Abstract
Newcastle disease (ND) and avian influenza (AI) are economically important infectious diseases of poultry. Sometime, concomitant secondary viral/or bacterial infections significantly alters the pathobiology of ND and AI in poultry. As of now, the disease patterns and dynamics of co-infections caused by ND virus (NDV, genotype XIII) and Low Pathogenic AI viruses (LPAI, H9N2) are explicitly elusive. Thus, we examined the clinicopathological disease conditions due to these two economically important viruses to understand the complex disease outcomes by virus–virus interactions in vaccinated flocks. The findings of clinicopathological and molecular investigations carried on 37 commercial ND vaccinated poultry flocks revealed simultaneous circulation of NDV and AIV in same flock/bird. Further, molecular characterization of hemagglutinin (HA) and neuraminidase (NA) genes confirmed that all the identified AIVs were of low pathogenicity H9N2 subtype and fusion (F) gene analysis of detected NDVs belong to NDV class II, genotype XIII, a virulent type. The NDV and H9N2 alone or co-infected flocks (NDV + LPAI) exhibit clinical signs and lesions similar to that of virulent NDV except the degree of severity, which was higher in H9N2–NDV co-infected flocks. Additionally, avian pathogenic E. coli and mycoplasma infections were detected in majority of the ailing/dead birds from the co-infected flocks during progression of the clinical disease. Overall, the findings highlight the multi-factorial disease complexity in commercial poultry and suggest the importance of NDV genotype XIII in intensifying the clinical disease in vaccinated birds.
Keywords: Clinicopathological patterns, Newcastle disease, Low pathogenic avian influenza, E. coli, Mycoplasma, Poultry
Introduction
Newcastle disease (ND) and avian influenza (AI) are the most devastating diseases of poultry and remain the foremost constraint to the growth of poultry industry around the globe. The ND is caused by the avian paramyxovirus serotype-1 (APMV-1), which is a member of the genus Avulavirus, family Paramyxoviridae under the order Mononegavirales [30, 44]. The virulent and avirulent ND viruses (NDVs) have the sequence of 112R/K–R–Q/K/R–K/R–R–F117 and 112G/E–K/R–Q–G/E–R–L117 in fusion protein (F) cleavage site, respectively [35]. All APMVs can be divided into 17 serotypes (APMV 1–17) [19] out of which APMV-1 is found to be associated with naturally occurring NDV infections with major economic consequences [46]. The APMV-1 can be divided into two distinct clades: class I and II, and class II is subdivided into 18 recognized genotypes [8]. The most recent fifth panzootic velogenic NDV (vNDV) belongs to a new sub-genotype VIIi, VIIh, XIIIa, XIIIb and XIIIc [30, 32].
Avian influenza is caused by various subtypes of influenza viruses of the family Orthomyxoviridae [25]. Two types of AI viruses (AIV) have been described based on their pathogenicity viz. high pathogenicity avian influenza viruses (HPAIV) that cause severe disease with high mortality, and low pathogenicity avian influenza virus (LPAIV) that generally causes asymptomatic infection or a mild disease [6]. ND and AI are the primary viral diseases of poultry worldwide and co-infection of NDV and AIV has been reported frequently [9, 10, 13, 38]. These days, reports are emerging on the detection of genotype XIII of NDV in vaccinated poultry flocks in several Asian countries [18, 21, 30, 41]. Additionally, NDV genotype XIII co-association with LPAIV–H9N2 may lead to severe clinical disease and results in high mortality compared to NDV alone [14]. The disease patterns during co-infections of genotype XIII NDV and LPAI are not well understood and information are limited on the impact of NDV–AIV co-infection on the mortality and severity of clinical disease especially in field conditions.
This study describes the clinicopathological disease caused by the genotype XIII of NDV and its exacerbation with the co-infecting LPAI–H9N2 subtype in the field conditions. The information presented will not only help to understand the virus–virus interactions and consequences on the bird’s health but would also highlight the multi-factorial disease complexity in the natural field conditions.
Materials and methods
Case history, pathological studies and sample collection
The study was carried out in 37 commercial multi aged poultry flocks vaccinated against NDV (LaSota at 7th day, NDV Killed at 14th day, LaSota at 28th day, Kumarov/R2B at 11th week and ND killed at the point of lay) from southern India including Tamil Nadu and Karnataka states with a history of respiratory/neurological signs and production drops during the period between January 2010–2012. Necropsy examination was carried out on dead and ailing birds. Pooled tissue samples such as brain, trachea, lungs, kidneys, spleen and intestines from 6 birds/flock were considered as a single pool for the virus isolation and histopathological studies. Heart blood and tracheal swabs were collected for bacterial and mycoplasma isolation. The clinical disease pattern was monitored until the mortality decreased to average rates up to 4 weeks.
Virus isolation
The pooled tissue samples were homogenized in phosphate buffered saline (PBS, pH 7.4) to obtain a 10% suspension, and clarified at 12,000× g for 10 min. After 1 h incubation with antibiotics, 100 µL of suspension was inoculated into three 11 day-old specific pathogen free (SPF) embryonated chicken eggs via allantoic route. The eggs were incubated at 37 °C till embryo death or up to 5 days, and haemagglutination (HA) test was carried out in amnioallantoic fluid (AAF) [35].
Histopathology
The collected tissues were processed and embedded in paraffin. Sections of 4 µm thickness were made and stained with Haematoxylin and Eosin [26].
Screening of NDV and AIV
Viral RNA from HA positive AAF was extracted using QIAamp Viral RNA Mini Kit (Qiagen, Germany). The cDNA was synthesized by random primer using SuperScript® First-Strand Synthesis System for RT-PCR (Invitrogen, Germany). The presence of AIV and NDV was confirmed by PCR targeting the M gene [36], and F gene [47], respectively. Typing of influenza A viruses and NDV was performed by further PCR amplification, sequencing of the HA, NA [17], and F genes of AIV and NDV, respectively by commercially.
Phylogenetic analysis
To ascertain the epidemiological aspects, the consensus sequences of F gene from each of the NDV positive isolate were aligned with representative virus strains of known genotype in the BioEdit software. The aligned sequence datasets were edited to equal length and were used to construct tree in MEGA7 using neighbor-joining method with 1000 bootstrap values [24].
Partial nucleotide sequences for the HA (616 bp) and NA (570) were aligned with CLC 6.6.2 (CLC Bio, Aarhus, Denmark). Maximum-likelihood analyses were performed using the software PhyML 3.0 [15] with the general time reversible (GTR) evolutionary model, an estimation of the proportion of invariable sites (I) and of the nucleotide heterogeneity of substitution rates (α), as selected by ModelGenerator 0.85 [20]. Nodal supports were assessed with 1000 bootstrap replicates.
Bacterial isolation and molecular detection
Standard methods for bacterial isolation as described by AAAP [12] were followed. Bacterial DNA from the culture was extracted using Wizard® Genomic DNA Purification Kit (Promega, USA). The presence of E. coli, Avibacterium paragallinarum and Ornithobacterium rhinotracheale (ORT) was screened by PCR targeting of 16S gene [29], followed by restriction fragment length polymorphism (RFLP) using restriction enzymes such as DdeI, RsaI, and EcoRI (M/s Fermentas, USA).
Screening for mycoplasma
Isolation of mycoplasmas from the tracheal swabs was carried out in Frey’s modified mycoplasma broth and agar medium [34]. The mycoplasmal DNA from all the cultures was isolated by using Wizard® Genomic DNA Purification Kit (Promega, USA). The presence of mycoplasma was confirmed by PCR targeting of 16S gene of Mycoplasma gallisepticum and Mycoplasma synoviae [34].
Statistical analysis
Data were analyzed using SPSS version 17.0 software. The duration of illness between LPAI and LPAI and ND co-infected farms were analyzed using student’s t test. Odds ratio (OR) and 95% confidence intervals (CIs) were calculated for risk for death (mortality rate) between ND and LPAI + ND co-infected flocks. The probability value (P) < 0.05 was considered as statistically significant.
Results
Virological and epidemiological findings
Out of 37 ND vaccinated poultry flocks screened, NDV of virulent pathotype was detected in 22 farms (59.5%) and LPAI virus (H9N2) in 11 farms (29.7%) by virus isolation, RT-PCR and sequencing. Notably, all AIV positive flocks were found to be simultaneously co-infected with NDV. Samples from all 22 flocks were further processed for genetic and phylogenetic analysis of the virus isolates. Sequence analysis of HA (CY099356, CY099357) and NA (CY099363, CY099347) genes of AIV revealed that all were belonging to low pathogenic H9N2 subtype (RSSR*G in HA gene). Putative protein sequence analysis of F protein of the NDV isolates revealed that presence of a motif, RRRKR*F, typical for the velogenic strains of NDV which complement the clinical picture of the disease.
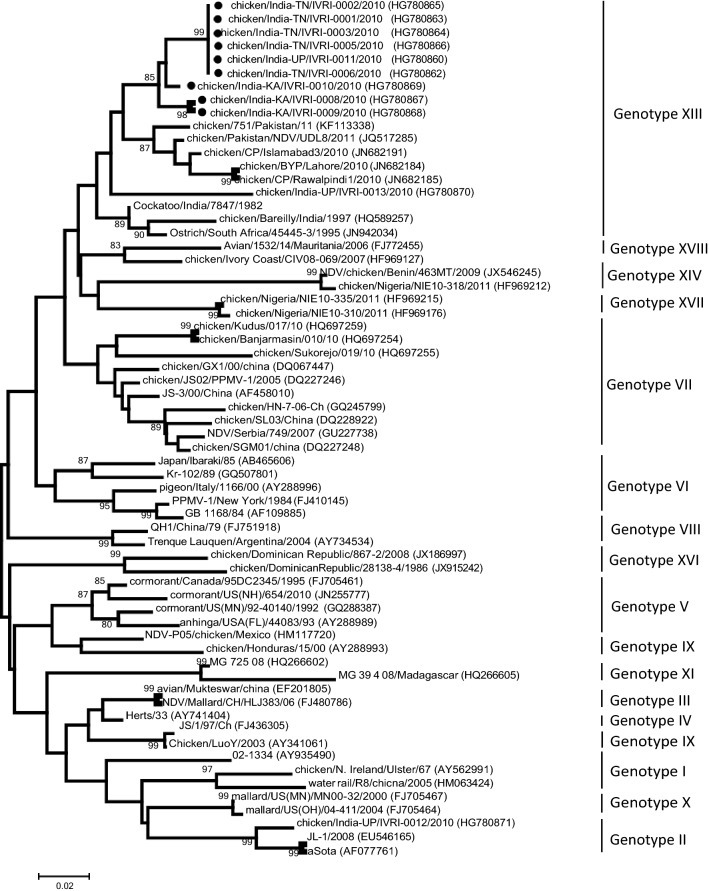
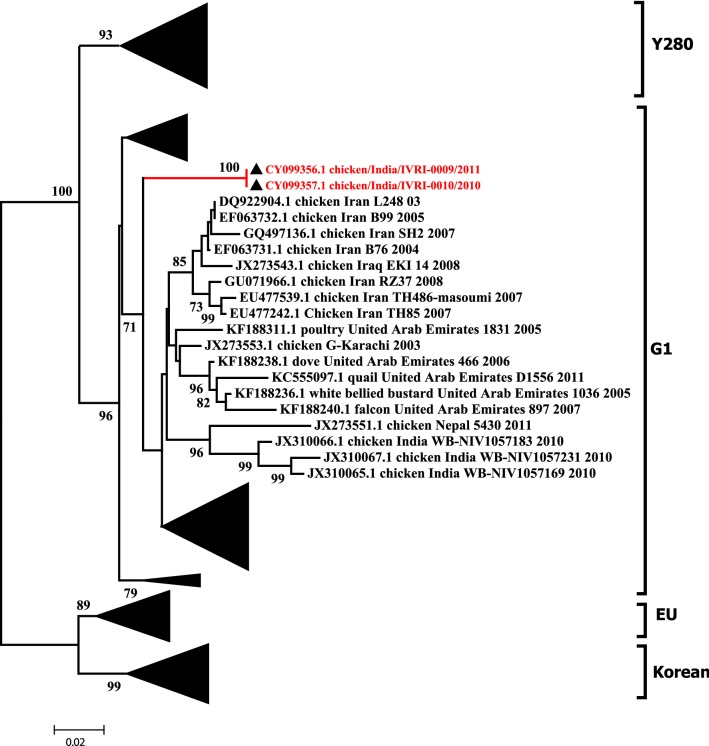
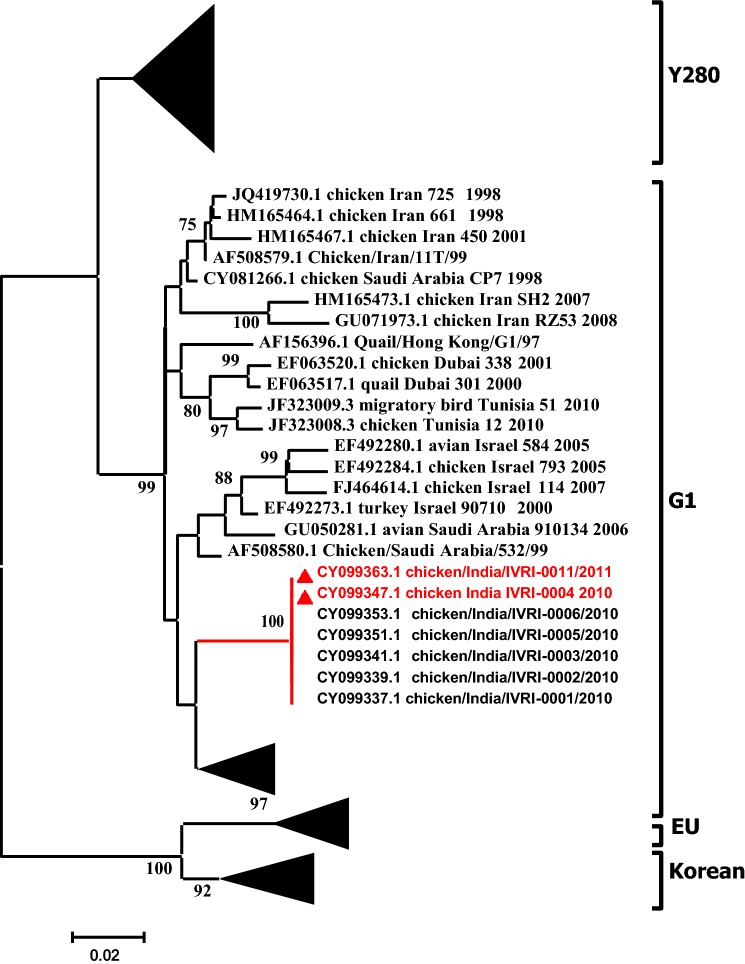
To understand the epidemiological clustering and distribution patterns of the NDV co-infecting the commercial poultry along with influenza, F gene based phylogenetic analysis was performed. The clustering pattern indicates the grouping of virus isolates into XIII genotype within Class II (Fig. 1). Phylogenetic analyses of the partial HA and NA genes revealed that multiple H9N2 genetic lineages are circulating in the poultry population of South Asia and in the Middle East during the last one decade. Although, the H9N2 subtype have been isolated from chicken and ducks in Eastern Asia, analysis of the HA gene suggests that it is different from the one documented in South Asia (India, Pakistan, Bangladesh). Nucleotide sequences obtained from the viruses isolated in this study were genetically related and formed independent genetic lineages within G1 clade, which is the most prevalent group in South East Asia (Fig. 2). Based on the NA gene analysis, a corresponding clustering pattern was observed where virus isolates reported in this study grouped with the AIV strains reported previously from Middle East and from India belonging to G1 clade (Fig. 3).
Fig. 1.
a Phylogenetic analysis of the partial F gene sequences representing different genotypes. Sequences analyzed in this study are marked with arrow. b A higher resolution clustering patterns of the NDV strains studied here and those that are available in public domains. Sequences presented here are marked with black filled circle. Boostraps lower than 80% are not shown
Fig. 2.
Maximum likelihood consensus tree derived from H9 sequences of influenza A virus hemagglutinin nucleotide sequences. Computations were realized with the GTR + I + G evolutionary model (I = 0.42; α = 1.13). Indian isolates are shown in red color. Bootstrap values lower than 50% were not shown. Labeling of branches is shown on the right side of the tree
Fig. 3.
Maximum likelihood consensus tree derived from influenza A virus neuraminidase nucleotide sequences. Computations were realized with the GTR + I + G evolutionary model (I = 0.29; α = 0.72). Sequences generated in this study and corresponding accession numbers are indicated by a filled triangle. Bootstrap values lower than 50% were not shown. Labeling of branches is shown on the right side of the tree
The mortality percentage was observed higher in flocks co-infected with NDV and AIV (2.6–44.40%) than NDV alone infected flocks (0.8–12%) and the difference was highly significant (P < 0.001) (Tables 1, 2). Flocks infected with only NDV were found to recover as early as 15–20 days post infection, whereas the recovery period was prolonged to 4–6 weeks in NDV and AIV co-infected flocks. The duration of illness in NDV and AIV co-infected flocks was significantly longer (P < 0.001) than the ND group (Table 2). Further, E. coli and mycoplasma were detected in all the ailing/dead birds from the above co-infected flocks during progression of the clinical disease, (after 7 days of initial appearance of clinical disease); whereas detection rate of E. coli and mycoplasma was only in seven NDV alone infected flocks. The mortality attained peak after 2–3 days of initial detection of mycoplasma and E. coli in both co-infection and NDV alone infected flocks, thereafter it declined rapidly in NDV alone infected flocks, whereas it extended up to one month in LPAI co-infected flocks.
Table 1.
Epidemiological findings of layer flocks affected with co-infection of LPAI, ND, E. coli, and mycoplasma
| S. no. | Flock code | Affected flock size | Age (weeks) | Mortality (%) | Duration of illness (days)a | Co-infections detected |
|---|---|---|---|---|---|---|
| 1. | LP1 | 100,000 | 76 | 4.90 | 40 | LPAI, ND, MG, MS and APEC |
| 2. | LP2 | 40,000 | 34 | 4.00 | 33 | LPAI, ND, MG, MS and APEC |
| 3. | LP3 | 6000 | 31 | 2.60 | 30 | LPAI, ND and APEC |
| 4. | LP4 | 7000 | 29 | 3.40 | 36 | LPAI, ND, MG and APEC |
| 5. | LP5 | 50,000 | 8 | 22.02 | 29 | LPAI, ND, MG, MS and APEC |
| 6. | LP6 | 25,000 | 7 | 15.41 | 38 | LPAI, ND, MS and APEC |
| 7. | LP7 | 15,000 | 8 | 20.00 | 27 | LPAI, ND, MG, MS and APEC |
| 8. | LP8 | 3800 | 6 | 40.82 | 45 | LPAI, ND and APEC |
| 9. | LP9 | 18,000 | 8 | 44.40 | 42 | LPAI, ND and APEC |
| 10. | LP10 | 5400 | 4 | 31.44 | 38 | LPAI, ND and APEC |
| 11. | LP11 | 4500 | 14 | 27.60 | 35 | LPAI, ND, MG and MS |
| 12. | N2 | 11,500 | 9 | 11.4 | 18 | ND, MG, MS and APEC |
| 13. | N4 | 4000 | 7 | 12.0 | 16 | ND, MG, MS and APEC |
| 14. | N8 | 22,000 | 25 | 1.4 | 11 | ND and MS |
| 15. | B9 | 7500 | 10 | 10.6 | 20 | ND, MG, MS and APEC |
| 16. | B10 | 12,000 | 8 | 10.2 | 15 | ND, MS and APEC |
| 17. | B11 | 9600 | 6 | 11.7 | 17 | ND, MG, MS and APEC |
| 18. | B12 | 10,000 | 35 | 1.2 | 7 | ND, MS and APEC |
| 19. | N1 | 8000 | 12 | 8.20 | 12 | ND |
| 20. | N3 | 10,000 | 34 | 0.8 | 8 | ND |
| 21. | B13 | 4800 | 8 | 9.0 | 14 | ND |
| 22. | B14 | 15,000 | 7 | 8.8 | 18 | ND |
ND Newcastle disease, LPAI low pathogenic avian influenza-H9N2, MG Mycoplasma gallisepticum, MS Mycoplasma synoviae, APEC avian pathogenic E. coli
aCalculated based on the farm mortality record after recovery
Table 2.
Statistical analysis of mortality and duration of illness between LPAI + ND co-infected and ND alone infected flocks
| LPAI + ND group | ND group | |
|---|---|---|
| Duration of illness | ||
| Mean ± SE (in days) | 35.73 ± 1.70 | 14.18 ± 1.28 |
| t Value | 10.15 | |
| P value | < 0.001 | |
| Mortality | ||
| Mean ± SE (in %) | 19.69 ± 4.58 | 7.75 ± 1.33 |
| OR (95% CI) | 2.94 (2.25–3.83) | |
| P value | < 0.001 | |
Clinical and pathological findings in young birds (growers) in LPAI co-infected flocks
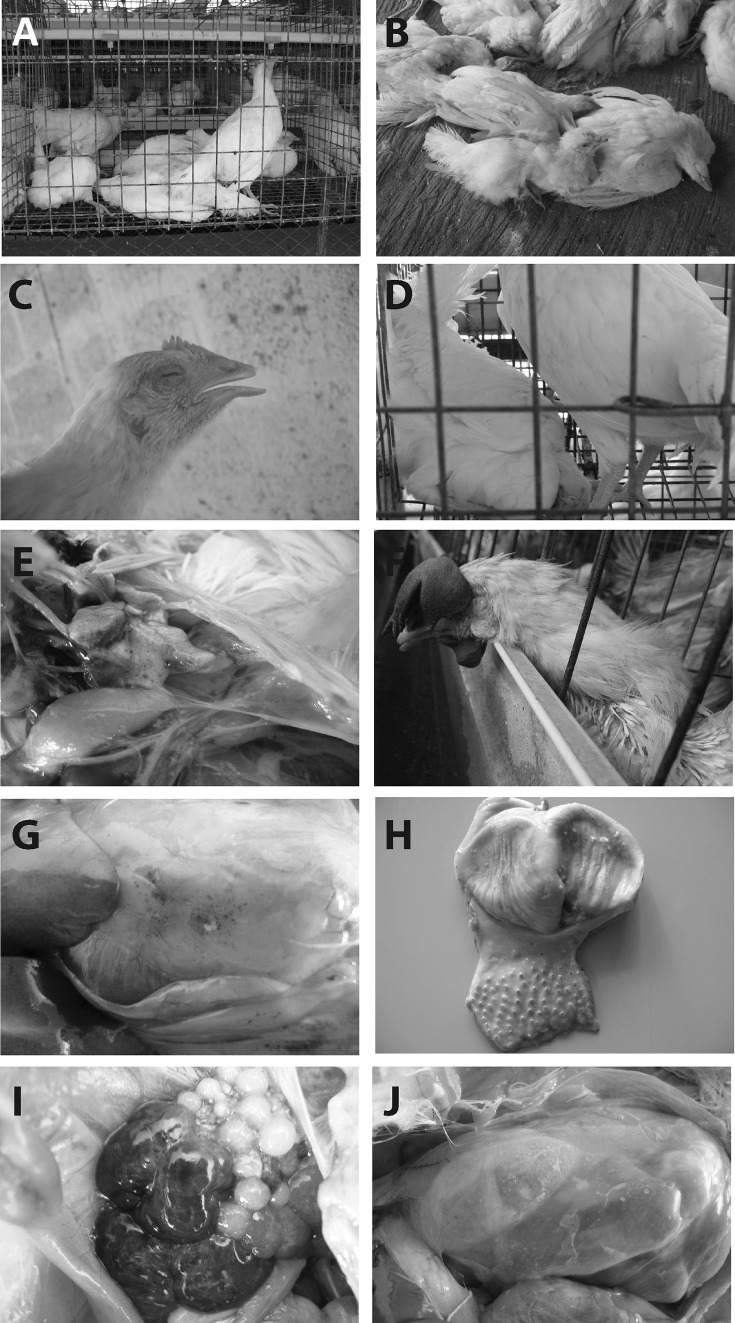
During initial stages, sudden death without any premonitory signs of the disease was observed (1–4 days). Later, the signs progressed as dullness, depression, somnolascence (Fig. 4a), ruffled feathers, reduced feed intake, water consumption, stunted growth (Fig. 4b), watery/greenish/white diarrhoea and prostration. Gasping (Fig. 4c), coryza and rales were suggestive of severe respiratory distress. Progressive neurological signs including of torticollis (Fig. 4d), head twitching, leg weakness, leg paralysis and loss of clutch reflex were consistently observed. The entire clinical course of the disease persisted for 3–4 weeks in all the complicated cases and the growers succumbed more rapidly than layers. Mortality rate was recorded between 15.4 and 44.4%.
Fig. 4.
Clinical picture of the virus infected birds. a Birds exhibiting depression and somnalescence, b birds exhibiting uneven growth in the affected flock, c the affected bird showing facial edema, lacrimal discharge, closed eye lids, severe respiratory distress with open mouth breathing, d affected bird displays torticollis, e lungs showing congestion, edema and frothy exudate, f dry, wrinkled, and cyanotic appearance of the comb, g multiple petechiae on the surface of gizzard fat, h moderate multifocal petechiae at the tips of proventricular papillae, i acute follicular haemorrhage results in purplish/tarry and enlarged follicles with liver-like appearance, j thickened and opaque abdominal air sacs
On postmortem examination, per-acutely died birds showed mild to marked congestion in the visceral organs. Birds died after 4–5 days of initial disease outbreak showed emaciated carcass, congestion of brain, haemorrhagic/catarrhal tracheitis, airsacculitis, diffuse pulmonary congestion and oedema (Fig. 4e), haemorrhagic proventriculitis, button ulcers in the small intestine, necrotic pancreatitis, spleenic atrophy and or mottling, nephritis-nephrosis complex and cloacal haemorrhages. The birds died 1 week after onset of clinical disease exhibited, thoracic airsacculitis, fibrinous adhesive pericarditis and fibrinous peri-hepatitis.
In histopathology, significant alterations were observed in brain, trachea, lung, proventriculus, intestine, pancreas, spleen, and kidney. Glial cell proliferation, neuronal shrinkage and neuronophagia were observed in the brain. Trachea showed extensive erosion of the epithelial layer, which exposed the stromal vessels leading to the initiation and progression of haemorrhages. The lesions in the lungs consisted of peribronchiolar oedema, and infiltration of mononuclear inflammatory cells. Marked haemorrhages and degeneration were observed in the mucosal folds of proventriculus. The predominant lesions in the intestines included degeneration and sloughing off of the epithelial cells covering tips of villi and mononuclear infiltrations. The lesions in the pancreas consisted of multifocal acute cellular swelling resulting in necrosis and shrunken acinar cells devoid of zymogen granules. Focal vacuolation and depletion of the follicular lymphocytes was observed in the spleen. In kidneys, the lesions consisted of tubular degeneration and necrosis with mild interstitial haemorrhages.
Clinicopathological findings in adult birds (layers) in LPAI co-infected flocks
Spread of the disease was comparatively slower in adult layers than young birds. The mortality rate ranged between 2.6 and 4.9%. Clinical signs consisted of depression, prostration, somnolascence, decreased feed intake and water consumption, facial swelling, cyanotic combs and wattles (Fig. 4f), drop/complete cessation of egg production, purulent conjunctivitis, watery diarrhea, gasping, loss of egg quality; including uneven sized and leathery eggs, small eggs etc. However, the respiratory signs were less severe in layers when compared to growers. Interestingly, the layers failed to exhibit any neurological signs, which were prominent in growers.
In necropsy examination, no lesions were detected form the birds that died during initial stage of the clinical disease (1–4 days), but the NDV and AIV could be detected from AAF. After 4 days, the lesions were largely confined to proventriculus, visceral and parietal peritoneum and abdominal cavity. Cachexia, caseous plugs in the larynx, haemorrhagic/catarrhal tracheitis/clear trachea, pulmonary congestion and edema, airsacculitis, petechiae in serosal layer of heart and abdominal fat (Fig. 4g), haemorrhagic proventriculitis (Fig. 4h), catarrhal enteritis, necrotic pancreatitis, atrophy of spleen, egg peritonitis, oophoritis, flaccid, misshapen/ruptured/haemorrhagic follicles (Fig. 4i), salpingitis nephritis-nephrosis complex and cloacal haemorrhages were observed in entire course of the disease. Fibrinous perihepatitis, pericarditis and abdominal airsacculitis (Fig. 4j) were observed after 1 week of the clinical onset. The clinical course of the disease continued for 2–3 weeks.
The prevailing histopathological alterations were restricted to the trachea, pancreas, spleen, peritoneal cavity, ovary and uterus. The lesions observed in respiratory, digestive tract, pancreas, spleen and kidneys were similar to that of HP lesions exhibited by growers. In addition to that diffuse serositis (peritonitis) including severe engorgement of vasculature, fibrous tissue proliferation and mononuclear cell infiltrations were observed in peritoneal cavity. Atretic follicles were characterized by the shrunken oocyte, separation of granulosa layer from thecal layer, and shrinkage of yolk spherules. Degeneration of granulosa and marked to severe haemorrhages in the follicular cavity of non-bursting type of follicles were also noticed. Oviduct showed atrophy and degeneration of tubular glands along with infiltrations of mononuclear inflammatory cells.
Clinical disease and pathological findings in NDV alone flocks
The young and adult birds exhibited similar clinical signs and lesions, but the severity of symptoms and lesions were less and recovery was faster when compared to LPAI co-infected flocks (Tables 3, 4).
Table 3.
Clinical findings of layer flocks affected with co-infection of LPAI/ND/E. coli/mycoplasma
| S. no. | Clinical finding | LPAI + NDa | NDa |
|---|---|---|---|
| 1. | Onset | Sudden | Gradual |
| 2. | Progress/spread of clinical disease | Rapid | Slow |
| 2. | Mortality | 15–44% | < 5% |
| 3. | Sudden death without any clinical signs | Observed | Not observed |
| 5. | Uniformity of the flock | Uneven | Even |
| 6. | Week birds | More prominent | Less prominent |
| 7. | Dullness and depression | More prominent | Varied between farms |
| 8. | Reduced feed intake and water consumption | More prominent | Less prominent |
| 9. | Greenish diarrhea | More prominent | Not observed |
| 10. | Ruffled feathers | More prominent | Absent |
| 11. | Respiratory distress | More prominent | Less prominent |
| 12. | Neurological signs | Very severe | Absent |
| 13. | Clinical course | 4–5 weeks | 2–3 weeks |
aBoth groups were complicated with MG, MS and E. coli in different combinations when the disease advanced
Table 4.
Postmortem findings observation in layer flocks affected with co-infection of LPAI/ND/E. coli/mycoplasma
| S. no. | Pathological finding | LPAI + ND | ND |
|---|---|---|---|
| 1. | No prominent lesion | Characteristic during initial 3–5 days | Occasional during initial 3–5 days |
| 2. | Brain congestion | Present | Absent |
| 3. | Tracheitis | Mucous/haemorrhagic | Mucous/haemorrhagic or no lesion |
| 4. | Lung alterations | Prominently congested, edematous and frothy | Occasional congestion and edema |
| 5. | Serosal haemorrhages | Present | Present |
| 6. | Proventriculus haemorrhages | Present | Present |
| 7. | Button ulcers in the intestine and caecal tonsils | Observed in young birds | Not observed |
| 8. | Egg peritonitis | Highly prominent | Prominent |
| 9. | Oophoritis | highly prominent | Prominent |
Both groups were complicated with MG, MS and E. coli in different combinations when the disease advanced
Discussion
Although, several reports are available on concurrent infections of poultry with various viral and bacterial agents, especially in respiratory disease complex (RDC) cases [1, 5, 9, 27, 42], studies on co-occurrence of LPAI and NDV in chicken is very limited [3, 38] and no description about the clinic-pathological patterns of these two agents during natural co-infection. This study demonstrated the natural co-infection of NDV, LPAI, mycoplasma and E. coli with severe clinical disease and high mortality when compared to pure form of NDV infection. These findings support the hypothesis that NDV infection makes the signs of LPAI–H9N2 infection more severe and predispose the animals for superinfection with E. coli and mycoplasma and vice versa. Newcastle disease and LPAI–H9N2 can produce severe disease depending on the type of secondary pathogen present [39, 43]. Mixed infections of H9N2 and other respiratory pathogens such as infectious bronchitis virus (IBV) and M. gallisepticum, can cause mortality between 20 and 60% in affected flocks [33, 38]. Previous experimental studies of [37] revealed that co-infection of the broilers with Ornithobacterium rhinotracheale (ORT) and H9N2 virus isolates induced higher mortality than infection with ORT or H9N2 virus alone. Co-infection of AIV and NDV demonstrated that co-infections can exacerbate clinical disease, affect virus replication, serological conversion and virus transmission [7, 11].
In the current study, the course of infection in NDV–LPAI co-infections extended up to 3–4 weeks, while compared to pure form of NDV infections, which lost up to 2 weeks. This could be due to potentiation of LPAI by NDV and its complicating agents like mycoplasma and E. coli. The mortality attained peak after 2–3 days of initial detection of mycoplasma and E. coli, suggesting that bacteria may play some role in the exhibition of the clinical syndrome. Co-infection of infectious bronchitis (IB) live vaccine and H9N2 avian influenza virus led to an extension of the shedding period of H9N2 virus, increasing the severity of clinical signs and mortality rates, causing macroscopic lesions in the embryos [16].
The clinic-pathological alterations were in concurrence with the previous studies on LPAI and/or NDV [31, 40]. Since, H9N2 produces minimal or no lesions when inoculated in to SPF chickens [31], other viruses and bacteria might have exacerbated LPAI in field conditions. The pathological alterations were almost similar (except degree of severity) in NDV–LPAI co-infected and NDV alone-infected flocks. This suggests that the NDV might play a triggering role in induction of clinic-pathological alterations. Protein cleavage site (HA0) of LPAI is only cleaved by trypsin-like proteases present in respiratory, gastrointestinal and reproductive tracts. When birds are co-infected with H9N2 and bacteria, the latter may release bacterial proteases into the other tissues which will cleave the HA0 and enhance its replication in multiple sites [2, 4, 16, 22]. For instance, it was demonstrated that the protease of S. aureus activated the HA of influenza virus, allowing multiple cycles of virus replication in the lungs of mice [45]. Alternatively, the stress of bacterial infection might have affected the immune system of chickens and exacerbated the pathogenicity of H9N2 influenza virus infection [22].
NDV cause severe lymphocytolysis leading to immuno-suppression, which paves way for replication of LPAI and both virus cause damage in respiratory epithelial cell lead to colonization of bacterial agent [23, 28]. Recent reports suggests that Genotype XIII NDV is emerging in India and it undergoes continuous evolutionary changes [18], leading to vaccine failures. Therefore, our results suggest that the genotype XIII NDV triggered a severe clinical disease in co-infection with LPAI. It is also assumed that mycoplasma and E. coli might have played a key role in this exacerbating effect. However, these finding needs to be evaluated carefully since no experimental attempts/demonstration of organisms in tissues were made. In order to fully prove the hypothesis about the existence of synergism after simultaneous infection with genotype XIII NDV, LPAI and other pathogens, an experimental study is highly desirable.
The fifth panzootic genotype XIII NDV is an emerging causes of disease in vaccinated birds. Clinical disease and mortality of genotype XIII NDV in vaccinated flocks were found to be exacerbated by LPAI–H9N2 co-infection or vice versa. The co-infection leads to severe disease in young grower than adult layer birds. Moreover, secondary bacterial infection with mycoplasma and E. coli caused exacerbation of clinical disease leading to enhanced mortality. The co-infection with LPAIV and NDV present an overlapping clinical and pathological picture often misleading the identification and diagnosis of both of these viruses. Several AIV infected countries including India not practicing vaccination against AIV, and they only impose culling followed by enhanced biosecurity and surveillance for control of AIV. In this context, prevention of genotype XIII NDV is most needed. Additional studies are needed to identify the mechanism in genotype XIII NDV evading protective antibody and formulate proper vaccination strategy. Further research is needed to ascertain the individual and cumulative possible contributions of co-infections in the disease intensity and how a virus–virus interaction influences the clinical outcome in these economically important diseases of poultry.
Acknowledgements
Authors are thankful to Indian Veterinary Research Institute and Indian Council of Agricultural Research for providing facilities and funds for carrying out the study.
Availability of data and materials
All reported data has been submitted to the public domain of NCBI and are accessible under accession numbers; HG 780865, HG 780863, HG 780864, HG 780866, HG 780860, HG780862, HG780869,HG 780867,HG 780868,CY 099356, CY 099357, CY 099363 and CY 099347.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed Z, Pandurang G, Acharya RS, Parihar NS. A report on outbreaks of respiratory disease in chicken in andhra pradesh with particular reference to infectious laryngotracheitis. Indian Vet J. 1969;46:646–650. [PubMed] [Google Scholar]
- 2.Akhtar S, Muneer M, Muhammad K, Tibu M, Anees M, Rasid I, Rehman R, Hussain I. Molecular characterization and epitope mapping of fusion (F) and hemagglutinin (HN) genes of avian paramyxovirus serotype I from peacocks in Pakistan. Pak J Zool. 2017;49:755–759. doi: 10.17582/journal.pjz/2017.49.2.sc9. [DOI] [Google Scholar]
- 3.Al-Mohana A, Kadhimv H, Al-Charrakh A, Al-Habubi Z, Nasir F, Al-Hilali A, Hadi Z. Molecular diagnosis of avian respiratory diseases in commercial broiler chicken flocks in province of Najaf, Iraq. Sci Res Essays. 2013;8:1191–1195. [Google Scholar]
- 4.Bano S, Naeem K, Malik SA. Evaluation of pathogenic potential of avian influenza virus serotype H9N2 in chickens. Avian Dis. 2003;47:817–822. doi: 10.1637/0005-2086-47.s3.817. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury JM. Avian mycoplasma infections: prototype of mixed infections with mycoplasmas, bacteria and viruses. Ann Microbiol (Paris) 1984;135A:83–89. doi: 10.1016/s0769-2609(84)80062-9. [DOI] [PubMed] [Google Scholar]
- 6.Capua I, Alexander DJ. Avian influenza: recent developments. Avian Pathol J WVPA. 2004;33:393–404. doi: 10.1080/03079450410001724085. [DOI] [PubMed] [Google Scholar]
- 7.Costa-Hurtado M, Afonso CL, Miller PJ, Spackman E, Kapczynski DR, Swayne DE, Shepherd E, Smith D, Zsak A, Pantin-Jackwood M. Virus interference between H7N2 low pathogenic avian influenza virus and lentogenic Newcastle disease virus in experimental co-infections in chickens and turkeys. Vet Res. 2014;45:1. doi: 10.1186/1297-9716-45-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diel G, da Silva LHA, Liu H, Wang Z, Miller PJ, Afonso CL. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2012;12:1770–1779. doi: 10.1016/j.meegid.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Dormitorio TV, Giambrone JJ, Guo K, Hepp GR. Detection and characterization of avian influenza and other avian paramyxoviruses from wild waterfowl in parts of the southeastern United States. Poult Sci. 2009;88:851–855. doi: 10.3382/ps.2008-00337. [DOI] [PubMed] [Google Scholar]
- 10.El Zowalaty ME, Chander Y, Redig PT, Abd El Latif HK, El Sayed MA, Goyal SM. Selective isolation of avian influenza virus (AIV) from cloacal samples containing AIV and Newcastle disease virus. J Vet Diagn Investig. 2011;23:330–332. doi: 10.1177/104063871102300222. [DOI] [PubMed] [Google Scholar]
- 11.França M, Howerth EW, Carter D, Byas A, Poulson R, Afonso CL, Stallknecht DE. Co-infection of mallards with low-virulence Newcastle disease virus and low-pathogenic avian influenza virus. Avian Pathol J WVPA. 2014;43:96–104. doi: 10.1080/03079457.2013.876530. [DOI] [PubMed] [Google Scholar]
- 12.Glisson J, Jackwood M, Pearson J, Reed W, Swayne D, Woolcock P. Isolation, identification, and characterization of avian pathogens. 5. Jacksonville: American Association of Avian Pathologists; 2008. [Google Scholar]
- 13.Goekjian VH, Smith JT, Howell DL, Senne DA, Swayne DE, Stallknecht DE. Avian influenza viruses and avian paramyxoviruses in wintering and breeding waterfowl populations in North Carolina, USA. J Wildl Dis. 2011;47:240–245. doi: 10.7589/0090-3558-47.1.240. [DOI] [PubMed] [Google Scholar]
- 14.Gowthaman V. 2011. Etio-pathology and differential diagnosis of low pathogenic avian influenza (LPAI) in poultry. Ph.D. thesis, Indian Veterinary Research Institute.
- 15.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 16.Haghighat-Jahromi M, Asasi K, Nili H, Dadras H, Shooshtari AH. Coinfection of avian influenza virus (H9N2 subtype) with infectious bronchitis live vaccine. Arch Virol. 2008;153:651–655. doi: 10.1007/s00705-008-0033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 18.Jakhesara SJ, Prasad VVSP, Pal JK, Jhala MK, Prajapati KS, Joshi CG. Pathotypic and sequence characterization of Newcastle disease viruses from vaccinated chickens reveals circulation of genotype II, IV and XIII and in India. Transbound Emerg Dis. 2014;63:523. doi: 10.1111/tbed.12294. [DOI] [PubMed] [Google Scholar]
- 19.Jeong J, Kim Y, An I, Wang S-J, Kim Y, Lee H-J, Choi K-S, Im S-P, Min W, Oem J-K, Jheong W. Complete genome sequence of a novel avian paramyxovirus isolated from wild birds in South Korea. Arch Virol. 2018;163:223–227. doi: 10.1007/s00705-017-3588-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khorajiya JH, Pandey S, Ghodasara PD, Joshi BP, Prajapati KS, Ghodasara DJ, Mathakiya RA. Patho-epidemiological study on Genotype-XIII Newcastle disease virus infection in commercial vaccinated layer farms. Vet World. 2015;8:372–381. doi: 10.14202/vetworld.2015.372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishida N, Sakoda Y, Eto M, Sunaga Y, Kida H. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch Virol. 2004;149:2095–2104. doi: 10.1007/s00705-004-0372-1. [DOI] [PubMed] [Google Scholar]
- 23.Kotani T, Odagiri Y, Nakamura J, Horiuchi T. Pathological changes of tracheal mucosa in chickens infected with lentogenic Newcastle disease virus. Avian Dis. 1987;31:491–497. doi: 10.2307/1590729. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D-H, Park J-K, Yuk S-S, Erdene-Ochir T-O, Kwon J-H, Lee J-B, Park S-Y, Choi I-S, Lee S-W, Song C-S. Complete genome sequence of a natural reassortant H9N2 avian influenza virus found in bean goose (Anser fabalis): direct evidence for virus exchange between Korea and China via wild birds. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2014;26:250–254. doi: 10.1016/j.meegid.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Luna L. Manual on histologic staining methods of the armed force institute of pathology. 3. New York City: McGraw-Hill Book Co; 1968. pp. 32–46. [Google Scholar]
- 27.Malik BS, Verma KC. Coexistence of antibodies against chronic respiratory disease, infectious laryngotracheitis, and infectious bronchitis on poultry farms of Uttar Pradesh, Andhra Pradesh, and Madras. Avian Dis. 1969;13:695–699. doi: 10.2307/1588577. [DOI] [PubMed] [Google Scholar]
- 28.Mast J, Nanbru C, van den Berg T, Meulemans G. Ultrastructural changes of the tracheal epithelium after vaccination of day-old chickens with the La Sota strain of Newcastle disease virus. Vet Pathol. 2005;42:559–565. doi: 10.1354/vp.42-5-559. [DOI] [PubMed] [Google Scholar]
- 29.Mendoza-Espinoza A, Koga Y, Zavaleta AI. Amplified 16S ribosomal DNA restriction analysis for identification of Avibacterium paragallinarum. Avian Dis. 2008;52:54–58. doi: 10.1637/8036-062507-Reg. [DOI] [PubMed] [Google Scholar]
- 30.Miller PJ, Haddas R, Simanov L, Lublin A, Rehmani SF, Wajid A, Bibi T, Khan TA, Yaqub T, Setiyaningsih S, Afonso CL. Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2015;29:216–229. doi: 10.1016/j.meegid.2014.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Mo IP, Brugh M, Fletcher OJ, Rowland GN, Swayne DE. Comparative pathology of chickens experimentally inoculated with avian influenza viruses of low and high pathogenicity. Avian Dis. 1997;41:125–136. doi: 10.2307/1592452. [DOI] [PubMed] [Google Scholar]
- 32.Nath B, Kumar S. Emerging variant of genotype XIII Newcastle disease virus from Northeast India. Acta Trop. 2017;172:64–69. doi: 10.1016/j.actatropica.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Nili H, Asasi K. Avian influenza (H9N2) outbreak in Iran. Avian Dis. 2003;47:828–831. doi: 10.1637/0005-2086-47.s3.828. [DOI] [PubMed] [Google Scholar]
- 34.OIE. Avian mycoplasmosis. Manual of diagnostic tests and vaccines for terrestrial animals; 2008. pp. 482–496.
- 35.OIE. Newcastle disease. Manual of diagnostic tests and vaccines for terrestrial animals, chapter 2314; 2012.
- 36.Ottiger H-P. Development, standardization and assessment of PCR systems for purity testing of avian viral vaccines. Biol J Int Assoc Biol Stand. 2010;38:381–388. doi: 10.1016/j.biologicals.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Pan Q, Liu A, Zhang F, Ling Y, Ou C, Hou N, He C. Co-infection of broilers with Ornithobacterium rhinotracheale and H9N2 avian influenza virus. BMC Vet Res. 2012;8:104. doi: 10.1186/1746-6148-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roussan DA, Haddad R, Khawaldeh G. Molecular survey of avian respiratory pathogens in commercial broiler chicken flocks with respiratory diseases in Jordan. Poult Sci. 2008;87:444–448. doi: 10.3382/ps.2007-00415. [DOI] [PubMed] [Google Scholar]
- 39.Seififi S, Asasi K, Mohammadi Ali. Natural co-infection caused by avian influenza H9 subtype and infectious bronchitis viruses in broiler chicken farms. Vet Arh. 2010;80:269–281. [Google Scholar]
- 40.Shalaby AA, Slemons RD, Swayne DE. Pathological studies of A/chicken/Alabama/7395/75 (H4N8) influenza virus in specific-pathogen-free laying hens. Avian Dis. 1994;38:22–32. doi: 10.2307/1591832. [DOI] [PubMed] [Google Scholar]
- 41.Siddique N, Naeem K, Abbas MA, Ali Malik A, Rashid F, Rafique S, Ghafar A, Rehman A. Sequence and phylogenetic analysis of virulent Newcastle disease virus isolates from Pakistan during 2009–2013 reveals circulation of new sub genotype. Virology. 2013;444:37–40. doi: 10.1016/j.virol.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 42.Smietanka K, Minta Z, Swiętoń E, Olszewska M, Jóźwiak M, Domańska-Blicharz K, Wyrostek K, Tomczyk G, Pikuła A. Avian influenza H9N2 subtype in Poland–characterization of the isolates and evidence of concomitant infections. Avian Pathol J WVPA. 2014;43:427–436. doi: 10.1080/03079457.2014.952221. [DOI] [PubMed] [Google Scholar]
- 43.Spickler A, Roth J. Emerging and exotic diseases of animals. Ames: CFSPH Iowa State University; 2008. pp. 203–204. [Google Scholar]
- 44.Susta L, Jones MEB, Cattoli G, Cardenas-Garcia S, Miller PJ, Brown CC, Afonso CL. Pathologic characterization of genotypes XIV and XVII Newcastle disease viruses and efficacy of classical vaccination on specific pathogen-free birds. Vet Pathol. 2015;52:120–131. doi: 10.1177/0300985814521247. [DOI] [PubMed] [Google Scholar]
- 45.Tashiro M, Ciborowski P, Klenk HD, Pulverer G, Rott R. Role of Staphylococcus protease in the development of influenza pneumonia. Nature. 1987;325:536–537. doi: 10.1038/325536a0. [DOI] [PubMed] [Google Scholar]
- 46.Terregino C, Aldous EW, Heidari A, Fuller CM, De Nardi R, Manvell RJ, Beato MS, Shell WM, Monne I, Brown IH, Alexander DJ, Capua I. Antigenic and genetic analyses of isolate APMV/wigeon/Italy/3920-1/2005 indicate that it represents a new avian paramyxovirus (APMV-12) Arch Virol. 2013;158:2233–2243. doi: 10.1007/s00705-013-1735-2. [DOI] [PubMed] [Google Scholar]
- 47.Toyoda T, Sakaguchi T, Hirota H, Gotoh B, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology. 1989;169:273–282. doi: 10.1016/0042-6822(89)90152-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All reported data has been submitted to the public domain of NCBI and are accessible under accession numbers; HG 780865, HG 780863, HG 780864, HG 780866, HG 780860, HG780862, HG780869,HG 780867,HG 780868,CY 099356, CY 099357, CY 099363 and CY 099347.