Abstract
Tomato (Lycopersicon esculentum) is one of the widely grown vegetables worldwide. Fusarium oxysporum f. sp. lycopersici (FOL) is the significant contributory pathogen of tomato vascular wilt. The initial symptoms of the disease appear in the lower leaves gradually, trail by wilting of the plants. It has been reported that FOL penetrates the tomato plant, colonizing and leaving the vascular tissue dark brown, and this discoloration extends to the apex, leading to the plants wilting, collapsing and dying. Therefore, it has been widely accepted that wilting caused by this fungus is the result of a combination of various physiological activities, including the accumulation of fungal mycelia in and around xylem, mycotoxin production, inactivation of host defense, and the production of tyloses; however, wilting symptoms are variable. Therefore, the selection of molecular markers may be a more effective means of screening tomato races. Several studies on the detection of FOL have been carried out and have suggested the potency of the technique for diagnosing FOL. This review focuses on biology and variability of FOL, understanding and presenting a holistic picture of the vascular wilt disease of tomato in relation to disease model, biology, virulence. We conclude that genomic and proteomic approachesare greater tools for identification of informative candidates involved in pathogenicity, which can be considered as one of the approaches in managing the disease.
Keywords: Fusarium oxysporum f. sp. lycopersici, Pathogenicity, Biology, Diversity, Lycopersicon esculentum, Vascular wilt
1. Introduction
The Fusarium genus is one of the utmost complex and adaptive species in the Eumycota and the Fusarium oxysporum (Fo) species complex includes plant, animal and human pathogens and a diverse range of non-pathogens (Gordon, 2017). Members of Fusarium species are ubiquitous soil-borne pathogens of a wide range of horticultural and food crops which cause destructive vascular wilts, rots, and damping-off diseases (Bodah, 2017). In addition to the losses caused before or during harvest, some Fusarium species are capable of producing mycotoxins in food and agricultural commodities (Nayaka et al., 2008, Nayaka et al., 2009, Mudili et al., 2014). Fusarium toxins are the most abundant natural contaminants of diets containing cereals and other grains (Venkataramana et al., 2014, Divakara et al., 2014, Kalagatur et al., 2015, Kumar et al., 2016) and suspected to be implicated in numerous diseases among mammals and other living beings (Nayaka et al., 2010, Venkataramana et al., 2014, Kalagatur et al., 2017, Kalagatur et al., 2018). The fumonisins, belong to the family of food-borne carcinogenic mycotoxins, with reports of toxic activity of Fo strains isolated from various products that exhibited different degrees of toxicity to experimental animals (Venkataramana et al., 2012). Members of Fusarium genus harbor biosynthetic machinery capable of producing interesting bioactive secondary metabolites, and produce antifungal, antibacterial and cytotoxic compounds, such as alkaloids, sesquiterpenes, polyketides, carotenoids, anthraquinone, cyclopentanone, and naphthoquinone derivatives (Manici et al., 2017). Fusarium oxysporumis an important, soil-inhabiting ubiquitous fungus, known for its phylogenetic diversity (Xiong and Zhan, 2018, Nicholas et al., 2017, Arpita et al., 2012). Strains of Foare saprophytic or non-pathogenic (Kumar et al., 2010). However, the phytopathogenic strains cause destructive vascular wilt disease and often limit the production of economically important crops (Servin et al., 2015, Shahzad et al., 2017). The species of Focause wilt disease in more than 150 hosts and range with specific formae speciales (Bertoldo et al., 2015). Asha et al., 2011, Nirmaladevi et al., 2016 reported Fusarium oxysporum Schlectend. Fr. f. sp. lycopersici (Sacc.) W.C. Snyder and H.N. Hansen (FOL) causes vascular wilt of tomato disease and reduced the yield to the maximum extent (Asha et al., 2011). The present review helps to alert recent progress in the application of molecular markers for understanding the diversity, biology and epidemiologyof FOL (Fig. 1).
Fig. 1.
Fusarium wilt caused by F. oxysporumf. sp. lycopersiciin field conditions.
2. Fusarium wilt of tomato
The family Solanaceae, includes more than 3000 species among them cultivated tomato, is the only vegetable crop cultivated throughout the world. This crop is a vital component of daily food and is consumed as unprocessed fresh fruits as well as invarious types of processed products (Brookie et al., 2018). Tomato wilt is one of the chief diseases of tomato caused by FOL (Borisade et al., 2017). The FOL enters the epidermis of root, later spreads through the vascular tissue and inhabits the plant xylem vessels, resulting in vessel clogging, and severe water stress as a result wilt like symptoms appear (Singh et al., 2017). The disease in morphologically identified by wilted plants bearing yellow colored leaves with minimal or absent crop yield. The dormant chlamydospore of FOL in infested soil can survive indefinitely in the absence of host (Khan et al., 2017, Cha et al., 2016). The progression of plant vascular infection by Fo is a complex phenomenon, and the sequential steps involved in the infection process are as follows: (1) root recognition through host-pathogen signals, (2) attachment to surface of root hairs and hyphal propagation, (3) invasion of the root cortex, and vascular tissue and differentiation within xylem vessels, (4) finally oozing of toxins and virulence factors. Colonization of the vessels leads to disease development and the characteristic wilting of the host plant (Di et al., 2016).
As a characteristic of soil-borne pathogen, FOL can survive extensively in soil as dormant propagules (chlamydospores). Host rootpresence triggers thegerminationof chlamydospores. The infection hyphae adhere to and then penetrate the root surface. The mycelium invades the root cortical cells intercellularly and enters vascular system through the xylem pits. Subsequently, the fungus displays a unique pathway of infection where it tends to colonize exclusively inside the vessels of xylem, further rapidly colonize the host. Within the vessels, the fungus starts to produce microconidia, which are transported to upwards through sap stream upon detachment. Further, germination of microconidia leads to mycelial penetration of the upper vessels. The characteristic wilt symptoms appear due to vessel blockage triggered by the gathering of fungal hypae and a combination of host-pathogen interaction such as, the release of toxins, gums, gels, and formation of tyloses. Typical disease indications, such as leaf epinasty, vein clearing, wilting and defoliation, appear and eventually precedes host plant death (Fig. 1, Fig. 2). During this phase the vascular wilt fungus, which stays limited to the xylem vessels, propagates through parenchymatous tissue and begins to sporulate abundantly on surface of the plant such as, leaf, steam etc. Dissemination of the pathogen can occur via seeds, transplants, soil or other means (McGovern, 2015, Joshi, 2018).
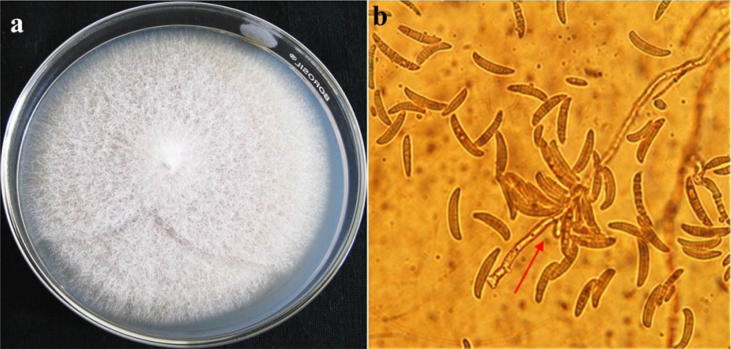
Fig. 2.
(a and b). Cultural and morphological features of Fusarium oxysporumf. sp. lycopersici. (a). F. oxysporumcolony of Fusarium sp. on PDA agar; (b). Microscopic view of macroconidia of F. oxysporumf. sp. Lycopersici, macroconidia abundant, commonly three septate and the attachment of the macroconidia to the mycelium is observed.
The ultra-structural aspect of the FOL and tomato plant interaction has been investigated based on light, fluorescence and electron microscopy. Scanning electron microscopy of transverse and longitudinal sections through the dried stems of tomato plants colonized by FOL revealed that microconidia were largely associated with the xylem vessels, which germinated, and the mycelium entered the cortex and vessels 10–14 days after inoculation. However, the hyphae within the vessels were thicker in diameter (1.5–2 µm) and propagated through the pits of vessels walls. No physical barricade within the vessels could control the spread of the microconidia. Uncolonized vessels appear granular, while the colonized vessels appear smooth. In tomato plants, when the vascular elements become infected with FOL, the contact parenchyma cells unsheathing the vessels develop calluses containing deposits. These contact parenchyma cells play a significant role in regulating storage, vascular contents, and the progress of defense-related functions. Light and transmission electron microscopy examination of tomato plant parenchyma cells shows the deposits of callose and the wall appositions associated with blabbing and vesiculation of the plasmalemma and usually contain globular bodies, that in later stages of development exhibit a striated or marbled appearance. Olivain et al. (2006) used confocal laser microscopy, green fluorescent protein (GFP) and expresser reporter genes DsRed2 to picturize the establishment of non-pathogenic and pathogenic strain on roots hairs of tomato by non-pathogenic strains. The hyphae that reached root surface created small networks. Fungal colonization was found be limited to the extent of the taproot and lateral roots and was never observed in the apical zones. The region ahead the apex is the core zone of root exudation (Di et al., 2016). At later stages, penetration of the epidermal cells was observed (see Fig. 3).
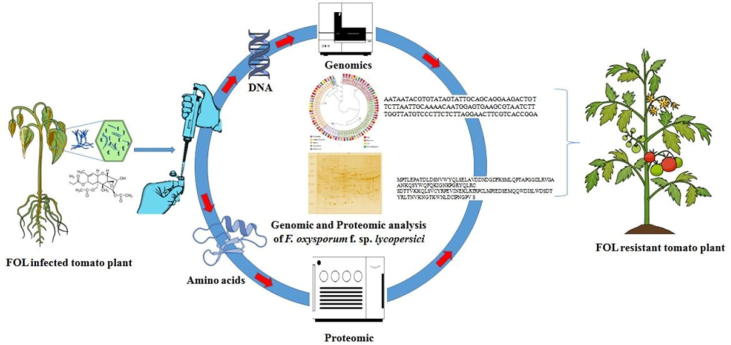
Fig. 3.
Schematic representation of application of multi-omics approaches for study F. oxysporumf. sp. lycopersicidiversity for developing FOL resistant tomato.
3. Mycotoxins from F. oxysporum
Certain molds produce toxic secondary metabolites called mycotoxins on a variedvariety of plants and agricultural commodities that are closely connected to animal and human food chains (Ramana et al., 2012). As a typical vascular wilt fungus, F. oxysporum produces the characteristic xylem vessel clogging and the wilting of infected plants. Colonization and clogging of vessels in addition to secretion of several toxins by the fungus includes fusaric acid, lycomarasmin, dehydrofusaric acid, etc., play amajor role in wilt symptoms development and progression. At least 11 species of Fusarium, such as plant pathogenic F. oxysporum produce the mycotoxinFumonisins (Desjardins, 2006). The toxigenic potential of F. oxysporum on plants and additional commodities and the extensivevariety and frequent presence of Fuminosintoxins has developed a major constraint in main food crops. There are findings of Fuminosin production by individual species of F. oxysporum and PCR based methods targeting the toxin genes of biosynthetic pathways have been studied (Proctor et al., 2008, Ramana et al., 2011). In our earlier study, Nirmaladevi et al. (2012), used a PCR based approach targeting the Fuminosin biosynthetic genes which allowed the detection of Fuminosin producing strains of FOL. Among the 45 strains tested, the primers has been used to detect 16 toxin producing strains of FOLindicating that some of the F. oxysporum strains causing tomato have the potential to produce Fumonisin (Nirmaladevi et al. (2012)). Fusaric acid is another potential toxin produced by F. oxysporum including FOL. The production of Fusaric acid has been connected with the virulence strains of Fusarium spp. Fusaric acid is a potent toxin natural contaminating infected plants and cereals causing typical wilt symptoms in plants and however, it has ill effects on humans and animals by enhancing the toxicity of trichothecenes (Wang and Ng, 1999). Singh et al. (2017) characterized the phytotoxic effects of Fusaric acid in tomato leaves which revealed reduced photosynthesis, leaf wilting and necrosis, enormous lipid peroxidation and intracellular reactive oxygen species and cell death. Further, the leaf proteome revealed differential expression of several proteins showing the potential role of Fusaric acid in decreasing cell viability and enhancing the fungal pathogenicity. In an attempt to demonstrate the role of Fusaric acid, Lopez-Diaz et al. (2018) found the gene fub1was vital for the synthesis of the toxin and its derivatives in F. oxysporum. Targeted deletion of fub1 and loss of Fusaric acid production in F. oxysporum led to reduced severity of wilt symptoms and virulence in host and the death of immunosuppressed mice.
4. Epidemiology
The overall distribution of FOL is known to be cosmopolitan and occurs predominantly as a soil saprophyte which standout amongst the most widely recognized and predominant fungi of cultivated soils. However, the different formae speciales (f. sp.) of Fooften have varying degrees of distribution. This disease affects the tomato grown at warm (28 °C) both in greenhouse and field condition (Bawa, 2016, Debbi et al., 2018). The disease is characterized by 70 to 60% of fruit yield loss with wilted plants containing yellowed leaves (Ravindra et al., 2015).
The three known FOL races (Races 1, 2 and 3) pathogens of tomato cultivars are distinguishable by their principle resistance genes. There are reports or Races 1 and 2 grownthrough the tomato growing regions of world whereas Race 3 has been reported in countriessuch as California, Australia, Southwestern Georgia and Mexico. Most commercial tomato varieties grown through the world are resistant to race 1 and 2, and a few are resistant to race 3 (Biju et al., 2017). FOL spread through short distance mainly through irrigation water and contaminated farm equipment’s and it can spread long distancesthrough infected transplants, soils etc., (Agrios, 2005). Certainly, once a region becomes contaminated with FOL, the fungus usually remains indefinitely (Animashaun et al., 2017, Prihatna et al., 2018).
5. Virulence genes requirements for the pathogenicity of FOL
Several research reports from the last decade, gain better insight into the molecular mechanisms involved in the pathogenesis FOL. Soil-borne phytopathogenic fungi must possess appropriate signaling mechanisms that enable them to respond by variations in geneexpression, leading to host recognition, root penetration and proliferation of hyphae withinthe host tissue leading to overcoming the host defense mechanism, and disease establishment (Rep and Kistler, 2010). The fungal growth and virulence factors are mainly governed by two pathways of signal transduction namely Mitogen activated protein kinase cascade (MAPK) and Cyclic adenosine monophosphate cAMP (Liu et al., 2016). The cAMP-PKA and MAPK cascades also function in FOL and may regulate a few key steps in infection process (Guo et al., 2016). Mutation generated by inactivation of gene encoding mitogen-activated protein kinase rendered the pathogen incapable of penetrating the tomato roots resulting in failure of appearance of disease symptoms. The inability of mutant strains to adhere to the root surface was detected byusing fluorescent microscopy expressing green fluorescent protein, whereas the wild-type strain was capable to firmly anchor and penetrate the root surface. Interestingly, pectate lyase and polygalacturonase enzymes secretion was reduced in mutant strain (Δfmk1), these two enzymes are involved in cell wall degradation during pathogenesis (Guo et al., 2016, Pareek and Rajam, 2017). The chitin synthase gene (chsV) encodes a chitin synthase (class V) an enzyme involved in membrane-associated chitin production; chitin is a vital component existing in cell wall of fungi (Liu et al., 2016). FOL resistant to secondary metabolites of plant was studied by DeConinck et al. (2015) using a non-pathogenic mutant of FOL obtained through random insertional mutagenesis. The mutant strain displayed comprehensive loss of virulence, and characterization of the insertion site revealed inactivation of the chsVgene. These findings suggest that the chsVgene is necessary to resist the defense compounds, a prerequisite for pathogenicity (Bharti et al., 2017).
Several genes FOL were identified whose protein products are released during infection into the host cells (Schmidt et al., 2013, Schmidt et al., 2016). The two protein produced in xylem are coded by genesSIX1 and SIX2 are positioned within 8 kb of each other and are on one of the smallest chromosomes (Boix-Ruíz et al., 2015, Rep et al., 2004). The SIX1 product is a small protein rich in cysteine that had revealed to be essential for FOL virulence (Selim et al., 2015). Eight fungal proteins from xylem sap of diseased plant was identified and the genetic material for these proteins are present in the similar region of chromosome (SIX1, SIX2) (Maldonado et al., 2018, Sasaki et al., 2015). Within the same chromosome a homolog of SIX1 and SIX1-H, are present which, encode for a salicylate hydroxylase homolog, and another gene, SIX3, encode for xylem-secreted protein. SIX1, SIX2, SIX3, and SHH1 were unique to FOL isolates. Despite their polyphyletic origin, all the FOL isolates had a genomic region containing of at least 8 kb identical genes comprising of SIX1, SIX2 and SHH1 that was lacking in other non-pathogenic isolates and formae speciales. The fungal virulence gene factors such as, SIX1, SIX2 and SIX3 encode a proteins, which is secreted into xylem sap may contribute to the wilting of plant by colonization of fungal hype. This genomic region initially existed in ancestral Foand subsequently vanished in all clonal lines except FOL (Jelinski et al., 2017, Maldonado et al., 2018, Debbi et al., 2018).
To identify the molecular necessities for the pathogenicity of FOL, Caroline et al. (2009) used the Agrobacterium facilitated insertional mutagenesis approach to generate more than 10,000 transformants of FOL and further screened them for loss of pathogenicity. Cellular processes involving lipid metabolism and amino acid, protein translocation, cell wall integrity, and degradation of protein seemed to be crucial for the pathogenicity of FOL based on the functional categorization of their pathogenic genes. Several genes, such as developmental regulator (flbA), phosphomannose isomerase, and chitin synthase V (chsV) were identified, which have a recognized role in virulence of Fo. In addition, gene knockout and complementation studies established that proteins involved in cell wall integrity, such as the glycosylphosphatidylinositol-anchored protein; proteins involved in peroxisome biogenesis; a transcriptional regulator and unknown function of protein are essential for pathogenicity and play a crucial role during the tomato infection by Fo.
6. Proteomics of F. oxysporum f. sp. lycopersici
Among the numerous promising advanced biotechnological approaches to get a well understanding of FOLconnected with its hostplant, is proteomics, a systems biology approach. Proteomics in combination with transcriptomics, genomics and other techniques it yields many valuable and informative data that can be used to understand plant pathogen interactions, virulence, the infection process, and downstream disease signaling mechanism. These insights in turn help design effective disease management strategies, possibilities for novel strategies for resistance breeding to overcome the huge crop losses (Kalita and Ram, 2018, deLamo et al., 2018). In an attempt to realize the mechanism of wilt caused by Folthe total proteome of 20 isolates were analyzed along with the cultural, morphological, virulence and molecular characteristics by Manikandanet al. (2018). The 17 different proteins showedby 2D analyses, among which 3 proteins were downregulated and 14 proteins were upregulated in Fol-8 in comparison to Fol-20. MALDI-TOF analysis and identification of these differentially expressed proteins exhibited the occurrence of the FAD binding domain containing protein, Cutinase-2, Chaperone, Cytochrome P450, sulfate anion transporter, Glycoside hydrolase family 85 protein, 60S ribosomal protein and, ATP-dependent RNA helicase. These are certainof the key proteins in virulence, symptom and wilt development. These proteins were also involved in sporulation, growth, maintenance of genome integrity and maximum penetration rate on host root tissues (Manikandanet al., 2018). Sun et al. (2014) report the comparative proteomics of F. oxysporum f. sp. cubense strains cultured inseveral conditions. These are mostly involved in post-translational modification, carbohydrate metabolism, inorganic ion transport, energy production, and enzymes includesgalactosidase, catalase-peroxidase, and chitinase which may be significant in the pathogenesis contribute to the high virulence of the wilt pathogen (Sun et al., 2014). deSain and Rep (2015) have reviewed the proteins secreted by pathogens, such as wilt fungus FOL during colonization to establish aeffective pathogen-host communication. The annotated genomic and proteomic analyses revealedFOLencodes 126 small, cysteine-rich and other potentially secreted proteins. A major subset of small, cysteine rich proteins such as Secreted in xylem (Six) 1–14 have been described in the xylem sap of infected host plants. Several of the SIX proteins play a serious role in colonization, disease symptom progressand the full virulence of FOLin tomato plants. Some of these proteins also known as Avirulence (Avr) 3, have also been implicated in plant immunity because they are known by the tomato resistance (R) protein immunity I-3 (Rep et al., 2004). Many enzymes such as Endopolygalacturonase (PG), exopolygalacturonase (PGX), tomatinase (TOM), metalloprotease (Mep) serine protease (Sep) produced by FOL also contribute to the pathogenicity (deSain and Rep (2015)). As a typical vascular wilt fungus, FOLenters roots and developmentsover epidermal and endodermal tissues and lastly colonize the xylem vessels. The molecular mechanism and interactions of FOL and tomato have been explored by investigating the composition of the xylem sap proteome of diseased plants and compared with the healthy plants. During colonization of tomato, SIX1 is one of the major fungal proteins that accumulate in xylem sap which is essential for virulence of FOLas well as its no virulence on host plants carrying the resistance gene I-3 (Rep et al., 2005, Rep et al., 2004). Other fungal proteins Six1, Six2, Six3, Six4, arabinanase, oxidoreductase, Serine protease are secreted by FOL into xylem sap through colonization of tomato (Houterman et al., 2007).
For FOLpathogenicity, required the Six 1, Six3, Six5, and Six6 and also confer avirulence to wilt fungus as these proteins are known by the tomato resistance (R) gene produces. They are also named as Avirulence (Avr) proteins; Six4 (Avr1), Six3 (Avr2), Six1 (Avr3), and as they trigger the I-1, I-2 and I-3 mediated resistance, respectively (Takken and Rep, 2010). FOLsecretes effector proteins during infection of tomato. The occurrence of Six5 and Avr2 in susceptible tomato plants confers virulence to the pathogen conversely it induces resistance in case of I-2 containing plants (Cao et al., 2018). In their effort to know the modifications in cellular protein expression in host leaves in response to Fusaric acid exposure, the total proteome of the leaves was analyzed by Singh et al. (2017). Difference expression in numerous proteins weredetected which fell into two categories such as up-accumulated proteins, and down-accumulated proteins and additional classified into five functional classes such as stress and defense; biosynthesis of protein, metabolism and processing and signal transduction, and transcription. Most of the down-regulated proteins were of the energy and metabolism class indicating the role of fusaric acid in decline of structure and breakdown of cells and the pathogenicity of FOL (Singh et al., 2017).
7. Genetics of host resistance
Chemical treatments and soil solarization infields usually fail to control the vascular wilt fungus. Planting Foresistant plant varieties is the most dependable method for disease inhibition. Cultivar resistance may vary by location; therefore, selection of an appropriate cultivar also need to be studied for the wilt pathogenicity in the field condition (Cheng et al., 2015, Yasushi and Tsutomu, 2006). Developing resistant varieties involves crossing resistant wild-type plants and existing cultivars for their properties, such as color, shape, and good taste. Resistance genes liked to molecular markers would be beneficial for tomato development programs (Hanson et al., 2016). The interaction between host and FOL is race and cultivar specific. Resistance to all FOL 3 races has been recognized among Lycopersicon spp., grown in wild, which has introgressed into commercial cultivars of tomato. The I and I-1 genes conferring resistance to FOL race 1 originate from accession LA716 of L. pennelliiand 160 of L. pimpinellifolium. The accession PI126915 (L. esculentum × L. pimpinelli folium hybrid) had resistance for races 1 and 2 (Cirulli and Alexander, 1966). The dominant gene (I2) in tomato, governing resistance against race 2 FOL, originates from the wild tomato species L. pimpinellifolium. The gene (I2) present in tomato (L. peruvianum) pocessedresistanceto both race1 and 2 (Neha et al., 2016). The gene (I-3) existing in L. pennelliiaccessions PI414773 and LA716 had resistance to race 3 (Zhao et al. (2015)). Gene to gene theory, the dominant race specific to resistance genes (R genes) present in anyspecies would respond to the secretion of dominant avirulence (Avr) genes of the pathogen (Pu et al., 2016).
I-2 gene, have resistance to FOL race 2, which will respond to avirulence gene (AvrI-2) present in race 2 of FOL and the activation of defense responses in plant (Essarioui et al., 2016). The Igenes are located and mapped on chromosomes 11 and 7 (Gonzalez-Cendales et al., 2016). I-2 is located within a similar bunch of 7 similar genes on chromosome 11 and the I-3 locus has been located on the chromosome 7 long arm (Gonzalez-Cendales et al., 2016). Five genomic positions were located on I2C family among them 2 genes are located on chromosome 11 which encode for cytoplasmic proteins comprising of nucleotide binding site and leucine rich repeats (LRRs). Few strain of I2 gene family revealed two important leucine rich repeat region which may contribute to resistance among Fusarium wilt with I2specificity (Ann-Maree et al., 2017).
8. Races and vegetative compatibility groups within FOL
Fusarium oxysporumspecies are grouped into formae speciales based on host specificity and additional subdivision within the formae speciales into races is on the basis of their pathogenicity to a specific group of cultivars of the host which may differ among resistant variety (Van Dam et al., 2016). The pathogenicity test to determine the forma speciales and the race of the pathogen, although time-consuming and subject to varying environmental conditions, is themost reliable method for categorizing pathogens based on host-specific within the Fo species complex (Ploetz, 2015). Further grouping within various formae speciales is based on vegetative compatibility, an approach of characterizing subspecific groups based on their genetics rather than the host-pathogen interaction.
There are three (race 1, race 2 and race 3) known physiological races within FOL that are differentiated between them based on their pathogenicity among diverse cultivars of tomato comprising of monogenic dominant resistance genes and race-specific. These resistance genes against FOL identified in wild tomato have been introduced into commercial varieties (Biju et al., 2017). Races 1 and 2 havebeen tested in most of the tomato cultivating areas across the world. Race 1, initially reported in 1886, severely affected and threatened commercial tomato production in Arkansas. The genes I and I-1 in tomato confer race 1 resistance (Petit-Houdenot and Fudal, 2017). The discovery and subsequent use of gene I led to the pathogen overcoming this resistance and consequently the emergence of race 2. Resistance to race 2FOL is governed by the dominant 1–2 gene in tomato (Catanzariti et al., 2015). Race 3 wasreported in Australia for first time had resistance to I-2 (Ann-Maree et al., 2015). In the early1980s, FOL race 3 caused significant yield losses and prevented land from being used for tomato cultivation in both continents. Most of the commercial tomato varieties resistant to races 1 and 2 of FOL and few cultivars resistant to race 3 are available. Races 1 and 2 are dispersed through most parts of the continents, however, race 3 has restricted dispersion throughout the world (Pena, 2005). The emergence of new races may be due to selection and mutation from pre-establishing races or avirulent isolates. Fusarium oxysporum lacks the sexual stage, and genetic exchange is therefore limited to parasexual cycle and genetic transformation, which requires heterokaryosis. Heterokaryon development in Fois regulated byasetofheterokaryonloci, whoseproducts maymediate eitherincompatibility/vegetative compatibility, leading to hyphal fusion followed by cell lysis (Shahi et al., 2016). Strains with the ability to procedure stable heterokaryon are assumed to be vegetatively compatible or much more likely genetically similar and belong to the same vegetative compatible group (VCG) (Strom and Bushley, 2016). The VCG experment is time consuming and laborious process, has assisted to characterize pathogenic strains and elucidate the population structure of FOL (Aguayo et al., 2017). Based on vegetative compatibility, FOL isolates are segregated into three VCGs (0030–0031 and 0035) (Chellappan et al., 2014). No correlation exists among the colony morphology, geographical origin, race or vegetative compatibility of FOL (Biju et al., 2017). This finding suggests that several genetic determinants for race specificity may exist within genetically isolated populations (VCGs). RFLP analysis of a worldwide collection of FOL revealed that isolates among VCG have a common ancestor; however, races within each VCG have developed independently (Gordon, 2017). Isozyme analysis and mtDNA, RFLPs of FOL showed that races within aVCG are closely related, although the races among different VCGs are diverse from each other. Vegetative compatibility grouping is an indication of evolutionary origin (Laurence et al., 2015). Micro-evolutionary events, such as changes in virulence within VCGs of FOL, may occur due to evolutionary and selection pressure triggered by the continuous use of resistant cultivars of tomato (Van Dam et al., 2016).
9. The relationship between FOL and non-pathogenic Fo
Studies on genetic diversity have primarily focused on the pathogenic strain of Fo, with lesser attention to the non-pathogenic Fo strains. Bao et al. (2002) examined non-pathogenic and pathogenic strains of Foisolated from roots of tomato, representing a wide range of geographical locations. Molecular markers such as AFLP, RAPD, ISSR and rDNA sequences have been used for analysis of study genetic diversity of all strains. The pathogenic and non-pathogenic strains segregated into dissimilar clusters based on ITS sequence analysis and AFLP. The pathogenic population exhibited less diversity than the non-pathogenic strains. Studies revealed that there is no connection between the geographic origin and genetic profiles among both the pathogenic and non-pathogenic Fostrains. Elias et al. (1991) isolated non-pathogenic Fo from roots of symptomless tomato, had no similarity with strains of FOL (VCGs 0030 and 0032).
Three non-pathogenic isolates from California were vegetatively compatible with the pathogenic strains of FOL belonging to VCG 0031. These non-pathogenic Foshared a common IGS haplotype, partial sequences and genomic DNA RFLPs with the pathogenicisolates of VCG 0031 (Sasaki et al., 2015). Mutations to virulence may occur in a non-pathogenic isolate that is in close proximity to the roots or vascular system of a susceptible host (Yadeta and Thomma, 2013). This isolate further may proliferate and lead to an epidemic. Further mutations altering virulence may occur within the VCG to combat the resistant cultivars of the host, leading to the emergence of new races (Biju et al., 2017).
10. Genetic variability between FOL and members of other formae speciales
The evolutionary lineages of Fospecies complex are monophyletic, with are a diverse complex (Singha et al., 2016). Phylogenetic analyses based on mitochondrial small subunit (mtSSU), rDNA intergenic spacer (IGS) region, ribosomal RNA gene and translation elongation factor (EF)-1α gene have aided to understand the evolutionary and genetic interactions among formae speciales of Fo (Czislowski et al., 2018). These experiments have revealed that a limited number of Fo formae speciales are monophyletic (Williams et al., 2016). Other formae speciales, cucumerinum, asparagi, gladioli, lini, cubense, dianthi, lycopersici, melonis, vasinfectum, lactucae, radicis-lycopersici, opuntiarum, and phaseoliwere found to be polyphyletic (van der Does et al., 2008), suggesting that virulence factor of pathogen have evolved several times independently towards a specific crop.
Relationships between isolates of FOL and formae speciales were observed by Nirmaladevi et al. (2016). The mitochondrial minor subunit rRNA gene and the α translation elongationfactor shown that FOL, melonis, radicis-lycopersici and batatasbelonged to the similar phylogenetic lineage. Based on IGS sequence analysis, Cai et al. (2003) showed that isolates in VCG 0035 had similarity to isolates of Fusarium oxysporumf. sp. radicis-lycopersici compared to isolates in other VCGs of FOL. Studies of Kawabe et al. (2005) based on phylogenetic studies of IGS sequences (rDNA) by NJ methods discovered A1, A2, and A3 well-supported clusterscontaining FOL isolates, the major group A2 composed of isolates of FOL along with a fewmembers of other formae speciales. Fusarium oxysporum f. sp. radicis-lycopersici, Fusarium oxysporumf. sp. melonisand Fusarium oxysporumf. sp. batataswere classified within the FOL large cluster in the IGS phylogeny. Few of formae speciales analyzed in their experiment were found phylogenetically dissimilar among A1, A2, and A3 groups (Kashiwa et al., 2016). Analysis of ITS sequences and AFLP of FOL and other formae speciales of Forevealed that specialized forms of Fo do not constitutemonophyletic lineages because they evolved in a divergent way.
11. Evolutionary relationships between VCGs and races of FOL
Various genetic tools are applied to analyze the evolutionary relationships and population structure among strains of FOL. A close relationship between VCGs has been confirmed bythe study of an array of markers. The genome of FOL is composed of a single copy, multiplecopies and repetitive DNA in the proportion of 68, 12 and 20%, respectively. When compared at the level of DNA, isolates from different VCGs and formae speciales revealed ahigh frequency of variation that did not correlate with the geographic origin or physiologicalrace of the isolates. However, in case of isolates within a VCG, less variation was observed, even though the isolates originated from diverse geographical locations and belonged to different races, suggesting that isolates within a VCG have arisen from a common ancestral progenitor. Races of FOL have arisen independently, and isolates within arace may differ genetically (Schmidt et al., 2016). Comparison of RAPD profiles of race 1 and race 2 isolates of FOL revealed two main groups that coincided with VCGs. In addition, there existed numerous single member of VCGs that might not be assigned to the two main RAPD clusters, suggesting the polyphyletic origin of FOL. The RFLP and RAPD analyses of FOL clearly indicate that the VCGs are diverse. Mutations altering virulence within eachVCG might have led to similar races in different VCGs (Schmidt et al., 2016). In FOL, it is a common assumption that race 2 emerged from race 1 and that race 3 was derived from race 2 (Sasaki et al., 2015, Schmidt et al., 2016).
Isolates of FOL characterize two genetically diverse evolutionary lineages. This hypothesis was supported by the interpretations of Elias et al. (1993) based on nuclear DNA RFLPs and based on mtDNA RFLPs and isozyme polymorphisms (Biju et al., 2017). Isolates of VCG 0032 and 0030 revealed a common mtDNA haplotype, whereas isolates in VCG 0033 shared a similar mtDNA haplotype with VCG 0031. The common mtDNA haplotypes of VCGs 0030 and 0032 indicate that they may share a common evolutionary lineage and the secondevolutionary lineage shared by VCGs 0031 and 0033. The association between molecular genotypes elucidated from two independent genetic markers, i.e., nuclear DNA RFLP and mtDNA RFLP, provides strong evidence of an asexual mode of reproduction on FOL as observed in other phytopathogenic fungi (Biju et al., 2017). Isolates representing VCGs 0030 and 0032 had identical IGS sequences, haplotypes, and genomic DNA grouped the twoVCGs with bootstrap (89%) support. This result revealed that these 2 VCGs have common ancestry origin (Cai et al., 2003). Similar results were testified by Balmas et al. (2005) basedon RAPD and microsatellite-primed PCR, VCGs 0030 and 0032 isolates shared few genetic markers, support the previous views of common ancestor origin. Observations made by Lievens et al. (2009) established that FOL contain three independent clonal lineages. The geographical and evolutionary linkages among isolates of FOL have been studied using partial sequences of MAT1, pg1 and IGS in combination with mating type (MAT) and VCG.
The first lineage consist of two isolates MAT1-1 and VCG 0031, the second lineage was shared by VCG 0030 and 0032 and MAT1-1, then the third lineage included MAT1-2 and VCG 0033 (Kashyap et al., 2015).
Phylogenetic studies of housekeeping gene by partial sequences of the α elongation factor (EF-1α) and a gene encoding exopolygalacturonase (pgx4), conducted on a worldwide collection of FOL strains demonstrated the most commonly observed vegetative compatibility groups showed multiple evolutionary lineages. At least 3 clonal lineages were represented by EF-1α grouping and by pgx4 clades (Giuseppe et al., 2015, Lievens et al., 2009). Although FOL is an asexually reproducing fungus, functional mating type genes (MAT1 and MAT2) have be identified with random distribution of alleles over the diverse clades of phylogenetic tree, regardless of the geographical origin. Evolutionary process other than recombination, such as, accumulation of mutations in loci and natural selection by positive selection pressure on the gene encoding virulence factors, may also result in multiple lineages (Vlaardingerbroek et al., 2016).
12. Molecular variability within FOL inferred from DNA fingerprints and markers
The development and evolutionary relationships among pathogenic and non-pathogenic strains in the regional population can be elucidated using phylogenetic and genetic diversityanalysis. This may also give information about dissemination of pathogen from other geographical areas (Chandra et al., 2011). The extent of genetic variability within the pathogenpopulations indicates the rate of evolution. Higher genetic variation signifies the expeditiousevolution in response to ecological changes leading to emergence of new species overcoming host resistance (Möller and Stukenbrock, 2017).
RFLP analysis of intergenic spacer region (IGS) of FOL isolates causing devastating disease of tomato greenhouse crops in Tanzania revealed high genetic diversity. Characterization based on IGS typing revealed six IGS types among 9 isolates of FOL (Lobna et al., 2017). Ahigh levels of genetic diversity was observed in FOL populations isolated from different geographical locations of India based on RAPD patterns. Further length variation in the ITS
region was observed in some isolates (Manikandan et al., 2018, Nirmaladevi et al., 2016). Phoutthasone et al. (2012) used AFLP markers to study genetic variation among FOL population in Thailand, which revealed correlation between pathogenicity among FOL population (Sharma et al., 2014). FOL isolates from some close geographical areas show high genetic relationships, suggesting the movement of the pathogen between these areas.Baysal et al. (2009) studied the molecular diversity of FOL isolates from the West Mediterranean region of Turkey. The pathogen hampers the production and is responsible forthe huge economic destabilization in tomato greenhouses. SRAP and ISSR markers used forthe genotyping of FOL races displayed significant differences among the pathogenic isolates. The hurdles in the management of the pathogen and its acquired resistance towards plant protecting chemicals may be linked to genetic diversity within the races. Molecular analyses showed diverse genetic variability among pathogenic isolates of r2 and r3 which may be linked to the pathogens exposed to abiotic stress (Aamir et al., 2018).
13. Protein and fatty acid analysis in the study of FOL
The evolutionary relationships within FOL and with other prokaryotes and eukaryotes havebeen elucidated using a wide array of DNA-based molecular approaches, which are versatile and highly informative methods. Other biomolecules, such as proteins and fatty acids, have also assisted in better understanding of the population structure of FOL (Al‐Sadi et al., 2015).
Isozyme assays, has also been used to identification of species, races or special forms of pathogenic fungi isolated form plant. It is an inexpensive and rapid method for analyses of alarge number of isolates performed by the electrophoresis of isozymes. Amino acid sequence differences may lead to changes in protein properties and thus altered the mobility of the proteins in a polyacrylamide gel (Sidaoui et al., 2017). Based on isozyme polymorphisms, Elias and Schneider (1992) found two distinct groups among the three FOL races. The first group contained VCG 0030 and 0032 isolates, and the second group included VCG 0031 isolates. Isolates within a VCG showed more similar isozyme profiles. The genetic similarity and distribution of isolates correlated with VCG rather than with their race, formae speciales or geographic origin. Isolates of VCG 0030 formed the first phenotype, and those belonging to VCG 0033 belonged to the second phenotype. Cellular fatty acid composition and analysis technique are used routinely to characterize, identify genera, species, and strains and differentiate of bacteria and yeasts. Although fewer types of fatty acids are secreted only by fungi which is absent in bacteria, it can be used for the characterization and identification of fungi (Willers et al., 2015). Fatty acid profiles have been used successfully to characterize Fusarium oxysporumf. sp. vasinfectum (Ellis et al., 2002). Matsumoto (2006) characterized and differentiated FOL strains based on their fatty acid methyl ester (FAME) profiles. The fatty acid 18:2 ω6, 9c was present in isolates of race 1 (38.2%), race 2 (43.1%), and race 3 (37.2%). Race 1 and race 2 isolates also dominantly contained 17.8% and 14.2% of the fatty acids 16:0 and 18:0. Principal component and cluster analysis showed that FAME profiles of the isolates connected with the similar vegetative compatibility groups (VCGs) compared to the similar races in FOL. Interestingly, isolates of VCGs0030 and VCG 0032 presented similar cluster grouping and FAME profiles. This finding is in agreement with reports of Kawabe et al. (2005), who observed a closephylogenetic relationship between isolates of VCG 0030 and of VCG 0032 based on DNA-based methods.
14. Concluding remarks and perspectives
Comparative phylogenetic studies of FOL is required to elucidate the diversity and origin of phylogenetically informative genes. Genomics will aid the discovery of additional informative genes needed to develop a highly resolved phylogenetic framework for FOL, resolve species boundaries, and develop robust molecular diagnostics in support of agricultural biosecurity. In this review, we discussed fusarium wilt of tomato, genetic variability, toxigenicity and pathogenicity of FOL and their link with other formae specialeswithin the Focomplex. The abovementioned information within FOL strains can be useful toplant breeders to have disease resistant plant breeding. The review will provide comprehensive insight in understanding host-pathogen interactions at the genetic level. Thisinformation essentially contributes to understanding the virulence pattern of the FOL, and assists development of molecular markers for the disease management strategies.
Acknowledgments
Acknowledgements
The authors acknowledge the recognition of University of Mysore as an Institution of Excellence and financial support from the Ministry of Human Resource Development, Government of India through the University Grants Commission, New Delhi, India. The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this research group NO (RGP-271).
Footnotes
Peer review under responsibility of King Saud University.
References
- M. Aamir, S.V. Kumar, D.M. Kumar, K.S. Pratap, A. Zehra, U.R. Sanmukh, S. Singh. Structural and functional dissection of differentially expressed tomato WRKY transcripts in host defense response against the vascular wilt pathogen (Fusarium oxysporum f. sp. lycopersici).https://doi.org/10.1371/ journal.pone.0193922; 2018. [DOI] [PMC free article] [PubMed]
- Agrios G.N. Elsevier Academic Press; Burlington, MA: 2005. Plant Pathology. [Google Scholar]
- Aguayo J., Mostert D., Fourrier-Jeandel C. Development of a hydrolysis probe-based real-time assay for the detection of tropical strains of Fusarium oxysporum f. sp. cubense race 4. PLoS ONE. 2017;12(2):e0171767. doi: 10.1371/journal.pone.0171767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sadi A.M., Al-Masoodi R.S., Al-Ismaili M., Al-Mahmooli I.H. Population structure and development of resistance to hymexazol among Fusarium solani populations from date palm, Citrus and cucumber. J. Phytopathol. 2015;163:947–955. [Google Scholar]
- Animashaun B.O., Popoola A.R., Enikuomehin O.A., Aiyelaagbe I.O.O., Imonmion J.E. Induced resistance to Fusarium wilt (Fusariumoxysporum) in tomato using plant growth activator. Acibenzolar-S-methyl. Nig. J. Biotech. 2017;32:83–90. [Google Scholar]
- Ann-Maree C., Ginny T.T., Lim Jones A. The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 2015;207:106–118. doi: 10.1111/nph.13348. [DOI] [PubMed] [Google Scholar]
- Ann-Maree C., Huong T.T.D., Pierrick B., de Sain Mara, Louise F.T., Rep Martijn, David A.J. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/ BAK1. Plant J. 2017;89:1195–1209. doi: 10.1111/tpj.13458. [DOI] [PubMed] [Google Scholar]
- Arpita D., Ramana V.M., Chandranayaka S., Murali H.S., Batra H.V. Molecular characterization of Fusarium oxysporum f. sp. cubense isolates from banana. Pest Manag. Horticul. Ecosys. 2012;18:171–178. [Google Scholar]
- Asha B.B., Nayaka S.C., Shankar A.C.U., Srinivas C., Niranjana S.R. Selection of effective bio–antagonistic bacteria for biological control of tomato wilt caused by Fusarium oxysporum f Sp. Lycopersici. BioscanIntquart. J. Life Sci. 2011;6:239–244. [Google Scholar]
- Balmas V., Scherm B., Di Primo P., Rau D., Marcello A., Migheli Q. Molecular characterisation of vegetative compatibility groups in Fusarium oxysporum f. spradicis–lycopersici and f. splycopersici by random amplification of polymorphic DNA and microsatellite–primed PCR. Eur. J. Plant Pathol. 2005;111:1–8. [Google Scholar]
- Bao J.R., Fravel D.R., O’Neill N.R., Lazarovits G., van Berkum P. Genetic analysis of pathogenic and nonpathogenicFusarium oxysporum from tomato plants. Can. J. Bot. 2002;80:271–279. [Google Scholar]
- Bawa I. Management strategies of Fusarium wilt disease of tomato incited by Fusarium oxysporum f. sp. lycopersici (Sacc.) A Review. Int. J. Adv. Acad. Res. 2016;2:5. [Google Scholar]
- Baysal O., Siragusa M., Ikten H., Polat I., Gumrukcu E., Yigit F., Carimi F., Teixeira da Silva J. Fusarium oxysporum f. sp. lycopersici races and their genetic discrimination by molecular markers in West Mediterranean region of Turkey. Physiol. Mol. Plant Pathol. 2009;74:68–75. [Google Scholar]
- Bertoldo C., Gilardi G., Spadaro D. Genetic diversity and virulence of Italian strains of Fusarium oxysporum isolated from Eustomagrandiflorum. Eur. J. Plant Pathol. 2015;141:83–97. [Google Scholar]
- Bharti P., Jyoti P., Kapoor P., Sharma V., Shanmugam V., Yadav S.K. Host-induced silencing of pathogenicity genes enhances resistance to Fusarium oxysporum wilt in tomato. Mol. Biotechnol. 2017;59(8):343–352. doi: 10.1007/s12033-017-0022-y. [DOI] [PubMed] [Google Scholar]
- Biju V.C., Fokkens L., Houterman P.M., Rep M., Cornelissen B.J.C. Multiple evolutionary trajectories have led to the emergence of races in Fusarium oxysporum f. sp. lycopersici. Appl. Environ. Microbiol. 2017;83:e02548–e2616. doi: 10.1128/AEM.02548-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodah E.T. Root rot diseases in plants: a review of common causal agents and management strategies. Agri. Res. Tech. 2017;5(3):555661. [Google Scholar]
- Boix-Ruíz A., Gálvez-Patón L., de Cara-García M., Palmero-Llamas D., Camacho-Ferre F., Tello-Marquina J.C. Comparison of analytical techniques used to identify tomato-pathogenic strains of Fusarium oxysporum. Phytoparasitica. 2015;43:471–483. [Google Scholar]
- Borisade O.A., Uwaidem Y.I., Salami A.E. Preliminary report on Fusarium oxysporum f.sp. Lycopersici (Sensulato) from some tomato producing agroecological areas in Southwestern Nigeria and susceptibility of F1-resistant tomato hybrid (F1-Lindo) to infection. Ann. Res. Rev. Biol. 2017;18(2):1–9. [Google Scholar]
- Brookie K.L., Best G.I., Conner T.S. Intake of raw fruits and vegetables is associated with better mental health than intake of processed fruits and vegetables. Front. Psychol. 2018;9:487. doi: 10.3389/fpsyg.2018.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Gale L.R., Schneider R.W., Kistler H.C., Davis R.M., Elias K.S., Miyao E.M. Origin of Race 3 of Fusarium oxysporum f. sp. lycopersici at a single site in California. Phytopathology. 2003;93:1014–1022. doi: 10.1094/PHYTO.2003.93.8.1014. [DOI] [PubMed] [Google Scholar]
- Cao L., Blekemolen M.C., Tintor N., Cornelissen B.J.C., Takken F.L.W. The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant. 2018;11:691–705. doi: 10.1016/j.molp.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Caroline B.M., van Wijk R., Reijnen L., Cornelissen B.J.C., Rep M. Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large–scale insertional mutagenesis. Genome Biol. 2009;10(1):R4. doi: 10.1186/gb-2009-10-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A.M., Lim G.T.T., Jones D.A. The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 2015;207:106–118. doi: 10.1111/nph.13348. [DOI] [PubMed] [Google Scholar]
- Cha J.Y., Han S., Hong H.J., Cho H., Kim D., Kwon Y., Kwon S.K., Crüsemann M., Lee Y.B., Kim J.F., Giaever G., Nislow C., Moore B.S., Thomashow L.S., Weller D.M., Kwak Y.S. Microbial and biochemical basis of a Fusarium wilt suppressive soil. ISME J. 2016;10:119–129. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra N.S., Wulff E.G., Udayashankar A.C., Nandini B.P., Niranjana S.R., Mortensen C.N., Prakash H.S. Prospects of molecular markers in Fusarium species diversity. Appl. Microbial. Biotechnol. 2011;90:1625–1639. doi: 10.1007/s00253-011-3209-3. [DOI] [PubMed] [Google Scholar]
- Chellappan, B.V., Houterman, P.M., Rep, M., Cornelissen, B.J.C. Evolution of races within Fusarium oxysporum f. sp. lycopersici. Molecular Plant Pathology, Swammerdam Institute for Life Sciences, University of Amsterdam, 1090 GB Amsterdam, The Netherlands; 2014.
- Cheng W., Song X.S., Li H.P., Cao L.H., Sun K., Qiu X.L., Xu Y.B., Yang P., Huang T., Zhang J.B. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015;13:1335–1345. doi: 10.1111/pbi.12352. [DOI] [PubMed] [Google Scholar]
- Cirulli M., Alexander L.J. A comparison of pathogenic isolates of Fusarium oxysporum f.sp. lycopersiciand different sources of resistance in tomato. Phytopathol. 1996;56:1301–1304. [Google Scholar]
- Czislowski E., Fraser-Smith S., Zander M., O'Neill W.T., Meldrum R.A., Tran-Nguyen L.T.T., Batley J., Aitken E.A.B. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant. Pathol. 2018;19:1155–1171. doi: 10.1111/mpp.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbi A., Boureghda H., Monte E., Hermosa R. Distribution and genetic variability of Fusarium oxysporum associated with tomato diseases in Algeria and a biocontrol strategy with indigenous Trichoderma spp. Front. Microbiol. 2018;9:282. doi: 10.3389/fmicb.2018.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeConinck B., Timmermans P., Vos C., Cammue B.P.A., Kazan K. What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 2015;20:91–101. doi: 10.1016/j.tplants.2014.09.007. [DOI] [PubMed] [Google Scholar]
- deLamo F.J., Constantin M.E., Fresno D.H., Boeren S., Rep M., Takken F.L.W. Xylem sap proteomics reveals distinct differences between r gene and endophyte-mediated resistance against Fusarium wilt disease in tomato. Front. Microbiol. 2018;9:2977. doi: 10.3389/fmicb.2018.02977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deSain M., Rep M. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 2015;16:23970–23993. doi: 10.3390/ijms161023970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins A.E. APS Press, St. Paul; 2006. Fusarium Mycotoxins Chemistry, Genetics and Biology. [Google Scholar]
- Di X., Takken F.L.W., Tintor N. How phytohormones shape interactions between plants and the soil-borne fungus Fusarium oxysporum. Front. Plant Sci. 2016;7:170. doi: 10.3389/fpls.2016.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakara S.T., Santosh P., Aiyaz M., Ramana M.V., Hariprasad P., Nayaka S.C., Niranjana S.R. Molecular identification and characterization of Fusarium spp. associated with sorghum seeds. J. Sci. Food Agric. 2014;94(6):1132–1139. doi: 10.1002/jsfa.6380. [DOI] [PubMed] [Google Scholar]
- Elias K.S., Schneider R.W., Lear M.M. Analysis of vegetative compatibility groups in nonpathogenic populations of Fusarium oxysporum isolated from symptomless tomato roots. Can. J. Bot. 1991;69:2089–2094. [Google Scholar]
- Elias K.S., Schneider R.W. Vegetative compatibility groups in Fusarium oxysporum f. sp. Lycopersici. Phytopathol. 1992;81:159–162. [Google Scholar]
- Elias K.S., Zamir D., Lichtmanpleban T., Katan T. Population–structure of Fusarium oxysporum f. splycopersici – restriction fragment length polymorphisms provide genetic evidence that vegetative compatibility group is an indicator of evolutionary origin. Mol. Plant Microbe Interact. 1993;6:565–572. [Google Scholar]
- Ellis R.J., Geuns J.M., Zarnowski R. Fatty acid composition from an epiphytic strain of Fusarium oxysporum associated with algal crusts. Acta Microbiol. Pol. 2002;51:391–394. [PubMed] [Google Scholar]
- Essarioui A., Mokrini F., Afechtal M. Molecular interactions between tomato and its wilt pathogen Fusarium oxysporum f. sp. Lycopersici– a review. Rev. Mar. Sci. Agron. Vét. 2016;4(1):66–74. [Google Scholar]
- Giuseppe O., Domenico B., Patrizia M., Maria Lodovica G., Angelo G. Fusarium oxysporum f. sp.papaveris: a new forma specialis isolated from Iceland poppy (Papavernudicaule) Phytopathol. Mediter. 2015;54:76–85. [Google Scholar]
- Gonzalez-Cendales Y., Catanzariti A.M., Baker B., Mcgrath D.J., Jones D.A. Identification of I-7 expands the repertoire of genes for resistance to Fusarium wilt in tomato to three resistance gene classes. Mol. Plant Pathol. 2016;17:448–463. doi: 10.1111/mpp.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T.R. Fusarium oxysporum and the Fusarium wilt syndrome. Ann. Rev. Phytopathol. 2017;55:23–39. doi: 10.1146/annurev-phyto-080615-095919. [DOI] [PubMed] [Google Scholar]
- Guo L., Yang L., Liang C., Wang J., Liu L., Huang J. The G-protein subunits FGA2 and FGB1 play distinct roles in development and pathogenicity in the banana fungal pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 2016;93:29–38. [Google Scholar]
- Hanson P., Lu S.F., Wang J.F., Chen W., Kenyon L., Tan C.W., Kwee L.T., Wang Y.Y., Hsu Y.C., Schafleitner R., Ledesma D., Yang R.Y. Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Scient. Horti. 2016;201:346–354. [Google Scholar]
- Houterman P.M., Speijer D., Dekker H.L., De Koster C.G., Cornelissen B.J.C., Rep M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 2007;8(2):215–221. doi: 10.1111/j.1364-3703.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- Jelinski N.A., Broz K., Jonkers W., Ma L.J., Kistler H.C. Effector gene suites in some soil isolates of Fusarium oxysporum are not sufficient predictors of vascular wilt in tomato. Phytopatholohy. 2017;107(7):842–851. doi: 10.1094/PHYTO-12-16-0437-R. [DOI] [PubMed] [Google Scholar]
- Joshi Renu. A review of Fusarium oxysporum on its plant interaction and industrial use. J. Med. Plants Stud. 2018;6(3):112–115. [Google Scholar]
- Kalagatur N.K., Mudili V., Siddaiah C., Gupta V.K., Natarajan G., Sreepathi M.H., Putcha V.L. Antagonistic activity of Ocimum sanctum L. essential oil on growth and zearalenone production by Fusarium graminearumin Maize grains. Front Microbiol. 2015;6:892. doi: 10.3389/fmicb.2015.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalagatur N.K., Karthick K., Allen J.A., Ghosh N., Sivaraman O., Chandranayaka S., Mudili V. Application of activated carbon derived from seed shells of Jatrophacurcasfor decontamination of zearalenonemycotoxin. Front Pharmacol. 2017;8:760. doi: 10.3389/fphar.2017.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalagatur N.K., Kamasani J.R., Mudili V. Assessment of detoxification efficacy of irradiation on zearalenonemycotoxin in various fruit juices by response surface methodology and elucidation of its in-vitro toxicity. Front Microbiol. 2018;9:2937. doi: 10.3389/fmicb.2018.02937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita P.J., Ram R.M. Proteomics in plant pathology: A tool for disease management. Int. J. Chem. Stu. 2018;6(4):1804–1813. [Google Scholar]
- Kashyap P.L., Rai S., Kumar S., Srivastava A.K., Anandaraj M., Sharma A.K. Mating type genes and genetic markers to decipher intraspecific variability among Fusarium isolates from pigeonpea. J. Basic Microbiol. 2015;55:846–856. doi: 10.1002/jobm.201400483. [DOI] [PubMed] [Google Scholar]
- Kawabe M., Kobayashi Y., Okada G., Yamaguchi I., Teraoka T., Arie T. Three evolutionary lineages of tomato wilt pathogen, Fusarium oxysporum f. sp. lycopersici, based on sequences of IGS, MAT1, and pg1, are each composed of isolates of a single mating type and a single or closely related vegetative compatibility group. J. Gen. Plant Pathol. 2005;71:263–272. [Google Scholar]
- Khan N., Maymon M., Hirsch A.M. Combating Fusarium infection using bacillus- based antimicrobials. Microorg. 2017;5(4):75. doi: 10.3390/microorganisms5040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H.B., Shankar A.C.U., Chandra Nayaka S., Kini S.R., Shekar Shetty H., Prakash H.S. Biochemical characterization of Fusarium oxysporum f. sp. Cubense isolates from India. African J. Biotechnol. 2010;9:523–530. [Google Scholar]
- Kumar K.N., Venkataramana M., Allen J.A., Chandranayaka S., Murali H.S., Batra H.V. Role of Curcuma longa L. essential oil in controlling the growth and zearalenone production of Fusarium graminearum. LWT-Food Sci. Technol. 2016;69:522–528. [Google Scholar]
- Laurence M.H., Summerell B.A., Liew E.C.Y. Fusarium oxysporum f. sp. canariensis: evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathol. 2015;64:1068–1075. [Google Scholar]
- Lievens B., Baarlen P.V., Verreth C., Kerckhove S.V., Rep M., Bart P.H.J. Thomma. Evolutionary relationships between Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis–lycopersici isolates inferred from mating type, elongation factor1a and exopolygalacturonase sequences”. Mycol. Res. 2009;113:1181–1191. doi: 10.1016/j.mycres.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Liu Z., Zhang X., Xin L., Chaoyu F., Xinyue H., Yanni Y., Zhonghua M. The chitin synthase FgChs2 and other FgChss co-regulate vegetative development and virulence in F. graminearum. Sci. Rep. 2016;6:34975. doi: 10.1038/srep34975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobna H., Hajer R., Noura C.H., Asma L., M'Hamdi-Boughalleb N., HorrigueRaouani N. Nematode virulence could affect interaction between Meloidogynejavanica (Nematoda: Heteroderidae) and Fusarium oxysporum f. sp. radicis lycopersici on tomato. J. Ent. Zool. Stu. 2017;5:1750–1754. [Google Scholar]
- Lopez-Diaz C., Rahjoo V., sulyok M., Ghionna V., Martin-Vicente A., Capilla J., di Pietro A., Lopez-berges M. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 2018;19(2):440–453. doi: 10.1111/mpp.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado B.L.D., Villarruel O.J.L., Calderón O.M.A., Sánchez E.A.C. Secreted in xylem (Six) genes in Fusarium oxysporum f.sp. cubense and their potential acquisition by horizontal transfer. Adv. Biotech. Micro. 2018;10(1):555779. [Google Scholar]
- Manici, L.M., Caputo, M.F., Saccà, L. Secondary metabolites released into the rhizosphere by Fusarium oxysporum and Fusarium spp. as underestimated component of nonspecific replant disease. 415, pp. 85–98; 2017.
- Manikandan R., Harish S., Karthikeyan G., Raguchander T. Comparative proteomic analysis of different isolates of Fusarium oxysporum f. sp. lycopersici to exploit the differentially expressed proteins responsible for virulence on tomato plants. Front. Microbiol. 2018;9:420. doi: 10.3389/fmicb.2018.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M. Comparison of two fatty acid analysis protocols to characterize and differentiate Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis- lycopersici. Mycoscience. 2006;47(4):190–197. [Google Scholar]
- McGovern R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015;73:78–92. [Google Scholar]
- Moller M., Stukenbrock E.H. Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 2017;15:771. doi: 10.1038/nrmicro.2017.143. [DOI] [PubMed] [Google Scholar]
- Mudili V., Siddaih C.N., Nagesh M., Garapati P., Naveen Kumar K., Murali H.S., Batra H.V. Mould incidence and mycotoxin contamination in freshly harvested maize kernels originated from India. J. Sci. Food Agric. 2014;94(13):2674–2683. doi: 10.1002/jsfa.6608. [DOI] [PubMed] [Google Scholar]
- Nayaka S.C., Shankar A.C.U., Niranjana S.R., Prakash H.S. Molecular detection and characterisation of Fusarium verticillioidesin maize (Zea mays. L) grown in southern India. Ann. Microbial. 2008;58:359–367. [Google Scholar]
- Nayaka S.C., Shankar A.C.U., Reddy M.S., Niranjana S.R., Prakash H.S., Shetty H.S., Mortensen C.N. Control of Fusarium verticillioides, cause of ear rot of maize, by Pseudomonas fluorescens. Pest Manag Sci. 2009;65:769–775. doi: 10.1002/ps.1751. [DOI] [PubMed] [Google Scholar]
- Nayaka S.C., Shankar A.C.U., Niranjana S.R., Wulff E.G., Mortensen C.N., Prakash H.S. Detection and quantification of fumonisins from Fusarium verticillioides in maize grown in southern India. World J. Microbiol. Biotechnol. 2010;26:71–78. [Google Scholar]
- Neha P., Shekhar S.S., Vati L., Chattopadhyay T. Molecular screening of tomato (Solanum lycopersicum L.) genotypes for resistance alleles against important biotic stresses. J. Appl Nat. Sci. 2016;8(3):1654–1658. [Google Scholar]
- Nicholas LeBlanc, AdilEssarioui Linda Kinkel, Corby Kistler H. Phylogeny, plant species, and plant diversity influence carbon use phenotypes among Fusarium populations in the rhizosphere microbiome. Phytobiomes. 2017;1:150–157. [Google Scholar]
- Nirmaladevi D., Srinivas C. Cultural, morphological, and pathogenicity variation in Fusarium oxysporum f. sp. lycopersici causing wilt of tomato. Batman ÜniversitesiYaşamBilimleriDergisi. 2012;2(1):1–16. [Google Scholar]
- Nirmaladevi D., Venkataramana M., Srivastava R.K., Uppalapati S.R., Gupta V.K., YliMattila T., Clement Tsui K.M., Srinivas C., Niranjana S.R., Chandra N.S. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f.sp.lycopersici. Sci. Rep. 2016;6:21367. doi: 10.1038/srep21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantal O., Claude H., Jarmila N., Jamshid F., Floriane L.H., Claude A. Colonization of tomato root by pathogenic and non-pathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl. Env. Microbiol. 2006;72:1523–1531. doi: 10.1128/AEM.72.2.1523-1531.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek M., Rajam M.V. RNAi-mediated silencing of MAP kinase signalling genes (Fmk1, Hog1, and Pbs2) in Fusarium oxysporum reduces pathogenesis on tomato plants. Fungal Biol. 2017;121:775–784. doi: 10.1016/j.funbio.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Pena R.J.H. Fusariumwilt of tomato caused by Fusarium oxysporum f. sp. lycopersici race 3 in Baja California Sur. Mexico. Plant Dis. 2005;12:1360. doi: 10.1094/PD-89-1360C. [DOI] [PubMed] [Google Scholar]
- Petit-Houdenot, Fudal Petit-Houdenot Y., Fudal I. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management Front. Plant. Sci. 2017;8:1072. doi: 10.3389/fpls.2017.01072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoutthasone S., Unartngam J., Soytong K. Genetic variation of Fusarium oxysporum f. sp. lycopersici isolated from tomatoes in Thailand using pathogenicity and AFLP markers. African J. Microbiol. Res. 2012;6:5636–5644. [Google Scholar]
- Ploetz R.C. Fusarium wilt of Banana. Phytopathol. 2015;105:1512–1521. doi: 10.1094/PHYTO-04-15-0101-RVW. [DOI] [PubMed] [Google Scholar]
- Prihatna C., Barbetti M.J., Barker S.J. A novel tomato Fusariumwilt tolerance gene. Front. Microbiol. 2018;9:1226. doi: 10.3389/fmicb.2018.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R.H., Busman M., Jeong-Ah S., Lee Y.W., Plattner R.D. A fumonisin biosynthetic gene cluster in Fusarium oxysporum strain O-1890 and the genetic basis for B versus C fumonisin production. Fungal Gene. Biol. 2008;45:1016–1026. doi: 10.1016/j.fgb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Pu Z., Ino Y., Kimura Y., Tago A., Shimizu M., Natsume S., Sano Y., Fujimoto R., Kaneko K., Shea D.J., Fukai E., Fuji S.I., Hirano H., Okazaki K. Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Front. Plant Sci. 2016;7:31. doi: 10.3389/fpls.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana M.V., Balakrishna K., Murali H.S., Batra H.V. Multiplex PCR-based strategy to detect contamination with mycotoxigenic Fusarium species in rice and finger millet collected from southern India. J. Sci. Food Agri. 2011;91:1666–1673. doi: 10.1002/jsfa.4365. [DOI] [PubMed] [Google Scholar]
- Ramana M.V., Nayaka S.C., Balakrishna K., Murali H.S., Batra H.V. A novel PCR-DNA probe for the detection of fumonisin producing Fusarium species from major food crops grown in southern India. Mycology. 2012;3(3) 167–164. [Google Scholar]
- Ravindra S., Biswas S.K., Nagar D., Singh J., Singh M., Kumar Mishra Yogesh. Sustainable integrated approach for management of Fusarium wilt of tomato caused by Fusarium oxysporiumsp. Lycopersici (Sacc.) Sander Hansen.Sus. Agri. Res. 2015;4(1):138–147. [Google Scholar]
- Rep M., Kistler H.C. The genomic organization of plant pathogenicity in Fusarium species. Curr. Opin. Plant Biol. 2010;13:420–426. doi: 10.1016/j.pbi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Rep M., van der Does H.C., Meijer M., van Wijk R., Houterman P.M., Dekker H.L., de Koster C.G., Cornelissen B.J. A small, cysteine–rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I–3– mediated resistance in tomato. Mol. Microbiol. 2004;53:1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- Rep M., Meijer M., Houterman P.M., van der Does H.C., Cornelissen B.J.C. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol. Plant-Microbe Interact. 2005;18:15–23. doi: 10.1094/MPMI-18-0015. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Nakahara K., Tanaka S., Shigyo M., Ito S.I. Genetic and pathogenicvariability of Fusarium oxysporum f. sp. cepaeisolated from onion and Welsh onion in Japan. Phytopathology. 2015;105:525–532. doi: 10.1094/PHYTO-06-14-0164-R. [DOI] [PubMed] [Google Scholar]
- Schmidt S.M., Houterman P.M., Schreiver I., Ma L., Amyotte S., Chellappan B., Boeren S., Takken F.L., Rep M. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Gen. 2013;14:119. doi: 10.1186/1471-2164-14-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S.M., Lukasiewicz J., Farrer R., van Dam P., Bertoldo C., Rep M. Comparative genomics of Fusarium oxysporum f. sp. Melonisreveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol. 2016;209(1):307–318. doi: 10.1111/nph.13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim E.M., El-Gammal N.A. Role of Fusaric acid mycotoxin in pathogensis process of tomato wilt disease caused by Fusarium oxysporum. J. Bioprocess. Biotech. 2015;5:255. [Google Scholar]
- Servin A., Elmer W., Mukherjee A., De la Torre-Roche R., Hamdi H., White J.C., Bindraban P., Dimkpa C. A review of the use of engineered nanomaterials to suppress plant disease and enhance crop yield. J. Nanopart. Res. 2015;17:92. [Google Scholar]
- Shahi S., Beerens B., Bosch M., Linmans J., Rep M. Nuclear dynamics and genetic rearrangement in heterokaryotic colonies of Fusarium oxysporum. Fungal Gen. Biol. 2016;91:20–31. doi: 10.1016/j.fgb.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Shahzad, R., Abdul Latif Khan., Saqib Bilal., SajjadAsaf., In-Jung Lee., 2017. Plant growth- promoting endophytic bacteria versus pathogenic infections: an example of Bacillus amyloliquefaciens RWL-1 and Fusarium oxysporum f. sp. lycopersici in tomato. Peer J. 5, e3107. [DOI] [PMC free article] [PubMed]
- Sharma M., Nagavardhini A., Mahendar T., RajuGhosh Suresh P., Rajeev K.V. Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (CicerarietinumL.) BMC Gen. 2014;15:454. doi: 10.1186/1471-2164-15-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidaoui A., Karkachi N., Bertella A., Boudeffeur S., Chhiba M., Tebyaoui A.El., Goumi Y., Kihal M. Morphological study and caracterisation of Fusarium oxysporumf.sp.albedinis by isozymes systems. Int. J. Sci. Engin. Res. 2017;8:1764–1768. [Google Scholar]
- Singh V.K., Singh H.B., Upadhyay R.S. Role of fusaric acid in the development of ‘Fusarium wilt’ symptoms in tomato: physiological, biochemical and proteomic perspectives. Plant Physiol. Biochem. 2017;118:320–332. doi: 10.1016/j.plaphy.2017.06.028. [DOI] [PubMed] [Google Scholar]
- Singha I.M., Yelena Kakoty, BalaGopalan Unni, Jayshree Das, Mohan C.K. Identification and characterization of Fusarium sp. using ITS and RAPD causing Fusarium wilt of tomato isolated from Assam. North East India. Genetic Eng. Biotechnol. 2016;14:99–105. doi: 10.1016/j.jgeb.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom N.B., Bushley K.E. Two genomes are better than one: history, genetics, and 483 biotechnological applications of fungal heterokaryons. Fungal Biol. Biotechnol. 2016;3:4. doi: 10.1186/s40694-016-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yi X., Peng M., Zeng H., Wang D., Li B., Tong Z., Chang L., Jin X., Wang X. Proteomics of Fusarium oxysporum race 1 and race 4 reveals enzymes involved in carbohydrate metabolism and ion transport that might play important roles in banana Fusarium wilt. PLoS ONE. 2014;9(12):e113818. doi: 10.1371/journal.pone.0113818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F., Rep M. The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 2010;11:309–314. doi: 10.1111/j.1364-3703.2009.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam P., Fokkens L., Linmans S.M., Schmidt J.H., Kistler H.C., Ma L.J., Rep M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016;18:4087–4102. doi: 10.1111/1462-2920.13445. [DOI] [PubMed] [Google Scholar]
- van der Does H.C., Lievens B., Claes L., Houterman P.M., Cornelissen B.J.C., Rep M. The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Env. Microbiol. 2008;10:1475–1485. doi: 10.1111/j.1462-2920.2007.01561.x. [DOI] [PubMed] [Google Scholar]
- Venkataramana M., Chandra Nayaka S., Balakrishna K., Murali H.S., Batra H.V. A novel PCR–DNA probe for the detection of fumonisin–producing Fusarium species from major food crops, grown in southern India. Mycol. Int. J. Fungal Biol. 2012;3:167–174. [Google Scholar]
- Venkataramana M., Navya K., Chandranayaka S., Priyanka S.R., Murali H.S., Batra H.V. Development and validation of an immunochromatographic assay for rapid detection of fumonisin B1 from cereal samples. J. Food Sci. Technol. 2014:1–9. doi: 10.1007/s13197-013-1254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaardingerbroek I., Beerens B., Rose L., Fokkens L., Cornelissen B.J., Rep M. Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Env.Microbiol. 2016;18:3702–3713. doi: 10.1111/1462-2920.13281. [DOI] [PubMed] [Google Scholar]
- Wang H., Ng T.B. Pharmacological activity of fusaric acid (5-butylpicolinic acid) Life Sci. 1999;65:849–856. doi: 10.1016/s0024-3205(99)00083-1. [DOI] [PubMed] [Google Scholar]
- Willers C., Jansen van Rensbur P.J., Claassens S. Phospholipid fatty acid profiling of microbial communities–a review of interpretations and recent applications. J. Appl. Microbiol. 2015;119:1207–1218. doi: 10.1111/jam.12902. [DOI] [PubMed] [Google Scholar]
- Williams A.H., Sharma M., Thatcher L.F., Azam S., Hane J.K., Sperschneider J., Kidd B.N., Anderson J.P., Ghosh R., Garg G. Comparative genomics andprediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Geno. 2016;17(1):191. doi: 10.1186/s12864-016-2486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Zhan A. Testing clustering strategies for metabarcoding-based investigation of community-environment interactions. Mol. Ecol. Res. 2018:1–13. doi: 10.1111/1755-0998.12922. [DOI] [PubMed] [Google Scholar]
- Yadeta K.A., Thomma J.B.P.H. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013;4:97. doi: 10.3389/fpls.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasushi H., Tsutomu A. PCR–based differentiation of Fusarium oxysporum f. sp.lycopersiciand radicis–lycopersici and races of F. oxysporum f. sp Lycopersici. J. Gen. Plant Pathol. 2006;72:273–283. [Google Scholar]
- Min Zhao., Hui-Min Ji., Yin Gao., XinXin Cao., Hui Yin Mao., Peng Liu., ShouQiangOuyang., 2015. Comparative transcriptome profiling conferring of resistance to Fusarium oxysporum infection between resistant and susceptible tomato.doi: http://dx.doi.org/ 10.1101/116988.
Further reading
- Staniaszek M., Sczechura W., Marczewski W. Identification of a new molecular marker C2–25 linked to the Fusarium oxysporum f. sp. radicis-lycopersici resistance Frl gene in tomato. Czech. J. Genet. Plant Breed. 2014;50:285–287. [Google Scholar]