., Koury S. T., Tan C. C., Schuster S. J., Kaissling B., Ratcliffe P. J. (1993). Distribution of erythropoietin producing cells in rat kidneys during hypoxic hypoxia. Kidney Int. 43, 815–823. 10.1038/ki.1993.115 [DOI] [PubMed] [Google Scholar]
- Ganz T. (2003). Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood 102, 783–788. 10.1182/blood-2003-03-0672 [DOI] [PubMed] [Google Scholar]
- Haase V. H. (2013). Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 27, 41–53. 10.1016/j.blre.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. L., Masson N., Dunne K., Flashman E., Kawamura A. (2017). The activity of JmjC histone lysine demethylase KDM4A is highly sensitive to oxygen concentrations. ACS Chem. Biol. 12, 1011–1019. 10.1021/acschembio.6b00958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill N. R., Fatoba S. T., Oke J. L., Hirst J. A., O’Callaghan C. A., Lasserson D. S. (2016). Global prevalence of chronic kidney disease - A systematic review and meta-analysis. PloS One 11, e0158765. 10.1371/journal.pone.0158765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano I., Suzuki N. (2019). The neural crest as the first production site of the erythroid growth factor erythropoietin. Front. Cell Dev. Biol. 7, 105. 10.3389/fcell.2019.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano I., Suzuki N., Yamazaki S., Sekine H., Minegishi N., Shimizu R. (2017). Renal anemia model mouse established by transgenic rescue with an erythropoietin gene lacking kidney-specific regulatory elements. Mol. Cell Biol. 37, e00451–e00416. 10.1128/MCB.00451-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. L. (1999). In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell Biol. 19, 8211–8218. 10.1128/MCB.19.12.8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys B. D., Lin S. L., Kobayashi A., Hudson T. E., Nowlin B. T., Bonventre J. V. (2010). Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am. J. Pathol. 176, 85–97. 10.2353/ajpath.2010.090517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai E., Horio M., Watanabe T., Iseki K., Yamagata K., Hara S. (2009). Prevalence of chronic kidney disease in the Japanese general population. Clin. Exp. Nephrol. 13, 621–630. 10.1007/s10157-009-0199-x [DOI] [PubMed] [Google Scholar]
- Inomata S., Itoh M., Imai H., Sato T. (1997). Serum levels of erythropoietin as a novel marker reflecting the severity of diabetic nephropathy. Nephron 75, 426–430. 10.1159/000189580 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Suzuki T., Miyata N. (2013). Small-molecular modulators of cancer-associated epigenetic mechanisms. Mol. Biosyst. 9, 873–896. 10.1039/c3mb25410k [DOI] [PubMed] [Google Scholar]
- Jeltsch A. (2006). Molecular enzymology of mammalian DNA methyltransferases. Curr. Top. Microbiol. Immunol. 301, 203–225. 10.1007/3-540-31390-7_7 [DOI] [PubMed] [Google Scholar]
- Jones M., Ibels L., Schenkel B., Zagari M. (2004). Impact of epoetin alfa on clinical end points in patients with chronic renal failure: a meta-analysis. Kidney Int. 65, 757–767. 10.1111/j.1523-1755.2004.00450.x [DOI] [PubMed] [Google Scholar]
- Jones P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- Kohli R. M., Zhang Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479. 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai N., Inoue C. N., Kondo Y., Iinuma K. (2000). Mitogenic action of lysophosphatidic acid in proximal tubular epithelial cells obtained from voided human urine. Clin. Sci. (Lond) 99, 561–567. 10.1042/cs0990561 [DOI] [PubMed] [Google Scholar]
- LeBleu V. S., Taduri G., O’Connell J., Teng Y., Cooke V. G., Woda C. (2013). Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 19, 1047–1053. 10.1038/nm.3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U., Lee K. L., Yang H., Poellinger L. (2009). Generating specificity and diversity in the transcriptional response to hypoxia. Nat. Rev. Genet. 10, 821–832. 10.1038/nrg2665 [DOI] [PubMed] [Google Scholar]
- Levery A. S., Eckardt K. U., Tsukamoto Y., Levin A., Coresh J., Rossert J. (2005). Definition and classification of chronic kidney disease: a position statement from kidney disease: improving global outcomes (KDIGO). Kidney Int. 67, 2089–2100. 10.1111/j.1523-1755.2005.00365.x [DOI] [PubMed] [Google Scholar]
- Martin E. R., Smith M. T., Maroni B. J., Zuraw Q. C., deGoma E. M. (2017). Clinical trial of vadadustat in patients with anemia secondary to stage 3 or 4 chronic kidney disease. Am. J. Nephrol. 45, 380–388. 10.1159/000464476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Osmond M. K., Pugh C. W., Heryet A., Nicholls L. G., Tan C. C. (1993). Identification of the renal erythropoietin-producing cells using transgenic mice. Kidney Int. 44, 1149–1162. 10.1038/ki.1993.362 [DOI] [PubMed] [Google Scholar]
- Mise K., Hoshino J., Ubara Y., Sumida K., Hiramatsu R., Hasegawa E. (2014). Renal prognosis a long time after renal biopsy on patients with diabetic nephropathy. Nephrol. Dial Transplant 29, 109–118. 10.1093/ndt/gft349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M., Eckardt K. U. (2006). Pathogenesis of renal anemia. Semin. Nephrol. 26, 261–268. 10.1016/j.semnephrol.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Obara N., Suzuki N., Kim K., Nagasawa T., Imagawa S., Yamamoto M. (2008). Repression via the GATA box is essential for tissue-specific erythropoietin gene expression. Blood 111, 5223–5232. 10.1182/blood-2007-10-115857 [DOI] [PubMed] [Google Scholar]
- Oliveira Arcolino F., Tort Piella A., Papadimitriou E., Bussolati B., Antonie D. J., Murray P. (2015). Human urine as a noninvasive sourse of kidney cells. Stem Cell Int. 2015, 362562. 10.1155/2015/362562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Suzuki N., Hirano I., Yamazaki S., Minegishi N., Yamamoto M. (2011). Isolation and characterization of renal erythropoietin-producing cells from genetically produced anemia mice. PloS One 6, e25839. 10.1371/journal.pone.0025839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrulienė K., Žiginskienė E., Kuzminskis V., Nedzelskienė I., Bumblytė I. A. (2017). Hepcidin serum levels and resistance to recombinant human erythropoietin therapy in hemodialysis patients. Medicina (Kaunas). 53, 90–100. 10.1016/j.medici.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Pfeffer M. A., Burdmann E. A., Chen C. Y., Cooper M. E., de Zeeuw D., Eckardt K. U. (2009). A trial of darbepoetin alfa in type 2 diabetes and chronic kidney disease. N Engl. J. Med. 361, 2019–2032. 10.1056/NEJMoa0907845 [DOI] [PubMed] [Google Scholar]
- Quaggin S. E., Kapus A. (2011). Scar wars: mapping the fate of epithelial-mesenchymal-myofibroblast transition. Kidney Int. 80, 41–50. 10.1038/ki.2011.77 [DOI] [PubMed] [Google Scholar]
- Rahmoune H., Thompson P. W., Ward J. M., Smith C. D., Hong G., Brown J. (2005). Glucose transporters in human renal proximal tubular cells isolated from the urine of patients with non-insulin-dependent diabetes. Diabetes 54, 3427–3434. 10.2337/diabetes.54.12.3427 [DOI] [PubMed] [Google Scholar]
- Sato K., Hirano I., Sekine H., Miyauchi K., Nakai T., Kato K. (2019). An immortalized cell line derived from renal erythropoietin-producing (REP) cells demonstrates their potential to transform into myofibroblasts. Sci. Rep. 9, 11254. 10.1038/s41598-019-47766-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller B., Doss S., Cock E., Del Aguila M. A., Nissenson A. R. (2008). Costs of managing anemia with erythropoiesis-stimulating agents during hemodialysis: a time and motion study. Hemodial Int. 12, 441–449. 10.1111/j.1542-4758.2008.00308.x [DOI] [PubMed] [Google Scholar]
- Schübeler D. (2015). Function and information content of DNA methylation. Nature 517, 321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Semenza G. L., Koury S. T., Nejfelt M. K., Gearhart J. D., Antonarakis S. E. (1991). Cell-type-specific and hypoxia-inducible expression of the human erythropoietin gene in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 88, 8725–8729. 10.1073/pnas.88.19.8725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Suzuki H., Nojima M., Kitamura H., Yamamoto E., Maruyama R. (2013). Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur. Urol. 63, 1091–1100. 10.1016/j.eururo.2012.11.030 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Szczech L., Tang K. L., Barnhart H., Sapp S., Wolfson M. (2006). Correction of anemia with epoetin alfa in chronic kidney disease. N Engl. J. Med. 355, 2085–2098. 10.1056/NEJMoa065485 [DOI] [PubMed] [Google Scholar]
- Smrzova J., Balla J., Bárány P. (2005). Inflammation and resistance to erythropoiesis-stimulating agents–what do we know and what needs to be clarified?. Nephrol. Dial Transplant 20 Suppl 8, viii2–viii7. 10.1093/ndt/gfh1109 [DOI] [PubMed] [Google Scholar]
- Souma T., Nezu M., Nakano D., Yamazaki S., Hirano I., Sekine H. (2016). Erythropoietin synthesis in renal myofibroblasts is restored by activation of hypoxia signaling. J. Am. Soc. Nephrol. 27, 428–438. 10.1681/ASN.2014121184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souma T., Suzuki N., Yamamoto M. (2015). Renal erythropoietin-producing cells in health and disease. Front. Physiol. 6, 167. 10.3389/fphys.2015.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souma T., Yamazaki S., Moriguchi T., Suzuki N., Hirano I., Pan X. (2013). Plasticity of renal erythropoietin-producing cells governs fibrosis. J. Am. Soc. Nephrol. 24, 1599–1616. 10.1681/ASN.2013010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N. (2015). Erythropoietin gene expression: developmental-stage specificity, cell-type specificity, and hypoxia inducibility. Tohoku J. Exp. Med. 235, 233–240. 10.1620/tjem.235.233 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Gradin K., Poellinger L., Yamamoto M. (2017). Regulation of hypoxia-inducible gene expression after HIF activation. Exp. Cell Res. 356, 182–186. 10.1016/j.yexcr.2017.03.013 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Obara N., Pan X., Watanabe M., Jishage K., Minegishi N. (2011). Specific contribution of the erythropoietin gene 3' enhancer to hepatic erythropoiesis after late embryonic stages. Mol. Cell Biol. 31, 3896–3905. 10.1128/MCB.05463-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Obara N., Yamamoto M. (2007). Use of gene-manipulated mice in the study of erythropoietin gene expression. Methods Enzymol. 435, 157–177. 10.1016/S0076-6879(07)35009-X [DOI] [PubMed] [Google Scholar]
- Suzuki N., Sasaki Y., Kato K., Yamazaki S., Kurasawa M., Yorozu K., et al. (2016). Efficacy estimation of erythropoiesis-stimulating agents using erythropoietin-deficient anemic mice. Haematologica 101, e356–e360. 10.3324/haematol.2015.140814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Matsuo-Tezuka Y., Sasaki Y., Sato K., Miyauchi K., Kato K. (2018. b). Iron attenuates erythropoietin production by decreasing hypoxia-inducible transcription factor 2α concentrations in renal interstitial fibroblasts. Kidney Int. 94, 900–911. 10.1016/j.kint.2018.06.028 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Vojnovic N., Lee K. L., Yang H., Gradin K., Poellinger L. (2018. a). HIF-dependent and reversible nucleosome disassembly in hypoxia-inducible gene promoters. Exp. Cell Res. 366, 181–191. 10.1016/j.yexcr.2018.03.020 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Yamamoto M. (2016). Roles of renal erythropoietin-producing (REP) cells in the maintenance of systemic oxygen homeostasis. Pflugers Arch. 468, 3–12. 10.1007/s00424-015-1740-2 [DOI] [PubMed] [Google Scholar]
- Tojo Y., Sekine H., Hirano I., Pan X., Souma T., Tsujita T. (2015). Hypoxia signaling cascade for erythropoietin production in hepatocytes. Mol. Cell Biol. 35, 2658–2572. 10.1128/MCB.00161-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubakihara Y., Akizawa T., Iwasaki M., Shimazaki R. (2015). High hemoglobin levels maintained by an erythropoiesis-stimulating agent improve renal survival in patients with severe renal impairment. Ther. Apher Dial 19, 457–465. 10.1111/1744-9987.12308 [DOI] [PubMed] [Google Scholar]
- Vogelmann S. U., Nelson W. J., Myers B. D., Lemley K. V. (2003). Urinary excretion of viable podocytes in health and renal disease. Am. J. Physiol. Renal Physiol. 285, F40–F48. 10.1152/ajprenal.00404.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L., Semenza G. L. (1993). General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. U. S. A. 90, 4304–4308. 10.1073/pnas.90.9.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Liu X., Jaenisch R., Lodish H. F. (1995). Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 83, 59–67. 10.1016/0092-8674(95)90234-1 [DOI] [PubMed] [Google Scholar]
- Wynn T. A., Ramalingam T. R. (2012). Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040. 10.1038/nm.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Souma T., Hirano I., Pan X., Minegishi N., Suzuki N. (2013). A mouse model of adult-onset anaemia due to erythropoietin deficiency. Nat. Commun. 4, 1950. 10.1038/ncomms2950 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang F., Wang L., Wang W., Liu B., Liu J. (2012). Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet 379, 815–822. 10.1016/S0140-6736(12)60033-6 [DOI] [PubMed] [Google Scholar]