Abstract
Caves are oligotrophic, dark and less-explored environments and are considered as sources of promising microbial strains in biotechnology. Hampoeil Cave is located in massive dolomite with thin bedded limestone in northwestern of Iran. In an isolation and screening program, various samples from soil, water, floor, wall and ceiling of Hampoeil cave and its invertebrates were collected. Four various treatments and 10 different isolation media were used for the isolation of the actinobacteria. Screening of the isolates for antimicrobial activity against 10 bacteria and fungi, 5 hydrolytic enzymes production and resistance to 5 heavy metals have been performed. Among 33 various samples, 76 actinobacteria from various genera, including Streptomyces, Micromonospora, Micrococcus, Kocuria and Corynebacterium were isolated. Eighty percent of the strains had one of the studied hydrolytic enzyme activity. At least one type of antimicrobial activity was seen in 25.3% of the isolates. Resistance to one metal or more was seen in 26.32% of the isolates. The ratio of rare-actinobacteria in the oligotrophic samples to enriched samples is 20% more than Streptomyces. Percentage of strains with the highest activity in esterase, amylase, DNase, protease or lipase activity that were isolated from organic-rich environmental samples were 100, 100, 100, 82 and 82%, respectively. Also, 26.32% of the actinobacterial isolates resisted to heavy metals. It was concluded that Hampoeil cave is a good source in finding cave-living actinobacteria potent in producing hydrolytic enzymes and bioremediation.
Keywords: Actinobacteria, Biodiversity, Cave, Enzyme, Metal
1. Introduction
Biodiversity is one of the bases of biotechnology development and is encouraged by OECD (Airoldi and Cinelli, 1997, Co-operation and Development, 2001). Discovering more genetic resources have resulted in an increased number of biotechnological products. Impacts of biodiversity can be shown by the fact that current global microbial biotechnology market is obtained by using potential of the cultivable microorganisms explored up to now. This market will reach 2244.20 US million dollars by 2023. This market is growing at the compound annual growth rate (CASR) of 6.1% between 2017 and 2023 (QYR-Research-Group, 2018). According to the EZbiolcloud, 14,381 valid bacterial taxa were published as of 28 May 2018 (Yoon et al., 2017), however, it was predicted that there are more than 1 trillion microbial species on Earth (Locey and Lennon, 2016). Rather than exploring new taxa of microorganisms, it is necessary to explore their potential in biotechnology. There are various strategies to reach this goal that is reviewed in Manual of Industrial Microbiology and Biotechnology (Baltz et al., 2010). By consideration of ∼80 years efforts and researches on rational and systematic screening of potent microorganisms in microbial biotechnology that have been started by René Dubos (Hopwood, 2007), it is now necessary to consider suitable strategy to avoid re-isolation of pre-isolated microbial strains.
Exploring less-known environments, such as caves, is one of the isolation and screening approaches in microbial biotechnology. Caves are defined as natural underground cavities with semi-darkness spaces, large enough for human exploration (Bates and Jackson, 1980). Carbonate caves are one of the widely distributed caves that are generated after dissolution of soluble rocks, e.g. limestone and dolomite during the million years (Wood, 1985). Normally, caves are ecosystems that has been considered as oligotrophic conditions (Airoldi and Cinelli, 1997) as well as less-explored microbial ecosystems (Barton and Jurado, 2007). Various microorganisms have been isolated from various caves worldwide, including Gram positive and Gram negative bacteria (Gulecal-Pektas, 2016, Yasir, 2018), protozoa (Kajtezović and Rubinić, 2013) and fungi (Belyagoubi et al., 2018, Cunningham et al., 1995).
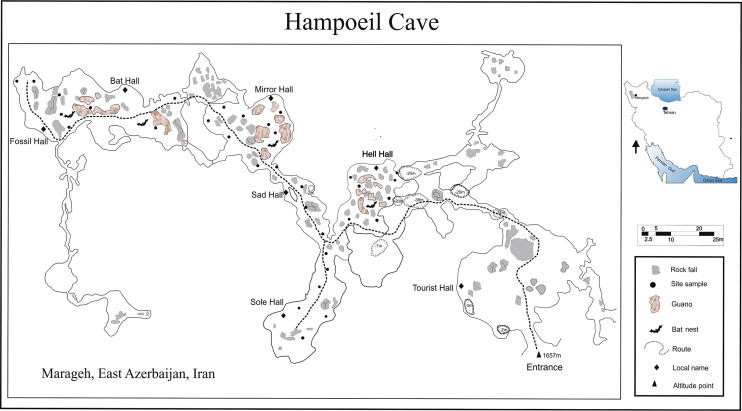
Iran, with 1,648,195 km2 area, lies between latitudes 24° and 40°N, and longitudes 44° and 64°E, is the 18th country in the world. Iran is a part of the Alpine–Himalayan orogenic belt that extends from Atlantic Ocean to Western Pacific, from Late Precambrian to Quaternary (Ghorbani, 2013). There is no report on the microbial diversity of the caves of Iran. Hampoeil Cave or Pigeon Cave is located 14 km southeast of Maragheh city, East Azerbaijan Province (Fig. 1) in northwestern Iran (46°18′29′'0.83E 37°18′42′'0.05N).
Fig. 1.
Geographical location of Hampoeil Cave (Maragheh, East Azerbaijan Province, Iran). The rout and site samples were shown in the map.
Actinobacteria are the most promising sources of valuable biological compounds and many commercial drugs have been driven by the members of this taxon, thus, finding new species of actinobacteria especially rare actinobacteria is a good strategy to provide valuable biochemicals used in medicine, agriculture and different industries (Hamedi et al., 2017, Hopwood, 2007). Actinobacteria are Gram positive, ubiquitous high G + C content bacteria that are found as free-living, symbiont of plants and animals and as their pathogens (Kurtbӧke, 2017). It was estimated that actinobacteria can be considered as a sources of one third of microbial biotechnology products and therefore has the third rank in the most sequenced genomes bacterial phyla (Yoon et al., 2017). Actinobacteria have been isolated from various terrestrial and water ecosystems, including caves (Riquelme et al., 2017). Oligotrophic properties of caves can result in coexistence of wide range of taxa and also stimulate inimitable strategies of indigenous microbiome to produce various biotechnological products (Bhullar et al., 2012).
By consideration of high biodiversity of actinobacteria in Iran and unexplored feature of Hampoeil cave as an oligotrophic environment, we were encouraged to find biodiversity of cultivable actinobacteria and their biological activities in the present study.
2. Materials and methods
2.1. Actinobacterial isolation media
The name and ingredients (g/l) of 10 isolation media were: Hickey Tresner agar (dextrin 10, yeast extract 1, extract 1, calcium chloride 0.2, N-Z-Amine 2), Soil extract agar (soil 4, yeast extract 1, Na2HPO4 0.5, KCl 1.7, MgSO4 0.5, CaCO3 0.2), Compost agar (compost 15, yeast extract 1), Rice bran agar (rice bran powder 15, yeast extract 1), Actinomycete agar (Sodium caseinate 2, L-arginine 0.1, sodium propionate 4, K2HPO4 0.5, MgSO4 0.1, FeSO4 0.001, glycerol 5), Starch casein nitrate agar (soluble starch 10, casein vitamin free 0.1, sodium nitrate 2, sodium chloride 2, K2HPO4 2, MgSO4 0.05, CaCO3 0.02, FeSO4 0.01), International Streptomyces Project (ISP2) (starch 10, yeast extract 4, peptone 2) and Nutrient agar (meat extract 1, meat peptone 5, sodium chloride 5, yeast extract 2) (Atlas, 2004). All media contains 15 g/l agar. The media ingredients were obtained from Merck Co. (Darmstadt, Germany), except for N-Z-Amine 2 that was obtained from Sigma-Aldrich (St. Louis, USA).
2.2. Biological activity screening media
ISP2 broth was used as seeding and fermentation medium for production of antimicrobial metabolites (Hamedi et al., 2015b). The name and ingredients (g/l) of the media used for screening biological activities were: Starch agar (starch 10, meat extract 1.5, yeast extract 1.5, NaCl 5) (Nakajima et al., 1985), Skim milk agar (skim milk powder 50, peptone 1, NaCl 5) (Zerdani et al., 2004), Tween 80 agar (peptone (10, NaCl 5), CaCl2·2H2O 0.1, tween 80 (1% (v/v) (Rai et al., 2014), Rhodamine-olive oil-agar (meat extract 1, meat peptone 5, NaCl 4, olive oil 31.25 ml, rhodamine B solution 10 mg/l, (Samad et al., 1989), DNA agar (tryptose 20, sodium chloride 5, deoxyribonucleic acid 2) (Jeffries et al., 1957), Minimal medium (glucose 10, L-asparagine 0.5, KH2PO4·7H2O 0.5, MgSO4·7H2O 0.5, FeSO4·7H2O 0.01 (Yadav et al., 2010). All media contain 18 g/l agar. The media ingredients were obtained from Merck Co. (Darmstadt, Germany), except for Rhodamine B and tryptose that were obtained from Sigma-Aldrich (St. Louis, USA).
2.3. Environmental sampling and pretreatments
Samples were obtained from soil and water from different sections of Hampoeil cave; floor, walls and ceiling of the cave and the live or dead invertebrate animals (Fig. 1). The samples were stored in zipped plastic bag (Zipkip Co., Karaj, Iran) or polyethylene terephthalate bottles (ParsPet Co., Tehran, Iran) and kept in a container that was cooled by dry-ice. The samples were studied in the lab, not later than 24 h. To minimize the growth of undesired microorganisms, the mud and soil samples were treated using four methods, including: (1) oven-drying at 120 °C for 10 min (2) putting in microwave for three min (3) suspending in sterile normal saline (0.9% (w/v) NaCl in distilled water) and centrifuging for 2500g, 15 min and, (4) adding chloramphenicol (50 µg/ml) to the isolation media (Hamedi et al., 2011). The outer surfaces of the invertebrate samples, including arthropods, were disinfected by three successive immersions in 70% ethanol to remove the microorganisms from the outer surfaces, then homogenized in sterile normal saline and cultured in the isolation media. The water samples were cultured in the isolation media without any pretreatments. Briefly, 1 ml of the water sample was spread on isolation media and incubated at appropriated conditions as described below.
2.4. Isolation and identification of the bacteria
The inoculated agar plates of the media were incubated at 28 °C, in a dark incubator for 14 days. Then, the plates were considered carefully, and the putative actinobacterial colonies were selected and isolated by subculture in the same medium. The purified isolates were cultured into Erlenmeyer flasks containing Brain heart infusion broth and incubated at 28 °C, 72 h, 180 rpm. The biomass was obtained by centrifugation and washed by 0.9% NaCl and poured in a sterile porcelain mortar. In an aseptic condition, the cells were disrupted by addition of liquid N2, pressed and rotated with the pestle it until liquid texture is achieved. The DNA was extracted by DNA extraction Kit according the instruction, and was subjected to PCR amplification of 16S rRNA gene (Hamedi et al., 2015b) with 9F (AAG AGT TTG ATC ATG GCT CAG) and 1541R (AGG AGG TGA TCC AAC CGC A) primers (Kumar et al., 2010). Briefly, each PCR reaction contained extracted DNA (2 μl), the primers (0.5 μM), MgCl2 (1.5 mM), dNTPs (0.15 μM), DMSO (5% v/v), standard Taq reaction buffer (1X), Taq DNA polymerase (2 units) and dH2O (up to 50 μl). PCR conditions were: 96 °C (3 min), 35 cycles of (96 °C for 30 s, 59–54 °C for 40 s, 68 °C for 140–190 s), 72 °C for 10 min. PCR products were observed through 1% agarose gel electrophoresis and SYBR Green I staining. The PCR products were sequenced by Macrogen (Seoul, Korea) and compared with that of other validated species in EzTaxon database (Kim et al., 2012). All molecular materials were purchased from (Pooya Gene Azma Co., Tehran, Iran).
2.5. Study of antibacterial activity of the isolates
The seeding material was prepared by inoculation of the isolates into ISP2 broth and incubated at 180 rpm, 28 °C, 48 h. The seeding material (10% v/v) was transferred into the 1000 ml Erlenmeyer containing 250 ml of fermentation medium and the fermentation flasks were incubated at 28 °C for 7 days, 180 rpm. After removing the biomass by centrifugation of the fermentation broth at 2500g for 10 min, the antimicrobial activity of the supernatant were assayed against several test strain microorganisms including, Escherichia coli UTMC 1465, E. coli TolC UTMC 1462, Pseudomonas aeruginosa UTMC 1463, Micrococcus luteus UTMC 1461, Mucor hiemalis UTMC 5057, Chromobacterium violaceum UTMC 1466, Candida albicans UTMC 5055, Staphylococcus aureus UTMC 1467, Pichia anomala UTMC 5056, and Bacillus subtilis UTMC 1464 by well agar diffusion method (Charousová et al., 2018). All test strains were obtained from University of Tehran Microorganisms Collection.
2.6. Hydrolytic enzyme production assay
2.6.1. Screening of amylase production
The isolates were screened for amylolytic activity according to their ability to hydrolysis of starch in Starch-agar. The inoculated plates were incubated on 28 °C for 5 days. After incubation period, to detect the amylase producing actinobacteria, the plates were flooded by iodine solution (1%). The amylase producing actinobacteria were identified by appearance of clear zone around their colonies which was surrounded by purple background (Nakajima et al., 1985).
2.6.2. Screening of protease activity
In order to screen the isolated actinobacteria for proteolytic activity, they were streaked on Skim milk agar plate. After incubation period (28 °C for 5 days), clear zones were appeared around the colonies of protease producing actinobacteria (Zerdani et al., 2004).
2.6.3. Screening of esterase activity
The esterase activity of actinobacterial isolates were screened according to their ability to hydrolysis Tween 80. The actinobacterial isolates were streaky cultured on Tween 80 agar plate. The inoculated plates were incubated at 28 °C for 5 days. The esterase producing actinobacteria isolates released fatty acid from Tween 80, the liberated fatty acids can bind to the calcium in the medium and produce insoluble crystals which were observed as a halo around the colonies (Rai et al., 2014).
2.6.4. Screening of lipase activity
To find lipase producing actinobacteria, the isolates were inoculated on Rhodamine-olive oil-agar. The inoculated plates were incubated on 28 °C for 5 days. Lipase producing actinobacteria were distinguished by appearing orange fluorescent halos around their streak culture under UV irradiation at 366 nm (Samad et al., 1989).
2.6.5. Screening of DNase activity
Ability of the isolates to produce DNase was assessed by inoculating the isolates on DNA agar plates. At the end of incubation (28 °C, 5 days), the plates were flooded with 1 N HCl. The clear zones were formed around the colonies are considered as degradation of DNA in the medium due to DNase activity of actinobacterial isolates (Jeffries et al., 1957).
2.6.6. Screening of heavy metal resistance
Resistant of actinobacterial isolates to several heavy metals including zinc, copper, cadmium, nickel and lead were primarily evaluated using growth test (Hamedi et al., 2015a). Briefly, the actinobacterial isolates were inoculated onto Minimal medium containing (Zn (35 mM), Cu (3 mM), Cd (2.5 mM), Ni (15 mM) and Pb (5 mM) heavy metal test media. The inoculated plates were incubated at 28 °C for 2 weeks. After incubation period, heavy metal resistant actinobacterial isolates, which can grow under the applied concentration of heavy metals, were further assessed in higher concentration of the desired heavy metal (Zn (70, 160 mM), Cu (7, 14 mM), Cd (5, 12 mM), Ni (30, 80 mM), Pb (9, 18 mM).
3. Results and discussion
Discovering new actinobacteria strains through conduction biodiversity studies is a strategic method towards finding new sources for current/new bioproducts. Poor investigated environments increase the likelihood of successful isolation of new gene pool. Harsh environment of the cave, including low nutrient and light caused to the low growth of plants and animals. However, a lot of microorganisms can be found in these oligotrophic and extreme environments (Lewin et al., 2016, Pedersen, 2000, Yasir, 2018).
3.1. Hampoeil cave and diversity of actinobacteria
Hampoeil cave is located in a rocky mountain and has about 1600 m height from the Mourdi Chay’s river bed. This area has semi-arid climate and the average annual rainfall is 300 ml. The cave entrance is 8 m and the height of it is about 25–40 m. Access to the sampling site of the cave requires suitable equipment, including cables and lighting facilities and experienced cave-guide. The first section of the cave consists of ∼1000 m2 square hall that was a historical residence of ancient cavemen and is currently a tourist attraction. However, due to a ∼14 m depth ravine, other sections of the cave are accessible only for professional cavemen and therefore significantly kept from pollution and human disturbance. Only cave-miners can access to the area that the samples obtained from there. These areas have enough oxygen for breathing, but had not any light. The cave is located in the upper vadose zone and development of karst system has occurred in the Triassic carbonate rocks. The cave has been developed and located in massive dolomite, with thin bedded limestone of the Elikah Formation. The cave is made entirely of dolomite and limestone and feature a wide variety of formations, including stalactites, stalagmites, soda straws, columns, and flowstone. Other sedimentary units around the cave are composed of sandstone and shale (Alavi and Shahrabi, 2009).
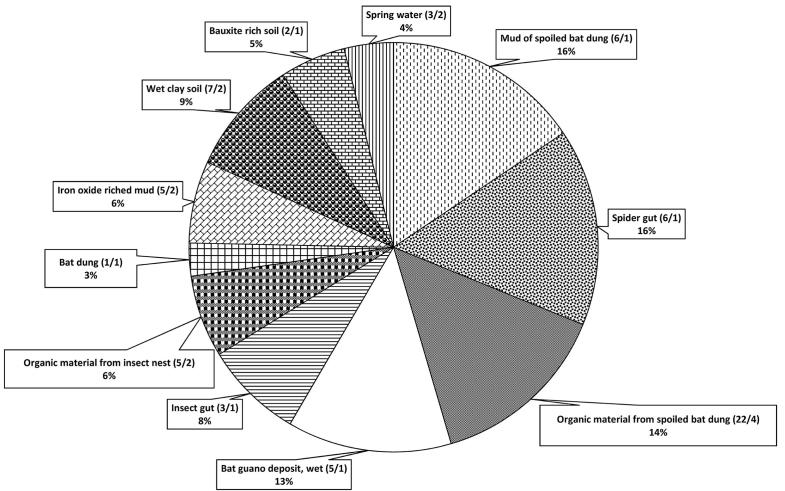
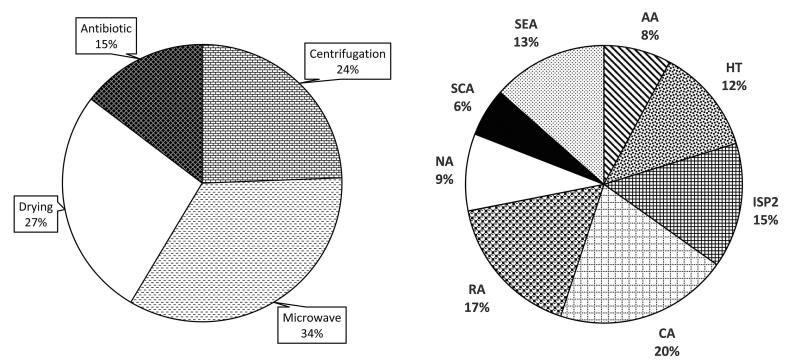
Among 33 various samples obtained from various sections of the Hampoeil cave at two steps, 76 actinobacteria were isolated from 22 samples and no actinobacteria were isolated from 11 samples, including 2 bat-feces eating arthropode, 1 iron-rich water, dead animals (2 scorpions, 1 worm and 1 insect), 1 water sample, 2 organic materials from insect nest dung and 1 bat skeleton. Some microorganisms rather than actinobacteria (specially Bacillus sp. and molds) were isolated, especially from organic rich samples, however, they are not further considered. Distribution of the actinobacterial isolates in various environmental samples was shown in Fig. 2. Maximum isolates per samples (82%) were seen in organic rich samples, including mud of spoiled bat dung, scorpion, and organic material from spoiled bat feces, bat guano deposit, insect, organic material from insect nest and bat dung. Distribution of the isolates according the pretreatments and the media used was shown in Fig. 3. Maximum and minimum amount of the isolates were obtained in microwave and antibiotic treatments, respectively. Among the media used, maximum and minimum number of the isolates was obtained in compost agar and Starch casein agar, respectively.
Fig. 2.
Distribution of actinobacteria from various environmental samples of Hampoeil cave. Percentage of the actinobacterial isolates and (numbers of the isolates actinobacteria/the numbers of the samples) in each environment were shown.
Fig. 3.
Distribution of the actinobacterial isolates according the treatments and the isolation media.
The result of molecular identification of the isolates according the similarity in 16SrRNA genes sequences are shown in Table 1. Among the isolates, 54% and 18.4% were Streptomyces and Micromonospora, respectively. After that, Micrococcus, Kocuria and Corynebacterium were 5% 4% and 5% of the isolates, respectively. All isolates were belonged to Actinobacteria class and there are no isolates that were belonged to other classes of Actinobacteria phylum. If the number of actinobacteria according their prevalence in the nature (Streptomyces/rare-actinobacteria) and related environments (oligotrophic/enriched) was considered, it is found that the numbers of the actinobacteria in enriched environments, such as bat guano deposit, organic materials from nest of arthropods or corpse of invertebrates is higher than that of non-enriched/oligotrophic samples. This finding broadly supports the work of other studies in this area linking bat guano deposit with more isolation of actinobacteria. It was reported that actinobacteria were 22.55% (second rank) of the common bacterial inhabitants of bat guano (De Mandal et al., 2015).
Table 1.
The actinobacterial strains isolated from Hampoeil cave.
| No. | UTMC code | Nearest neighborhood | Similarity % |
|---|---|---|---|
| 1 | 3161 | Micromonospora aurantiaca | 100 |
| 2 | 3162 | Micromonospora vinacea | 99.8 |
| 3 | 3163 | Kocuria gwangalliensis | 99.82 |
| 4 | 3164 | Streptomyces flavoviridis | 99.80 |
| 5 | 3165 | Streptomyces tendae | 100 |
| 6 | 3166 | Kocuria flava | 99.45 |
| 7 | 3167 | Micrococcus flavus | 100 |
| 8 | 3168 | Micromonospora soli | 100 |
| 9 | 3169 | Promicromonospora iranensis | 99.82 |
| 10 | 3170 | Micromonospora chalcea | 99.61 |
| 11 | 3171 | Rhodococcus cerastii | 99.80 |
| 12 | 3172 | Micromonospora krabiensis | 98.21 |
| 13 | 3173 | Streptomonospora halotolerans | 98.85 |
| 14 | 3174 | Micromonospora aloeverae | 100 |
| 15 | 3175 | Micromonospora soli | 100 |
| 16 | 3176 | Micromonospora chersina | 99.62 |
| 17 | 3177 | Micrococcus aloeverae | 100 |
| 18 | 3178 | Streptomyces brevispora | 99.61 |
| 19 | 3180 | Micromonospora echinofusca | 99.60 |
| 20 | 3181 | Micromonospora chalcea | 99.63 |
| 21 | 3182 | Actinomadura apis | 99.81 |
| 22 | 3183 | Nonomuraea ceibae | 99.28 |
| 23 | 3185 | Nocardia pigrifrangens | 99.81 |
| 24 | 3186 | Micromonospora chalyaphumensis | 99.64 |
| 25 | 3187 | Micromonospora soli | 100 |
| 26 | 3188 | Saccharomonospora azurea | 100 |
| 27 | 3189 | Micrococcus yunnanensis | 99.64 |
| 28 | 3190 | Streptomyces pratensis | 100 |
| 29 | 3191 | Nocardia jinanensis | 99.19 |
| 30 | 3192 | Micrococcus yunnanensis | 99.74 |
| 31 | 3193 | Arthrobacter citraus | 99.13 |
| 32 | 3194 | Corynebacterium mucifaciens | 100 |
| 33 | 3195 | Corynebacterium mucifaciens | 100 |
| 34 | 3196 | Corynebacterium xerosis | 100 |
| 35 | 3197 | Micromonospora chalcea | 99.71 |
| 36 | 3199 | Kocuria rhizophila | 99.73 |
| 37 | 3201 | Streptosporangium jiaoheense | 100 |
| 38 | 3202 | Streptomyces cinereoruber subsp. fructofermentans | 99.47 |
| 39 | 3204 | Micromonospora vinacea | 99.82 |
| 40 | 3224 | Streptomyces radiopugnans | 99.87 |
| 41 | 3226 | Streptomyces djakartensis | 99.47 |
| 42 | 3227 | Streptomyces ambofaciens | 99.87 |
| 43 | 3228 | Streptomyces violaceorubidis | 99.88 |
| 44 | 3229 | Streptomyces violascens | 99.62 |
| 45 | 3230 | Streptomyces thermocarboxydus | 100 |
| 46 | 3231 | Streptomyces albogriseolus | 100 |
| 47 | 3232 | Streptomyces qinglanensis | 99.01 |
| 48 | 3233 | Streptomyces thermocarboxydus | 100 |
| 49 | 3234 | Streptomyces djakartensis | 99.47 |
| 50 | 3235 | Streptomyces erythrogriseus | 100 |
| 51 | 3236 | Streptomyces cellulosae | 100 |
| 52 | 3237 | Streptomyces spiroverticillatus | 99.61 |
| 53 | 3238 | Streptomyces helimycini | 99.67 |
| 54 | 3239 | Streptomyces luteus | 99.62 |
| 55 | 3240 | Saccharothrix texasensis | 99.87 |
| 56 | 3241 | Streptomyces ramulosus | 99.60 |
| 57 | 3242 | Streptomyces spiroverticillatus | 99.80 |
| 58 | 3243 | Streptomyces arenae | 99.72 |
| 59 | 3244 | Streptomyces ochraceiscleroticus | 98.74 |
| 60 | 3245 | Streptomyces polyantibioticus | 99.55 |
| 61 | 3246 | Streptomyces spiroverticillatus | 99.61 |
| 62 | 3247 | Streptomyces heliomycini | 100 |
| 63 | 3249 | Streptomyces viridochromogenes | 98.47 |
| 64 | 3252 | Streptomyces leeuwenhoekii | 99.21 |
| 65 | 3253 | Streptomyces pratensis | 99.80 |
| 66 | 3254 | Streptomyces pratensis | 100 |
| 67 | 3255 | Streptomyces spiroverticillatus | 99.80 |
| 68 | 3256 | Streptomyces viridochromogenes | 99.75 |
| 69 | 3257 | Streptomyces djakartensis | 99.47 |
| 70 | 3258 | Streptomyces cinereoruber subsp. cinereoruber | 99.87 |
| 71 | 3259 | Streptomyces glomeroaurantiacus | 99.54 |
| 72 | 3260 | Streptomyces ambofaciens | 99.88 |
| 73 | 3261 | Streptomyces albogriseolus | 99.87 |
| 74 | 3262 | Streptomyces griseus subsp. griseus | 99.82 |
| 75 | 3263 | Streptomyces pratensis | 100 |
| 76 | 3264 | Streptomyces spiroverticillatus | 99.49 |
It is interesting to note that in this study, the ratio of rare-actinobacteria in the oligotrophic samples to enriched samples is 20% more than that of Streptomyces. Rare-actinobacteria were isolated/identified in oligotrophic samples collected from caves in various countries. For example, Rhodoccocus (17%), Pseudonocardia (16%) were identified from moon milk samples of Collemboles cave in Belgium (Maciejewska et al., 2018). Also, 3 Nocardia sp. and 7 Streptomyces sp. were isolated from the water and moon milk speleothem, from Bolshaya Oreshnaya cave, Siberia, Russia (Axenov-Gibanov et al., 2016). From the sediments and microbial mats developing on cave walls and ceilings of the volcanic caves (12 caves in Portugal and 2 caves from Canada), 2 Arthrobacter and 12 Streptomyces were isolated (Riquelme et al., 2017).
3.2. Hydrolytic activity of the actinobacteria
The results of the enzyme activities studied (amylase, DNase, protease, lipase or esterase) between the isolates were shown in Table 2. Among 76 isolates, 5.26, 21.05, 21.05, 23.68 and 9.21% of the isolates, had 5, 4, 3, 2, 1 enzyme activity(ies), respectively and 19.74 of the isolates did not show any hydrolytic enzyme activity. It seems that there is a relationship between the enzyme activity and environmental sources of the strains. Percentages of the strains with the highest activity in esterase, amylase, DNase, protease or lipase activity that were isolated from organic-rich environmental samples were 100, 100, 100, 82 and 82% of the strains, respectively. But, percentages of the strains with no esterase, amylase, DNase, protease or lipase activity that were isolated from these environmental samples were 66, 54, 61, 53 and 63%, respectively. It seems that living in organic-rich environments caused to selection of higher enzyme producer actinobacteria. Saccharomonospora azurea UTMC 3188, Streptomyces sp. UTMC 3246 and Streptomyces sp. UTMC 3262 had the maximum hydrolytic enzyme activities among the isolates. Streptomyces sp. UTMC 3246 that isolated from an arthropod (spider), had the highest activity in esterase and lipase activities and can be considered for further study. Streptomyces spiroverticillatus is the closest strain (99.61%) to it and is a producer of tautomycin, a polyketide with antifungal and anticancer activities (Chen et al., 2010). Three isolates with similarity between (97.8%-98.7%) in 16S rRNA gene to Streptomyces spiroverticillatus have been isolated from volcanic caves in Portugal (Riquelme et al., 2017).
Table 2.
Hydrolytic enzyme activities in the strains isolated from Hampoeil cave. + and – are used for positive and negative results, respectively. More activity was shown by more +.
| UTMC code | Esterase | Amylase | DNase | Protease | Lipase |
|---|---|---|---|---|---|
| Micromonospora aurantiaca UTMC 3161 | + | ++ | + | ++ | – |
| Micromonospora sp. UTMC 3162 | + | ++ | – | ++ | – |
| Kocuria sp. UTMC 3163 | – | ++ | – | – | + |
| Streptomyces sp. UTMC 3164 | + | +++ | + | ++ | – |
| Streptomyces tendae UTMC 3165 | ++ | ++ | + | – | – |
| Kocuria sp. UTMC 3166 | – | +++ | – | – | + |
| Micrococcus flavus UTMC 3167 | – | +++ | – | – | + |
| Micromonospora soli UTMC 3168 | + | ++ | – | – | |
| Promicromonospora sp. UTMC 3169 | + | ++++ | + | ++ | + |
| Micromonospora sp. UTMC 3170 | ++ | + | ++ | ++ | + |
| Rhodococcus sp. UTMC 3171 | + | ++ | – | ++ | – |
| Micromonospora sp. UTMC 3172 | + | ++ | – | – | – |
| Micromonospora aloeverae UTMC 3174 | + | + | – | – | – |
| Micromonospora soli UTMC 3175 | + | ++ | + | – | – |
| Micrococcus aloeverae UTMC 3177 | + | – | + | + | + |
| Streptomyces sp. UTMC 3178 | – | +++ | + | + | |
| Micromonospora sp. UTMC 3180 | – | + | – | – | – |
| Micromonospora sp. UTMC 3181 | + | + | + | +++ | – |
| Actinomadura sp. UTMC 3182 | + | – | – | + | – |
| Nonomuraea sp. UTMC 3183 | – | + | – | – | – |
| Micromonospora sp. UTMC 3186 | + | ++ | + | ++ | – |
| Saccharomonospora azurea UTMC 3188 | +++ | +++ | ++ | ++ | – |
| Micrococcus sp. UTMC 3189 | + | – | ++ | ++ | + |
| Nocardia sp. UTMC 3191 | – | – | + | – | – |
| Micrococcus sp. UTMC 3192 | – | – | + | +++ | + |
| Corynebacterium mucifaciens UTMC 3194 | – | ++ | – | – | + |
| Corynebacterium xerosis UTMC 3196 | – | ++ | – | + | – |
| Micromonospora sp. UTMC 3197 | + | ++ | – | – | + |
| Kocuria sp. UTMC 3199 | + | ++ | – | – | – |
| Streptosporangium jiaoheense UTMC 3201 | – | + | – | – | – |
| Micromonospora sp. UTMC 3204 | – | +++ | – | + | – |
| Streptomyces sp. UTMC 3224 | + | +++ | – | + | + |
| Streptomyces sp. UTMC 3226 | + | ++ | – | – | – |
| Streptomyces sp. UTMC 3227 | + | – | – | – | – |
| Streptomyces sp. UTMC 3228 | + | ++ | – | – | – |
| Streptomyces sp. UTMC 3229 | + | ++ | + | – | – |
| Streptomyces thermocarboxydus UTMC 3230 | + | ++ | + | – | – |
| Streptomyces sp. UTMC 3231 | + | ++ | – | – | – |
| Streptomyces thermocarboxydus UTMC 3233 | + | ++ | + | – | – |
| Streptomyces erythrogriseus UTMC 3235 | + | ++ | – | – | – |
| Streptomyces cellulosae UTMC 3236 | + | + | + | – | – |
| Streptomyces sp. UTMC 3237 | + | + | – | – | – |
| Streptomyces sp. UTMC 3238 | – | + | – | – | – |
| Streptomyces sp. UTMC 3239 | + | ++ | – | ++ | + |
| Streptomyces sp. UTMC 3241 | ++ | – | – | – | – |
| Streptomyces sp. UTMC 3243 | + | ++ | – | – | – |
| Streptomyces sp. UTMC 3244 | +++ | +++ | – | + | – |
| Streptomyces sp. UTMC 3245 | + | +++ | – | +++ | – |
| Streptomyces sp. UTMC 3246 | +++ | +++ | – | +++ | + |
| Streptomyces heliomycini UTMC 3247 | ++ | ++ | – | + | – |
| Streptomyces sp. UTMC 3249 | ++ | ++ | + | – | – |
| Streptomyces sp. UTMC 3252 | + | ++ | + | + | – |
| Streptomyces sp. UTMC 3253 | + | ++ | – | +++ | – |
| Streptomyces pratensis UTMC 3254 | ++ | +++ | – | – | – |
| Streptomyces sp. UTMC 3255 | +++ | ++ | + | +++ | – |
| Streptomyces sp. UTMC 3257 | + | +++ | + | +++ | – |
| Streptomyces sp. UTMC 3258 | + | +++ | + | +++ | – |
| Streptomyces sp. UTMC 3260 | ++ | +++ | + | ++ | – |
| Streptomyces sp. UTMC 3261 | + | +++ | + | +++ | – |
| Streptomyces sp. UTMC 3262 | ++ | +++ | + | +++ | + |
| Streptomyces sp. UTMC 3264 | + | ++ | + | +++ | + |
3.3. Antibiotic activity of the actinobacteria
The results of antibiotic activities among the isolates were shown in Table 3. As seen, at least one type of antimicrobial activity against the microbial test strains was seen in 25.3% of the isolates. However, there was no antibiotic activity against P. aeruginosa, E. coli, C. violaceum, S. aureus and P. anomola. Among the isolates, 25.3%, 7.3%, 3.16%. 8.4% and 3.61% had antibiotic activity against M. luteus, E. coli TolC, B. subtilis, S. aureus and P. anomola, respectively. maximum antibiotic activity was seen in Streptomyces sp. 3202 that was effect on M. luteus, E. coli TolC, B. subtilis and S. aureus.
Table 3.
The strains isolated from Hampoeil cave with antimicrobial activity. 1: Micrococcus luteus UTMC1461, 2: Escherichia coli TolC UTMC1462, 3: Pseudomonas aeruginosa UTMC1463, 4: Bacillus subtilis UTMC1464, 5: Escherichia coli UTMC1465, 6: Chromobacterium violaceum UTMC1466, 7: Staphylococcus aureus UTMC1467, 8: Candida albicans UTMC5055, 9: Pichia anomola UTMC5056 and 10: Mucor hiemalis UTMC5057.
| Actinobacterial isolate | 1 | 2 | 4 | 7 | 9 |
|---|---|---|---|---|---|
| Streptomyces tendae UTMC 3165 | 30 | – | – | 18 | – |
| Kocuria sp. UTMC 3166 | 14 | – | – | – | – |
| Micromonospora soli UTMC 3168 | 15 | – | – | – | – |
| Micromonospora sp. UTMC 3172 | 14 | – | – | – | – |
| Micromonospora soli UTMC 3175 | 13 | – | – | – | – |
| Streptomyces sp. UTMC 3178 | 25 | – | – | 12 | – |
| Micromonospora sp. UTMC 3181 | 20 | 20 | – | 22 | – |
| Saccharomonospora azurea UTMC 3188 | 14 | – | – | – | – |
| Streptosporangium jiaoheense UTMC 3201 | 30 | – | – | – | – |
| Streptomyces sp. UTMC 3202 | 23 | 17 | 17 | 22 | – |
| Streptomyces sp. UTMC 3227 | 18 | – | – | – | – |
| Streptomyces sp. UTMC 3229 | 23 | – | 27 | – | – |
| Streptomyces albogriseolus UTMC 3231 | 29 | – | – | – | – |
| Streptomyces thermocarboxydus UTMC 3233 | 23 | – | – | – | – |
| Streptomyces erythrogriseus UTMC 3235 | 13 | – | – | – | – |
| Streptomyces cellulosae UTMC 3236 | 16 | – | – | – | – |
| Streptomyces sp. UTMC 3237 | – | 23 | – | – | 28 |
| Streptomyces sp. UTMC 3242 | 27 | – | – | – | 11 |
| Streptomyces sp. UTMC 3244 | – | – | 23 | 17 | – |
| Streptomyces sp. UTMC 3245 | 20 | 16 | – | – | 12 |
| Streptomyces sp. UTMC 3246 | 25 | 24 | – | – | – |
| Streptomyces pratensis UTMC 3254 | 17 | – | – | – | – |
| Streptomyces sp. UTMC 3255 | 28 | 20 | – | – | – |
What is surprising is that antimicrobial activity of the isolates is not high. Although, there are some antimicrobial activity observed against some test strains, including M. luteus, E. coli TolC and B. subtilis, but all of these test strains are sensitive to antimicrobial substance, rather than antibiotics, e.g. fatty acids (Chandrasekaran et al., 2011, Vakharia et al., 2001). By consideration of 91% esterase activity in the strains containing antibacterial activity against M. luteus, as a highly sensitive test strain to fatty acids, it seems, fatty acids liberated from esterase activity caused to antimicrobial activity that seen for many of the isolates. Among the isolates, only Streptomyces sp. UTMC 3237 may be considered for farther studies. This strain has 99.61% taxonomical similarity to Streptomyces spiroverticillatus, a known antifungal producer (Chen et al., 2010).
3.4. Metal resistance of the actinobacteria
The effect of the metals on the actinobacteria isolates was shown in table 4. Among the isolates, 73.68% were sensitive to all metal studied, including Pb (5 mM), Ni (15 mM), Cd (2.5 mM), Cu (3 mM) and Zn (35 mM). But, 26.32% of the isolates were resisted one metal or more. The strains designated Nocardia sp. UTMC 3191 and Streptomyces sp. UTMC 3261 were resisted to all five metals studied and Micromonospora soli UTMC 3168, Streptomyces sp. UTMC 3178 and Streptomyces pratensis UTMC 3254 were resisted to three metals studied.
Table 4.
Metal resistance in the strains isolated from Hampoeil cave with antimicrobial activity. R: resistant, S: sensitive.
| UTMC code | Zn (35 mM) | Cu (3 mM) | Cd (2.5 mM) | Ni (15 mM) | Pb (5 mM) |
|---|---|---|---|---|---|
| Micromonospora aurantiaca UTMC 3161 | R | S | S | S | S |
| Micromonospora sp. UTMC 3162 | S | S | S | R | R |
| Streptomyces sp. UTMC 3164 | R | S | R | R | S |
| Micromonospora soli UTMC 3168 | S | R | R | R | R |
| Micromonospora sp. UTMC 3170 | R | R | S | S | R |
| Rhodococcus sp. UTMC 3171 | R | S | S | R | S |
| Streptomyces sp. UTMC 3178 | R | R | S | R | R |
| Micromonospora sp. UTMC 3181 | R | S | S | S | S |
| Nocardia sp. UTMC 3191 | R | R | R | R | R |
| Corynebacterium xerosis UTMC 3196 | R | S | S | S | S |
| Micromonospora sp. UTMC 3197 | S | R | S | S | S |
| Streptomyces sp. UTMC 3224 | S | S | S | R | S |
| Streptomyces sp. UTMC 3234 | S | S | S | R | S |
| Streptomyces sp. UTMC 3245 | R | R | S | S | S |
| Streptomyces sp. UTMC 3253 | S | S | R | S | S |
| Streptomyces pratensis UTMC 3254 | R | R | R | R | S |
| Streptomyces sp. UTMC 3261 | R | R | R | R | R |
| Streptomyces sp. UTMC 3262 | R | S | S | R | R |
Maximum tolerance to Zn was 70 mM that were seen in Micromonospora aurantiaca UTMC 3161, Streptomyces sp. UTMC 3178, Nocardia sp. UTMC 3191 and Streptomyces pratensis UTMC 3254. It was 35 mM for other metal resistant isolates. The strains Streptomyces sp. UTMC 3178 and Micromonospora aurantiaca UTMC 3261 had maximum tolerance to Cu that was 7 mM. In the other metal resistant actinobacteria, maximum resistant to Cu, was 3 mM. Maximum resistance to Cd was 5 mM that was seen in the Nocardia sp. UTMC 3191 and Streptomyces pratensis UTMC 3254, it was 2.5 mM for the others metal resistant actinobacterial isolates. Maximum resistance to nickel was 30 mM that were seen in Micromonospora sp. UTMC 3162, Streptomyces sp. UTMC 3164, Micromonospora soli UTMC 3168, Rhodococcus sp. UTMC 3171, Streptomyces sp. UTMC 3178 and Nocardia sp. UTMC 3191. Maximum resistance to Pb was 9 mM that were in Micromonospora soli UTMC 3168 and Nocardia sp. UTMC 3191.
The most interesting finding was that potential of 26.32% of the actinobacterial isolates in resistance to heavy metals and therefore can be considered as candidates in study to use in gray biotechnology. In accordance with the present results, previous studies have demonstrated that some neighborhoods of the metal resistant actinobacteria isolates have potential in bioremediation. Streptomyces sp. UTMC 3231 and Streptomyces sp. UTMC 3164 had 99.87% and 99.80% similarities to Streptomyces albogriseolus and Streptomyces flavoviridis, respectively. Streptomyces albogriseolus HUT6045 and Streptomyces flavoviridis HUT 6147 has potential for the selective bioaccumulation of thorium and uranium (Nakajima and Tsuruta, 2002, Tsuruta, 2006). Also, S. flavoviridis HUT 6147 have potential in biosorption and recycling of gold (Tsuruta, 2004). Rhodococcus sp. UTMC 3171 is 99.80% similarity to Rhodococcus cerastii, an actinobacterium with potential for degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), a high explosive used in weapons manufacturing (Wang et al., 2017).
4. Conclusion
The result of current study showed the impact of caves as good sources to find various actinobacteria with versatile potential in biotechnology. In Hampoeil cave, if there is need to find more actinobacteria, enriched environmental samples are preferred, however, more rare-actinobacteria were obtained from oligotrophic samples. Also, it was concluded that actinobacteria isolated from Hampoeil cave are significant in at least two major aspects of hydrolytic enzyme production and bioremediation. Future studies on the current topics are recommended.
5. Authors’ contribution
Javad Hamedi introduced the conception of the study and interpreted the data. Maghsoud Kafshnouchi had contribution in doing the experiments. All authors had participation in sampling according biological (Hamedi and Kafshnoochi) and geological characteristics (Mohsen Ranjbaran). The map of the cave was drawn by M. Ranjaran.
Acknowledgments
Acknowledgement
The authors are grateful to Behzad Ghomi, Fardin Hassandoost and Fereydoon Vahdano-Bonab (from Maragheh Caving Club, Maragheh Etehad Caving Group) for their kind guidance in Hampoeil Cave. Also, the authors are thankful to Leila Parvizi and Fahimeh Mohammadnia from University of Tehran Microorganisms Collection for their kindly technical assistances.
Declarations of interest
None.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Javad Hamedi, Email: jhamedi@ut.ac.ir.
Mohsen Ranjbaran, Email: m.ranjbaran@ut.ac.ir.
References
- Airoldi L., Cinelli F. Sources and biochemical composition of suspended particulate material in a submarine cave with sulphur water springs. Mar. Biol. 1997;128:537–545. [Google Scholar]
- Alavi M., Shahrabi M. The map unit; Tehran, Iran: 2009. GSI Maragheh geological map Geological Survey of Iran. [Google Scholar]
- Atlas R.M. CRC Press; 2004. Handbook of microbiological media. [Google Scholar]
- Axenov-Gibanov D.V., Voytsekhovskaya I.V., Tokovenko B.T., Protasov E.S., Gamaiunov S.V., Rebets Y.V., Luzhetskyy A.N., Timofeyev M.A. Actinobacteria isolated from an underground lake and moonmilk speleothem from the biggest conglomeratic karstic cave in Siberia as sources of novel biologically active compounds. PloS one. 2016;11:e0149216. doi: 10.1371/journal.pone.0149216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R.H., Demain A.L., Davies J.E. American Society for Microbiology Press; 2010. Manual of industrial microbiology and biotechnology. [Google Scholar]
- Barton H.A., Jurado V. What's up down there? Microbial diversity in caves. Microbe-Am. Soc. Microbiol. 2007;2:132. [Google Scholar]
- Bates R., Jackson J. Second ed. American Falls Church; Virginia: 1980. Glossary of geology. [Google Scholar]
- Belyagoubi L., Belyagoubi-Benhammou N., Jurado V., Dupont J., Lacoste S., Djebbah F., Ounadjela F.Z., Benaissa S., Habi S., Abdelouahid D.E. Antimicrobial activities of culturable microorganisms (actinomycetes and fungi) isolated from Chaabe Cave. Algeria. Int. J. Speleol. 2018;47:8. [Google Scholar]
- Bhullar K., Waglechner N., Pawlowski A., Koteva K., Banks E.D., Johnston M.D., Barton H.A., Wright G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PloS one. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran M., Senthilkumar A., Venkatesalu V. Antibacterial and antifungal efficacy of fatty acid methyl esters from the leaves of Sesuvium portulacastrum L. Eur. Rev. Med. Pharmacol. Sci. 2011;15:775–780. [PubMed] [Google Scholar]
- Charousová I., Medo J., Hleba L., Javoreková S. Streptomyces globosus DK15 and Streptomyces ederensis ST13 as new producers of factumycin and tetrangomycin antibiotics. Braz. J. Microbiol. 2018 doi: 10.1016/j.bjm.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-L., Xu Y.-H., Zheng Y.-G., Shen Y.-C. Improvement of tautomycin production in Streptomyces spiroverticillatus by feeding glucose and maleic anhydride. Biotechnol. Bioprocess. Eng. 2010;15:969–974. [Google Scholar]
- Co-operation, O.f.E., Development . OECD Publishing; 2001. Biological resource centres: underpinning the future of life sciences and biotechnology. [Google Scholar]
- Cunningham K., Northup D., Pollastro R., Wright W., LaRock E. Bacteria, fungi and Biokarst in Lechuguilla cave, Carlsbad caverns national park, new Mexico. Environ. Geol. 1995;25:2–8. [Google Scholar]
- De Mandal S., Panda A.K., Bisht S.S., Kumar N.S. First report of bacterial community from a bat guano using illumina next-generation sequencing. Genom. data. 2015;4:99–101. doi: 10.1016/j.gdata.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani M. The Economic Geology Iran. Springer; 2013. A summary of geology of Iran; pp. 45–64. [Google Scholar]
- Gulecal-Pektas Y. Bacterial diversity and composition in oylat cave (Turkey) with combined sanger/pyrosequencing approach. Pol. J. Microbiol. 2016;65:69–75. doi: 10.5604/17331331.1197277. [DOI] [PubMed] [Google Scholar]
- Hamedi J., Dehhaghi M., Mohammdipanah F. Isolation of extremely heavy metal resistant strains of rare actinomycetes from high metal content soils in Iran. Int. J. Environ. 2015;Res:9. [Google Scholar]
- Hamedi J., Imanparast S., Mohammadipanah F. Molecular, chemical and biological screening of soil actinomycete isolates in seeking bioactive peptide metabolites. Iran. J. Microbiol. 2015;7:23. [PMC free article] [PubMed] [Google Scholar]
- Hamedi J., Mohammadipanah F., Pötter G., Spröer C., Schumann P., Göker M., Klenk H.-P. Nocardiopsis arvandica sp. nov., isolated from sandy soil. Int. J. Syst. Evol. Microbiol. 2011;61:1189–1194. doi: 10.1099/ijs.0.022756-0. [DOI] [PubMed] [Google Scholar]
- Hamedi J., Poorinmohammad N., Wink J. Biology and Biotechnology of Actinobacteria. Springer; 2017. The role of actinobacteria in biotechnology; pp. 269–328. [Google Scholar]
- Hopwood D.A. Oxford University Press; 2007. Streptomyces in nature and medicine: the antibiotic makers. [Google Scholar]
- Jeffries C.D., Holtman D.F., Guse D.G. Rapid method for determining the activity of microorgan-isms on nucleic acids. J. Bacteriol. 1957;73:590. doi: 10.1128/jb.73.4.590-591.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajtezović N., Rubinić J. Proceeding of the 3rd International Conference “Waters in Sensitive & Protected Areas”. 2013. p. 15. [Google Scholar]
- Kim O.-S., Cho Y.-J., Lee K., Yoon S.-H., Kim M., Na H., Park S.-C., Jeon Y.S., Lee J.-H., Yi H. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. System. Evolution. Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kumar V., Bisht G.S., Institu S.B.S.P.G. An improved method for isolation of genomic DNA from filamentous actinomycetes. Int. J. Eng. Technol. Manage. Appl. Sci. 2010;2:2. [Google Scholar]
- Kurtbӧke D. Biology and Biotechnology of Actinobacteria. Springer; 2017. Ecology and habitat distribution of actinobacteria; pp. 123–149. [Google Scholar]
- Lewin G.R., Carlos C., Chevrette M.G., Horn H.A., McDonald B.R., Stankey R.J., Fox B.G., Currie C.R. Evolution and ecology of actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016;70:235–254. doi: 10.1146/annurev-micro-102215-095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locey K.J., Lennon J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. 2016;113:5970–5975. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska M., Całusińska M., Cornet L., Adam D., Pessi I.S., Malchair S., Delfosse P., Baurain D., Barton H.A., Carnol M. High-throughput sequencing analysis of the actinobacterial spatial diversity in moonmilk deposits. Antibiotics. 2018;7:27. doi: 10.3390/antibiotics7020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima A., Tsuruta T. Competitive biosorption of thorium and uranium by actinomycetes. J. Nucl. Sci. Technol. 2002;39:528–531. [Google Scholar]
- Nakajima R., Imanaka T., Aiba S. Nucleotide sequence of the Bacillus stearothermophilus alpha-amylase gene. J. Bacteriol. 1985;163:401–406. doi: 10.1128/jb.163.1.401-406.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K. Exploration of deep intraterrestrial microbial life: current perspectives. FEMS. Microbiol. Lett. 2000;185:9–16. doi: 10.1111/j.1574-6968.2000.tb09033.x. [DOI] [PubMed] [Google Scholar]
- QYR-Research-Group, 2018. Global Microbial Fermentation Technology Market size, Status and Forecast 2025. Report Code: WGR3042315.
- Rai B., Shrestha A., Sharma S., Joshi J. Screening, optimization and process scale up for pilot scale production of lipase by Aspergillus niger. Biomed. Biotechnol. 2014;2:54–59. [Google Scholar]
- Riquelme C., Dapkevicius M.D.L.E., Miller A.Z., Charlop-Powers Z., Brady S., Mason C., Cheeptham N. Biotechnological potential of actinobacteria from Canadian and Azorean volcanic caves. Appl. Microbiol. Biotechnol. 2017;101:843–857. doi: 10.1007/s00253-016-7932-7. [DOI] [PubMed] [Google Scholar]
- Samad M.Y.A., Razak C.N.A., Salleh A.B., Yunus W.Z.W., Ampon K., Basri M. A plate assay for primary screening of lipase activity. J. Microbiol. Methods. 1989;9:51–56. [Google Scholar]
- Tsuruta T. Biosorption and recycling of gold using various microorganisms. J. Gen. Appl. Microbiol. 2004;50:221–228. doi: 10.2323/jgam.50.221. [DOI] [PubMed] [Google Scholar]
- Tsuruta T. Bioaccumulation of uranium and thorium from the solution containing both elements using various microorganisms. J. Alloy. Comp. 2006;408:1312–1315. [Google Scholar]
- Vakharia H., German G.J., Misra R. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active alpha-hemolysin. J. Bacteriol. 2001;183:6908–6916. doi: 10.1128/JB.183.23.6908-6916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Boukhalfa H., Marina O., Ware D.S., Goering T.J., Sun F., Daligault H.E., Lo C.C., Vuyisich M., Starkenburg S.R. Biostimulation and microbial community profiling reveal insights on RDX transformation in groundwater. Microbiol. Open. 2017:6. doi: 10.1002/mbo3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W.W. Origin of caves and other solution openings in the unsaturated (vadose) zone of carbonate rocks: a model for CO2 generation. Geology. 1985;13:822–824. [Google Scholar]
- Yadav A.K., Srivastava A.K., Yandigeri M.S., Kashyap S.K., Modi D.R., Arora D.K. Characterization of indigenous copper-resistant Streptomycetes from chickpea (Cicer arietinum L.) fields. Ann. Microbiol. 2010;60:605–614. [Google Scholar]
- Yasir M. Analysis of bacterial communities and characterization of antimicrobial strains from cave microbiota. Braz. J. Microbiol. 2018;49:248–257. doi: 10.1016/j.bjm.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerdani I., Faid M., Malki A. Feather wastes digestion by new isolated strains Bacillus sp. in Morocco. Afr. J. Biotechnol. 2004;3:67–70. [Google Scholar]