Abstract
Cancer treatment decisions are increasingly guided by which specific genes are mutated within each patient’s tumor. For example, agents inhibiting the epidermal growth factor receptor (EGFR) benefit many colorectal cancer (CRC) patients, with the general exception of those whose tumor includes a KRAS mutation. However, among the various KRAS mutations, that which encodes the G13D mutant protein (KRASG13D) behaves differently; for unknown reasons, KRASG13D CRC patients benefit from the EGFR-blocking antibody cetuximab. Controversy surrounds this observation, because it contradicts the well-established mechanisms of EGFR signaling with regard to RAS mutations. Here, we identified a systems-level, mechanistic explanation for why KRASG13D cancers respond to EGFR inhibition. A computational model of RAS signaling revealed that the biophysical differences between the three most common KRAS mutants was sufficient to generate different sensitivities to EGFR inhibition. Integrated computation with experimentation then revealed a non-intuitive, mutant-specific dependency of wild-type RAS activation by EGFR that is determined by the interaction strength between KRAS and the tumor suppressor neurofibromin (NF1). KRAS mutants that strongly interacted with and competitively inhibited NF1 drove wild-type RAS activation in an EGFR-independent manner, whereas KRASG13D weakly interacted with and could not competitively inhibit NF1 and, thus, KRASG13D cells remained dependent upon EGFR for wild-type RAS activity. Overall, our work demonstrates how systems approaches enable mechanism-based inference in genomic medicine and can help identify patients for selective therapeutic strategies.
Introduction
Cancer treatment decisions are increasingly influenced by which specific genes are mutated within each patient. This has been referred to as personalized medicine, precision medicine, and genomic medicine. One example of personalized medicine in cancer involves the use of epidermal growth factor receptor (EGFR)-blocking antibodies and inhibitors in colorectal cancer (CRC) patients. Clinical trials have shown that humanized therapeutic antibodies that target EGFR, like cetuximab and panitumumab, provide a survival benefit to CRC patients (1, 2). These drugs are now approved for CRC patients, except for those with KRAS mutations.
Approximately 40% of patients with CRC have an acquired KRAS mutation within their tumor (3). The Ras family of guanosine triphosphatases (GTPases) – HRAS, NRAS, and KRAS – serve as key nodes in the EGFR signaling network (Fig. 1A). The signals that propagate from Ras to its effectors, like the RAF family of kinases, during the course of EGFR signaling can also be initiated by constitutively active mutant KRAS proteins. These mutant KRAS proteins are not dependent upon EGFR for their activation (4). Thus, it seemed logical that the presence of a constitutively active mutant KRAS would indicate resistance to anti-EGFR agents. Clinical trials concluded that CRC patients with constitutively active mutant KRAS do not benefit from anti-EGFR agents (5, 6). This relationship between EGFR inhibitors, KRAS mutations, and CRC appears consistent with the conventional understanding of EGFR signaling.
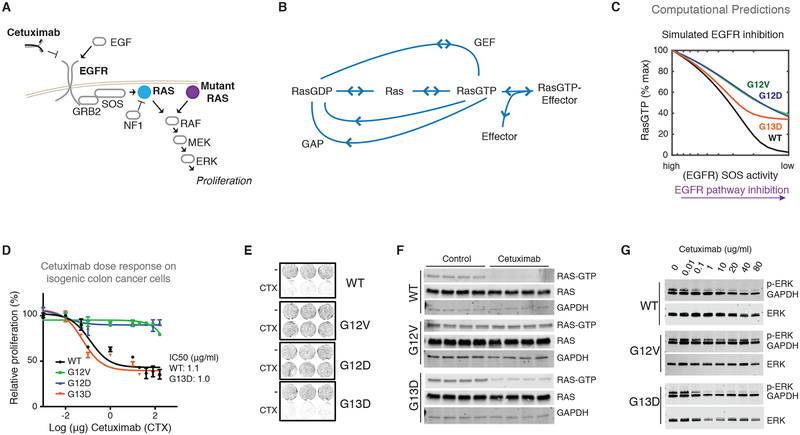
Fig. 1. The KRAS mutant-specific response to anti-EGFR agent cetuximab in CRC.
(A) EGFR signals through the RAS GTPases to drive proliferation. Constitutively active Ras mutants are active in an EGFR-independent manner and are known to cause resistance to EGFR inhibitors. (B) The biochemical processes that influence Ras nucleotide binding for both wild-type and mutant Ras proteins and that are the focus of the mathematical model. (C) Simulated anti-EGFR dose response from the computational Ras model. (D) MTT proliferation assays to assess dose responses of KRAS WT SW48 (WT) colon cancer cells and three derivative isogenic cell lines, each with one of the three most common KRAS mutants in colon cancer (G12D, G12V, and G13D), to the EGFR-blocking antibody cetuximab (CTX at dose indicated for 48 hours). Data are means ± SD of seven biological replicates and are representative of three experiments. (E) Two-dimensional colony formation assay for each cell line in the isogenic panel treated without or with cetuximab (CTX; 20μg/ml) for seven days. Images are representative of six independent experiments. (F) Ras binding domain (RBD) pull-down Ras activation assays for isogenic SW48 cells cultured without and with cetuximab (as in E). Four biological replicates for each condition were included in each of three independent experiments. (G) Immunoblots of ERK phosphorylation in whole-cell lysates from isogenic SW48 cells cultured in the presence of increasing concentrations of cetuximab. Blots are representative of three independent experiments.
However, multiple studies that evaluated whether there were differences between the common, constitutively active KRAS mutants suggest that the relationship between oncogenic KRAS mutants and the response to EGFR inhibitors is more complicated. Initially, a retrospective analysis of phase III clinical trial data found that the anti-EGFR agent cetuximab benefited CRC patients with a KRAS G13D mutation, but not patients with any other KRAS mutation (7). Although this claim has been further supported with additional clinical trials and experimental model systems (7–9), the finding remains controversial because it is difficult to reconcile known principles of Ras biology with KRAS G13D patients responding differently (4, 10–12). Without a mechanism, expert opinion has been to consider the KRAS G13D mutation equivalent to other KRAS mutations and to assume that patients with the KRAS G13D mutation would not benefit from anti-EGFR agents, despite the evidence to the contrary. Resolving this problem has the potential to benefit a large number of cancer patients. For example, there are approximately 10,000 new cases of KRAS G13D CRC in the United States alone (13, 14).
Here, we performed a computational and experimental investigation of this problem. Applying our previously described computational systems biology methods for studying Ras mutant proteins (15) revealed that the controversial KRAS G13D behavior that has been interpreted to be inconsistent with known mechanisms of Ras biology is actually fully consistent with known mechanisms of Ras biology. Our model suggests that cancers with the G13D mutant are more sensitive to EGFR inhibition because the amount of active, cellular, wild-type GTP-bound Ras (RasGTP) decreases in G13D cancers much more than in cancers with other Ras mutations. The model also suggests that the key difference between G13D and the other common Ras mutants is that G13D does not bind well to the tumor suppressor neurofibromin (NF1), and that a strong interaction with NF1 leads to the competitive inhibition of its GAP activity and increased wild-type Ras GTP levels in an EGFR-independent manner, whereas a weak interaction with NF1 does not result in competitive inhibition of NF1 and wild-type Ras GTP levels remain EGFR dependent. Our experiments confirm these model predictions. Overall, this work demonstrates the power of computational systems biology approaches to address problems in personalized medicine, and it also highlights the necessity of mathematical models based on fundamental biochemistry as a tool for understanding the behaviors of biological networks that are important to disease.
Results
Systems Modeling of Oncogenic KRAS mutants
We previously developed a mathematical model of the processes that regulate Ras signaling (15). This model is based upon the well-established architecture of the Ras signaling module and the readily available biochemical rate constants of wild-type and mutant proteins (text S1 and fig. S1). These processes, nucleotide exchange, GTP hydrolysis, and effector binding, can be considered the “central dogma of Ras signaling.” A Ras mutant is incorporated into the computational model through the inclusion of its specific biochemical rate constants. We then simulate the reactions between Ras and its interaction partners in accordance with the accepted biochemical understanding of these processes. In other words, we simulate Ras signaling in silico at the level of chemical mass-action kinetics. We use model simulations to find the behaviors that logically follow from this well-accepted information, but may nevertheless be non-obvious due to the complexity and scope of the system (15, 16).
Here, we utilize our mathematical model to computationally investigate how Ras mutations should influence the response to EGFR inhibition. The three most common Ras mutants in CRC are G12D, G12V, and G13D (3). We updated our model, which already included G12D and G12V mutants (15), to also include the G13D mutant by incorporating the known biochemical differences between each mutant and wild-type Ras, as has been previously measured experimentally (17, 18) (fig. S1). We found that the available data for the G13D mutant were sufficient to result in its constitutive activation, just as the available data for G12D and G12V have been shown to be sufficient to explain these mutants’ constitutive activation (figs. S2 and S3, A and B).
We then used the model to investigate how Ras signaling networks containing each mutant would respond to EGFR inhibition. We did this by using the computational model to find the abundance of total cellular, active RasGTP that should occur for conditions of high EGFR activation [which leads to Ras activation through the Ras guanine exchange factors (GEFs) son of sevenless 1 (SOS1) and son of sevenless 2 (SOS2)] to conditions of low EGFR activation (where low amounts of Ras activation by Ras GEFs would occur). Unexpectedly, our simulations of EGFR inhibition, which were based on the biochemical properties of these mutants, found the G13D-containing network displayed larger reductions in Ras signals than the G12D and G12V-containing networks (Fig. 1B and fig. S3C). This was notable, because expert opinion had been that it did not make sense for different Ras mutants to respond differently to EGFR inhibition. Our analysis revealed that it is fully consistent with Ras central dogma for some mutants to respond more strongly to EGFR inhibition. Moreover, our analysis suggests that the available biochemical data are sufficient to explain a mechanism by which G13D would be the most sensitive of the most common KRAS mutants in CRC.
Evaluation of an Experimental Model System for this Phenomenon
To experimentally study KRAS allele-specific differences and model predictions, we obtained a panel of isogenic CRC cells that was previously derived from the SW48 CRC cell line and was used to study the KRAS G13D response to cetuximab (7). We obtained isogenic cells with the following KRAS genotypes: G12D/wild-type (WT) (G12D cells), G12V/WT (G12V cells), G13D/WT (G13D cells), and WT/WT (WT cells) (Fig. S4A). The mutant isogenic cells display constitutively increased amounts of active RasGTP when compared to the parental WT cells (fig. S4B), consistent with all three of these mutants being constitutively active. No significant changes in ERK phosphorylation were noted with the Ras mutant isogenic lines, which is consistent with other recent work on signaling within Ras mutant isogenic SW48 cells (19).
We performed dose-response experiments with the EGFR-blocking antibody cetuximab to evaluate the described difference for these cells. When treated with increasing doses of cetuximab, both the G13D cells and WT cells displayed reduced proliferation (Fig. 1C) and reduced colony formation (Fig. 1D and fig. S4C), whereas each in the G12D and G12V cells were not noticeably affected. We also evaluated dose responses to mitogen-activated protein kinase kinase (MEK) inhibitors to evaluate whether these cells were more sensitive to any inhibition of the pathway. We observed that all cell lines responded similarly to MEK inhibition (fig. S5A), suggesting that the G13D cells are not simply more sensitive to all agents that target the EGFR-RAS-extracellular signal-regulated kinase (ERK) pathway.
We hypothesized that if there was a difference in how these cells depended upon EGFR signals, then we should be able to detect net proliferation differences when these cells are cultured in medium containing low amounts of serum. Consistent with our hypothesis, we observed that G13D and WT cells proliferated more slowly than G12D and G12V cells when grown in low-serum media, but that all cells proliferated at a similar rate when supplemental EGF was added to the media (fig. S4D). This further suggests that G13D cells display an increased dependency upon EGFR signaling compared to G12D and G12V cells. Also, because the patterns of response appeared analogous to the clinical observations regarding KRAS genotype and response (7), the data suggest this cell line would be useful to test our experimental model.
An Alternative Experimental Model for this Phenomenon
We desired an additional experimental system for comparing mutant-specific responses to treatment. We hypothesized that the introduction of mutant KRAS G12D or G12V into the WT Ras cells should reduce sensitivity to cetuximab, whereas introduction of KRAS G13D would have a minimal effect on sensitivity. In our experiments, we observed that transfected KRAS G12D or G12V, but not G13D or WT, promoted resistance to cetuximab, consistent with our hypothesis and consistent with the G13D mutant being comparably more sensitive to EGFR inhibition (fig. S6).
Experimental Evaluation of Predicted Signaling Differences
Our model suggests that there should be signaling differences between G13D cells and cells with one of the other common KRAS mutations (G12D cells and G12V cells). We measured the abundance of active, GTP-bound Ras (RasGTP) in cells treated with or without cetuximab, and we detected a reduction in RasGTP only in G13D and WT cells, but not in G12V cells (Fig. 1E). As RasGTP signals are transmitted downstream through the ERK MAPK cascade (Fig. 1A), we also measured phosphorylated ERK for cells treated with different doses of cetuximab. We detected reductions in the abundance of phosphorylated ERK in both the sensitive G13D and WT cells upon treatment with cetuximab but not in the resistant G12V cells (Fig. 1F). All cells displayed reductions in phosphorylated ERK when treated with a MEK inhibitor (fig. S5B). In contrast to the observed changes in ERK phosphorylation, we did not detect changes in AKT phosphorylation under treatment with cetuximab (fig. S7), consistent with little to no change in AKT phosphorylation after EGFR inhibition in nine other colorectal cancer cell lines (20).
Experimental Confirmation of EGFR Dependence
We performed additional experiments to confirm that the response of these isogenic SW48 cells to cetuximab was EGFR dependent. First, we used siRNA to knock down EGFR expression in these four different isogenic cell lines (fig. S8A). We observed reduced ERK phosphorylation and reduced proliferation of the WT cells and of the G13D cells with EGFR knockdown, but no difference in the G12V cells or the G12D cells (fig. S8A,B). We then performed dose-response experiments with the EGFR-blocking antibody panitumumab to complement the studies with the EGFR-blocking antibody cetuximab. As with cetuximab, we observed that G13D and WT cells both displayed reduced proliferation when treated with panitumumab whereas G12V and G12D cells were insensitive to panitumumab (fig. S9A). Immunoblots similarly observed reduced ERK phosphorylation for WT and G13D cells, but not for G12V cells (fig. S9B). To more broadly evaluate the response to agents that target EGFR, we also performed dose-response experiments using the tyrosine kinase inhibitor erlotinib which is a small molecule compound that targets the kinase domain of EGFR. We found G13D and WT cells to be more sensitive to erlotinib than G12V and G12D cells (fig. S10A). In contrast to our experiments with cetuximab and panitumumab (Fig. 1C and fig. S9A), G12V cells and G12D cells here appeared to be slightly sensitive to erlotinib (fig. S10A). Immunoblots observed reduced ERK phosphorylation for WT and G13D cells, but not G12V cells (fig. S10B), raising the possibility that the partial sensitivity of G12V (and G12D) cells may come from off-target effects.
Model Prediction of Differences in Wild-Type Ras Activation
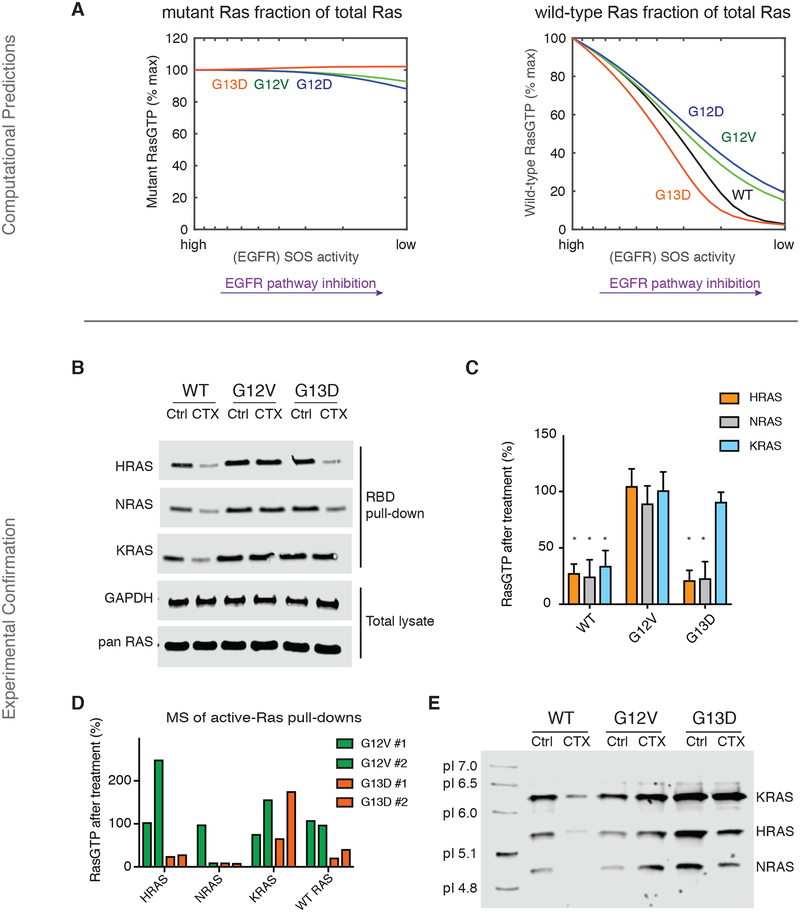
Our computational model includes both mutant (KRAS) and wild-type (KRAS, NRAS, and HRAS) pools of Ras because CRC cells express all three Ras proteins (21). The differences in total RasGTP that our model predicts, (as in Fig. 1B) are accordingly distributed between GTP bound mutant Ras proteins and GTP-bound wild-type Ras proteins. We queried our model to determine whether the predicted changes in signal were coming from mutant Ras, wild-type Ras, or both. Our simulations suggest that EGFR inhibition should cause no appreciable changes in the amount of mutant Ras bound to GTP (Fig. 2A and fig. S3D). This is consistent with the conventional wisdom that anti-EGFR agents should not influence mutant Ras signaling. However, our simulations predicted that EGFR inhibition should result in large changes in wild-type RasGTP (Fig. 2A and fig. S3D). This suggests that the non-obvious response to anti-EGFR agents may have a basis in wild-type Ras signaling.
Fig. 2. The Ras model predicts, and experiments confirm, that wild-type Ras activation distinguishes sensitive from non-sensitive cancer cells.
(A) Simulated anti-EGFR dose response for the Ras model, further subdivided to reveal the change in active, GTP-bound mutant Ras (left) and the change in active, GTP-bound wild-type Ras (right), within each modeled genotype. (B) Ras binding domain (RBD) pull-down Ras activation assays for isogenic SW48 cells (WT, KRAS G12V, and KRAS G13D) cultured without or with cetuximab. CTX; 20μg/ml) or without cetuximab for 48 hours. Blots are representative of four independent experiments. (C) Densitometry-based quantification of the ratio of RasGTP between cetuximab-treated and untreated cells from three independent assays represented in (B). The quantified data are means ± SD. *P<0.05, one-way ANOVA (F=35.22) with post-hoc Tukey’s test for multiple comparisons between WT or G13D cells vs. G12V cells for each RAS isoform. (D) Mass spectrometry-based quantification of the GTP-bound wild-type HRAS, wild-type NRAS, total (both wild-type and mutant) KRAS, and wild-type H/N/KRAS in cetuximab-treated KRASG12V or KRASG13D cells relative to untreated counterparts. (CTX; 20μg/ml for 48 hours). Data from two independent experiments are presented. (E) Isoelectric focusing of excised gel bands from RBD pull-down lysates, performed upon excised gel bands. Lysates are from isogenic SW48 cells (WT, KRAS G12V, and KRAS G13D) cultured without or with cetuximab (CTX; 20μg/ml for 48 hours). Blot is representative of three independent experiments.
Experimental Confirmation of Differences in Wild-Type Ras Activation
We returned to our experimental system to test the model-based hypothesis that EGFR inhibition causes a larger drop in wild-type RasGTP in G13D cells than in cells with one of the other common Ras mutants. We measured Ras activation in the presence and absence of cetuximab for each of the Ras proteins (HRAS, NRAS, and KRAS) (Fig. 2, B and C) by using antibodies specific for each form of Ras (fig. S11). We observed a large reduction in GTP-bound wild-type HRAS and GTP-bound wild-type NRAS after cetuximab treatment only in G13D and WT cells, consistent with our model’s predictions. We also observed a larger reduction in GTP-bound KRAS in WT cells than in G12V and G13D cells, consistent with the presence of one constitutively active KRAS allele for the two mutant cell lines.
To complement these studies, we developed a mass-spectrometry assay that could quantify the amount of active HRAS, NRAS, KRAS, and wild-type H/N/KRAS through the use of isotopically labeled peptides unique to HRAS, NRAS, KRAS, wild-type H/N/KRAS, and the G12V and G13D Ras mutants. This approach revealed greater reductions in active (GTP-bound) wild-type HRAS, wild-type NRAS, and wild-type H/N/KRAS in G13D cells treated with cetuximab than in G12V cells treated with cetuximab (Fig. 2D; data file S1). Additionally, total (wild-type and mutant) KRAS in G13D cells displayed a partial reduction in GTP binding, consistent with one KRAS allele being wild-type and one KRAS allele being mutant.
We also developed an approach to differentiate between HRAS, KRAS, and NRAS through isoelectric focusing (IEF). Analysis of Ras binding domain (RBD) lysates that were further separated by IEF prior to immunoblotting revealed decreased relative levels of active NRAS and HRAS in isogenic WT and G13D cells treated with cetuximab, but not in G12V cells treated with cetuximab (Fig. 2E). Additionally, active KRAS levels were most strongly reduced in WT cells treated with cetuximab, but did not demonstrate any reduction in the (KRAS) G12V and (KRAS) G13D cells treated with cetuximab. We also note that in several experiments there was an increased level of RasGTP detected within G12V cells treated with cetuximab. We hypothesize this may be a temporary, rebound, increase in signal after a partial loss of negative feedback (22, 23). Altogether, the immunoblots, mass spectrometry, and IEF demonstrate reductions in wild-type HRAS and NRAS GTP levels in both WT and G13D cells, but not the G12V cells.
Model-based Identification of a Determinative Interaction with NF1
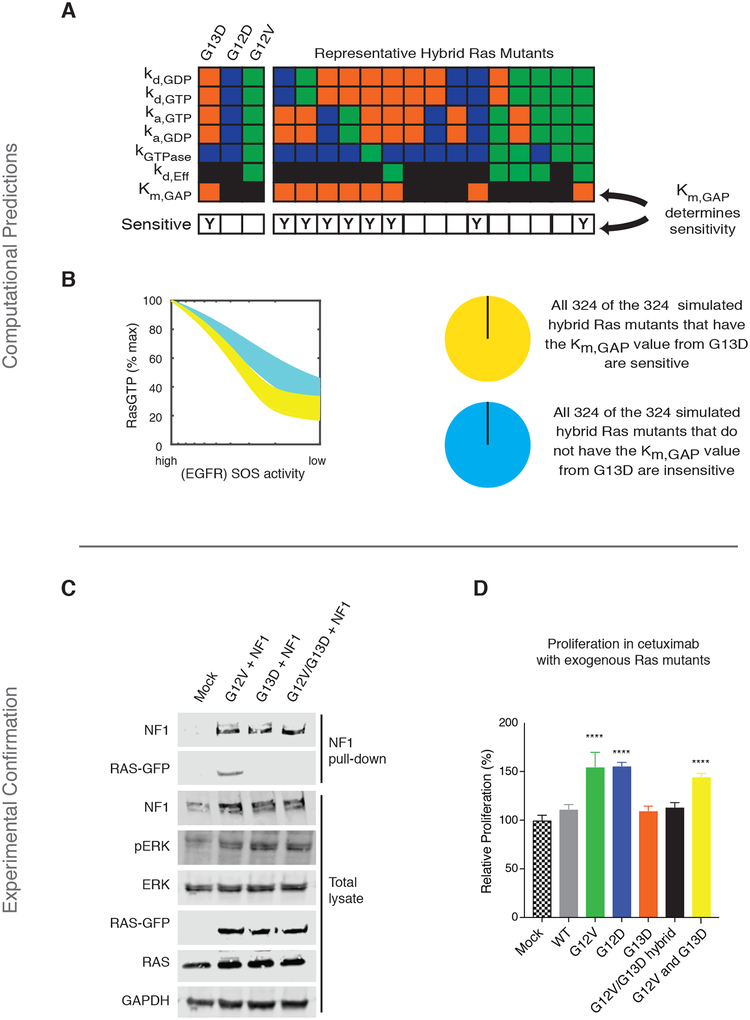
The behavior of each mutant in our computational model is determined by its parameter values (fig. S1). We set out to determine which specific parameter value(s) are responsible for the G13D mutant being more sensitive to modeled EGFR inhibition. We reasoned that it may be possible to determine which parameter(s) are responsible for sensitivity by systematically considering synthetic Ras mutants that were built from combinations of the G12V, G12D, and G13D parameter values. We therefore created 648 such computational Ras mutants by considering all combinations of the parameters from the G13D, G12V, and G12D mutants, effectively creating computational hybrid Ras mutants (Fig. 3A). We used our model to simulate dose responses to EGFR inhibition for each of 648 different computational hybrids, and then we determined whether any single parameter could distinguish between the sensitive and resistant hybrid mutant networks. Our analysis found that all hybrids that were sensitive to simulated EGFR inhibition contained the Km (or the enzymatic Michaelis constant) that characterizes the interaction between KRAS G13D and the Ras GTPase-activating protein (Ras GAP) neurofibromin (NF1), and also that all mutants that were insensitive to simulated EGFR inhibition had the Km value that applied to the G12D and G12V mutants (Fig. 3B and fig. S3E). Thus, this demonstrates that that this parameter is necessary and sufficient for sensitivity to EGFR inhibition in our systems model of Ras signaling.
Fig. 3. The Ras model predicts, and experiments confirm, that mutant-specific interactions with tumor suppressor NF1 determine whether or not cells respond to anti-EGFR agents.
(A) Schematic to explain computational Ras hybrid mutants. G13D, G12D, and G12V have been described to differ in seven biochemical parameters. 648 different computational hybrids were generated by considering all of the possible combinations of these differentiating parameters. For each mutant, the model was evaluated to determine whether the computational hybrid was sensitive (“Y” in bottom row) or resistant to simulated EGFR inhibition. Orange indicates a parameter value specific to KRAS G13D, blue a parameter value specific to G12D, green a value specific to G12V, and black a value specific to KRAS WT that is also used for a mutant when do specific data is available. Fifteen representative hybrid mutants are shown from the 648 total hybrid mutants to visualize how they hybrid mutants contain combinations of the individual parameters used to model a G13D, G12D, or G12V mutant. (B) Simulated dose responses for all 648 hybrids, color coded on the basis of whether the hybrid had the Ras/NF1 Km value of the G13D mutant or that of the G12V or G12D mutant. (C) Co-immunoprecipitation of NF1 with KRAS G12V, G13D, and G12V/G13D (GG/VD) from mixtures of lysates from NF1-transfected cells with lysates from RAS-transfected cells. Blots are representative of three independent experiments. (D) MTT proliferation assays of cetuximab-treated KRAS WT SW48 cells transfected with WT, G12V, G12D, G13D, G12V/G13D double mutant (GG/VD), or both G12V and G13D KRAS. (CTX; 20μg/ml for 48 hours). Data are means ± SD of eight biological replicates and are representative of three experiments. Significance was determined with a cutoff of 25% induced growth and ****P<0.001 when compared to mock transfection by one-way ANOVA (F=90.69) with post-hoc Tukey’s test for multiple comparisons.
Experimental Confirmation of a Determinative Interaction with NF1
Ras GAPs like NF1 facilitate the inactivation of wild-type RasGTP to RasGDP, and oncogenic Ras mutants are insensitive to Ras GAPs. An increased Km essentially indicates that the GAP cannot bind well to the mutant Ras protein. It was initially unclear to us why reduced binding to GAP would influence the response to anti-EGFR agents as we modeled all three Ras mutants to have no increase in GTP hydrolysis once bound, so binding to GAP would intuitively be inconsequential.
We set out to test this computational results that suggest the strength of the interaction with NF1 can determine whether a cell line with a given mutation is sensitive or resistant to cetuximab. It has previously been reported G13D Ras binds much less well to NF1 (17). We reproduced this impaired binding with a co-immunoprecipitation study (fig. S12A) and with Bioluminescence Resonance Energy Transfer (BRET) (fig. S12B). We hypothesized that a KRAS G12V/G13D hybrid mutant (GG/VD), where the glycine residues at codons 12 and 13 were replaced with a valine and aspartic acid, respectively, would be constitutively active and bind poorly to NF1. We created this mutant and, when it was transfected into parental SW48 cells, we found it to be constitutively active, as demonstrated by to the presence of increased ERK phosphorylation (Fig. 3C and fig. S12C). We also found that this GG/VD combination mutant bound much less well to NF1 than KRAS G12V (Fig. 3C and fig. S12B).
If the ability to bind NF1 is the critical factor that determines whether or not a mutant promotes resistance to cetuximab, as suggested by our model, we reasoned that the KRAS G12V/G13D mutant would not promote resistance to cetuximab. We used our transfection-based assay (as shown in fig. S6) to evaluate the ability of transfected Ras mutants to alter WT cells sensitivity to cetuximab. Consistent with our hypothesis, we observed that the G12V/G12D double mutant did not promote resistance, despite being constitutively active (Fig. 3D).
A Mechanism for KRAS Mutant Allele-Specific Responses to EGFR Inhibition
We considered how differences in the interaction between KRAS and NF1 might result in differences in network signal output. Our previous systems analysis of oncogenic Ras found that the reversible and non-productive binding interaction between a Ras mutant and a Ras GAP can promote wild-type Ras activation (15), as the GAP-insensitive Ras mutant can effectively behave as a competitive inhibitor of Ras GAPs (24). Several other studies have also observed increased wild-type Ras when mutant Ras is present (25–27). Our new study suggests that G13D is an exception to this process because it binds much less well to NF1 and therefore cannot lead to wild-type Ras activation through the competitive inhibition of NF1 Ras GAP activity.
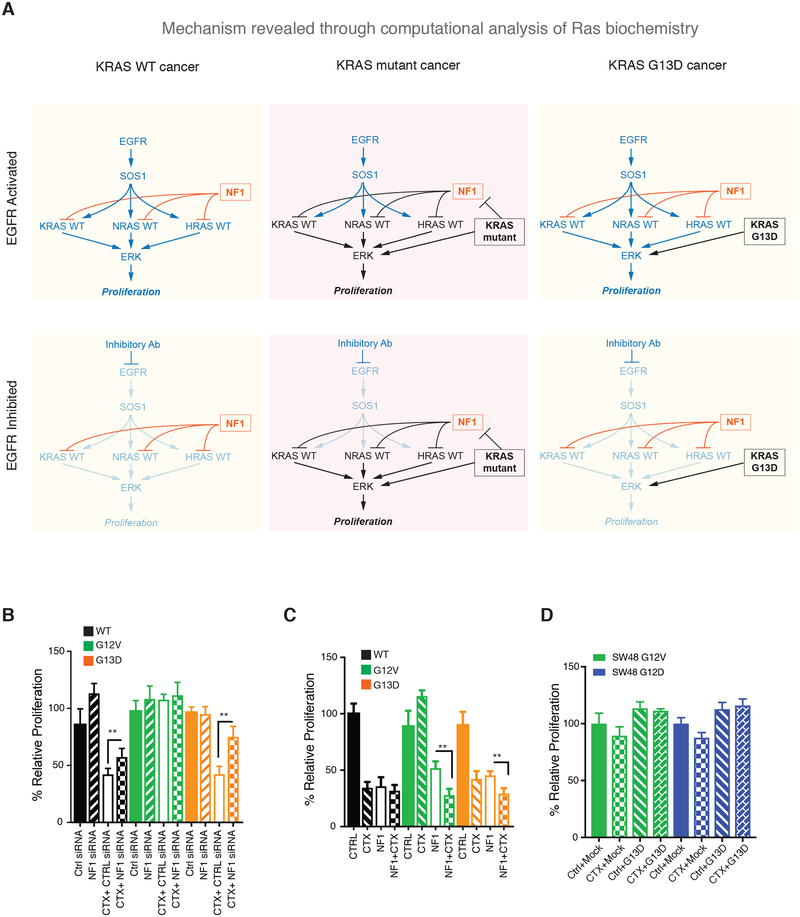
We therefore propose a mechanism that explains why KRAS G13D, but not other common KRAS mutants like G12D and G12V, responds to cetuximab (Fig. 4A). In a WT cell, Ras activation is dependent upon EGFR and can be counteracted with EGFR inhibitors. In a G12D or G12V cell, the mutant KRAS is constitutively active. Through the competitive inhibition of the Ras GAP NF1, wild-type Ras is also active in an EGFR-independent manner and the cells will be insensitive to therapeutic EGFR inhibition. In a G13D cell, the mutant KRAS is constitutively active and wild-type Ras activation is dependent on EGFR because the G13D mutant cannot drive wild-type Ras activation through the competitive inhibition of NF1. Assuming that the activation of proliferative signals downstream from Ras requires a quantity of Ras signal that is greater than the mutant alone can typically provide, inhibition of wild-type Ras through EGFR inhibition should negatively impact proliferation signals within the G13D cell. This assumption that wild-type Ras signaling is required in addition to mutant Ras signaling is consistent with emerging data that cancer promotion requires both wild-type and mutant Ras signals (15, 22, 25–27).
Fig. 4. A mechanism for KRAS-allele specific response to anti-EGFR agents with experimental validation.
(A) In a KRAS WT cancer, NF1 ensures there are low levels of RasGTP when EGFR is not active (or is inhibited). In KRAS G12D and KRAS G12V cancers, mutant Ras is active. Wild-type Ras is also active through the competitive inhibition of NF1 through the non-productive interaction between these Ras mutants and NF1. In a KRAS G13D cancer, mutant Ras is active but wild-type Ras remains dependent on EGFR for activation due to the inability of KRAS G13D to bind NF1. (B) MTT proliferation assays for isogenic SW48 cells with siRNA knock down of NF1 and/or with cetuximab treatment. (C) MTT proliferation assays for isogenic SW48 cells with neurofibromin transfection and/or with cetuximab treatment. (D) MTT proliferation assays of cetuximab treated KRAS G12D SW48 cells (left) and KRAS G13D SW48 cells (right) transfected with KRAS WT, G12V, G12D, or G13D. For B-D, (CTX; 20μg/ml for 72 hours). Data are means ± SD of eight biological replicates and are representative of 3 experiments. *P<0.05 and **P<0.01 by one-way ANOVA (F>27 for all three graphs) with post-hoc Tukey’s test.
Experimental Confirmation of Our Mechanism
We desired to test and confirm this proposed mechanism. We hypothesized that reduced expression of NF1 would make both G13D cells and WT cells less sensitive to cetuximab but would not largely affect G12V cells. This is because we reasoned reduced NF1 should result in increased wild-type RasGTP, thereby making these cells less dependent upon EGFR for wild-type Ras activation. We performed small interfering RNA (siRNA)-mediated knockdown experiments of NF1 in WT, G13D, and G12V cells and compared proliferation in the presence and absence of cetuximab. As hypothesized, NF1 knockdown reduced the sensitivity of G13D cells and WT cells to cetuximab with minimal effect on G12V cells (Fig. 4B). We also hypothesized that increased expression of NF1 should make G12V cells more sensitive to cetuximab. This is because we reasoned that these cells would become more dependent upon EGFR for wild-type Ras activation as NF1 levels increased. To test, we transfected WT, G13D, and G12V cells with NF1 and then treated with cetuximab. As hypothesized, increased NF1 expression made the G12V cells significantly more sensitive to cetuximab (Fig. 4C). Lastly, we reasoned that this mechanism also suggests that the introduction of KRAS G13D into a G12V or G12D cell would not cause the G12V or G12D to become sensitive to cetuximab, as the codon 12 KRAS mutant can still competitively inhibit NF1. We experimentally tested this hypothesis by transfecting G12V and G12D cells with KRAS G13D and found that the introduction of the KRAS G13D mutant did not cause the cells to become sensitive to cetuximab (Fig. 4D), consistent with our proposed mechanism.
Validation of Our Mechanism in Additional CRC Cell Lines
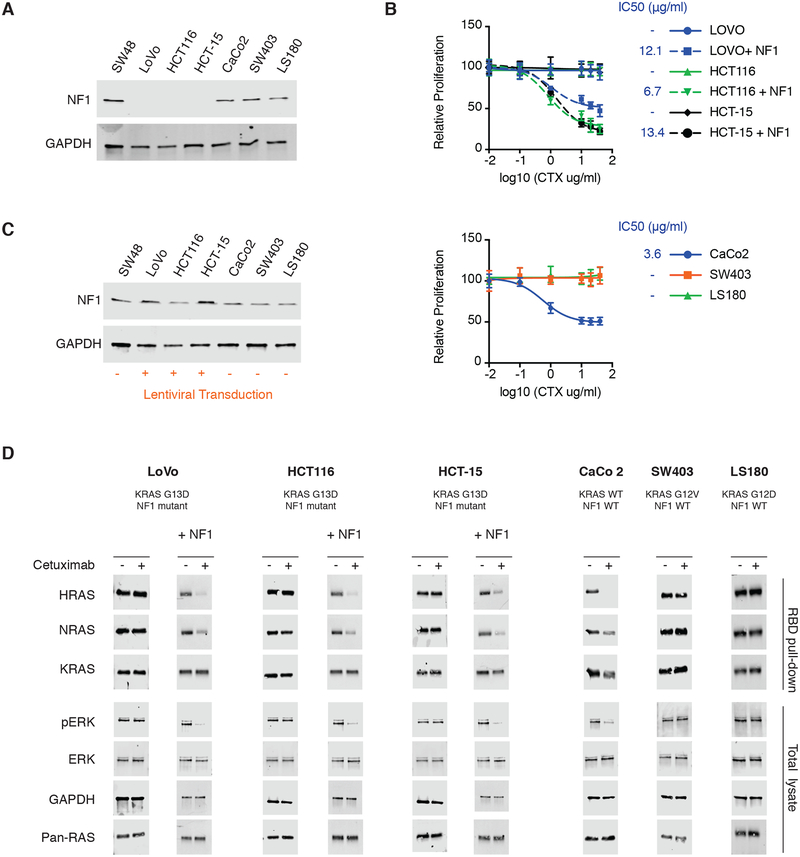
We lastly set out to determine whether the patterns of EGFR sensitivity and wild-type RAS activation in relation to KRAS mutation status that we first predicted with a mathematical model and then observed in an isogenic panel of SW48 CRC cell lines would be more generally observable in other CRC cell lines. We obtained three hemizygous KRAS G13D CRC cell lines (LoVo, HCT116, and HCT-15), one CRC cell line for each of KRAS WT (CaCo2), KRAS G12V (SW403), and KRAS G12D (LS180). Of note, all three of these KRAS G13D CRC cell lines have an NF1 mutation, whereas the other three CRC cell lines do not. We evaluated whether the NF1 mutations result in reduced NF1 protein expression with immunoblots and we did not detect NF1 expression in the three KRAS G13D cell lines (Fig. 5A). Consistent with our finding that reduced NF1 expression can convert a KRAS G13D SW48 cell line from being sensitive to cetuximab to insensitive to cetuximab (Fig. 4D), we observed that the cell lines that both had a NF1 mutant and the KRAS G13D mutant (LoVo, HCT116, and HCT-15) were insensitive to cetuximab (Fig. 5B). We also observed the KRAS WT (CaCo2) CRC cell line was sensitive to cetuximab and that the KRAS G12V (SW403) and KRAS G12D (LS180) CRC cell lines were insensitive to cetuximab, consistent with the data from the isogenic SW48 cell lines.
Fig. 5. Evaluation of proposed mechanism in additional CRC cell lines.
(A) Immunoblot of NF1 expression in an extended panel of CRC cell lines that includes three KRAS G13D, NF1 mutant, CRC cell lines (LoVo, HCT116, and HCT-15), KRAS WT, NF1 WT, CaCo2 cells, KRAS G12V, NF1 WT, SW403 cells, and KRAS G12D, NF1 WT, LS180 cells. Parental, KRAS WT, NF1 WT, SW48 cells are included for comparison. Blots are representative of three independent experiments. (B) MTT proliferation assays to assess dose responses of the extended panel of CRC cell lines. (top) Dose responses for the three KRAS G13D, NF1 mutant, cell lines and for the same three cell lines that have been transduced to express NF1. (bottom) Dose responses from the three NF1 WT cell lines. (CTX at dose indicated for 48 hours). Data are means ± SD of eight biological replicates and are representative of three experiments. IC50 values are presented for sensitive cell lines; (−) indicates a resistant cell line. (C) Immunoblot of NF1 expression in the KRAS G13D, NF1 mutant, CRC cell lines (LoVo, HCT116, and HCT-15) after lentiviral transduction with NF1. Non-transduced SW48, CaCo2, SW403, and LS180 cells are included for comparison. Blots are representative of 3 independent experiments. (D) Ras binding domain (RBD) pull-down Ras activation assays and ERK phosphorylation immunoblots for CRC cell lines LoVo, HCT116, HCT-15, CaCo2, SW403, and LS180, cultured without or with cetuximab (CTX; 20μg/ml for 48 hours). The NF1 mutant cell lines were investigated both in native form and after transduction with NF1 (+NF1). Blots are representative of 3 independent experiments.
The mechanism we have proposed (Fig. 4A) assumes that NF1 is present. We therefore hypothesized that if we reintroduced NF1 protein expression to the KRAS G13D, NF1 mutant, CRC cell lines that they would gain sensitivity to cetuximab. We used lentiviral transduction to express full length NF1 in these three cell lines (Fig. 5C). We observed that the reintroduction of NF1 caused a reduction in proliferation for these cells (fig. S13). Cetuximab dose responses found that these three KRAS G13D mutant with exogenous NF1 expression (LoVo + NF1, HCT116 + NF1, and HCT-16 + NF1) were now sensitive to cetuximab (Fig. 5B), fully consistent with our proposed mechanism. Additionally, we performed immunoblots on RBD lysates and whole cell lysates to evaluate RAS-GTP levels and ERK phosphorylation. We observed changes in RasGTP levels and in ERK phosphorylation with cetuximab treatment for KRAS WT CaCo2 cells, but not in the CRC cells with a KRAS G12D mutation, a KRAS G12V mutation, or a KRAS G13D mutation with a co-occurring NF1 mutation (Fig. 5D). Additionally, we observed reductions in RAS GTP and ERK phosphorylation in the KRAS G13D cells when NF1 protein had been reintroduced, but not in their NF1 mutant state (Fig. 5D). Altogether, these experiments suggest that the mechanism we identified through computational modeling and with SW48 isogenic cells is more general. It also highlights that NF1 mutations that co-occur with a KRAS G13D mutation may confer resistance to cetuximab.
Discussion
Every year, there are approximately ten-thousand new cases of KRAS G13D CRC in the United States alone. Despite the Phase III clinical trial evidence that these patients would benefit from Food and Drug Administration (FDA)-approved EGFR inhibitors, the apparent discrepancy between the known mechanisms of Ras signaling and these clinical effects has been seen as problematic. The field has chosen to favor intuition over empirical data, and has considered KRAS G13D equivalent to other codon 12, 13, and 61 KRAS mutations. This practice should be reconsidered; our mathematical work has identified a mechanism that is fully consistent with fundamental Ras biology and the idea that the KRAS G13D mutant can be more sensitive to EGFR inhibition.
Our mathematical model describes the “central dogma” of Ras biology. That is, Ras GEFs activate Ras, Ras GAPs inactivate Ras, active Ras binds to Ras effectors, and Ras has very slow GTPase activity and very slow GEF-independent nucleotide exchange activity. Our model is based upon peer-reviewed data that biochemically and biophysically characterize each of these reactions. Our simulations find that Ras central dogma permits different mutations to respond differently to the same upstream inhibitor. Additionally, our computational analysis finds the available biochemical data for the KRAS G13D mutant is sufficient to provide a mechanistic explanation for why KRAS G13D patients benefit from EGFR inhibition.
Differences in wild-type Ras activation between these KRAS mutant cells as they are treated with EGFR inhibitors are the critical aspect we uncovered with our model. Our experiments tested and confirmed this mechanism. Of note, the mathematical model, its analysis, and these hypotheses were posted to bioRxiv before the experimental work in this study began (28). This helps demonstrate that these were true, prospective, predictions.
Our study also suggests one mechanism by which KRAS G13D cancers may become resistant to EGFR agents. We demonstrated that decreased NF1 expression makes KRAS G13D mutant cancer cells more resistant to EGFR inhibition. Accordingly, we would hypothesize that CRC patients who have both NF1 and KRAS G13D mutations will be less likely to receive benefit and/or will receive a smaller benefit. We queried colorectal genomics studies to ask how often NF1 and KRAS G13D mutations co-occur (13). We found that KRAS G13D CRC patients had a co-occurring NF1 mutation less than 4% of the time, suggesting that a very large proportion of KRAS G13D CRC patients may be able to benefit from anti-EGFR agents that have been approved for CRC like cetuximab and panitumumab.
That KRAS G13D CRC cell lines commonly have NF1 mutations while patients do not is intriguing. We hypothesize that a requirement for wild-type Ras activation in CRC limits the KRAS G13D CRC that can yield cell lines to any cells that harbor an additional mutation that promotes an increase in wild-type RasGTP. Within actual cancer patients, we hypothesize that wild-type Ras in KRAS G13D CRC patients is promoted by extracellular signals that activate receptor tyrosine kinases like EGFR.
There are likely additional mechanisms that can lead to resistance to EGFR inhibition, just as there are multiple mechanisms for resistance to other targeted therapies. Future work will attempt to uncover these relationships. It is also possible that there are additional Ras mutants that respond to EGFR inhibition through similar mechanisms involving reduced binding to NF1. Future work will attempt to identify additional exceptional responder Ras mutants.
Our focus on the “central dogma” of Ras signal regulation has allowed us to construct a model for which there are readily available, high-quality, biochemical data for wild-type and mutant Ras. Although our model is limited in scope, it has been able to uncover multiple unappreciated aspects of Ras biology (15, 16). Many systems models extend to larger considerations of networks. There is a clear appreciation that features beyond the scope of our model, such as positive and negative feedback (19, 22, 26, 27, 29) play important contributions to Ras signaling. Once processes like the positive feedback of RasGTP on SOS1 (29) and the differences in the regulation of SOS1 and SOS2 (30) are biophysically characterized to the level of the different KRAS mutant alleles, it would be possible to determine whether there are additional differences between KRAS G13D, G12D, and G12V that further contribute to cancers with the KRAS G13D mutant being more sensitive to EGFR inhibition.
Our work demonstrates how systems approaches can uncover non-obvious, mechanistic bases for clinical observations that otherwise defy expert-level explanation. Many genes associated with cancer and other diseases have multiple pathological variants. Our work is significant to these other genes and diseases, as we have demonstrated how apparently similar variants can exhibit different responses to the same pharmacological treatment. As clinical genomics becomes more common, and as the number of targeted therapies approved and in development continues to grow, we believe that it will be increasingly necessary to perform integrated mathematical analysis of biomolecular systems to understand how mutant allele-specific behaviors emerge and influence response to treatment.
Materials and Methods
Mathematical Model and Analysis
Details of the model and its development have been published previously (15, 31–34), and are summarized here and further described in the Supplementary Text. The model focuses on Ras and the types of proteins that directly interact with Ras to regulate Ras-GTP levels: Ras GEFs (such as SOS1), Ras GAPs (such as NF1), and Ras effector proteins (such as the RAF kinases). The model includes 1) GEF mediated nucleotide exchange, 2) intrinsic nucleotide exchange, 3) GAP mediated nucleotide hydrolysis, 4) intrinsic nucleotide hydrolysis, and 5) effector binding. GEF and GAP reactions, 1 and 3 above, are described mathematically with reversible and irreversible Michaelis-Menten kinetics, respectively. We consider only the subset of total GEFs and GAPs that are active within our model. The other reactions are described with first- and/or second-order mass-action kinetics. It is assumed that wild-type and Ras mutant proteins have identical reaction mechanisms as indicated above, and that differences in rate constants (or enzymatic parameters) for the reactions account for described differences. For example, Ras mutant protein G12V hydrolyzes GTP more slowly than does wild-type Ras. In this case, the rate constant for this reaction kGTPase,G12V is smaller than the rate constant for the same reaction with wild-type Ras, kGTPase,WT. All reactions are grouped into a set of differential equations and the steady-state quantity of RasGTP-effector complexes (and RasGTP) is solved for the specified conditions.
Parameters of the model for proteins correspond to biochemically observable properties. Rate constants, enzymatic properties (Vmax and Km), and protein abundances for wild-type Ras proteins have been previously obtained, utilized, and published (listed in Supplementary Text) (15). Mutant proteins can be characterized by their difference from wild-type proteins in terms of a multiplicative factor, α. Values for α are determined from previous experimental studies that measured the desired property for both wild-type and mutant Ras proteins (35, 36). For G12V and G12D, we use the same α values that were previously obtained and utilized in our model (15). For G13D previous experiments described this mutant to have an elevated nucleotide dissociation rate compared to wild-type Ras (α = 3.6625) (18). Previous studies have also described Ras G13D to be insensitivity to Ras GAP (37), and to have no appreciable binding to the Ras GAP NF1 (17). A 100-fold increase in the Km of GAP on Ras G13D is used to model the immeasurable binding to the Ras GAP NF1. We estimated the change must be at least 100 times large as changes of approximately 50-fold have previously been measured for other Ras mutants (38), so we assumed that the difference must be larger to be undetectable. The decreased GTPase activity of the G12D mutant is used for the G13D mutant because we could not find an α factor at the time we began our; using the same value as G12D allowed us to introduce impaired GTPase activity while also allowing us to focus on the known biochemical differences.
Computational “hybrid” mutants are modeled mutants that have properties of two distinct Ras mutants. For example, a hybrid Ras mutant may be modeled with all of the properties of Ras G12D, except for the faster intrinsic nucleotide dissociation properties of G13D. Such a hybrid could be used to evaluate how faster nucleotide dissociation would influence signaling through the comparison of this hybrid’s behavior with that of the G12D mutant.
The Ras network within the CRC context is assumed to be EGFR driven, and EGFR is assumed to activate Ras through increased activation of Ras GEFs like SOS1 and SOS2. We use a ten-fold increase in Vmax for GEF reactions to indicate EGFR activation, just as we have done previously to model receptor tyrosine kinase mediated Ras activation (15). To simulate an EGFR inhibition dose response, levels of GEF activity between the “high” (10× increase) case and the basal “low” (1×) level were considered and the resulting level of RasGTP determined by model simulation. We assume that the three Ras proteins, HRAS, NRAS, and KRAS, share similar biochemistry and can be modeled with the same set of biochemical properties; such an assumption is consistent with measurements of the three Ras proteins (39, 40). We assume that measurements that provide α for one Ras protein are good approximations for the same mutant to the other Ras proteins. We assume that more than one Ras gene is expressed in CRC cells. This is consistent with many data (21, 41). We here model Ras mutants as being heterozygous, such that for a KRAS mutant, one half of total KRAS will be mutant and one half of total KRAS will be wild-type. Here, we assume that 50% of total Ras is KRAS (and that 25% of total Ras is mutant). This assumption is consistent with mass spectrometric quantification of KRAS, NRAS, and HRAS levels (21).
RasGTP and RasGTP-effector complex are considered as measures of Ras pathway activation. Model simulations are used to determine steady-state levels of RasGTP and RasGTP-effector. Simulations and analysis are performed in MATLAB (9.1.0.441655, MathWorks).
Cell Line Models and Culture Method
SW48 cells and isogenic counterparts were cultured in RPMI media supplemented with FBS (10%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). SW403 cells were culture in L-15 Media with FBS (10%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). LS-180 cells were cultured in DMEM with FBS (10%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). LoVo cells were cultured in F12-K media with FBS (10%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). HCT116 cells were grown in McCoys-5a media with FBS (10%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). HCT-15 cells were grown in RPMI media supplemented with FBS (10%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). CaCo2 cells were grown in DMEM with with FBS (20%), penicillin (100 unit/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). All cells were grown in indicated media and incubated at 37 °C in 5% CO2 unless indicated otherwise in experimental methods. SW48 cells were obtained from Horizon Discovery. SW403, HCT116, HCT-15, CaCo2, SW48, and LoVo were obtained from ATCC.
Western Blot Analysis
Cell lysates were generated using radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% nonyl phenoxypolyethoxylethanol (NP-40), 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris of pH 8.0) containing protease inhibitor cocktail (Cell Signaling Technologies) and incubated on ice for 1 hour. Total protein concentration was determined by Pierce-Protein assay (Thermo Fisher Scientific,). Protein samples (20 μg) were resolved by electrophoresis on 10–12% sodium dodecyl sulfate–polyacrylamide gels and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corporation) for 20 minutes at 25V. The blots were probed with the appropriate primary antibody and the appropriate fluorophore conjugated secondary antibody. The protein bands were visualized using the Licor Clx Odyssey imaging station (Licor Biosystems). Comparative changes were measured with Licor Image Studio software.
Cell-Proliferation Assay
Cells (5000 per well) were seeded in 96-well plates in phenol-red-free medium supplemented with charcoal-stripped FBS. Treatments were initiated after the cells were attached. At the appropriate time points, cell viability was determined by MTT assay; 10 μl of MTT (5mg/ml in phosphate-buffered saline) was added to each well followed by incubation at 37°C for 2 hours. The formazan crystal sediments were dissolved in 100 μl of dimethyl sulfoxide and absorbance was measured at 590 nm using the Tecan Infinite 200 Pro-plate reader. Each treatment was performed in seven replicate wells and repeated three times.
siRNA-mediated Gene Knockdown
The appropriate recombinant SW48 cells were plated in a 10 cm plate in DMEM supplemented with 10% FBS 24 h before transfection. The following day, cells were transfected with siRNAs against NF1 (2 μg) or control siRNA (2 μg) using Lipofectamine 2000. For EGFR knockdown, cells were plated 96 well format in 100 μl of OptiMEM (10%FBS) with 0.1μg of siRNA mixed with 0.5 μl Lipofectamine 2000 per well. 24hours following EGFR siRNA delivery, cells were treated with Cetuximab for 48 hours, and proliferation was measured by MTT assay. Silencer Select Small interfering RNAs were purchased from Thermo Fisher. Silencer Select-NF1 (s56534) was comprised of pooled RNAs targeting exons 2, 10, 16, 18 and 19 in the NF1 mRNA. Silencer Select-EGFR (s565) was comprised of pooled siRNA targeting five unique sequences within exon 2 of the EGFR mRNA. Silencer Select Control siRNA (4390843) was used as negative control. All siRNAs were reconstituted in RNase-free molecular grade water upon arrival from vendor at concentration of 5mM.
Expression Plasmid Transfection
Cells were plated in 96 well plate at 5000 cells per well in antibiotic free media. 24 h later cells were transfected with expression plasmids with duplex containing 0.2 μg of DNA and 0.25ul of Lipofectamine 2000 per well. Cell proliferation was assayed within at least 48 h.
Ras expression constructs from the NCI Ras Initiative clone collection for KRAS4B-WT (Addgene #83129), NRAS-WT (Addgene #83173), HRAS-WT (Addgene #83181), KRAS-G13D (Addgene #83133), KRAS-G12V (Addgene #83132), and KRAS-G12D (Addgene #83131) were Gateway cloned into EGFP expression vector pEZYegfp (Addgene #18671). KRAS G13D was used to create the G12V/G13D (GG/VD) through site directed mutagenesis. NF1 expression construct (Addgene #70423) was Gateway cloned into pEZYflag (Addgene #18700) and NF1 expression construct (Addgene #70424) was Gateway cloned into pcDNA3.1-ccdB-Nanoluc (Addgene #87067) and pLenti6.2-ccdB-Nanoluc (Addgene #87075). The RAS Clone Collection was a gift from Dominic Esposito (Addgene kit # 1000000070 and kit # 1000000089). pEZYegfp and pEZYflag were gifts from Yu-Zhu Zhang (Addgene plasmid # 18671 ; http://n2t.net/addgene:18671 ; RRID:Addgene_18671 and Addgene plasmid # 18700 ; http://n2t.net/addgene:18700 ; RRID:Addgene_18700). pLenti6.2-ccdB-Nanoluc and pcDNA3.1-ccdB-Nanoluc were gifts from Mikko Taipale (Addgene plasmid # 87075 ; http://n2t.net/addgene:87075 ; RRID:Addgene_87075 and Addgene plasmid # 87067 ; http://n2t.net/addgene:87067 ; RRID:Addgene_87067).
Colony-Formation Assay
Cells were trypsinized and 4000 cells per well were plated in triplicate 6-well plates in DMEM supplemented with FBS. Colonies were formed after 7 days. The cells were fixed with ice-cold methanol and stained with crystal violet. Images were obtained using the Licor Clx Odyssey imaging station (Licor Biosystems). Colony formation was quantified by measuring absorbance per well. Comparison were made by normalizing to control wells. A total of five experimental replicates were performed with each containing three biological replicates.
Active Ras Pull-down Assay
Isolation of active GTP bound Ras was performed using the Active Ras Pull-Down and Detection Kit (Thermo Fisher Scientific,) following manufacturers protocol. Ras abundance was measured by Western blot and/or by mass spectrometry. Western blot analysis of RBD pull-down lysates was performed with mouse anti-KRAS antibody (WH0003841, Sigma), Rabbit anti-NRAS (ab16713, Abcam), Rabbit anti-HRAS (18295, Protein Tech), mouse anti-pan-RAS antibody (1862335, Thermo Scientific) and mouse anti-GAPDH (sc-4772, Santa Cruz Biotechnology).
Mass Spectrometry
RBD lysates from cetuximab treated and non-treated cells (CTX; 20μg/ml for 48h) were precipitated using Methanol-Chloroform. Dried pellets were dissolved in 8 M urea, reduced with 5 mM tris (2-carboxyethyl) phosphine hydrochloride (TCEP), and alkylated with 50 mM chloroacetamide. Proteins were then trypsin digested overnight at 37° C. Samples were digested at 50 μl final volume. Heavy labeled peptides were spiked-in to the digested samples at appropriate concentrations so that a single LCMS injection contained 10 μl of digested sample with 500 fmol of heavy labeled peptides. Peptides used were: SFEDIHQYR for HRAS; SFADINLYR for NRAS; SFEDIHHYR for KRAS; LVVVGAGGVGK for wild-type H/N/KRAS; LVVVGAGDVGK for G13D mutant H/N/KRAS, and LVVVGAVGVGK for G12V mutant H/N/KRAS and the same peptide sequences were previously used for quantification of endogenous RAS and mutant RAS genes in similar isogenic SW48 cells (21).
The samples were analyzed on a Fusion mass spectrometer (Thermo Fisher Scientific). Samples were injected directly onto a 25 cm, 100 μm ID column packed with BEH 1.7 μm C18 resin (Waters). Samples were separated at a flow rate of 300 nL/min on a nLC 1200 (Thermo Fisher Scientific). Buffer A and B were 0.1% formic acid in water and 90% acetonitrile, respectively. A gradient of 1–25% B over 110 min, an increase to 40% B over 10 min, an increase to 100% B over another 10 min and held at 90% B for a final 10 min of washing was used for 140 min total run time. Peptides were eluted directly from the tip of the column and nanosprayed directly into the mass spectrometer by application of 2.8 kV voltage at the back of the column. The Fusion was operated in a data dependent mode. Full MS1 scans were collected in the Orbitrap at 120k resolution. The cycle time was set to 3 s, and within this 3s the most abundant ions per scan were selected for CID MS/MS in the ion trap. Monoisotopic precursor selection was enabled and dynamic exclusion was used with exclusion duration of 5 s
Peak area quantitation of the heavy peptides and corresponding light peptides from the samples were extracted by Skyline (42). Within each sample, we used mutant Ras as a standard to normalize against. We then compared the ratio of normalized wild-type peptide levels in cetuximab treated conditions to normalized wild-type peptide levels in non-cetuximab treated conditions.
Isoelectric Focusing of Active RAS Isoforms
SW48-WT RAS, SW48-KRAS G12V and SW48 KRAS G13D cells were cultured in T-75 adherent culture flasks. Cells were grown in growth media alone or growth media with Cetuximab (20 μg/ml of ) for 48 hours. Media was removed and cells were washed with ice cold TBS. Cells were scraped in 1 ml of lysis wash buffer (25mM Tris-HCl, pH 7.2, 150mM NaCl, 5mM MgCl2, 1% NP-40 and 5% glycerol). Cells were lysed on ice and were vortexed every 10 seconds. Cell lysates were subjugated to RBD co-immunoprecipitation as previously described above. RBD Co-Immunoprecipitation product was resolved by SDS PAGE in a 12% polyacrylamide gel. Bands were excised from the 21kD region of the gel. Gel products were liquified at 95°C for 5 minutes. Protein was extracted and purified utilizing the ReadyPrep 2-D cleanup Kit (BioRad Laboratories) following manufacturers protocol. Protein samples were added to 50% glycerol loading buffer and incubated at room temperature for 20 minutes. Samples and IEF Ladder were resolved on Criterion Bio-Lyte IEF Gel with a 3–10 pH range (BioRad Laboratories). Gels were run at the following power conditions with constant voltage: 100V for 60 minutes, 250V for 60 minutes, and 500 V for 30 minutes in a stepwise fashion with a total run time of 150 minutes. The IEF gel was then soaked in 5% SDS buffer for 24 hours with gentle rocking at 4°C. Protein was electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corporation) for 1hr at a constant 25V. The PVDF blots were probed with the Anti-pan-RAS primary antibody from the Active Ras Pull-Down and Detection Kit (Thermo Fisher Scientific) and the anti-mouse DyLight 800 fluorophore conjugated secondary antibody (Invitrogen). The protein bands were visualized using the Licor Clx Odyssey imaging station (Licor Biosystems). Comparative changes were measured with Licor Image Studio software.
Co-immunoprecipitation
H293T cells were individually transfected with the expression plasmid for NF1-Flag, WT KRAS-GFP, G12V KRAS-GFP, G12D KRAS-GFP or KRAS G13D-GFP. Cells were harvested in IP Lysis/Wash Buffer (0.025M Tris, 0.15M NaCl, 0.001M EDTA, 1% NP-40, 5% glycerol; pH 7.4 and 1× protease inhibitor) 24 h post-transfection. Whole cell lysates (500 μg) were pre-cleared for 0.5 h using Control Agarose Resin slurry (Thermo Fisher Scientific,). Immunoprecipitation was performed by first incubating 800 μl of H293T NF1-Flag pre-cleared lysate with 200 μl of either WT KRAS-GFP, G12V KRAS-GFP, G12D KRAS-GFP or G13D KRAS -GFP pre-cleared cell lysate. Each cell lysate mixture had EDTA (pH 8.0) added to make a final concentration of 10mM. GTP-gamma-S was added to the solution to a final concentration of 100nM. This solution was incubated at room temperature for 20 minutes with gentle rocking. The reaction was terminated by adding MgCl2 to the solution at a final concentration of 50mM. The final steps of the Co-IP were performed using the Pierce immunoprecipitation Kit (Thermo Fisher Scientific) with immobilized anti-NF1 Ab (Santa Cruz Biotechnologies, CA). 500 μg of the cell lysate was added and incubated at room temperature under rotary agitation for 2 h. At the end of the incubation, the complexes were washed five times with Lysis buffer. The Western blotting was probed with mouse monoclonal NF1 antibody (Santa Cruz Biotechnologies) and mouse monoclonal RAS antibody (Thermo Fisher Scientific).
Lentiviral Transduction
293FT cells were used to generate lentiviral particles by transfection using lipofectamine 2000 (Life Technologies Corporation). Packaging plasmids pMD2G, PMDLg/RRE, and pRSV/Rev were cotransfected with pCDH NF1-NanoLuc C-term expression plasmid. Lentivirus containing supernatant was harvested at 48 h and 72 h after transfection. LoVo, HCT116 and HCT-15 cells were plated in respective media with heat inactivated FBS (10%) and 2 mM L-Glutamine two days before infection. For infection, LoVo, HCT116 and HCT-15 cells were transduced with pCDH NF1-nanoLuc lentivirus with polybrene (8 μg/ml) for 10 hours. The cells were washed and media was replenished and cells were incubated for 48 h. Following this, cells were placed in 1μg/ml puromycin selection for seven days. Cells were harvested for western blots and MTT assays as described previously.
AKT phosphorylation
The pAKT antibodies were validated by starving WT SW48 cells in RPMI pen/strp media for 12 hours, cells were stimulated with EGF (50ng/ml) for 5 minutes. Whole cell lysates were prepared and resolved on 12% polyacrylamide gel. Gels were transferred to PDF membrane, and probed with anti-phospho-T308 AKT1 rabbit antibody (AB13038, Cell Signaling Technology), anti-phospho-S473 AKT1 rabbit monoclonal antibody (AB4060, Cell Signaling Technology) and anti-pan AKT mouse monoclonal antibody (AB2920, Cell Signaling Technology) in 3% Bovine Serum Albumin solution. Cell lines indicated were either treated with vehicle-control (Ctrl) or 20ug/ml of Cetuximab for 48 hours. Whole cell lysates were prepared and analyzed by western blot analysis as previously described.
Bioluminescence Resonance Energy Transfer (BRET) Assay
HEK-293T cells were grown in DMEM 10% FBS without antibiotic. Cells were seeded at 5×103 cells per well in a 96 well white opaque Perkin Elmer microplate. 24 hours after seeding, cells were co-transfected with a constant concentration 0.1 μg of NF1-NanoLuc pcDNA expression plasmid and increasing concentrations RAS-EGFP pcDNA expression plasmid (0, 0.05, 0.1, 0.2, 0.4, 0.6, 0.8 and 1.2 μg) with 0.25ul of Lipofectamine 2000 per well following manufacturers protocol (Thermo Fisher). 24 hours later, media was aspirated from each well and 25μl of Nano-Glo Live Cell Reagent was added to each well per manufacturer’s protocol (Promega). Plates were placed on orbital shaker for 1 minute at 300 RPM. Following incubation, the plate was read on the Tecan Infinite M200 Pro with LumiColor Dual Setting with an integration time of 1000ms. BRET ratio was calculated from the dual emission readings. BRET ratio was plotted as a function of the RAS-GFP/NF1-NanoLuc plasmid ratio. BRET assays were repeated five times, each with 8 biological replicates.
Statistical Analysis
Significant differences amongst sample groups of greater than or equal to three were determined by one-way ANOVA followed by post-hoc Tukey’s test for multiple comparisons with GraphPad Prism7 software. Significant differences amongst two sample groups was determined by one-tailed unpaired t-test. Mass spectrometry was performed twice. Every other experiment was performed at least three times, and P values are indicated in each figure.
Supplementary Material
Data file S1. Mass spectrometry data and analysis of untreated and cetuximab-treated isogenic SW48 cells.
Text S1. Extended description of the Ras model
Fig. S1. Schematic of the reactions of the Ras model.
Fig. S2. Available parameters for the Ras G13D mutant are sufficient to account for its constitutive activation.
Fig. S3. Ras model predictions when level of RasGTP-Effector complex is used as a readout of signal strength instead of RasGTP.
Fig. S4. Isogenic colon cancer cells display a KRAS mutant-specific response to cetuximab.
Fig. S5. Evaluation of the response of the isogenic panel to MEK inhibitor.
Fig. S6. Transfection-based cetuximab sensitivity assay.
Fig. S7. Evaluation of AKT phosphorylation upon cetuximab treatment.
Fig. S8. Evaluation of EGFR knockdown in isogenic SW48 cells.
Fig. S9. Evaluation of panitumumab treatment of isogenic SW48 cells.
Fig. S10. Evaluation of erlotinib treatment of isogenic SW48 cells.
Fig. S11. Evaluation of Ras antibodies.
Fig. S12. Detection of impaired binding between KRAS G13D and NF1.
Fig. S13. Evaluation of KRAS G13D CRC cell lines that have been transduced with NF1.
Acknowledgments:
We thank Shumei Kato for providing cetuximab, and Michael Elliott, Craig Wilen, Alan Saghatelian, and members of the Salk Institute Cancer Center for helpful conversations and feedback.
Funding: Support for this work was provided by NIH K22CA216318, NIH T32CA009370, and by the Mass Spectrometry Core of the Salk Institute with funding from NIH P30CA014195 and the Helmsley Center for Genomic Medicine.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository with the dataset identifier PXD014755. All materials are available upon request from the corresponding author.
References and Notes:
- 1.Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, van Hazel G, Wierzbicki R, Langer C, Moore MJ, Cetuximab for the treatment of colorectal cancer. N Engl J Med 357, 2040–2048 (2007); published online EpubNov 15 (10.1056/NEJMoa071834). [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, Wolf M, Amado RG, Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 25, 1658–1664 (2007); published online EpubMay 1 (10.1200/JCO.2006.08.1620). [DOI] [PubMed] [Google Scholar]
- 3.N. Cancer Genome Atlas, Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012); published online EpubJul 18 (10.1038/nature11252). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephen AG, Esposito D, Bagni RK, McCormick F, Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014); [DOI] [PubMed] [Google Scholar]
- 5.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR, K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359, 1757–1765 (2008); published online EpubOct 23 (10.1056/NEJMoa0804385). [DOI] [PubMed] [Google Scholar]
- 6.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD, Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26, 1626–1634 (2008); published online EpubApr 1 (10.1200/JCO.2007.14.7116). [DOI] [PubMed] [Google Scholar]
- 7.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O’Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S, Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. Jama 304, 1812–1820 (2010); published online EpubOct 27 (10.1001/jama.2010.1535). [DOI] [PubMed] [Google Scholar]
- 8.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E, Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol 30, 3570–3577 (2012); published online EpubOct 10 (10.1200/JCO.2012.42.2592). [DOI] [PubMed] [Google Scholar]
- 9.Nakamura M, Aoyama T, Ishibashi K, Tsuji A, Takinishi Y, Shindo Y, Sakamoto J, Oba K, Mishima H, Randomized phase II study of cetuximab versus irinotecan and cetuximab in patients with chemo-refractory KRAS codon G13D metastatic colorectal cancer (G13D-study). Cancer Chemother Pharmacol 79, 29–36 (2017); published online EpubJan (10.1007/s00280-016-3203-7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segelov E, Thavaneswaran S, Waring PM, Desai J, Robledo KP, Gebski VJ, Elez E, Nott LM, Karapetis CS, Lunke S, Chantrill LA, Pavlakis N, Khasraw M, Underhill C, Ciardiello F, Jefford M, Wasan H, Haydon A, Price TJ, van Hazel G, Wilson K, Simes J, Shapiro JD, Response to Cetuximab With or Without Irinotecan in Patients With Refractory Metastatic Colorectal Cancer Harboring the KRAS G13D Mutation: Australasian Gastro-Intestinal Trials Group ICECREAM Study. J Clin Oncol 34, 2258–2264 (2016); published online EpubJul 1 (10.1200/JCO.2015.65.6843). [DOI] [PubMed] [Google Scholar]
- 11.Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R, Wiezorek J, Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol 31, 759–765 (2013); published online EpubFeb 20 (10.1200/JCO.2012.45.1492). [DOI] [PubMed] [Google Scholar]
- 12.Morelli MP, Kopetz S, Hurdles and complexities of codon 13 KRAS mutations. J Clin Oncol 30, 3565–3567 (2012); published online EpubOct 10 (10.1200/JCO.2012.43.6535). [DOI] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013); published online EpubApr 2 (10.1126/scisignal.2004088). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018. CA Cancer J Clin 68, 7–30 (2018); published online EpubJan (10.3322/caac.21442). [DOI] [PubMed] [Google Scholar]
- 15.Stites EC, Trampont PC, Ma Z, Ravichandran KS, Network analysis of oncogenic Ras activation in cancer. Science 318, 463–467 (2007); published online EpubOct 19 (10.1126/science.1144642). [DOI] [PubMed] [Google Scholar]
- 16.Stites EC, Trampont PC, Haney LB, Walk SF, Ravichandran KS, Cooperation between Noncanonical Ras Network Mutations. Cell Rep, (2015); published online EpubJan 15 (10.1016/j.celrep.2014.12.035). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gremer L, Gilsbach B, Ahmadian MR, Wittinghofer A, Fluoride complexes of oncogenic Ras mutants to study the Ras-RasGap interaction. Biol Chem 389, 1163–1171 (2008); published online EpubSep (10.1515/BC.2008.132). [DOI] [PubMed] [Google Scholar]
- 18.Palmioli A, Sacco E, Airoldi C, Di Nicolantonio F, D’Urzo A, Shirasawa S, Sasazuki T, Di Domizio A, De Gioia L, Martegani E, Bardelli A, Peri F, Vanoni M, Selective cytotoxicity of a bicyclic Ras inhibitor in cancer cells expressing K-Ras(G13D). Biochem Biophys Res Commun 386, 593–597 (2009); published online EpubSep 4 (10.1016/j.bbrc.2009.06.069). [DOI] [PubMed] [Google Scholar]
- 19.Hood FE, Klinger B, Newlaczyl AU, Sieber A, Dorel M, Oliver SP, Coulson JM, Bluthgen N, Prior IA, Isoform-specific Ras signaling is growth factor dependent. Mol Biol Cell 30, 1108–1117 (2019); published online EpubApr 15 (10.1091/mbc.E18-10-0676). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, Wong KK, Cantley LC, Engelman JA, Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest 121, 4311–4321 (2011); published online EpubNov (10.1172/JCI57909). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mageean CJ, Griffiths JR, Smith DL, Clague MJ, Prior IA, Absolute Quantification of Endogenous Ras Isoform Abundance. PLoS One 10, e0142674 (2015)10.1371/journal.pone.0142674). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young A, Lou D, McCormick F, Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov 3, 112–123 (2013); published online EpubJan (10.1158/2159–8290.CD-12–0231). [DOI] [PubMed] [Google Scholar]
- 23.Lito P, Rosen N, Solit DB, Tumor adaptation and resistance to RAF inhibitors. Nat Med 19, 1401–1409 (2013); published online EpubNov (10.1038/nm.3392). [DOI] [PubMed] [Google Scholar]
- 24.Bollag G, McCormick F, Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351, 576–579 (1991); published online EpubJun 13 (10.1038/351576a0). [DOI] [PubMed] [Google Scholar]
- 25.Keller JW, Haigis KM, Franklin JL, Whitehead RH, Jacks T, Coffey RJ, Oncogenic K-RAS subverts the antiapoptotic role of N-RAS and alters modulation of the N-RAS:gelsolin complex. Oncogene 26, 3051–3059 (2007); published online EpubMay 10 (10.1038/sj.onc.1210103). [DOI] [PubMed] [Google Scholar]
- 26.Lim KH, Ancrile BB, Kashatus DF, Counter CM, Tumour maintenance is mediated by eNOS. Nature 452, 646–649 (2008); published online EpubApr 3 (10.1038/nature06778). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D, Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 25, 243–256 (2014); [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stites EC, Differences in sensitivity to EGFR inhibitors could be explained by described biochemical differences between oncogenic Ras mutants. bioRxiv 10.1101/005397, (2014). [DOI] [Google Scholar]
- 29.Jeng HH, Taylor LJ, Bar-Sagi D, Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat Commun 3, 1168 (2012)10.1038/ncomms2173). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corbalan-Garcia S, Yang SS, Degenhardt KR, Bar-Sagi D, Identification of the mitogen-activated protein kinase phosphorylation sites on human Sos1 that regulate interaction with Grb2. Mol Cell Biol 16, 5674–5682 (1996); published online EpubOct (10.1128/mcb.16.10.5674). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stites EC, Modeling membrane localization: case study of a Ras signaling model. Adv Exp Med Biol 680, 661–667 (2010)10.1007/978-1-4419-5913-3_73). [DOI] [PubMed] [Google Scholar]
- 32.Stites EC, Ravichandran KS, Mathematical investigation of how oncogenic ras mutants promote ras signaling. Methods Mol Biol 880, 69–85 (2012)10.1007/978-1-61779-833-7_5). [DOI] [PubMed] [Google Scholar]
- 33.Stites EC, Ravichandran KS, Mechanistic modeling to investigate signaling by oncogenic Ras mutants. Wiley Interdiscip Rev Syst Biol Med 4, 117–127 (2012); published online EpubJan-Feb (10.1002/wsbm.156). [DOI] [PubMed] [Google Scholar]
- 34.Stites EC, Shaw AS, Quantitative Systems Pharmacology Analysis of KRAS G12C Covalent Inhibitors. CPT Pharmacometrics Syst Pharmacol 7, 342–351 (2018); published online EpubMay (10.1002/psp4.12291). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eccleston JF, Moore KJ, Brownbridge GG, Webb MR, Lowe PN, Fluorescence approaches to the study of the p21ras GTPase mechanism. Biochem Soc Trans 19, 432–437 (1991); published online EpubApr ( [DOI] [PubMed] [Google Scholar]
- 36.Chuang E, Barnard D, Hettich L, Zhang XF, Avruch J, Marshall MS, Critical binding and regulatory interactions between Ras and Raf occur through a small, stable N-terminal domain of Raf and specific Ras effector residues. Mol Cell Biol 14, 5318–5325 (1994); published online EpubAug ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischbach MA, Settleman J, Specific biochemical inactivation of oncogenic Ras proteins by nucleoside diphosphate kinase. Cancer Res 63, 4089–4094 (2003); published online EpubJul 15 ( [PubMed] [Google Scholar]
- 38.Donovan S, Shannon KM, Bollag G, GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta 1602, 23–45 (2002); published online EpubMar 14 ( [DOI] [PubMed] [Google Scholar]
- 39.Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A, Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry 37, 7420–7430 (1998); published online EpubMay 19 (10.1021/bi972621j). [DOI] [PubMed] [Google Scholar]
- 40.Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A, Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry 36, 4535–4541 (1997); published online EpubApr 15 (10.1021/bi962556y). [DOI] [PubMed] [Google Scholar]
- 41.Omerovic J, Hammond DE, Clague MJ, Prior IA, Ras isoform abundance and signalling in human cancer cell lines. Oncogene 27, 2754–2762 (2008); published online EpubApr 24 (10.1038/sj.onc.1210925). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010); published online EpubApr 1 (10.1093/bioinformatics/btq054). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sydor JR, Engelhard M, Wittinghofer A, Goody RS, Herrmann C, Transient kinetic studies on the interaction of Ras and the Ras-binding domain of c-Raf-1 reveal rapid equilibration of the complex. Biochemistry 37, 14292–14299 (1998); published online EpubOct 6 (10.1021/bi980764f). [DOI] [PubMed] [Google Scholar]
- 44.Rudolph MG, Linnemann T, Grunewald P, Wittinghofer A, Vetter IR, Herrmann C, Thermodynamics of Ras/effector and Cdc42/effector interactions probed by isothermal titration calorimetry. J Biol Chem 276, 23914–23921 (2001); published online EpubJun 29 (10.1074/jbc.M011600200). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. Mass spectrometry data and analysis of untreated and cetuximab-treated isogenic SW48 cells.
Text S1. Extended description of the Ras model
Fig. S1. Schematic of the reactions of the Ras model.
Fig. S2. Available parameters for the Ras G13D mutant are sufficient to account for its constitutive activation.
Fig. S3. Ras model predictions when level of RasGTP-Effector complex is used as a readout of signal strength instead of RasGTP.
Fig. S4. Isogenic colon cancer cells display a KRAS mutant-specific response to cetuximab.
Fig. S5. Evaluation of the response of the isogenic panel to MEK inhibitor.
Fig. S6. Transfection-based cetuximab sensitivity assay.
Fig. S7. Evaluation of AKT phosphorylation upon cetuximab treatment.
Fig. S8. Evaluation of EGFR knockdown in isogenic SW48 cells.
Fig. S9. Evaluation of panitumumab treatment of isogenic SW48 cells.
Fig. S10. Evaluation of erlotinib treatment of isogenic SW48 cells.
Fig. S11. Evaluation of Ras antibodies.
Fig. S12. Detection of impaired binding between KRAS G13D and NF1.
Fig. S13. Evaluation of KRAS G13D CRC cell lines that have been transduced with NF1.