Abstract
Nosema apis and Nosema ceranae are microsporidian parasite worldwide spread causing an emerging infectious disease of European honeybee Apis mellifera. The Nosema presence was deeply investigated in several countries but low information are presents about islands. In this investigation was evaluated the presence N. ceranae and N. apis in apiaries located in Tuscanian Archipelago islands (Central Italy). For N. ceranae detection, two different Real-Time PCR (qPCR) methods, the 16S rRNA and Hsp70 gene amplification qPCR, were performed on honey bee samples; while, for N. apis only the 16S rRNA qPCR amplification was performed. On all islands, only N. ceranae was present, while N. apis was not found in the samples. The two qPCR showed significant difference (p < 0.040) in N. ceranae spores quantification. The single-copy Hsp70 gene method qPCR assay systematically detected a lower amount of N. ceranae copies compared to the multi-copy 16S rRNA gene method.
Keywords: Nosema ceranae, Nosemosis, qPCR, Hsp70 and 16S rRNA gene, Apis mellifera, Tuscanian Archipelago
1. Introduction
Microsporidia are unicellular eukaryotes infecting vertebrate and invertebrate animal hosts. So far, three microsporidia species have been described associated to the honey bees, all belonging to the Nosema genus. The infections are known as nosemosis irrespective they are caused by Nosema apis Zander (Fries, 1993), Nosema ceranae (Fries et al., 1996) or Nosema neumanni (Chemurot et al., 2017). The first two species are spread worldwide and responsible for known symptoms (Fries et al., 2006, Higes et al., 2006, Klee et al., 2007, Martin-Hernandez et al., 2007). However, N. neumanni was recently detected only in Ugandan bees and its consequences for the host bees have not been described yet.
N. apis and N. ceranae are generally considered original parasites of European and Asian honey bees, respectively (Botías et al., 2012). However, the second showed capable to effectively infect Apis mellifera individuals also, which allowed its spread far beyond the Asian continent (Fries et al., 2006).
It was suggested that N. ceranae may have replaced N. apis in Italy (Ferroglio et al., 2013) and in other countries (Ansari et al., 2017, Calderón et al., 2008, Fries et al., 2006, Giersch et al., 2009, Higes et al., 2009b, Invernizzi et al., 2009, Klee et al., 2007, Pacini et al., 2016, Tapaszti et al., 2009), although the possibility of a direct competition between the two species is still debated.
In Italy, N. ceranae was found in honey bee colonies reared in all regions (Ferroglio et al., 2013, Klee et al., 2007, Maiolino et al., 2014, Mutinelli et al., 2010, Porrini et al., 2016), but small islands have never been investigated in this respect. Thus, no data are available for the islands of Tuscanian Archipelago: Giannutri (area: 26 ha), Montecristo (103 ha), Pianosa (104 ha), Capraia (193 ha), Giglio (204 ha), Gorgona (223 ha) and Elba (22.350 ha).
Both Nosema species do infect the epithelial cells of the honey bee ventriculum (Fries et al., 1996, Higes et al., 2007) with effects recognizable both at individual and colony level, like reduced lifespan, lethargic behaviour and poor honey and pollen harvest (Eiri et al., 2015, Higes et al., 2009c, Higes et al., 2008).
Diarrhoeic faeces, high mortality at the hive entrance and swollen bee abdomens are commonly seen in N. apis infected colonies (Bourgeois et al., 2010), but N. ceranae infections tend to produce subtle symptoms that are difficult to spot in the field. Nevertheless, damage may be severe and result into adult population decline until colony collapse (Fries et al., 1996, Giersch et al., 2009, Higes et al., 2006).
Nosema spp. spores can be detected by light microscopy in the honey bee ventriculum, but reliable species determination requires advanced techniques. Although individual spore morphology may be examined by transmission electron microscope with this purpose (Chen et al., 2009, Forsgren and Fries, 2010, Fries et al., 1996), biomolecular bioassays are generally preferred for practical reasons and because they allow accurate quantification by the Real-Time PCR (qPCR) approach.
The 16S rRNA gene is commonly used in qPCR assays where species discrimination and quantification are required (Chen et al., 2008, Forsgren and Fries, 2010, Higes et al., 2006, Klee et al., 2007, Martin-Hernandez et al., 2007). However, this is a multi-copy gene characterized by a variable number of sequences in the N. ceranae genome. This suggested to develop a new qPCR method, which is based on the single-copy N. ceranae gene Hsp70 (Cilia et al., 2018).
The aim of this investigation was to seek the presence of Nosema spp. in managed colonies from the Tuscanian Archipelago islands and, concurrently, compare the results of the qPCR methods based on the 16S rRNA and Hsp70 genes.
2. Materials and methods
In July and August of 2017, honey bee workers were sampled on the islands of Tuscanian Archipelago in order to determine the Nosema infection.
Samples were taken on asymptomatic apiaries from the island of Pianosa (42°35′05.2″N 10°05′22.3″E), Capraia (43°01′54.4″N 9°29′59.8″E), Gorgona (43°26′00.1″N 9°54′18.0″E), and Elba (42°51′43.3″N 10°25′03.1″E) islands (Fig. 1). On Giannutri and Montecristo, no managed apiaries are known. On Giglio, the contacted beekeepers refused to take samples for this research.
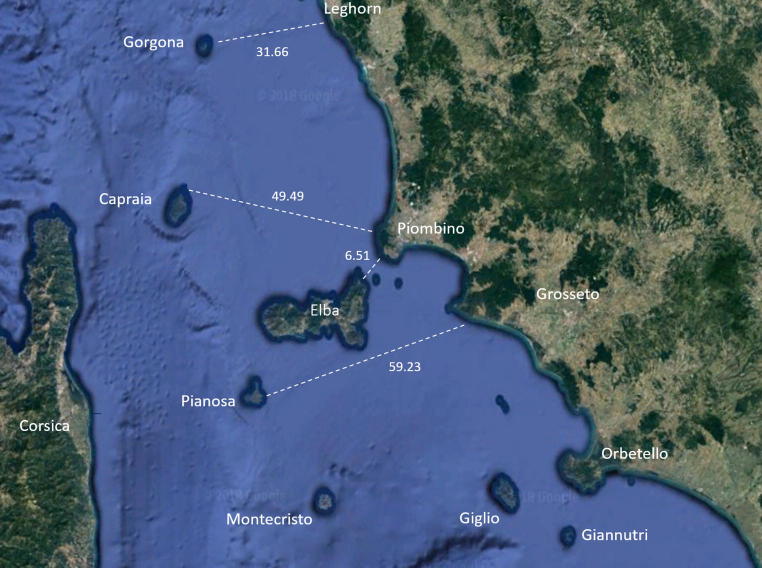
Fig. 1.
Map of Tuscanian Archipelago (Central Italy) with the distances in kilometres (km) between each island and the nearest land.
On each island, two colonies from the same apiary were sampled. Each sample consisted on 50 forager bees, which were refrigerated until analysis. The samples were divided in two subsamples of 25 bees, which were extracted separately. For extraction, the gastrum (later referred to as abdomen as of common use) was carefully dissected with tweezers. The abdomens of each subsample were pooled and homogenized (in 1 ml of DNA free water) with a Tissue Lyser II (Qiagen, Hilden, Germany) operated for 3 min at 30 Hz.
Total DNA was extracted with High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany) following the manufacturer’s instructions.
Two aliquots from each DNA extract were taken and analysed by different qPCR methods. In one case, primers and probes for N. apis and N. ceranae designed on a 16S rRNA gene sequence were used as described by Forsgren and Fries (2010). The optimized amplification and quantitation protocol was: 15 min at 98 °C followed by 40 cycles of denaturation for 5 s at 98 °C, annealing/extension for 10 s at 63 °C and melt curve analysis from 65 to 95 °C (in 0.5 °C increments) 10 sec/step. The other aliquot served to quantify N. ceranae copies in each sample using the Hsp70 gene-based primers and probes described by Cilia et al. (2018). In this case, amplification and quantitation were performed according to the following cycling conditions: initial activation step 95 °C for 10 min, PCR cycling (40 cycles of 95 °C for 15 s, 56 °C for 60 s).
All the analyses were conducted in duplicate and the final infection data reported as average of Nosema number copies per honey bee.
Data obtained were statistically analysed through a three-way ANOVA. Number of N. ceranae copies were tested against colony, qPCR method and replicate. Statistical significance threshold was set at P = 0.05. Where applicable, multiple comparison of means was made by using Newman-Keuls test.
Regression was calculated on the averages of the replicate analyses from each colony to compare the two qPCR methods.
All the statistics were calculated with the package STATISTICA (Sistema software di analisi dei dati), ver. 7.1, StatSoft Italia srl (2005).
3. Results
All of the samples resulted in DNA amplification below the detection limit for N. apis. Table 1 reports the number of N. ceranae copies resulting from each replicate analysis performed with the two methods. No detectable N. ceranae infections were found in the samples from Elba Island, whereas the samples from Gorgona, Capraia and Pianosa samples were positive.
Table 1.
Replicates of Nosema ceranae gene copies detected with 16S rRNA and Hsp70 qPCR assay in honey bee samples from Tuscanian Archipelago.
| Island | Colony | Replicate | 16S gene copies/bee | Hsp70 gene copies/bee |
|---|---|---|---|---|
| Capraia | C1 | 1 | 3,16E+05 | 2,28E+04 |
| 2 | 3,28E+05 | 2,44E+04 | ||
| C2 | 1 | 1,16E+07 | 3,12E+06 | |
| 2 | 1,28E+07 | 3,24E+06 | ||
| Gorgona | G1 | 1 | 4,52E+04 | 3,36E+04 |
| 2 | 4,96E+04 | 3,64E+04 | ||
| G2 | 1 | 5,88E+06 | 3,96E+05 | |
| 2 | 6,12E+06 | 4,16E+05 | ||
| Pianosa | P1 | 1 | 5,52E+05 | 3,80E+05 |
| 2 | 5,80E+05 | 4,28E+05 | ||
| P2 | 1 | 6,36E+07 | 3,88E+06 | |
| 2 | 6,60E+07 | 3,96E+06 | ||
| Elba | E1 | 1 | <LOD | <LOD |
| 2 | <LOD | <LOD | ||
| E2 | 1 | <LOD | <LOD | |
| 2 | <LOD | <LOD | ||
Samples positive for N. ceranae contained an average of 13.99 (±10.34 s.e.; s.d. = 25.34) * 10^6 copies per bee with the 16S rRNA-based method, but only 1.33 (±0.71 s.e.; s.d. = 1.74) * 10^6 by using the Hsp70 sequences.
A principal effects ANOVA conducted on the data of Table 1 resulted in a significant effect of colony (F(5, 16) = 4.020, p = 0.015) and analytical method (F(1, 16) = 5.380, p = 0.034) but not of replicate (F(1, 16) = 0.004, p = 0.950).
On average, the 16S rRNA-based technique detected 8.59 (±2.91 s.e.) times the N. ceranae copies found with the Hsp70 method, and the ratio between the copies detected with the two methods (16S rRNA/Hsp70) was in the range 1.35–16.53.
Results show both high between-sample variability of this ratio (s.d. = 7.12) and insufficient correlation between the paired replicate averages to make direct conversion between the two methods possible (Pearson r = 0.832 (F(1, 4) = 9.020, p = 0.040)).
In a regression analysis of data (y = b0 + b1 * x), the number of copies detected with 16S rRNA and Hsp70 methods were respectively considered dependent and independent variables, with good fit to the linear model (F(1,4) = 9.02p < 0.040), b0 = −2,061,789 (p = 0.817) and b1 = 12,09 (p = 0.040). However, low determination index (Adjusted R2 = 0.616) confirms that major factors other than Hsp70 estimate should be considered to account for the variability in the number of N. ceranae copies detected with the 16S rRNA-based method.
4. Discussion
The results of this investigation allow to include Tuscanian Archipelago among the Italian areas where N. ceranae can be found in honey bee colonies (Ferroglio et al., 2013, Klee et al., 2007, Maiolino et al., 2014, Mutinelli et al., 2010, Porrini et al., 2016). This confirms that, despite the likely infrequent exchanges with the mainland populations, this pathogen may find suitable conditions to thrive in small Mediterranean islands. In this respect, samples collected from the comparatively larger Elba island Elba should be considered more occasional finding than a realistic representation of the overall situation.
N. apis was not detected in any of the investigated colonies. Although, the results of this investigation do not clarify this aspect, seasonality (Martin-Hernandez et al., 2007, Ondrus, 2017), different plasticity to temperature (Fries et al., 2006, Higes et al., 2009a, Higes et al., 2009b, Klee et al., 2007, Paxton et al., 2007) and/or competition between the two investigated species (Chen et al., 2012, Forsgren and Fries, 2010, Martín-Hernández et al., 2012) may account for the difference in prevalence between the two Nosema species.
In the three islands where positive samples could be found, the number of N. ceranae copies detected whit both qPCR methods was in the order of magnitude of 10^4 or higher, which is compatible with fully developed infections and suggestive of consolidated pathogen populations in those islands.
As resulted by the three-way ANOVA test, colony was a significant source of variability, suggesting heterogeneous levels of infection in the considered hives. Conversely, the replicate did not result into significant effect. This is compatible with high repeatability of both qPCR methods (Cilia et al., 2018, Martin-Hernandez et al., 2007). However, Hsp70 qPCR assay systematically detected less N. ceranae copies than the alternate method, whose results were one or two orders of magnitude higher. This and the unsteady 16S rRNA/Hsp70 ratio calculated at sample level agrees with the variable number of sequences of the 16S rRNA gene of N. ceranae (Sagastume et al., 2016). Molecular diagnostics may take advantage of multi-copy genes in that they allow designing high sensitivity tests (more possibility to detect the target gene), but the fluctuations in the number of 16S rRNA sequences in the N. ceranae genome may ultimately affect the quantification reliability (Sagastume et al., 2011). Conversely, the Hsp70 method bases on a highly-conserved region of the N. ceranae genome (Gomez-Moracho et al., 2014, Wang et al., 2017), making it suitable to accurate quantification.
The results of the 16S rRNA amplification may be correlated to the highly variable number of copies of multi-copy genes. The PCR primers may affect their capability to anneal to the template influencing the assay sensitivity. On the other hand, the results of the Hsp70 gene amplification, detecting a lower number of copies, may be correlated to its highly conserved sequence. The designed PCR primers anneal to the template once, with high sensitivity and specificity.
In conclusion, the results of this investigation provide evidence that, as well known, N. ceranae may successfully proliferate in asymptomatic honey bee colonies living in small Mediterranean islands and indicate that N. apis might not find similarly suitable conditions to thrive. In this respect, further research is required to elucidate the long-term host-parasite relationship in limited environments characterized by genetic isolation. A comparison between different q-PCR assays showed profound differences in the results when the primers are designed on single-copy or multi-copy sequences. The choice of analytical method may then be critical when reliable quantification is required.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ansari M.J., Al-Ghamdi A., Nuru A., Khan K.A., Alattal Y. Geographical distribution and molecular detection of Nosema ceranae from indigenous honey bees of Saudi Arabia. Saudi J. Biol. Sci. 2017;24:983–991. doi: 10.1016/j.sjbs.2017.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botías C., Anderson D.L., Meana A., Garrido-Bailón E., Martín-Hernández R., Higes M. Further evidence of an oriental origin for Nosema ceranae (Microsporidia: Nosematidae) J. Invertebr. Pathol. 2012;110:108–113. doi: 10.1016/j.jip.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Bourgeois A.L., Rinderer T.E., Beaman L.D., Danka R.G. Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. J. Invertebr. Pathol. 2010;103:53–58. doi: 10.1016/j.jip.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Calderón R.A., Sanchez L.A., Yañez O., Fallas N. Presence of Nosema ceranae in Africanized honey bee colonies in Costa Rica. J. Apic. Res. 2008;47:328–329. [Google Scholar]
- Chemurot M., De Smet L., Brunain M., De Rycke R., de Graaf D.C. Nosema neumanni n. sp. (Microsporidia, Nosematidae), a new microsporidian parasite of honeybees, Apis mellifera in Uganda. Eur. J. Protistol. 2017;61:13–19. doi: 10.1016/j.ejop.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Chen Y., Evans J.D., Smith I.B., Pettis J.S. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008;97:186–188. doi: 10.1016/j.jip.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Chen Y.P., Evans J.D., Murphy C., Gutell R., Zuker M., Gundensen-Rindal D., Pettis J.S. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J. Eukaryot. Microbiol. 2009;56:142–147. doi: 10.1111/j.1550-7408.2008.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Chung W.P., Wang C.H., Solter L.F., Huang W.F. Nosema ceranae infection intensity highly correlates with temperature. J. Invertebr. Pathol. 2012;111:264–267. doi: 10.1016/j.jip.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Cilia G., Cabbri R., Maiorana G., Cardaio I., Dall’Olio R., Nanetti A. A novel TaqMan ® assay for Nosema ceranae quantification in honey bee, based on the protein coding gene Hsp70. Eur. J. Protistol. 2018;63:44–50. doi: 10.1016/j.ejop.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Eiri D.M., Suwannapong G., Endler M., Nieh J.C. Nosema ceranae can infect honey bee larvae and reduces subsequent adult longevity. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferroglio E., Zanet S., Peraldo N., Tachis E., Trisciuoglio A., Laurino D., Porporato M. Nosema ceranae has been infecting honey bees Apis mellifera in Italy since at least 1993. J. Apic. Res. 2013;52:60–61. [Google Scholar]
- Forsgren E., Fries I. Comparative virulence of Nosema ceranae and Nosema apis in individual European honey bees. Vet. Parasitol. 2010;170:212–217. doi: 10.1016/j.vetpar.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Fries I. Nosema Apis - a parasite in the honey bee colony. Bee World. 1993;74:5–19. [Google Scholar]
- Fries I., Feng F., da Silva A., Slemenda S.B., Pieniazek N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae) Eur. J. Protistol. 1996;32:356–365. [Google Scholar]
- Fries I., Martín R., Meana A., García-Palencia P., Higes M. Natural infections of Nosema ceranae in European honey bees. J. Apic. Res. 2006;45:230–233. [Google Scholar]
- Giersch T., Berg T., Galea F., Hornitzky M. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie. 2009;40:117–123. [Google Scholar]
- Gomez-Moracho T., Maside X., Martin-Hernandez R., Higes M., Bartolomé C. High levels of genetic diversity in Nosema ceranae within Apis mellifera colonies. Parasitology. 2014;141:475–481. doi: 10.1017/S0031182013001790. [DOI] [PubMed] [Google Scholar]
- Higes M., García-Palencia P., Martín-Hernández R., Meana A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia) J. Invertebr. Pathol. 2007;94:211–217. doi: 10.1016/j.jip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Higes M., Martín-Hernández R., Botías C., Bailón E.G., González-Porto A.V., Barrios L., Del Nozal M.J., Bernal J.L., Jiménez J.J., Palencia P.G., Meana A. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008;10:2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- Higes M., Martin-Hernandez R., Garci-a-Palencia P., Mari-n P., Meana A. Horizontal transmission of Nosema ceranae (Microsporidia) from worker honeybees to queens (Apis mellifera) Environ. Microbiol. Rep. 2009;1:495–498. doi: 10.1111/j.1758-2229.2009.00052.x. [DOI] [PubMed] [Google Scholar]
- Higes M., Martín-Hernández R., Garrido-Bailón E., Botías C., Meana A. The presence of Nosema ceranae (Microsporidia) in North African honey bees (Apis mellifera intermissa) J. Apic. Res. 2009;48:217–219. [Google Scholar]
- Higes M., Martín-Hernández R., Garrido-Bailón E., González-Porto A.V., García-Palencia P., Meana A., del Nozal M.J., Mayo R., Bernal J.L., Kamer Y., Klinberg E., Zioni N., Inbar S., Chejanovsky N., Alaux C. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ. Microbiol. Rep. 2009;1:110–113. doi: 10.1111/j.1758-2229.2009.00014.x. [DOI] [PubMed] [Google Scholar]
- Higes M., Martín R., Meana A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006;92:93–95. doi: 10.1016/j.jip.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Invernizzi C., Abud C., Tomasco I.H., Harriet J., Ramallo G., Campá J., Katz H., Gardiol G., Mendoza Y. Presence of Nosema ceranae in honeybees (Apis mellifera) in Uruguay. J. Invertebr. Pathol. 2009;101:150–153. doi: 10.1016/j.jip.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Klee J., Besana A.M., Genersch E., Gisder S., Nanetti A., Tam D.Q., Chinh T.X., Puerta F., Ruz J.M., Kryger P., Message D., Hatjina F., Korpela S., Fries I., Paxton R.J. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007;96:1–10. doi: 10.1016/j.jip.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Maiolino P., Iafigliola L., Rinaldi L., De Leva G., Restucci B., Martano M. Histopathological findings of the midgut in European honey bee (Apis Mellifera L.) naturally infected by Nosema spp. Vet. Med. Anim. Sci. 2014;2:4. [Google Scholar]
- Martín-Hernández R., Botías C., Bailón E.G., Martínez-Salvador A., Prieto L., Meana A., Higes M. Microsporidia infecting Apis mellifera: coexistence or competition. Is Nosema ceranae replacing Nosema apis? Environ. Microbiol. 2012;14:2127–2138. doi: 10.1111/j.1462-2920.2011.02645.x. [DOI] [PubMed] [Google Scholar]
- Martin-Hernandez R., Meana A., Prieto L., Salvador A.M., Garrido-Bailon E., Higes M. Outcome of colonization of apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007;73:6331–6338. doi: 10.1128/AEM.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutinelli F., Costa C., Lodesani M., Baggio A., Medrzycki P., Formato G., Porrini C. Honey bee colony losses in Italy. J. Apic. Res. 2010;49:119–120. [Google Scholar]
- Ondrus P. The seasonality, thermal dependence, and spread of Nosema ceranae in new locales. Bee World. 2017;94:105–109. [Google Scholar]
- Pacini A., Mira A., Molineri A., Giacobino A., Bulacio Cagnolo N., Aignasse A., Zago L., Izaguirre M., Merke J., Orellano E., Bertozzi E., Pietronave H., Russo R., Scannapieco A., Lanzavecchia S., Schnittger L., Signorini M. Distribution and prevalence of Nosema apis and N. ceranae in temperate and subtropical eco-regions of Argentina. J. Invertebr. Pathol. 2016;141:34–37. doi: 10.1016/j.jip.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Paxton R.J., Klee J., Korpela S., Fries I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie. 2007;38:558–565. [Google Scholar]
- Porrini C., Mutinelli F., Bortolotti L., Granato A., Laurenson L., Roberts K., Gallina A., Silvester N., Medrzycki P., Renzi T., Sgolastra F., Lodesani M. The status of honey bee health in Italy: results from the nationwide bee monitoring network. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagastume S., del Águila C., Martín-Hernández R., Higes M., Henriques-Gil N. Polymorphism and recombination for rDNA in the putatively asexual microsporidian Nosema ceranae, a pathogen of honeybees. Environ. Microbiol. 2011;13:84–95. doi: 10.1111/j.1462-2920.2010.02311.x. [DOI] [PubMed] [Google Scholar]
- Sagastume S., Martín-Hernández R., Higes M., Henriques-Gil N. Genotype diversity in the honey bee parasite Nosema ceranae: multi-strain isolates, cryptic sex or both? BMC Evol. Biol. 2016;16:216. doi: 10.1186/s12862-016-0797-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapaszti Z., Forgách P., Kővágó C., Békési L., Bakonyi T., Rusvai M. First detection and dominance of Nosema ceranae in Hungarian honeybee colonies. Acta Vet. Hung. 2009;57:383–388. doi: 10.1556/AVet.57.2009.3.4. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ru Y., Liu W., Wang D., Zhou J., Jiang Y., Shi S., Qin L. Morphological characterization and HSP70-, IGS-based phylogenetic analysis of two microsporidian parasites isolated from Antheraea pernyi. Parasitol. Res. 2017;116:971–977. doi: 10.1007/s00436-017-5373-6. [DOI] [PubMed] [Google Scholar]