Abstract
Almost all organisms exhibit ~24-h rhythms, or circadian rhythms, in a plentitude of biological processes. These rhythms are driven by endogenous molecular clocks consisting of a series of transcriptional and translational feedback loops. Previously, we have shown that the inner nuclear membrane protein MAN1 regulates this clock and thus the locomotor rhythm in flies, but the mechanism remains unclear. Here, we further confirmed the previous findings and found that knocking down MAN1 in the pacemaker neurons of adult flies is sufficient to lengthen the period of the locomotor rhythm. Molecular analysis revealed that knocking down MAN1 led to reduced mRNA and protein levels of the core clock gene period (per), likely by reducing its transcription. Over-expressing per rescued the long period phenotype caused by MAN1 deficiency whereas per mutation had an epistatic effect on MAN1, indicating that MAN1 sets the pace of the clock by targeting per.
Electronic supplementary material
The online version of this article (10.1007/s12264-019-00404-6) contains supplementary material, which is available to authorized users.
Keywords: Circadian clock, Drosophila, Nuclear envelope, MAN1, Period
Introduction
Circadian rhythms, or ~24-h rhythms in various behavioral and physiological processes, are a fundamental property of life. These rhythms exist across phylogeny from prokaryotes to humans, and are driven by a relatively conserved molecular clockwork consisting of transcriptional and translational feedback loops [1]. In Drosophila, two transcription factors CLOCK (CLK) and CYCLE (CYC) heterodimerize and activate the transcription of period (per) and timeless (tim) via E-box elements located in the promoter/enhancer regions of these genes [2]. PER and TIM proteins accumulate in the cytoplasm, bind each other, and enter the nucleus where they repress the transcriptional activity of CLK/CYC, thus inhibiting their own transcription and forming the major feedback loop of the clockwork. PER/TIM undergo post-translational modifications that ultimately lead to their degradation. Once they are degraded, CLK/CYC can activate transcription again and start a new cycle. This serves as the basis of the overt 24-h rhythms. CLK/CYC also drive the transcription of two additional transcription factors, vrille (vri) and PAR-domain protein 1ε/δ (Pdp1ε/δ). The former represses while the latter activates clk transcription, forming accessory loops of the clockwork.
The nuclear envelope (NE) has long been considered simply as a barrier that separates genetic material from the rest of the cell and controls the entry/exit of molecules into/out of the nucleus [3]. However, in the past 20 years or so, emerging studies have demonstrated that NE interacts extensively with chromatin and DNA, influencing the spatial organization of the genome and modulating gene expression [3, 4]. Recent work has reported that circadian-regulated genes are rhythmically recruited to the nuclear periphery where they interact with chromosomal regions that associate with nuclear lamins (lamin-associated domains) and transcription is attenuated via epigenetic modifications [5]. Once they move away from the nuclear envelope, transcription is re-activated and thus cyclic transcription can be achieved. However, the detailed mechanisms underlying this process remain largely unclear. Previously, we found three NE proteins, MAN1, lamin B1, and lamin B receptor, to be involved in determining the circadian period length in both human cell lines and Drosophila, but little is known regarding how they exert effects on the clock in vivo [6]. Therefore, the goal of this study is to investigate the mechanism underlying how MAN1 regulates the clock in flies.
Materials and Methods
Fly Strains
We used the following fly strains: w1118, yw, genetic background control lines for the RNAi lines (VDRC:60100), UASMAN1RNAi-1 (NIG:3167R-1), UASMAN1RNAi-2 (NIG:3167R-2), UASMAN1RNAi-3 (VDRC:108906), MAN1GS2297 [6], timGAL4;UASdcr2 [6], cryG4-16 [6], UASdcr2;cryGAL4-16 [6], cryG4-39;UASdcr2 [6], pdfG4-GS [7], UASper [8], perL [9], UASclk [10], and clkjrk [11]. Dicer2 (dcr2) was co-expressed to enhance the effects of RNAi [12].
Drosophila Activity-Monitoring and Behavioral Analysis
Flies were reared on standard cornmeal-yeast-sucrose medium and kept in 12 h light:12 h dark (LD) cycles at 25°C. Male flies 3–4 days old were used to monitor locomotor activity levels using the Drosophila Activity Monitor system (Trikinetics, Waltham, MA, USA) for 4 days of LD followed by 7 days of constant darkness (DD). To monitor the locomotor activity of strains containing the pdfG4-GeneSwitch (pdf-GS) driver, flies were raised on standard cornmeal-yeast-sucrose medium until pupation [7]. After eclosion, flies were transferred to tubes with standard food containing 250 µmol/L RU486 (Sigma-Aldrich, St. Louis, MO, USA) for 2–3 days before monitoring locomotor activity. The food used during behavioral assays also contained 250 µmol/L RU486. Ethanol used to solubilize RU486 was added to the food as vehicle control.
Analyses of period, power, and rhythmicity during DD were carried out using ClockLab software (Trikinetics). For DD rhythmicity, rhythmic flies were defined as those with χ2 power-significance ≥10. Period calculations considered all flies with power-significance ≥10. One-way ANOVA and Tukey’s multiple comparison test (Prism Graphpad, La Jolla, CA, USA) were used to compare the differences between genotypes.
RNA Extraction and Quantitative Real-Time PCR
Flies were entrained in LD for 3 days, collected on DD1 at the indicated circadian time points (CTs) and frozen immediately on dry ice. Fly heads were isolated and homogenized in TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Chloroform was subsequently added and the mixture centrifuged at 12,000g for 15 min at 4°C. Aqueous top layer was collected and ethanol (Sigma-Aldrich) was added to precipitate RNA. The precipitates were collected by centrifuging at 12,000g for 10 min at 4°C. The RNA pellets were washed with 75% ethanol. After air drying, the pellets were dissolved in RNAse-free water. Contaminating genomic DNA was removed by RQ1 DNase (Promega, Madison, WI, USA) digestion, and total RNA was directly amplified with TransScript Green One-Step qRT-PCR SuperMix (Transgen Biotechnology, Beijing, China). All qPCR reactions were carried out on a Step One Plus Real-Time PCR System (Life Technologies). The templates were reverse-transcribed at 45°C for 5 min, denatured at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C, 15 s at 60°C, 30 s at 72°C, and 15 s at 75°C for data acquisition. The primers used for expression analysis were as follows: MAN1 forward, 5′-GACTCACGGGGAAATCTGC-3′ and reverse, 5′-GCAGATTGGGTTCAGAAAGC-3′; tim forward, 5′-CTGGGGAGTGACCATGG-3′ and reverse, 5′-GCTGGAATCGCCACTG-3′; per forward, 5′-CAGCAGCAGCCTAATCG-3′ and reverse, 5′-GAGTCGGACACCTTGG-3′; clk forward, 5′-TACTGCGTGAGGATATCG-3′ and reverse, 5′-GTTGTTGTTCTGGTTGC-3′; Pdp1ε forward, 5′-GAACCCAAGTGTAAAGACAATGCG-3′ and reverse, 5′-CTGGAAATACTGCGACAATGTGG-3′; vri forward, 5′-TGTTTTTTGCCGCTTCGGTCA-3′ and reverse, 5′-TTACGACACCAAACGATCGA-3′; beta-Actin forward, 5′-CTAACCTCGCCCTCTCCTCT-3′ and reverse, 5′-GCAGCCAAGTGTGAGTGTGT-3′; rp49 forward, 5′-TACAGGCCCAAGATCGTGAA-3′ and reverse, 5′-GCACTCTGTTGTCGATACCC-3′; pre-per forward, 5′-TAGCTCAACGTTCCCATTCG-3′, and reverse 5′-AGATAATGCAGCACAGGAAGC-3′; and Cbp20 forward, 5′-AGATCCACGAGCTCTTCTCC-3′, and reverse 5′-CCGATCGGACATAGTACTCC-3′.
Beta-actin or rp49 (for mRNA analysis) and cbp20 (for pre-mRNA analysis) were used as normalization controls. The delta-delta CT method was used for quantification. The value of the control genotype was set to 1. Student’s t test (Microsoft Excel, Seattle, WA, USA) was used to compare the differences between genotypes.
Western Blot
Flies were entrained in LD for 3 days, collected during DD1 at the indicated CTs, and frozen immediately on dry ice. Heads were separated and homogenized in RIPA buffer (20 mmol/L Tris at pH 7.5, 150 mmol/L NaCl, 1 mmol/L EDTA, 0.05 mmol/L EGTA, 10% glycerol, 1% Triton X-100, 0.4% sodium deoxycholate, 0.1% SDS) containing a protease inhibitor mixture (Roche, Basel, Switzerland) and phosphatase inhibitor mixture (Roche). This homogenate was sonicated 3 to 5 times for 8 s each time, and then centrifuged at 13,000 rpm for 10 min at 4°C to remove cell debris. The supernatant was collected, transferred to new tubes, and centrifuged again at 13,000 rpm for 10 min at 4°C. The supernatant was collected and protein lysates were prepared in SDS-PAGE loading buffer. Equal amounts of protein were run on 7.5% SDS-PAGE gels and then transferred to nitrocellulose membranes. After incubation with primary antibodies at 4°C overnight, the membranes were incubated with secondary antibodies at room temperature for 1 h. The primary antibodies used were rat anti-TIM (1:1000; a gift from Dr. Joanna Chiu, University of California, Davis, CA, USA), guinea pig anti-PER (1:1000; a gift from Dr. Joanna Chiu), and mouse anti-HSP70 (1:5000; Sigma-Aldrich). Secondary antibodies were conjugated with IRDye 800 (LI-COR Biosciences, Lincoln, NE, USA) and incubated at 1:10000. Blots were visualized with the Odyssey Infrared Imaging System (LI-COR Biosciences).
Protein levels of TIM, PER, and HSP70 were quantified by the software image studio for Odyssey. TIM and PER levels were normalized to HSP70. The value of the control genotype was set to 1. Student’s t test (Microsoft Excel, Seattle, WA, USA) was used to compare the differences between genotypes.
Immunostaining
Male flies were entrained for 3 days in LD and collected on DD1. Flies were anesthetized with CO2 and dissected in 3.7% formaldehyde diluted in phosphate-buffered saline (PBS). After fixing for 30 min at room temperature, the brains were rinsed 3 times in PBS and incubated in PBS with 1% Triton for 15 min at room temperature. The brains were then incubated with 5% goat serum diluted in PBT (PBS with 0.3% Triton) for 1 h at room temperature, followed by overnight incubation with 1:500 rabbit anti-PER (a gift from Dr. Michael Rosbash, Brandeis University, Waltham, MA, USA) and 1:50 mouse anti-PDF (Developmental Studies Hybridoma Bank, Iowa City, IA, USA) in PBT at 4°C. After 6 × 20-min PBT rinses, the brains were incubated with the secondary antibodies donkey anti-mouse AlexaFluor-594 (1:1000; Life Technologies) and donkey anti-rabbit AlexaFluor-488 (1:1000; Abcam, Cambridge, UK). After incubating overnight at 4°C, the brains were rinsed 6 × 20-min in PBS, and then mounted and imaged using an Olympus FV1000 confocal microscope with a 60× oil-immersion lens (Olympus, Tokyo, Japan). Images were acquired using the same settings (power, gain, and offset) for each experiment. The intensity of PER signals was quantified by ImageJ software. Student’s t test (Microsoft Excel) was used to compare the average intensity values between different genotypes.
Results
Knocking Down MAN1 in Adult Pacemaker Neurons Lengthens the Period of the Locomotor Rhythm
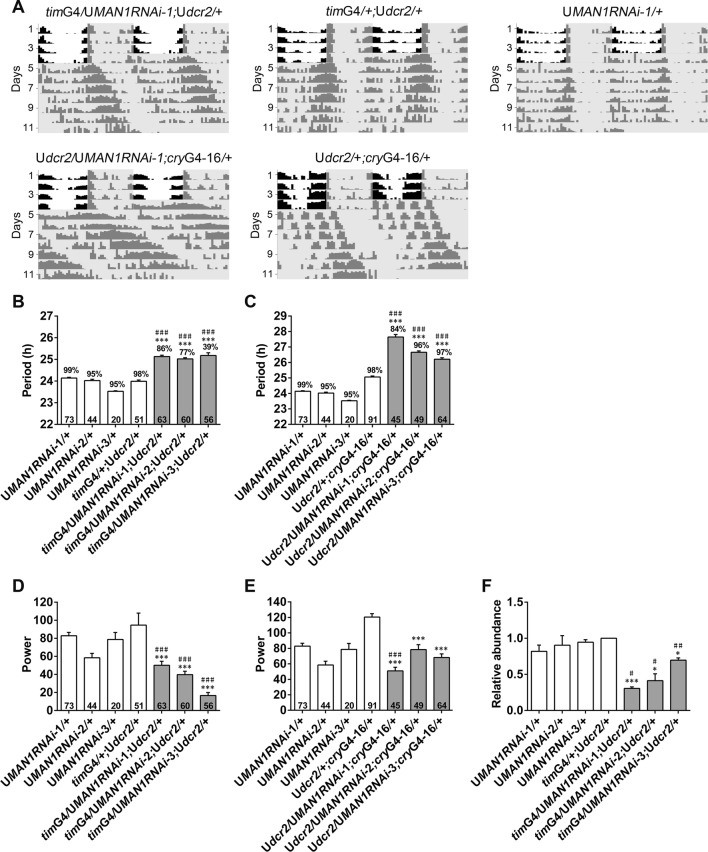
In a previous study, we demonstrated that both knocking down and over-expressing MAN1 in circadian neurons significantly lengthens the period of the locomotor rhythm under constant darkness (DD) [6]. Here, we used three RNAi lines (including two lines that were not tested in our previous study) to knock down MAN1 in all clock cells using a timGAL4 driver or mainly in circadian neurons using a cryptochrome (cry)GAL4-16 driver (Fig. S1) [13, 14]. We found a ~1 to > 2 h longer period of the DD locomotor rhythm in the RNAi flies, accompanied by a modest yet significant reduction in the power of the rhythm (Fig. 1A–E and Table 1). We then verified the knockdown efficiency of the RNAi (Fig. 1F). We also over-expressed MAN1 using cryGAL4-16 and found a > 1 h longer period as well as substantially reduced power of the rhythm, confirming our previous findings (Table S1) [6].
Fig. 1.
Knocking down MAN1 lengthens the period of the locomotor rhythm. A Double-plotted representative actograms of the indicated genotypes. Flies were monitored in LD for 4 days and then DD for 7 days. Gray shades indicate dark phases. B–E The period (B, C) and power (D, E) of the DD locomotor rhythm of flies with MAN1 knocked down and controls. Digits on the bars are the number of flies tested. Percentage of rhythmicity is indicated above the bars (P < 0.001, one-way ANOVA; ***/###P < 0.001, Tukey’s multiple comparison test). F Plots of relative mRNA abundance of MAN1 in whole head extracts of flies with MAN1 knocked down and controls determined by qRT-PCR (n = 3; */#P < 0.05, **/##P < 0.01, ***P < 0.001 * vs GAL4 controls, #vs UAS controls, Student’s t test). Error bars represent the standard error of the mean (SEM). G4, GAL4; U, UAS.
Table 1.
Knocking down MAN1 lengthens the period of the locomotor rhythm.
| Genotype | Period (h) ± SEM | Power ± SEM | %Rhythmic | n |
|---|---|---|---|---|
| timG4/UMAN1RNAi-1;Udcr2/+ | 25.13 ± 0.06***### | 50.17 ± 4.39***### | 86 | 63 |
| timG4/UMAN1RNAi-2;Udcr2/+ | 25.02 ± 0.05***### | 39.71 ± 3.88***### | 77 | 60 |
| timG4/UMAN1RNAi-3;Udcr2/+ | 25.18 ± 0.13***### | 16.69 ± 3.24***### | 39 | 56 |
| timG4/+;Udcr2/+ | 23.99 ± 0.06 | 94.69 ± 13.26 | 98 | 51 |
| Udcr2/UMAN1RNAi-1;cryG4-16/+ | 27.64 ± 0.16***### | 50.88 ± 4.75***### | 84 | 45 |
| Udcr2/UMAN1RNAi-2;cryG4-16/+ | 26.66 ± 0.09***### | 78.41 ± 6.37*** | 96 | 49 |
| Udcr2/UMAN1RNAi-3;cryG4-16/+ | 26.21 ± 0.10***### | 68.20 ± 4.63*** | 97 | 64 |
| Udcr2/+;cryG4-16/+ | 25.06 ± 0.06 | 120.49 ± 4.41 | 98 | 91 |
| UMAN1RNAi-1/+ | 24.14 ± 0.03 | 82.89 ± 3.68 | 99 | 73 |
| UMAN1RNAi-2/+ | 24.02 ± 0.06 | 58.46 ± 4.91 | 95 | 44 |
| UMAN1RNAi-3/+ | 23.53 ± 0.03 | 78.75 ± 7.69 | 95 | 20 |
One-way ANOVA, P < 0.001, Tukey’s multiple comparison test, ***P < 0.001 vs G4 and Udcr2; ###P < 0.001 vs URNAi. G4, GAL4; U, UAS.
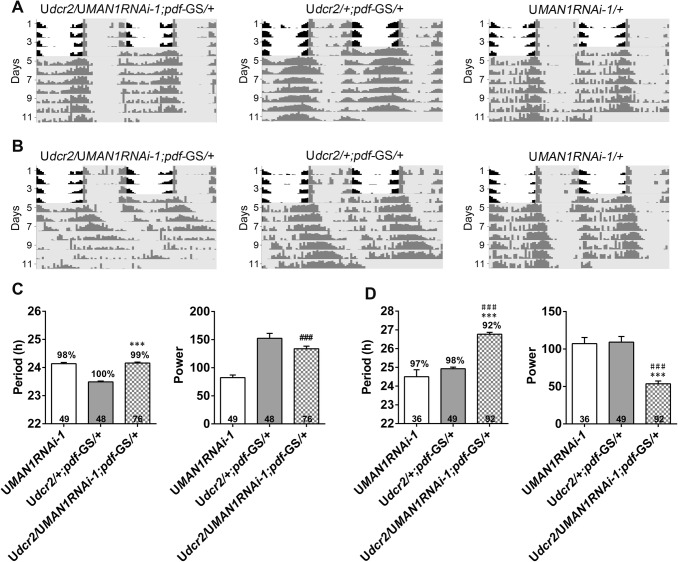
To test whether MAN1 functions in the adult circadian system, we used a gene-switch system to turn on RNAi expression specifically in the pigment-dispersing factor (PDF)-expressing pacemaker neurons of adult flies a few days before the start of behavioral monitoring [7]. We found that the period in drug-treated flies with RNAi expression activated was lengthened by close to 2 h, whereas the period in the vehicle-treated controls was not significantly altered (Fig. 2 and Table 2). Knocking down MAN1 in the pacemaker neurons of adults modestly reduced the power of the rhythm as well. We also over-expressed MAN1 in the adult pacemaker neurons but found no significant effect on the period or power (Table S2). Taken together, these results showed that knocking down MAN1 in the adult pacemaker neurons is sufficient to lengthen the period of the clock. Given that over-expressing MAN1 in adults did not appear to alter the locomotor rhythm, we suspected that the behavioral phenotypes observed in UASMAN1/+;cryGAL4-16/+ were due to developmental defects caused by over-expression. Therefore, for further investigations we focused on MAN1RNAi flies.
Fig. 2.
Knocking down MAN1 in adults lengthens the period of the locomotor rhythm. A, B Double-plotted representative actograms of the indicated genotypes. Flies treated with vehicle control (A) or RU486 (B) to activate the pdfG4-geneswitch (pdf-GS) driver. C, D The period and power of DD locomotor rhythms of flies with MAN1 knocked down and controls. Flies treated with vehicle control (C) or RU486 (D). Flies are monitored in LD for 4 days and then DD for 7 days. Gray shades indicate dark phases. Error bars represent SEM. Digits on the bars are the number of flies tested. Percentage of rhythmicity is indicated above the bars (P < 0.001, one-way ANOVA; ***/###P < 0.001, * vs GAL4 controls; # vs UAS controls, Tukey’s multiple comparison test). G4, GAL4; U, UAS.
Table 2.
Knocking down MAN1 in adults lengthens the period of the locomotor rhythm.
| Genotype | Period (h) ± SEM | Power ± SEM | %Rhythmic | n |
|---|---|---|---|---|
| Ethanol-treated | ||||
| Udcr2/+;pdf-GS/+ | 23.49 ± 0.03 | 152.41 ± 8.75 | 100 | 48 |
| UMAN1RNAi-1/+ | 24.14 ± 0.04 | 82.40 ± 4.53 | 98 | 49 |
| Udcr2/UMAN1RNAi-1;pdf-GS/+ | 24.16 ± 0.03*** | 133.76 ± 4.63### | 99 | 76 |
| pdf-GS/+ | 24.53 ± 0.05 | 134.92 ± 7.82 | 100 | 31 |
| UMAN1/+ | 23.56 ± 0.03 | 76.89 ± 6.23 | 96 | 45 |
| UMAN1/+;pdf-GS/+ | 23.50 ± 0.07 | 150.68 ± 11.94### | 100 | 11 |
| Drug-treated | ||||
| Udcr2/+;pdf-GS/+ | 24.93 ± 0.09 | 109.22 ± 7.52 | 98 | 49 |
| UMAN1RNAi-1/+ | 24.50 ± 0.37 | 107.24 ± 8.09 | 97 | 36 |
| Udcr2/UMAN1RNAi-1;pdf-GS/+ | 26.77 ± 0.10***### | 53.59 ± 3.65***### | 92 | 92 |
| pdf-GS/+ | 24.94 ± 0.08 | 80.53 ± 5.49 | 96 | 50 |
| UMAN1/+ | 23.74 ± 0.05 | 101.28 ± 5.79 | 98 | 52 |
| UMAN1/+;pdf-GS/+ | 24.92 ± 0.08### | 88.54 ± 5.35 | 91 | 87 |
One-way ANOVA, P < 0.001, Tukey’s multiple comparison test, ***P < 0.001 vs G4, ###P < 0.001 vs UAS. Drug-treated, flies treated with RU486 after eclosion; ethanol-treated, flies treated with ethanol instead of RU486 after eclosion. G4, GAL4; U, UAS.
Knocking Down MAN1 Reduces per mRNA and Protein Levels
We next sought to characterize the mechanism by which MAN1 sets the pace of the clock by examining the effects of knocking down MAN1 on clock gene expression. Previously, we showed that silencing MAN1 reduces the transcription of BMAL1 in human cell lines but does not alter the mRNA level of cyc (the Drosophila ortholog of BMAL1) in fly heads [6]. Here, we found that knocking down MAN1 substantially reduced the per mRNA level and increased clk mRNA level (Fig. 3A). To further confirm this, we measured the protein level of PER and found it to be decreased, consistent with reduction in the mRNA level (Fig. 3B). On the other hand, TIM, the binding partner of PER, was not significantly changed (Fig. 3C) [15]. Because the period of the locomotor rhythm under DD is believed to be determined by the small ventral lateral neurons that express PDF (sLNvs), we assessed PER in these cells and found a significant reduction of the PER protein level at least at Circadian Time 20 (CT20; CT0 was defined as subjective lights-on time) (Fig. 3D,E) [16].
Fig. 3.
Knocking down MAN1 reduces the mRNA and protein levels of per.A Plots of relative mRNA abundance versus CT for clock genes determined by qRT-PCR in whole head extracts of MAN1 RNAi (timG4/UMAN1RNAi;Udcr2/+) and control (timG4/+;Udcr2/+) flies collected on DD1 (n ≥ 3). For each time series, the value of the lowest time point was set to 1. B, C Western blots of proteins from whole head extracts of MAN1RNAi and control flies collected on DD1 and probed with PER and TIM antibodies (left). Quantification of PER and TIM protein levels (right, n = 4). PER and TIM protein levels were normalized to that of HSP70. For each time series, the value of the control at the peak time point was set to 1. D Brains from Udcr2/+;cryG4-16/+ and Udcr2/UMAN1RNAi;cryG4-16/+ flies collected at CT 0 and 20 on DD1 immunostained with PER (green) and PDF (red) antisera. Scale bar, 10 µm. E Quantification of PER protein levels in the s-LNvs of images as in (D) (n = 72–80). Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. G4, GAL4; U, UAS.
per has an Epistatic Effect on MAN1 in Period Length Determination
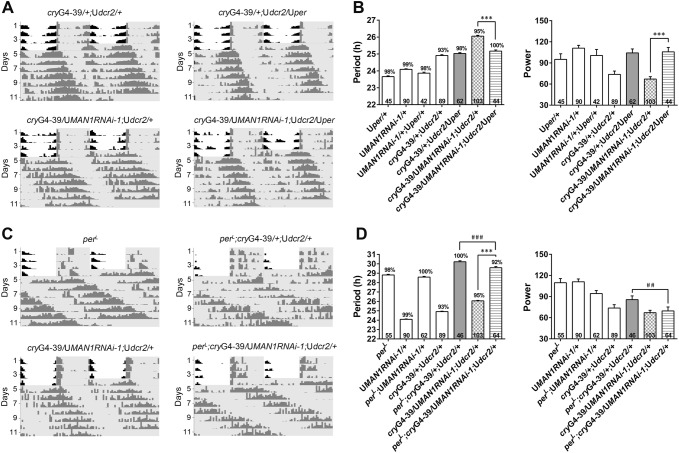
Based on our findings, we proposed that MAN1 times the clock by regulating per and/or clk mRNA levels. To test this hypothesis, we first over-expressed per in flies with MAN1 knocked down and found that this significantly reverted the long period phenotype, whereas over-expressing per in control flies did not significantly affect the period (Fig. 4A, B and Table 3) [8]. On the other hand, knocking down MAN1 on a perL mutant background did not lead to further lengthening of the period (Fig. 4C, D and Table 3) [9]. These results demonstrated that when per is either over-expressed or mutated, the influence of MAN1 on period length is blocked, indicating an epistatic effect of per on MAN1. In combination with the molecular changes, we propose that MAN1 sets the pace of the clock via per.
Fig. 4.
per has an epistatic effect on MAN1 in period length determination. A, C Double-plotted representative actograms of the indicated genotypes. Flies were monitored in LD for 4 days and then DD for 7 days. Gray shades indicate dark phases. B The period and power of the DD locomotor rhythm of MAN1RNAi flies over-expressing per. D The period and power of the DD locomotor rhythm of MAN1RNAi flies carrying the perL mutation. Error bars represent SEM. Digits on the bar are the number of flies tested. Percentage of rhythmicity is indicated above the bars (P < 0.001, one-way ANOVA, **/##P < 0.01, ***/###P < 0.001, * vs MAN1-RNAi flies, #vs over-expression or mutant flies, Tukey’s multiple comparison test). G4, GAL4; U, UAS.
Table 3.
Genetic interaction between MAN1 and per.
| Genotype | Period (h) ± SEM | Power ± SEM | %Rhythmic | n |
|---|---|---|---|---|
| Uper/+ | 23.66 ± 0.05 | 95.10 ± 7.92 | 98 | 45 |
| UMAN1RNAi-1/+ | 24.09 ± 0.03 | 111.00 ± 3.99 | 99 | 90 |
| UMAN1RNAi-1/+;Uper/+ | 23.87 ± 0.06 | 100.63 ± 8.51 | 98 | 42 |
| cryG4-39/+;Udcr2/+ | 24.91 ± 0.05 | 73.76 ± 4.68 | 93 | 89 |
| cryG4-39/+;Udcr2/Uper | 25.02 ± 0.05 | 104.29 ± 5.58 | 98 | 62 |
| cryG4-39/UMAN1RNAi-1;Udcr2/+ | 26.06 ± 0.05+++††† | 67.12 ± 3.37††† | 95 | 103 |
| cryG4-39/UMAN1RNAi-1;Udcr2/Uper | 25.17 ± 0.07*** | 105.72 ± 6.29*** | 100 | 44 |
| perL | 28.81 ± 0.07 | 109.66 ± 5.96 | 98 | 55 |
| perL;UMAN1RNAi-1/+ | 28.59 ± 0.08 | 94.23 ± 4.37 | 100 | 62 |
| perL;cryG4-39/+;Udcr2/+ | 30.22 ± 0.11 | 85.63 ± 5.37 | 100 | 46 |
| perL;cryG4-39/UMAN1RNAi-1;Udcr2/+ | 29.58 ± 0.12***### | 69.64 ± 5.58## | 92 | 64 |
One-way ANOVA, P < 0.001, Tukey’s multiple comparison test, ***P < 0.001 vs MAN1RNAi-1 flies; ##P < 0.01, ###P < 0.001 vs perL;cryG4-39/+;Udcr2/+ flies; +++P < 0.001 vs cryG4-39/+;Udcr2/+ flies; †††P < 0.001 vs UMAN1RNAi-1/+ flies. G4, GAL4; U, UAS.
We also tested for genetic interactions between MAN1 and clk. Over-expressing clk slightly shortened the period, and knocking down MAN1 in flies over-expressing clk did not substantially lengthen the period, indicating that over-expressing clk also has an epistatic effect on MAN1 (Table S3) [10]. Given that CLK activates the transcription of per, it is possible this epistatic effect is caused by enhanced per [11, 17]. The clkjrk/+ mutation, on the other hand, leads to a very poor rhythm and thus it is not possible to determine whether a genetic interaction exists between clkjrk and MAN1 (Table S3) [11].
Knocking Down MAN1 Reduces per Pre-mRNA Levels
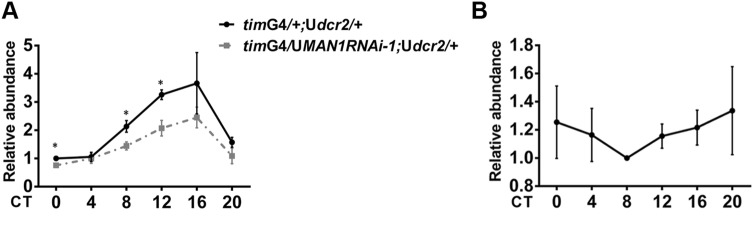
Since genetic interaction analysis strongly suggested that MAN1 determines the period by regulating per, we next addressed how MAN1 acts on per. As knocking down MAN1 decreased the per mRNA level, we assayed the pre-mRNA level of per in these flies, which reflects the status of per transcription. We found that the per pre-mRNA level was significantly reduced in MAN1RNAi flies, indicating that MAN1 functions to promote per transcription (Fig. 5A). Furthermore, to test whether MAN1 serves as a permissive or instructive signal, we examined the temporal expression profile of MAN1. We did not find significant changes in MAN1 mRNA levels throughout the day, demonstrating that MAN1 is not circadian-regulated at the transcription level (Fig. 5B).
Fig. 5.
Knocking down MAN1 reduces the pre-mRNA level of per. A Plots of relative pre-mRNA abundance of per determined by qRT-PCR in whole head extracts of MAN1RNAi (timG4/MAN1RNAi;Udcr2/+) and control (timG4/+;Udcr2/+) flies during DD1 (n = 4; *P < 0.05, Student’s t test). B Plots of relative mRNA abundance of MAN1 in whole head extracts of w1118 flies in DD1 determined by qRT-PCR (n = 3; P > 0.05, one-way ANOVA).
Discussion
Previously, we showed that MAN1 binds to and promotes the transcription of the core clock gene BMAL1 (the mammalian ortholog of Drosophila cyc) in human U2OS cells, while over-expressing MAN1 in flies enhances cyc mRNA levels and knocking down MAN1 does not alter the cyc level [6]. We proposed that MAN1 uses a conserved mechanism to regulate the clock by promoting the transcription of BMAL1/cyc. However, we found that the circadian phenotypes associated with MAN1 over-expression were likely due to developmental defects rather than a relatively specific and direct effect of MAN1 on the clock, as over-expressing MAN1 in adult flies did not result in significant changes in circadian behavior. On the other hand, knocking down MAN1 in adults led to a substantially lengthened period, which means that MAN1RNAi flies are an appropriate model in which to study the role of MAN1 in the clock.
Molecular analysis has revealed that knocking down MAN1 reduces per while increasing the clk mRNA levels. We confirmed the altered expression of per at the protein level and intended to do the same for clk, but unfortunately we do not have a CLK antibody that works well. However, we believe MAN1 is more likely to act directly on per rather than clk to determine period length. First of all, over-expressing MAN1 leads to a trend of increased per mRNA, opposite to the effect of knocking down MAN1 that we found here [6]. As for clk, both over-expressing and knocking down MAN1 led to elevated levels, implying an indirect modulation. Second, downregulation of PER levels are associated with a lengthened period, similar to the phenotype of MAN1RNAi, whereas upregulated clk shortens the period [10, 18]. Third, based on our current understanding of how the clock operates, if an increase in clk is able to influence the period length, it should lead to an increase of per/tim transcription and subsequently their mRNA levels [2]. However, we found significantly less per mRNA in MAN1RNAi flies, which would be unexpected if CLK function is indeed enhanced. What is more, over-expressing per specifically rescued the long-period phenotype of MAN1RNAi, while perL mutation had an epistatic effect on MAN1. It is also possible that knocking down MAN1 does not further lengthen the period of perL flies is the result of a “ceiling effect”, but in our opinion this is less likely because we have unpublished results demonstrating that genetically modulating certain genes can lengthen the period of perL by an additional ~3 h. Therefore, we believe knocking down MAN1 in the presence of perL does not reach the limit of the clock timing mechanism, and the failure to further lengthen the period indeed reflects true genetic interactions. Based on these findings, we propose that MAN1 sets the pace of the clock by promoting per expression.
Previous studies have demonstrated an important role for MAN1 in transcriptional regulation [19]. In line with this, we found that knocking down MAN1 reduced per transcription, although it remains to be further determined whether MAN1 binds directly to the per locus to regulate transcription or indirectly by modulating other factors that in turn control per transcription. It has been shown that circadian genes are rhythmically recruited to the NE where they interact with lamin-associated domains and their transcription is attenuated by histone methylation events [5]. Given the role of MAN1 in clock regulation, it is possible that MAN1 participates in modulating transcription when circadian genes such as per move to the nuclear periphery.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31471125 and 31671215) and “1000 Talents” Program of China. We thank Dr. Joanna Chiu, Dr. Ravi Allada, Dr. Michael Rosbash, Dr. Yong Zhang, and Dr. Fang Guo for kindly providing fly stocks and reagents.
Conflict of interest
The authors declare no competing interests.
References
- 1.Li S, Zhang L. Circadian Control of Global Transcription. Biomed Res Int. 2015;2015:187809. doi: 10.1155/2015/187809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stancheva I, Schirmer EC. Nuclear envelope: connecting structural genome organization to regulation of gene expression. Adv Exp Med Biol. 2014;773:209–244. doi: 10.1007/978-1-4899-8032-8_10. [DOI] [PubMed] [Google Scholar]
- 4.Mattout-Drubezki A, Gruenbaum Y. Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell Mol Life Sci. 2003;60:2053–2063. doi: 10.1007/s00018-003-3038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Sifakis EG, Sumida N, Millan-Arino L, Scholz BA, Svensson JP, et al. PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell. 2015;59:984–997. doi: 10.1016/j.molcel.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Lin ST, Zhang L, Lin X, Zhang LC, Garcia VE, Tsai CW, et al. Nuclear envelope protein MAN1 regulates clock through BMAL1. Elife. 2014;3:e02981. doi: 10.7554/eLife.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol. 2011;21:1783–1793. doi: 10.1016/j.cub.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(SICI)1096-9861(20000619)422:1<66::AID-CNE5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, et al. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/S0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 11.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/S0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 12.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 13.Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/S0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 14.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/S0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 15.Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, et al. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 16.Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 17.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 18.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, et al. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bengtsson L. What MAN1 does to the Smads TGFbeta/BMP signaling and the nuclear envelope. Febs J. 2007;274:1374–1382. doi: 10.1111/j.1742-4658.2007.05696.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.