Abstract
Non-structural protein 4 (nsP4) polymerase of chikungunya virus (CHIKV) has a crucial role in genome replication and hence could act as a promising target for novel therapeutics. Though, nsP4 is important in viral life cycle, but it is less explored as therapeutic target. The catalytic core of nsP4 Polymerase includes conserved GDD motif which is present not only across different CHIKV strains but also across other Alphaviruses. This emphasizes the uniqueness and importance of this motif in the functioning of nsP4 polymerase and hence, we focused on GDD motif for docking of drug molecules. Herein, a model of nsP4 polymerase was developed using Swiss Model, validated by Ramachandran plot and molecular dynamic simulation. Molecular docking was performed using LeadIT FlexX flexible docking module with FDA approved drug molecule library. On the basis of flexX score, top 5 leads with flexX scores − 33.7588, − 30.2555, − 29.6043, − 28.916 and − 28.5042 were selected. The bonding pattern of these leads were analysed in discovery studio and were further screened on the basis of molecular dynamic simulation studies. Simulation analysis revealed that only the top lead, Mitoxantrone Hydrochloride which is an anticancer drug and is currently indicated in leukemias and lymphomas interacted favourably and stably with nsP4. Our findings suggest that Mitoxantrone Hydrochloride can be a potential novel inhibitor of CHIKV polymerase and should be further validated by in vitro assays.
Electronic supplementary material
The online version of this article (10.1007/s13337-019-00547-0) contains supplementary material, which is available to authorized users.
Keywords: Catalytic triad, Chikungunya virus (CHIKV), Molecular docking, RNA dependent RNA polymerase
Introduction
Of late, Chikungunya fever’s devastating outbreaks have been reported worldwide. The Chikungunya virus (CHIKV) belongs to genus Alphavirus of Togaviridae family and is transmitted through Aedes aegypti [1]. The first outbreak of CHIKV occurred in Tanzania in 1953 and later on reached to Asian countries including Bangladesh, India, China, Indonesia, Sri Lanka, Thailand and Bhutan [7]. It also reached Europe, where outbreaks were reported in Italy, France, Croatia and Madeira [19] and outbreaks were also reported from African countries most likely due to travelling and shipping [7]. A sudden onset of fever (lasting from few days to 2 weeks), swelling and polyarthralgia which may last as long as a year, are the elementary manifestation of CHIKV fever [24].
CHIKV genome codes for four non-structural proteins (nsPs) i.e. nsP1, nsP2, nsP3, nsP4 and five structural proteins i.e. capsid (C), envelope glycoprotein E1, E2, E3 and 6k [7, 11, 17, 21]. nsPs play a key role in the intracellular replication of CHIKV [17]. Inside the host cell, nascent single polypeptide P1234 is formed which is then cleaved to mature nsP4 and precursor P123 replication complex, which is further processed to produce nsp1 and P23 complex and finally the cleavage of P23 results in mature nsP1, nsP2, nsP3, nsP4 [11, 22, 25]. nsP1 possess viral capping, methyltransferase and guanyltransferase activity. Multifunctional nsP2 protein has protease activity, RNA helicase activity and RNA triphosphatase activity. C-terminal of macrodomian of nsP3 is mainly involved in binding with host cellular proteins. nsP4 exhibits RNA dependent RNA polymerase (RdRp) activity which is responsible for transcription and genome replication of CHIKV [2, 7].
Approximately 100 residues from N-terminal of nsP4 are variable [25] while C-terminal (approximately 500 amino acids) form highly conserved polymerase domain all through alphaviruses. The catalytic core of polymerase constitutes GDD motif C (Gly314, Asp315 and Asp316), responsible for the polymerase activity of nsP4 [15, 25]. Additionally, divalent metal ions like magnesium cations which act in catalysis and are required for the polymerase activity are coordinated by conserved aspartate residues of GDD motif [3, 25]. Mutation in GDD motif to SDD in influenza virus, to GDN in Mononegavirales and in poliovirus polymerase completely abolish the polymerase activity indicating the significance of this domain [25]. Along with GDD motif, Ser279, Gly280, Thr284, and Asn288 are highly conserved among different genotypes of CHIKV and even other alphaviruses like Semliki forest (SFV), Sindbis virus (SINV) [9] etc. demonstrating significance of these residues in structural stability and/or functioning of nsP4 polymerase.
Several molecules having the inhibitory effect on CHIKV replication have been reported but few studies were documented on nsP4 as a target. Benzimidazole-related compound [26], ribavirin, diketo acid derivative [15], hesperetin-a flavanoid derivatives [17], compound A [2] and favipiravir [9] are some compounds already reported in the literature exhibiting anti-CHIKV nsP4 activity. But all of these molecules are still at the experimental level as far as their use in CHIKV intervention is concerned. Some of these molecules are reported to have activity against other viruses also suggesting repurposing of these drugs as anti CHIKV may become a possibility in near future. Current indications of some of these reported CHIKV-nsP4 inhibitors are: (1) favipiravir is reported to inhibit replication of influenza virus. In Japan favipiravir is approved for use under certain conditions i.e. either if the virus become resistant to other antiviral or if the patient has pandemic influenza virus infection. In USA favipiravir is currently undergoing late phase III clinical trials where two phase III studies have been completed. The results of these clinical trials are not available in public domain [14]. Different classes of flavonoids possessing antiviral activity have been reported against Dengue Virus (DENV), Herpes Simplex Virus (HSV), Human Cytomegalovirus (HCMV) etc. (2) hesperetin has antioxidant, anti-inflammatory, anti-allergic and anti-carcinogenic activity and (3) ribavirinhas been shown to inhibit the replication of SFV [2] and its combinations with other compounds and/or interferons have been used to treat hepatitis C and viral hemorrhagic fevers [4] including lassa and hanta fever. Drug repositioning therefore has a great advantage over traditional approaches to drug discovery, as it significantly reduces the cost and time of drug development [5]. The existing drugs have well approved and established formulations, manufacturing methods, extensive absorption distribution, metabolism, excretion and toxicity (ADMET) data as well as clinical trial safety data. Lots of drugs have been repurposed like colesevelam was originally used to reduce low-density lipoprotein cholesterol but now repurposed to treat type 2 diabetes mellitus [18]. Another example of drug repurposing is aspirin, initially discovered as an analgesic and later on indicated for the treatment of cardiovascular disease but recent studies demonstrated its anti-tumor activity [13]. The number of repositioned drugs has steady increased over last few years.
As nsP4 is a largely less explored but important protein for the replication of CHIKV, which has become a global threat in recent times. Therefore in the current study we focused on the in silico screening of potential inhibitors of nsP4 combined with drug repurposing strategy in order to move towards effective CHIKV therapeutics promptly.
Material methods
Sequence retrieval, homology modelling and validation of nsP4
The polyprotein sequence of nsP4 of CHIKV IND-06-Guj strain was retrieved from NCBI GenBankID ADZ47898.1 in FASTA format. The polymerase part constitutes 460 amino acids from 152 to 611 amino acids of complete nsP4. It was submitted for homology modelling using Swiss model, as no PDB structure is available for this protein. The model was generated based on template identification, then aligning the target sequence and the template sequence followed by model building. The quality of model was evaluated by Q mean Z-score. The top best 3 models of nsP4 polymerase were generated and each model was validated by Ramachandran Plot. The best fit model was selected based on Q mean Z-score and Ramachandran plot validation for further study. Furthermore, the quality of selected model was also checked by ProCheck, ProQ and the ERRAT.
Sequence alignments, receptor preparation and molecular docking
Through exhaustive literature search crucial and conserved residues were delineated in the region of interest of nsP4. The conservation of selected residues was marked in different genotypes of CHIKV through multiple sequence alignments using clustal omega and also the selected CHIKV genotype was aligned with human polymerases to look for exclusive presence of these residues in viral polymerases. The generated nsP4 model was uploaded in the LeadIT FlexX flexible docking module which is fully automated tool used for the molecular docking studies. Pocket 1 and pocket 2 were generated around the GDD motif and other conserved residues respectively within a sphere of 8 Å. Computational docking of 2924 FDA approved molecules which is one of the subset of ZINC drug database (http://zinc.docking.org/) [12] was performed on the selected pockets. The module executes high throughput virtual screening process in which it predicts the geometry of the complex with their corresponding binding energies and flexX scores. It completely relies on the principle of incremental construction algorithm and also provides 2D protein–ligand interaction map using Pose View tool present in the module. During docking, the flexible ligand is first disintegrated into fragments and then incrementally built into the active site using different placement strategies. It generates a number of poses for a particular ligand and selects upto 10 best poses. In this software, the flexX score was calculated according to their binding pose and potential interactions with key residues [6, 23]. After docking, the top leads were selected according to the flexX scores and the interactions were analyzed using pose view. Further detailed analysis of protein–ligand interaction for selected leads was done by using PyMol, Discovery Studio and molecular dynamic simulations.
Molecular dynamic simulation
MD simulation (MDS) allows exploring in-depth analysis of protein. Stability of modelled protein and its interaction with top selected leads were validated using MDS studies in Gromacs. GROMAC 5.0.7 package with OPLS force filed was used to perform MDS to stabilize the modelled protein as well as the docked protein complex by minimizing their energy. The structures were solvated using spc216.gro in which 11 chloride ions were added.
Results and discussion
Homology modelling of nsP4 polymerase and validation of structure
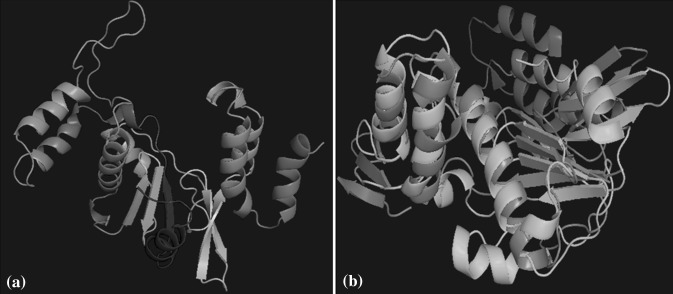
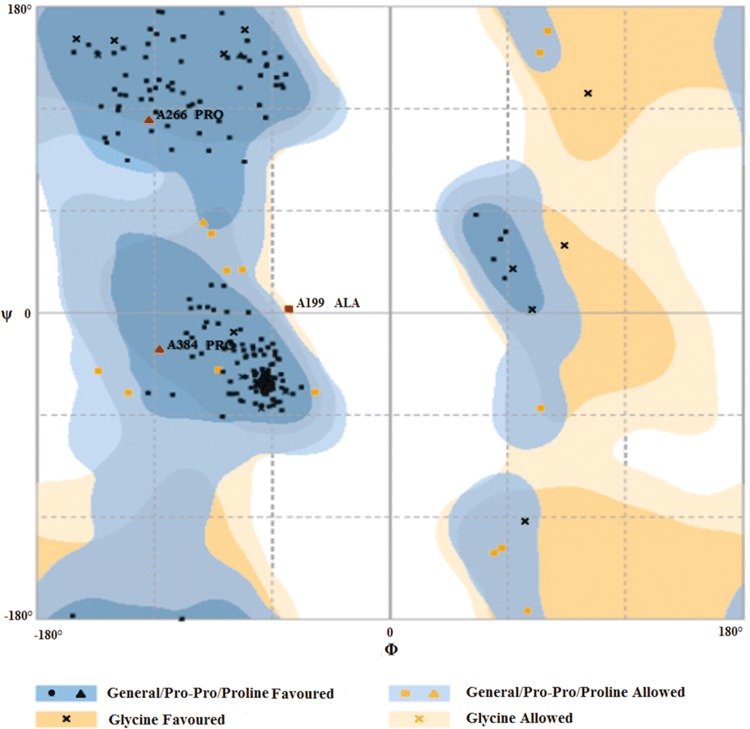
Structure of nsP4 polymerase of CHIKV was developed using I304 V polymerase mutant of Encephalomyocarditis virus (EMCV) as a template on Swiss model. The template and model were selected based on the Qmean Z-score. The generated model (Fig. 1a) has the highest Z-score among all other models and was validated by Ramachandran plot (using Rampage, PDBSum and Procheck). 92.4% residues fell into favoured region, 6.2% residues in allowed region and 1.3% residues fell into outlier region (Fig. 2). According to Rampage, 92.4% residues fell into favoured region, 6.2% residues in allowed region and 1.3% residues fell into outlier region. Moreover according to Procheck, 87.8% residues were present in most favoured regions and 1.5% residues were in disallowed regions. The quality of protein model was evaluated by ProQ software, which predicts the generated models as a fairly good model. Additionally, ERRAT was also performed in which the overall quality factor is 67.757. Although the result of ERRAT program was not favourable but still the model was considered for study as rest of the parameters from other software tools were considered promising good quality model.
Fig. 1.
Structure of nsP4 polymerase. a Swiss model generated tertiary structure of nsP4 polymerase b Superimposition of template (I304 V polymerase mutant of EMCV) with the CHIKV nsP4 polymerase. Cyan colour indicates the sequence of CHIKV polymerase and the green colour denotes the template sequence (colour figure online)
Fig. 2.
Ramachandran plot analysis of the nsp4 model using RAMPAGE
The modelled structure was then superimposed with the template structure. Though the entire structure did not superimpose completely with the template but there was a perfect superimposition of the region containing selected pockets (Fig. 1b) suggesting that this region can be safely used for docking studies. MDS demonstrates that RMSD of protein structure is about 0.3 nm, indicating stable nsP4 model structure and hence can be considered for docking studies.
Identification of pocket and molecular docking
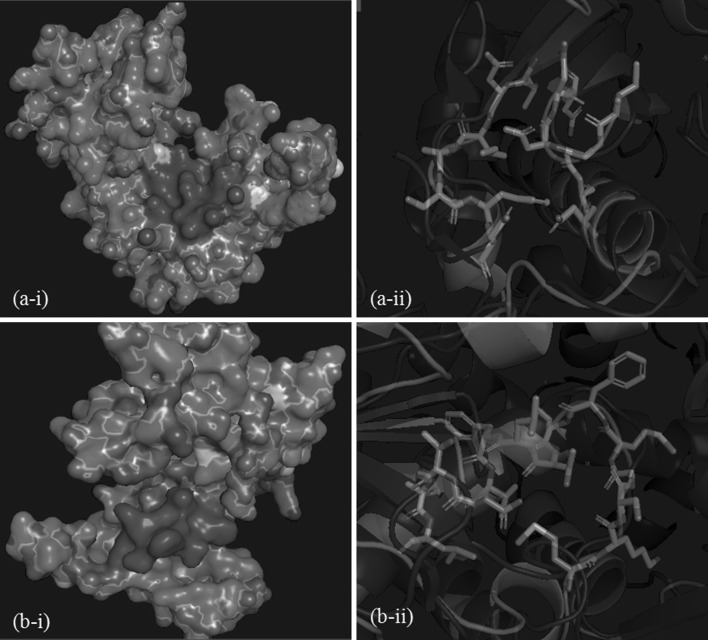
Pocket 1 comprises of 14 residues which include GDD motif at the position 314, 315 and 316 (Fig. 3a; Table 1) while pocket 2 comprises of 15 residues including Ser279, Gly280, Thr284 and Asn288 that are conserved among CHIKV, SFV, SINV, hepatitis C virus (HCV), West Nile virus (WNV), murine norovirus (MNV) and poliovirus(Fig. 3b; Table 1). Asn288 being present in both the pockets. Docking was performed onto generated pockets and the lowest flexX score for pocket 1 and pocket 2 was − 33.7588 and − 20.8165 respectively. Lower the flexX scores, strong and stable is the ligand-receptor interaction. We observed good flexX scores for pocket 1 as it is deeply seated with free side chains but pocket 2 did not show good flexX scores as it is shallow and has less interacting free side chain. Therefore, we selected top leads only from pocket 1. Mitoxantrone Hydrochloride, Spectinomycin, diminazene aceturate, Imatinib and Cefadroxil are the top selected leads from pocket 1 with flexX scores − 33.7588, − 30.2555, − 29.6043, − 28.916 and − 28.5042 respectively. None of these molecules are reported as potential inhibitors of CHIKV particularly nsp4 in the available literature [8, 15]. Mitoxantroneis an anticancer drug widely used to treat leukemias and lymphomas through inhibition of the activity of topoisomerase II enzyme which is essential for the repair of damaged DNA [10, 16]. Mitoxantrone hydrochloride is an FDA approved drug for multiple sclerosis and is also used for leukemia and prostate cancer in combination with other drugs. Spectinomycin and Cefadroxil has antibacterial activity while diminazene aceturate is used for the treatment of protozoal diseases. Imatinib is an anticancer drug, used to treat certain tumors and act as an inhibitor of tyrosin kinase enzyme. Imatinib is already under patent in US for its second medical use in the treatment of hepatitis and other liver disorders and is also shown to inhibit replication of herpes virus, poxvirus, influenza virus, parainfluenza virus, respiratory syncytial virus, rhinovirus, yellow fever virus, west nile virus and encephalitis virus [20]. Current therapeutic indication of each lead with their ZINC ID is given in Table 2.
Fig. 3.
Representation of generatedpockets. a-i and b-i are surface view (magenta colour) of pocket 1 and pocket 2 respectively. a-ii and b-ii are stick view of pocket 1 and pocket 2 respectively against cartoon view of superimposed protein structures from Fig. 1b
Table 1.
Residues of pocket 1 and pocket 2 of nsp4 polymerase
| Pockets for docking | Amino acids |
|---|---|
| Pocket 1 | THR 219, ASP220, ILE221, ALA222, SER223, PHE224, ASP225, ASN288, ASN292, ILE313, GLY314, ASP315, ASP316, ASN317 |
| Pocket 2 | MET277, LYS278, SER279, GLY280, MET281, PHE282, LEU283, THR284, LEU285, PHE286, VAL287, ASN288, THR289, LEU290, LEU291 |
Table 2.
Selected leads with their ZINC ID and current therapeutic use
| S. no. | ZINC I.D | Compound name | Molecular formula | Therapeutic use |
|---|---|---|---|---|
| 1. | ZINC03794794 | Mitoxantrone hydrochloride | C22H30Cl2N4O6 | Antineoplastic |
| 2. | ZINC03831468 | Spectinomycin | C14H24N2O7 | Antibacterial |
| 3. | ZINC03830706 | Diminazeneaceturate | C22H29N9O6 | Antiparasitic |
| 4. | ZINC19632618 | Imatinib | C29H31N7O | Anticancer |
| 5. | ZINC33943584 | Cefadroxil (Sumacef) | C16H17N3O5S | Antibacterial |
The protein–ligand interaction patterns focuses on the importance of the selected conserved residues as all of them are mainly involved in making strong salt bridge, electrostatic and hydrogen bonds with the ligands. Asp315 and Asp316 of GDD domain mainly involved in making strong bonds with the leads (Table 3). Mitoxantrone hydrochloride formed one salt bridge, three electrostatic bonds, five conventional hydrogen bonds, eight carbon hydrogen bonds and two other bonds with different residues within the docking site, primarily interacting with Asp315 of GDD domain (Table 3a; supplementary Fig. 1). Spectinomycin exhibits two salt bridge and 4 hydrogen bonds with the residues within the pocket and bonded with Asp315 and Asp316 of GDD motif with one salt bridge and one conventional hydrogen bond each (Table 3b; supplementary Fig. 2), Diminazene aceturate formed six polar interactions in all including one electrostatic bond with Asp 316 and one Pi-anion electrostatic bond with Asp315 of GDD motif (Table 3c; supplementary Fig. 3), whereas, Imatinib, another potential lead docked successfully within the pocket and interacted with Asp 316 and Asp315 of GDD forming salt bridge and electrostatic bond within the docking site (Table 3d; supplementary Fig. 4). The last lead cefadroxil exhibits salt bridge and conventional hydrogen bond with Asp315 and Asp316 (Table 3e; supplementary Fig. 5). Moreover, it is observed that the bond length between the leads and the residues are mostly short (less than 3 Å) indicating strong bond strength. Asp315 and Asp316 of GDD domain are most likely available residues which exhibits favourable interaction with the leads. It is clearly evident in our study that the residues of GDD motif play an important role as most of the leads form strong stable bonds with DD in the pocket as is also reported earlier [15]. Moreover since we used a library of FDA approved drugs for which the toxicity data is already approved, we do not expect harsh side effects unless effective dose are calculated to be too high after in vitro and in vivo validations of selected leads.
Table 3.
Analysis of type of bond and bond length between the selected lead and pocket residues
| Residues | Bond type | Bond length (Å) | Residues | Bond type | Bond length (Å) |
|---|---|---|---|---|---|
| (a) Mitoxantrone Hydrochloride | (b) Spectinomycin | ||||
| ASP315 | Salt bridge hydrogen bond | 1.99022 | ASP316 | Salt bridge hydrogen bond | 2.04442 |
| ASP220 | Electrostatic bond | 4.53851 | ASP315 | Salt bridge hydrogen bond | 1.87122 |
| ASP225 | Electrostatic bond | 2.94717 | ASP315 | Conventional hydrogen bond | 2.10764 |
| ASP315 | Electrostatic bond | 4.55126 | ASP316 | Conventional hydrogen bond | 1.78343 |
| ASP315 | Conventional hydrogen bond | 2.04657 | ILE221 | Conventional hydrogen bond | 3.2643 |
| ASP315 | Conventional hydrogen bond | 1.77048 | ASP220 | Carbon hydrogen bond | 2.61277 |
| ILE221 | Conventional hydrogen bond | 3.20613 | (c) Diminazeneaceturate | ||
| SER223 | Conventional hydrogen bond | 2.8178 | ASP220 | Salt bridge hydrogen bond | 1.60495 |
| ASP225 | Conventional hydrogen bond | 2.97774 | GLU218 | Electrostatic bond | 5.42497 |
| ILE221 | Carbon hydrogen bond | 3.03815 | ASP316 | Electrostatic bond | 3.01854 |
| ASP315 | Carbon hydrogen bond | 3.09365 | ASP225 | Electrostatic bond | 5.34922 |
| ILE221 | Carbon hydrogen bond | 3.07513 | ILE221 | Conventional hydrogen bond | 2.18196 |
| ASP220 | Carbon hydrogen bond | 3.05238 | ASP315 | Pi-anion electrostatic bond | 2.5895 |
| ASP225 | Carbon hydrogen bond | 1.81343 | ILE221 | Pi-alkyl hydrophobic bond | 5.16513 |
| ASP315 | Carbon hydrogen bond | 2.50143 | (d) Imatinib | ||
| GLY314 | Carbon hydrogen bond | 2.76843 | ASP316 | Salt bridge hydrogen bond | 1.72328 |
| SER223 | Pi-donor hydrogen bond | 3.23261 | ASP316 | Salt bridge hydrogen bond | 2.03976 |
| LYS226 | Pi-alkyl hydrophobic bond | 5.1176 | ASP315 | Electrostatic bond | 4.91715 |
| (e) Cefadroxil | ASP220 | Conventional hydrogen bond | 1.62601 | ||
| ASP315 | Salt bridge hydrogen bond | 1.91205 | SER223 | Conventional hydrogen bond | 2.70859 |
| ASP220 | Electrostatic bond | 4.31123 | ASP225 | Conventional hydrogen bond | 2.94526 |
| ASP315 | Conventional hydrogen bond | 1.88675 | ILE313 | Carbon hydrogen bond | 2.22606 |
| ASP316 | Conventional hydrogen bond | 2.13024 | ASP315 | Pi-anion electrostatic bond | 4.5074 |
| ILE221 | Conventional hydrogen bond | 2.10114 | SER223 | Pi-donor hydrogen bond | 4.1704 |
| SER223 | Conventional hydrogen bond | 3.16906 | SER223 | Pi-sigma hydrophobic bond | 3.16013 |
| ASP225 | Conventional hydrogen bond | 2.87312 | ALA222 | Alkyl hydrophobic bond | 3.22803 |
| ASP315 | Pi-anion electrostatic bond | 3.51659 | ALA222 | Pi-alkyl hydrophobic bond | 5.15777 |
| LYS226 | Alkyl hydrophobic bond | 4.23487 | LYS341 | Pi-alkyl hydrophobic bond | 5.1248 |
| LYS226 | Pi-alkyl hydrophobic bond | 5.13147 | |||
Molecular dynamic simulation
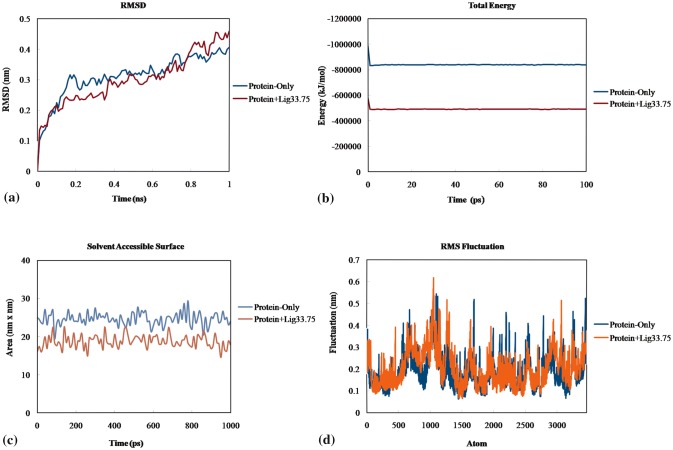
Root mean square deviation (RMSD), root mean square fluctuation (RMSF), total energy and solvent accessible surface area (SASA) of the protein and docked protein complex were calculated and analysed to explore the stability and specific binding of docked protein complex. RMSD value of only protein first increase and then stabilize around 0.3 nm at the end of 0.4 ns. On the basis of MDS only mitoxantrone hydrochloride exhibited stable binding with the nsP4 polymerase. The trajectory of docked protein complex (i.e. protein with bound mitoxantrone hydrochloride) stabilizes at 2.4 nm that demonstrates stable and precise binding of protein with mitoxantrone hydrochloride (Fig. 4a). Total energy was calculated which indicate the stability of docked protein (Fig. 4b). SASA of protein and docked protein complex also signify the stability as there were low fluctuations over the time of simulation (Fig. 4c). Furthermore, fluctuation of each atom (amino acid) were analysed by RMSF values during the time of simulation. RMSF value of protein and protein complex were calculated and showed lower fluctuations indicating the less changes on protein upon binding to mitoxantrone hydrochloride (Fig. 4d).
Fig. 4.
Molecular dynamic simulation. a RMSD b Total energy c SASA d RMSF of modelled protein only and mitoxantrone hydrocholoride bound protein complex i.e. protein + lig33.75 during the time of simulations
In the drug discovery, high throughput computing has taken the era to new heights where the complete information about the 3D structure of protein has been exploited along with the in silico screening of large libraries against the targeted proteins. Drug repositioning has further improved the whole process of drug discovery as the leads selected have already been approved by FDA so these drugs can be repurposed for the inhibition of CHIKV replication as the ADMET details are already identified for them. Although, further substantiation of the selected lead via in vitro or in vivo experiments including cell culture assays and animal studies could be done to determine efficacy and optimum dose for human application.
Polymerase activity of CHIKV nsp4 is important for viral replication but is still an insufficiently explored protein for inhibitors. Relevance of selected inhibitor namely Mitoxantrone hydrochloridethat exhibited strong and stable bond with the residues of docking pocket was focussed upon. As mitoxantrone hydrochloride is an FDA approved drug so its ADMET evaluation is already determined. Although its side effects are linked to cardio-myopathy at high doses but the drug has demonstrated improved therapeutics with fewer side effects and is approved in combination for cancer therapy. Scientists have also tried to increase its bioavailability by novel drug delivery methods. Zang et al. [27] reported the enhanced stability, sustained release kinetics of encapsulated mitroxantrone along with improved therapeutic efficiency.
Since Mitoxantrone Hydrochloride, Spectinomycin, diminazene aceturate, Imatinib and Cefadroxil interacted with the residues of interest of nsP4 polymerase with low score and stable polar interactions. Mitoxantrone hydrochloride and imatinib are anti-cancer drugs and generally have severe side effects, therefore their benefits in CHIKV treatment remains to be weighed over risk. Further lead optimizations through scaffold hopping to increase the therapeutic index of these compounds, ADMET verification and in vitro and in vivo validations of the selected leads might show the way to the development of effective antiviral therapy of CHIKV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to Jaypee Institute of Information Technology (JIIT), NOIDA and Ram Lal Anand College, University of Delhi for providing infrastructural resources and Ritu Ghildiyal acknowledges Indian Council of Medical Research (ICMR), for providing financial support [Project Sanction order: VIR/24/20l2-ECD-I].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abdelnabi R, Neyts J, Delang L. Chikungunya virus infections: time to act, time to treat. Curr Opin Virol. 2017;24:25–30. doi: 10.1016/j.coviro.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Abu Bakar F, Ng L. Nonstructural proteins of alphavirus—potential targets for drug development. Viruses. 2018;10:71. doi: 10.3390/v10020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P. A sequence motif in many polymerases. Nucl Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babusis D, Curry MP, Kirby B, Park Y, Murakami E, Wang T, et al. Sofosbuvir and ribavirin liver pharmacokinetics in patients infected with hepatitis C virus. Antimicrob Agents Chemother. 2018;62:e02587-17. doi: 10.1128/AAC.02587-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown AS, Patel CJ. A review of validation strategies for computational drug repositioning. Briefings Bioinform. 2016;19(1):174–177. doi: 10.1093/bib/bbw110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bursulaya BD, Totrov M, Abagyan R, Brooks CL. Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des. 2003;17:755–763. doi: 10.1023/B:JCAM.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 7.Burt FJ, Chen W, Miner JJ, Lenschow DJ, Merits A, Schnettler E, et al. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect Dis. 2017;17:e107–e117. doi: 10.1016/S1473-3099(16)30385-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen MW, Tan YB, Zheng J, Zhao Y, Lim BT, Cornvik T, et al. Chikungunya virus nsP4 RNA-dependent RNA polymerase core domain displays detergent-sensitive primer extension and terminal adenylyltransferase activities. Antiviral Res. 2017;143:38–47. doi: 10.1016/j.antiviral.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Delang L, Guerrero NS, Tas A, Quérat G, Pastorino B, Froeyen M, et al. Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J Antimicrob Chemother. 2014;69:2770–2784. doi: 10.1093/jac/dku209. [DOI] [PubMed] [Google Scholar]
- 10.Hajihassan Z, Rabbani-Chadegani A. The effect of mitoxantrone as an anticancer drug on hepatocytes nuclei and chromatin: selective release of histone proteins. Indian J Pharmacol. 2011;43:187–191. doi: 10.4103/0253-7613.77362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoornweg TE, van Duijl-Richter MKS, Ayala Nuñez NV, Albulescu IC, van Hemert MJ, Smit JM. Dynamics of Chikungunya virus cell entry unraveled by single-virus tracking in living cells. J Virol. 2016;90:4745–4756. doi: 10.1128/JVI.03184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishida J, Konishi M, Ebner N, Springer J. Repurposing of approved cardiovascular drugs. J Transl Med BioMed Cent. 2016;14:1–15. doi: 10.1186/s12967-016-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koszalka P, Tilmanis D, Hurt AC. Influenza antivirals currently in late-phase clinical trial. Influenza Other Respi Viruses. 2017;11:240–246. doi: 10.1111/irv.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar SP, Kapopara RG, Patni MI, Pandya HA, Jasrai YT, Patel SK. Exploring the polymerase activity of chikungunya viral non structural protein 4 (nsP4) using molecular modeling, e-pharmacophore and docking studies. Int J Pharm Life Sci. 2012;3:1752–1765. [Google Scholar]
- 16.Nitiss J. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oo A, Hassandarvish P, Chin SP, Lee VS, Abu Bakar S, Zandi K. In silico study on anti-Chikungunya virus activity of hesperetin. PeerJ. 2016;4:e2602. doi: 10.7717/peerj.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasheed S, Sánchez SS, Yousuf S, Honoré SM, Choudhary MI. Drug repurposing: in-vitro anti-glycation properties of 18 common drugs. PLoS One. 2018;13:1–9. doi: 10.1371/journal.pone.0190509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezza G. Dengue and chikungunya: long-distance spread and outbreaks in naïve areas. Pathog Glob Health. 2014;108:349–355. doi: 10.1179/2047773214Y.0000000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riviere P. Use of Imatinib to treat liver disorders and viral infections. United States Patent; 2012. https://patentimages.storage.googleapis.com/7b/65/30/6f8461cfb7ed85/US8293745.pdfv. Accessed 2 Feb 2019.
- 21.Shah PS, Wojcechowskyj JA, Eckhardt M, Krogan NJ. Comparative mapping of host-pathogen protein-protein interactions. Curr Opin Microbiol. 2015;27:62–68. doi: 10.1016/j.mib.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A, Kumar A, Yadav R, Uversky VN, Giri R. Deciphering the dark proteome of Chikungunya virus. Sci Rep. 2018;8:5822. doi: 10.1038/s41598-018-23969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sousa SF, Ribeiro JM, Coimbra JTS, Neves RPP, Martins SA, Moorthy NSHN, et al. Protein-ligand docking in the new millennium—a retrospective of 10 years in the field. Curr Med Chem. 2013;20:2296–2314. doi: 10.2174/0929867311320180002. [DOI] [PubMed] [Google Scholar]
- 24.Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 25.Tomar S, Hardy RW, Smith JL, Kuhn RJ. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol. 2006;80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada Y, Orba Y, Sasaki M, Kobayashi S, Carr MJ, Nobori H, et al. Discovery of a novel antiviral agent targeting the nonstructural protein 4 (nsP4) of chikungunya virus. Virology. 2017;505:102–112. doi: 10.1016/j.virol.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Ren Y, Wang Y, He Y, Feng W, Song C. Pharmacokinetics, distribution and anti-tumor efficacy of liposomal mitoxantrone modified with a luteinizing hormone-releasing hormone receptor-specific peptide. Int J Nanomed. 2018;13:1097. doi: 10.2147/IJN.S150512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.