Abstract
The possible correlation between HPV16 and HPV18 genomic copies with the grade of cervical lesions needs more investigations. The aim of this study was to quantify genomic copies of HPV16 and 18 simultaneously and to find out the correlation between genomic copies numbers and different grades of lesions. Therefore, a total of 102 formalin-fixed and paraffin-embedded tissue specimens, 33 LSILI, 43 HSIL, and 26 squamous/adenocarcinoma were subjected to DNA extraction. The β-globin gene was selected to qualify the extracted DNA as well as normalization of viral titers using Taq-Man real-time PCR. The presence HPV16 and/or 18 were screened in tissue samples by nested PCR method, then an in- house Taq-Man Duplex real-time PCR assay was employed to quantify their genomic copies. The mean age of participants was 43 ± 13. Out of 102, 80 samples were positive for HPV16 and/or 18 DNA. There was a statistically significant association between HPV16 genomic copies and progression of cervical lesions (P < 0.001). In contrast, no such an association was found in the case of HPV18 (P = 0.51). Moreover, with 95% confidence intervals, 2.3–4 genomic copies of HPV16 genome/cell could be applicable to distinguish LSIL from HSIL and SCC. In conclusion, quantification of HPV16 genomic copy number showed a close association with progression of cervical lesion. Furthermore, HPV16 genomic copies of 4 copies/cell could be a set point to differentiate LSIL from HSIL.
Keywords: HPV16 and HPV18, LSIL, HSIL, SCC, Taq-Man real-time PCR, Genomic copies
Introduction
Cervical cancer is the fourth most prevalent and the fifth lethal cancer amongst women worldwide [16]. In Iran, with an incidence rate of 2.5 case per 100,000, it ranks 12th amongst women [6, 16]. Almost 20% of all cancers were established by viruses such as human papillomavirus (HPV) [1]. HPV usually infects the basal layer of the epithelial cells, and then establishes a persistent infection, which might develop to gynecological, esophageal, breast, and colorectal malignancy over time [18, 25, 29]. Persistence infections with High-Risk papillomaviruses (HR-HPVs) are the major risk factor for developing both high-grade cervical lesions and invasive cervical cancer [3]. Worldwide, 70% of cervical cancers are caused by oncogenic HPV16 and 18 [14]. The behavior of HPVs in various grade of lesions was reported to be biologically different, which is influenced by viral and host factors [5]. In order to improve assessment of suspected individuals, a molecular test might be beneficial. Meanwhile, to reduce the number of women with normal cytology for further histologic investigation and to avoid unnecessary follow-up regarding their suspected state of HR-HPV infections, a reliable molecular marker would be of great value. It is suggested that a molecular quantitative assay, could be employed as a potential discriminative of lesions grades. [17]. HPV replication can be quantified and used as a predictor of viral persistence risk and the malignancy progression [12].
Previous studies implied that HR-HPV genome quantification by molecular assays strongly enhances the sensitivity of the negative predictive value of the histology screening when compared to cytology method [20, 26]. The majority of cross-sectional studies reported the association between HPV genomic copies and the disease progression. Wu et al. [31] reported that such association is more significant in the case of HPV16 than other types, when investigated in low and high grade and high intraepithelial lesions. Ramirez-Flores et al. also reported that HPV16 genomic copies were higher than type 18 in cervical lesion neoplasia. They showed that HPV16 genomic copy number had a moderate positive association with cervical lesion progression that was not detected in the case of HPV type 18 [28]. In practice, establishing a reliable quantification assay in addition to the common histological examinations, not only improves the power of screening test for differentiating cervical lesions, but also reduces misconception by histological errors. Herein, to clarify a possible correlation between the grade of confirmed cervical intraepithelial lesions with HPV16 and 18 genomic copies, an in-house Taq-Man real-time PCR was developed and determined a discriminative value in order to differentiate low grade from high grade lesions.
Materials and methods
Study design and samples
In this cross-sectional study, 102 formalin-fixed paraffin embedded biopsy (FFPE) samples including 33 ISIL cases with CIN I; 23 HSIL cases with CIN II; 20 HSIL cases with CIN III; 23 cases with SCC and 3 other cases with adenocarcinoma were collected. All samples were selected from archive of department of pathology of Motahari Clinic affiliated to Shiraz University of Medical Sciences, 2014–2015. The study was approved by the local Ethics Committee of Shiraz University of Medical Sciences (IR.SUMS.REC. 1395. S301). The Hematoxylin and eosin (H&E) stained slides were reclassified into different histologic grade of cervix CINI, II, III and SCC by the pathologist).
DNA extraction and qualification
10 μm-thick sections of FFPE tissue blocks were cut and collected in autoclaved Eppendorf micro-centrifuge tubes (1.5 ml) and were then subjected to deparafinazation step as described in a previous study [24]. Genomic DNA was extracted using the QIAamp FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted DNA was stored at −20 °C until further analysis. In order to assess the quality of extracted DNA and to normalize the copy of DNA in tissue samples; all extracted DNAs were subjected to an in house Taq-Man real-time PCR for β-globin gene quantification.
Cloning and standard curve
In order to prepare standard fragment of related genes for quantification purpose, HPV E7 gene fragment and β-globin gene were cloned into plasmid constructs. Hence, to detect HPV16 and 18 as well as human β-globin gene fragment using Real-Time PCR, three pairs of primer were designed by AlleleID software (Table 1). Amplified PCR products were run on 2% agarose gel, excised and purified by GF-1 PCR Clean-up Kit (Vivantis Technologies, Malaysia), and thereafter inserted into pTZ57R/T plasmid using InsTAclone PCR Cloning kit (Thermo Scientific). The constructs were transformed in DH5alpha E. coli and subsequently plasmid was purified using GF-1 Plasmid DNA Extraction Kit (Vivantis Technologies, Malaysia). The optical density (OD) of purified plasmids were measured by a NanoDrop™ spectrophotometer (Nano Drop™ 2000; Thermo Scientific) and then used to calculate the absolute DNA copy number. Standard curve of β-globin gene plasmid ranging from 3 × 105 to 30 copies/reaction in 1:10 serial dilutions was used for cell quantification of each sample. HPV16 and HPV18 standard curves with ten-fold serial dilutions (ranged from 3 × 107 to 30 copies for HPV16 and 3 × 106 to 30 copies of HPV18) were used for HPV genome quantification in each sample in a background of 200 ng/reaction human genome. HPV genomic copies was normalized by calculating the ratio of virus copy numbers to half of β-globin gene copy numbers, which measures the absolute copy number of HPV DNA per cell.
Table 1.
Sequences and concentration of HPV16, HPV18 and β-globin primers and probes used in real-time PCR
| Oligonucleotide | Sequence | Concentration (µM) | Product size (bp) |
|---|---|---|---|
| HPV16 E7 F | 5′-AGGAGGATGAAATAGATGG-3′ | 0.4 | |
| HPV16 E7 R | 5′-CTTCCAAAGTACGAATGTC-3 | 0.4 | 141 |
| HPV16 E7 probe | FAM-CACAACCGAAGCGTAGAGTCA-BHQ | 0.16 | |
| HPV18 E7 F | 5′-CGATGAAATAGATGGAGTTAA-3′ | 0.4 | |
| HPV18 E7 R | 5′-CACACTTACAACACATACA-3′ | 0.4 | 92 |
| HPV18 E7 probe | JOE-ACATTTACCAGCCCGACGAG-BHQ | 0.16 | |
| β-globin F | 5′-TGTGTTCACTAGCAACCTCAA-3′ | 0.4 | |
| β-globin R | 5′-CTCACCACCAACTTCATCCA-3′ | 0.4 | 111 |
| β-globin probe | FAM-CCTGAGGAGAAGTCTGCCGTTACTGCC-BHQ | 0.16 |
Determination of the specificity and detection limit of Real-time PCR
Specificity of each HPV genotype primers and probe were checked, using HPV16 and HPV18 positive control in Tag-Man Real time assay. In order to find copy number limit of our test; OD (optical density) of HPV16 and HPV18 E7 gene and β-globin, cloned into plasmid was measured and used in the formula to find out each gene copy number. The formula calculates the number of dsDNA copies by entering OD amount of DNA (ng) and length of PCR product (bp). After calculating ten-fold serial dilutions were made and used as templates of Real-Time PCR. The final dilution in which Ct was determined was set as detection limit of the test. The specificity of the test was also evaluated by the use of some negative as well as positive controls.
Quantification of β-globin gene and samples normalization
β-globin gene quantified in all positive samples using Taq-Man real-time PCR and samples with negative β-globin were excluded. The sequences of β-globin specific primers and Taq-Man probe are shown in Table 1. Reaction ingredient included Ampliqon RealQ Plus 2x Master Mix for probe High Rox, 0.16 µM of each HPV genotype probes and 0.4 µM of each primer in 25 µl final volume. Cycling conditions consisted of a 95 °C denaturation step for 15 min, followed by 35 cycles consisting of alternating 95 °C for 20 s, and 60 °C annealing for 20 s and 72 °C elongation for 20 s.
HPV type 16 and 18 screening by Nested-PCR assay
In an attempt to find HPV16/18 infected samples, a Nested-PCR assay was used. Type specific HPV16 and 18 primers were employed in a nested PCR assay on samples which were positive for β-globin gene to assure that only suitable types be included for next duplex HPV DNA quantification. Primers sequences that target HPV L1 gene, each PCR run components and test conditions were previously explained in details by Farhadi et al. [15].
Duplex Taq-Man real time-PCR assay for detection of HPV16 and 18
In the next step, 80 samples that were positive for β-globin and HPV16 and/or HPV18 were processed for quantification of viral genome, using an in-house duplex Real-Time PCR. Primers and probe sequences used for HPV16 and 18 quantifications are shown in Table 1. Reaction components were Ampliqon real Q Plus 2x Master Mix for probe High Rox as mix and 0.16 µM of each HPV genotype probes and 0.4 µM of each primer in 25 µl final volume. Real-Time PCR was performed using Qiagen Inc.-Rotor-Gene Q. Cycling conditions consisting of a 95 °C early denaturation step for 15 min, followed by 35 cycles of 95 °C for 30 s, and 58 °C annealing for 30 s, and 72 °C elongation for 30 s. In each run a standard plasmid, containing HPV16 or HPV18 E7 gene, was included. genomic copies were determined based on cycle threshold (Ct) values above their corresponding standards in a comparison to β-globin gene.
Statistical analysis
SPSS 21 was used for statistical analysis. Graph Pad 7.0 was used to generate a scatter dot plot of the genomic copies data. One-way ANOVA, paired t test, Kruskal–Wallis and T Test were used to compare different parameter. P values less than 0.05 were considered to be statistically significant.
Results
Demographic and pathological result of the participants
The mean age of patients was 42 ± 12 years (Table 2). A statistically significant correlation was observed between older age and increase of/higher lesions grade (P < 0.001). Out of 101 samples 80 specimens were positive for HPV16 and 18, of which 25 samples were LSIL, 32 samples were HSIL and 20 samples were SCC and 3 samples were Adenocarcinoma, based on pathological reevaluation.
Table 2.
Proportion of the histological classifications by HPV infection status
| HPV16 | HPV18 | HPV16 and 18 co-infection | Total | Age mean | |
|---|---|---|---|---|---|
| LSIL | 5 (20%) | 4 (16%) | 16 (64%) | 25 | 38 ± 10 |
| HSIL | 11 (34%) | 3 (9.4%) | 18 (56%) | 32 | 39 ± 10 |
| SCC | 5 (25%) | 3 (15%) | 12 (60%) | 20 | 50 ± 10 |
| ADC | 0 | 0 | 3 (10%) | 3 | 67 ± 7 |
| Total | 21 (26.3%) | 10 (12.5%) | 49 (61.3%) | 80 | 49 ± 9 |
LSIL low grade squamous cell intraepithelial lesions, HSIL high grade squamous cell intraepithelial lesions, SCC squamous cell carcinoma, ADC adenocarcinoma of cervix
Nested PCR for HPV16/18 screening
To screen for the presence of HPV16 or 18 in cervical lesions, the nested-PCR was performed. The nested-PCR results showed that out of 101 positive samples for β-globin gene, 91 (90%) were either positive for HPV16 and/or 18. To be exact, out of 33 LSIL samples, 26 were positive, out of 42 HSIL samples, 39 were positive, and out of 23 SCC samples, all cases were harboring HPVs genome inside.
Determination of the specificity and detection limit of Real-time PCR
The specificity test showed no cross-reactivity for HPV16 and HPV18 quantification assays when positive controls were included as templates into reactions. In addition, HPV16 and 18 plasmids in ten-fold serial dilutions showed a detection limit of 30 copies of DNA, which is equivalent to 10−7 ng DNA/µl for both HPV genotypes.
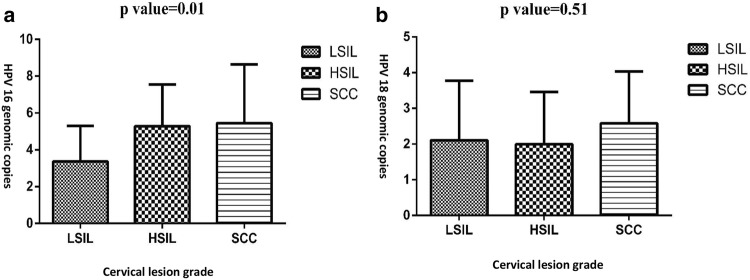
Figure 1 illustrates the distribution of the logarithmically transformed absolute virus copy number per cell in a determined histological classification. Mean (95% CI) log of HPV16 and 18 genomic copies was 4.5 (3.8–5.2) and 2.1 (1.8–2.6) copies per cell (log10-transformed), respectively.
Fig. 1.
Distribution of the logarithmically transformed absolute virus copy number of HPV16 (a) and HPV18 (b) per cell by cervical histological classification. In (a), P value was 0.02 between LSIL/HSIL and 0.029 between LSIL/SCC
Duplex Taq-Man real time-PCR assay for quantification of HPV16 and 18
To find a correlation between genomic copies and grading as well as disease progression, copy number of HPV16 and 18 genomes amongst LSIL, HSIL and SCC were compared between mono and co-infection cases. In this regard, it was shown that while a significant correlation was found for HPV16 genomic copies and grade of the cervical lesions (P < 0.001), such a relationship was not observed for HPV18 (P = 0.5). HPV16 mean genomic copies was significantly higher in samples with HSIL or higher cervical intraepithelial lesion than in women with LSIL (P = 0.02) (Table 3). Overall, there was a linear increasing association between HPV16 genomic copies and disease progression (P < 0.05). Moreover, 61% (n = 49) of the samples had co-infection of HPV16 and 18, whose HPV16 genomic copies was significantly higher than HPV18 (P = 0.01). Also, there was a significant difference between HPV16 and HPV18 genomic copies in different cervical lesions grade (P = 0.004). Although HPV16 genomic copies in mono-infected patients was higher compared to HPV16/18 co-infections and increased from LSIL to SCC, it was not statistically significant (P = 0.36). Furthermore, HPV18 copies was not increased in different histological lesions in both mono- and co-infected patients (P = 0.5 and P = 0.8). HPV18 genomic copies in patients with mono-infection appeared to be lower than those with confection, but then again it was not statistically significant (P = 0.2).
Table 3.
DNA viral loads in mono-HPV and multiple HPV16/18 infections
| HPV16 | HPV18 | |
|---|---|---|
| Total samples | 70 | 59 |
| Mean viral load of total samples (95% CI) | 4.5 (3.8–5.2) | 2.1 (1.8–2.6) |
| HPV mono-infection samples, n (%) | 21 (26.3) | 10 (12.5) |
| Mean viral load of mono-infection (95% CI) | 4.9 (3.8–6.7) | 1.8 (0.5–3.3) |
| HPV co-infection, n (%) | 49 (61%) | |
| Mean viral load of co-infection (95% CI) | 4.3 (3.4–5.2) | 2.3 (1.8–2.8) |
| P value of viral load between mono-infection and co-infection | 0.36 | 0.2 |
Viral loads are expressed in logarithmic viral load per human cell
Discrimination between different lesion grades
Finally, regarding significant association of genomic copies and grading of lesion, a discriminative value by 95% confidence intervals was determined for HPV16 virus copies in LSIL cervical lesions with a mean of 2.3–4 copies per cell. Mean of HPV16 copies with 95% confidence interval for HSIL grade was determined in a range of 4.5–6.3 copies per cell and 4.7–7 copies per cell for SCC. This value was statistically different between LSIL and HSIL (P = 0.003) and LSIL and SCC (P = 0.007); hence, could be used to discriminate LSIL from other grades.
Discussion
Infection with high risk HPV types is the leading cause of cervical intraepithelial neoplasia [17]. The risk of cervical lesion progression from low to high-grade appears to be greater in patients with persistent oncogenic HPV infection as well as higher genomic copies [27]. Studies have suggested that HR-HPVs genome quantification might enhance the power of histology test’s predictive value in screening HSIL. Moreover, some studies showed that this correlation was type-dependent and could not be adjusted to all the HPV genotypes [7, 10, 17, 31].
The results of our study showed a significant association between HPV16 genomic copies and cervical lesion progression from LSIL to SCC (P < 0.001). Also, HPV16 genomic copies were significantly higher in HSIL (CINII and III) than LSIL (CINI) (P = 0.02) (Table 3). In agreement, Wu et al. found a strong correlation between HPV16 genomic copies and disease severity, which was in line with Carcopino et al. [7, 31]. Also, Broccolo et al. and Dong et al. in separate studies showed that HPV16 and HPV-31 genomic copies were associated with increasing the grade of related lesions [4, 13]. In addition, Mittal et al. [27] showed that higher genomic copies at LSIL grade reduced the risk of developing HSIL or SCC in these women by four-fold, which was compatible with our finding.
Interestingly, in the case of HPV18, we could not find such a relationship between genomic copies and grade of cervical lesions (P = 0.5). In agreement with this result, Wu et al., Broccolo et al. and Dong et al. reported no association between HPV18 genomic copies and the progression of cervical lesion from LSIL to SCC [4, 13, 31]. The reason behind the lower genomic copies of HPV18 in more invasive lesion; however, is not clear. Nonetheless, significant difference between HPV16 and HPV18 genomic copies in cervical lesions can be related to different mechanism of pathogenesis of each type. In this accordance Barbosa and Schlegel [2] reported that HPV-18 DNA was five-fold more efficient than HPV-16 for the transformation of keratinocytes in vitro. Moreover Villa and Schlegel [30] reported that HPV18 induce immortalization of epithelial cells which making it more aggressive than those immortalize with HPV16. Moreover, Dong et al. reported low HPV18 genomic copies in HSIL grade might be due to genome integration. These finding indicated that cancerous lesions containing HPV18, is more likely to be associated with HPV genome integration rather than episomal status [13]. They also mentioned that increasing HPV transcript levels can be associated with higher lesion grades rather than viral copy number [13, 21].
The comparison of mono and co-infection of HPV16/18 also showed that genomic copies varied among these types as the higher amount was detected for specimens infected only with HPV16. In this accordance, Wu et al. and Flores et al. also reported higher HPV16 genomic copies in mono-infected cases [17, 31]. In the case of HPV16/18 co-infection; however, HPV16 genomic copies appeared to be lower than mono-infection with no statistically differences (P = 0.3). Besides, HPV16 had higher genomic copies in co-infections.
Comparing the HPV genomic copies at different stages showed that HPV18 mean genomic copies was lower in HSIL grade than LSIL in both mono and co-infection, but again with the progression toward SCC, it had an increasing trend with no statistically significant association. In a study by Wu et al. [31] a statistically significant decrease was reported for HPV18 genomic copies in HSIL in comparison with other grades.
With respect to previous study, women with multiple HPV genotypes infection including HPV16 were more likely to have cervical lesions and abnormal cytology with the highest OR in the HSIL group [11]. In this study, 70 (68%) of the samples were infected with HPV16 (20% HPV16 mono-infection and 48% HPV16/18 co-infection). Moreover, 59 (73%) patients were infected with HPV18 (9% HPV18 mono-infection). In this regard, Khorsandizadeh et al. [22] reported the most commonly detected viruses HPV16 (56.4% of all HPV types) and HPV18 (15.6% of all HPV types). Another study by Kiani et al. [23] showed that 69.7% of cervical lesions were infected with HPV16 and/or 18. The rate of HPV16 and 18 co-infection was reported to be various (16.3–55%) in malignant lesions [19]. Interestingly, in this study it was found that 61% (49 out of 80) of positive samples harbored both HPV16 and 18 genomes. De Jesus, et al. [9] reported that 75.4% of all grades of cervical lesion amongst Brazilian patients had co-infection with HPV 16 and 18. Carrillo-García et al. [8] demonstrated that co-infections were common in both high-grade and low-grade cervical lesions, and if multiple HR-HPV types caused this co-infection, the risk of progression to higher grade lesion or CC increased.
The exact grading to discriminate between various stages of malignant disease remains to be a drawback of this methodology. Therefore, an alternative molecular method such as quantitative PCR to improve or even surrogate it, seems to be beneficial. In our study, we found that 2.3 to 4 genomic copies of HPV16 per cell can reliably discriminate the LSIL grade from HSIL and SCC. In the study by Carcopino et al., which evaluated the clinical significance of HPV16 and 18 genomic copies, the correlation of genomic copies and lesion stage was demonstrated, and it was determined that 3.0 × 106 copies/million cells threshold could be a suitable discriminative value for highly specific discrimination of grade 2 cervical lesion from other grades (21). Another research by Dong et al. [13] also indicated that the discriminative value level for the log 10-transformed genomic copies of HPV16, is 4.26 copies per 10,000 cells. To date, the clinical utility of HPV genomic copies quantitation remains questionable as it has often yielded conflicting results. One of the main reasons for this drawback could be the lack of standardized procedures as well as suitable controls, leading to an unreliable comparison between the findings from different studies.
The mean age of SCC was significantly higher than LSIL and HSIL (p < 0.001). All cases diagnosed with SCC were up to 40 years old (95% CI 46–55). The highest HPV16 genomic copies was seen in SCC grade and the oldest cases were seen in SCC too; thus, we could say that older age and higher genomic copies could promote the severity of lesions. This result was consistent with Briolat’s study [3] that reported a significant association between the patients’ age and increasing severity of the cervical lesions. Another study by Mittal et al. showed that women aged 50–60 were twice at greater risk of persisting HPV infection in comparison with women aged 40–49.
Using the formalin-fixed paraffin-embedded tissue as specimen, small sample size, as well as the few cases of SCC and adenocarcinoma can be noted as the limitations of this study.
In conclusion The results of this study showed that 90% of cervical lesions are infected with two important high risk HPVs including HPV16 and 18. Moreover, this study highlights the relation between HPV16 genomic copies and the severity of the cervical lesions, whereas an HPV18 genomic copy appears to be of no clinical utility. Furthermore, HPV16 genomic copies as a differentiating factor of LSIL from HSIL grades could be used in clinical diagnosis, with a special attention to women with HPV16 genomic copies higher than 4 copies per cells. Also determination of genomic copies as well as co-infection of HPV16/18 might be more helpful for prediction of prognosis than determination of sole HPV genotyping.
Acknowledgements
The present study was extracted from the thesis written by Negar Joharinia, and was financially supported by Shiraz University of Medical Sciences (Grant No: 94-10934). Special thanks to Prof. Abbas Behzad-Behbahani and the staff of Diagnostic Laboratory Sciences and Technology Research Center for their assistance. The authors wish to thank Mr. H. Argasi at the Research Consultation Center (RCC) of Shiraz University of Medical Sciences for his invaluable assistance in editing this manuscript.
Compliance with ethical standards
Conflict of interest
All the authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bakhtiarizadeh S, et al. Almost complete lack of human cytomegalovirus and human papillomaviruses genome in benign and malignant breast lesions in Shiraz, Southwest of Iran. Asian Pac J Cancer Prev. 2017;9(12):3319–3324. doi: 10.22034/APJCP.2017.18.12.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa M, Schlegel RJO. The E6 and E7 genes of HPV-18 are sufficient for inducing two-stage in vitro transformation of human keratinocytes. Oncogene. 1989;4(12):1529–1532. [PubMed] [Google Scholar]
- 3.Briolat J, et al. HPV prevalence, viral load and physical state of HPV-16 in cervical smears of patients with different grades of CIN. Int J Cancer. 2007;121(10):2198–2204. doi: 10.1002/ijc.22959. [DOI] [PubMed] [Google Scholar]
- 4.Broccolo F, et al. Prevalence and viral load of oncogenic human papillomavirus types associated with cervical carcinoma in a population of North Italy. J Med Virol. 2009;81(2):278–287. doi: 10.1002/jmv.21395. [DOI] [PubMed] [Google Scholar]
- 5.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16(1):1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer, I.I.C.o.H.a. Human papillomavirus and related diseases report. ICO Information Centre on HPV and Cancer, 2015.
- 7.Carcopino X, et al. Significance of HPV 16 and 18 viral load quantitation in women referred for colposcopy. J Med Virol. 2012;84(2):306–313. doi: 10.1002/jmv.23190. [DOI] [PubMed] [Google Scholar]
- 8.Carrillo-García A, et al. Impact of human papillomavirus coinfections on the risk of high-grade squamous intraepithelial lesion and cervical cancer. Gynecol Oncol. 2014;134(3):534–539. doi: 10.1016/j.ygyno.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 9.de Jesus SP, et al. A high prevalence of human papillomavirus 16 and 18 co-infections in cervical biopsies from southern Brazil. Braz J Microbiol. 2018;49:220–223. doi: 10.1016/j.bjm.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Río-Ospina L, et al. The DNA load of six high-risk human papillomavirus types and its association with cervical lesions. BMC Cancer. 2015;15(1):100. doi: 10.1186/s12885-015-1126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson EL, et al. Cervical cytology and multiple type HPV infection: a study of 8182 women ages 31–65. Gynecol Oncol. 2014;133(3):405–408. doi: 10.1016/j.ygyno.2014.03.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong L, et al. Human papillomavirus viral load as a useful triage tool for non-16/18 high-risk human papillomavirus positive women: a prospective screening cohort study. Gynecol Oncol. 2018;148(1):103–110. doi: 10.1016/j.ygyno.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Dong B, et al. Type-specific high-risk human papillomavirus viral load as a viable triage indicator for high-grade squamous intraepithelial lesion: a nested case- control study. Cancer Manag Res. 2018;10:4839–4851. doi: 10.2147/CMAR.S179724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eghbali SS, et al. Oncogenic human papillomavirus genital infection in southern Iranian women: population-based study versus clinic-based data. Virol J. 2012;9(1):194. doi: 10.1186/1743-422X-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farhadi A, et al. High-risk human papillomavirus infection in different histological subtypes of renal cell carcinoma. J Med Virol. 2014;86(7):1134–1144. doi: 10.1002/jmv.23945. [DOI] [PubMed] [Google Scholar]
- 16.Fitzmaurice C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores R, et al. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118(5):1187–1193. doi: 10.1002/ijc.21477. [DOI] [PubMed] [Google Scholar]
- 18.Gao G, Smith DI. Human papillomavirus and the development of different cancers. Cytogenet Genome Res. 2017;150:185–193. doi: 10.1159/000458166. [DOI] [PubMed] [Google Scholar]
- 19.Guan P, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer. 2012;131(10):2349–2359. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 20.Hesselink AT, et al. High-risk human papillomavirus DNA load in a population-based cervical screening cohort in relation to the detection of high-grade cervical intraepithelial neoplasia and cervical cancer. Int J Cancer. 2009;124(2):381–386. doi: 10.1002/ijc.23940. [DOI] [PubMed] [Google Scholar]
- 21.Ho CM, et al. Type-specific human papillomavirus oncogene messenger RNA levels correlate with the severity of cervical neoplasia. Int J Cancer. 2010;127(3):622–632. doi: 10.1002/ijc.25078. [DOI] [PubMed] [Google Scholar]
- 22.Khorasanizadeh F, et al. Epidemiology of cervical cancer and human papilloma virus infection among Iranian women—analyses of national data and systematic review of the literature. Gynecol Oncol. 2013;128(2):277–281. doi: 10.1016/j.ygyno.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 23.Kiani SJ, et al. Detection and typing of human papilloma viruses by nested multiplex polymerase chain reaction assay in cervical cancer. Jundishapur J Microbiol. 2015;8(12):e26441. doi: 10.5812/jjm.26441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoodvan S, et al. No detection of Streptococcus g allolyticus and Helicobacter pylori in colorectal cancer tissue samples in Shiraz. Iran. Iran J Cancer Prev. 2017;10(1):e6337. [Google Scholar]
- 25.Mahmoudvand S, et al. Presence of human papillomavirus DNA in colorectal cancer tissues in Shiraz, Southwest Iran. Asian Pac J Cancer Prev APJCP. 2014;16(17):7883–7887. doi: 10.7314/APJCP.2015.16.17.7883. [DOI] [PubMed] [Google Scholar]
- 26.Mayrand M-H, et al. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med. 2007;357(16):1579–1588. doi: 10.1056/NEJMoa071430. [DOI] [PubMed] [Google Scholar]
- 27.Mittal S, et al. Risk of high-grade precancerous lesions and invasive cancers in high-risk HPV-positive women with normal cervix or CIN 1 at baseline—a population-based cohort study. Int J Cancer. 2017;140(8):1850–1859. doi: 10.1002/ijc.30609. [DOI] [PubMed] [Google Scholar]
- 28.Ramírez-Flores M, et al. HPV 16 and 18 viral loads are greater in patients with high-grade cervical epithelial lesions. Eur J Gynaecol Oncol. 2016;37(5):644–648. [PubMed] [Google Scholar]
- 29.Serrano B, et al. Epidemiology and burden of HPV-related disease. Best Pract Res Clin Obstet Gynaecol. 2017;47:14–26. doi: 10.1016/j.bpobgyn.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Villa LL, Schlegel RJV. Differences in transformation activity between HPV-18 and HPV-16 map to the viral LCR-E6-E7 region. Virology. 1991;181(1):374–377. doi: 10.1016/0042-6822(91)90507-8. [DOI] [PubMed] [Google Scholar]
- 31.Wu Z, et al. Association between human papillomavirus (HPV) 16, HPV18, and other HR-HPV viral load and the histological classification of cervical lesions: results from a large-scale cross-sectional study. J Med Virol. 2017;89(3):535–541. doi: 10.1002/jmv.24645. [DOI] [PubMed] [Google Scholar]