Abstract
Noncovalent interactions between biomolecules are critical to their activity. Native mass spectrometry (MS) has enabled characterization of these interactions by preserving noncovalent assemblies for mass analysis, including protein-ligand and protein-protein complexes for a wide range of soluble and membrane proteins. Recent advances in native MS of lipoprotein nanodiscs have also allowed characterization of antimicrobial peptides and membrane proteins embedded in intact lipid bilayers. However, conventional native electrospray ionization (ESI) can disrupt labile interactions. To stabilize macromolecular complexes for native MS, charge reducing reagents can be added to the solution prior to ESI, such as triethylamine, trimethylamine oxide, and imidazole. Lowering the charge acquired during ESI reduces Coulombic repulsion that leads to dissociation, and charge reduction reagents may also lower the internal energy of the ions through evaporative cooling. Here, we tested a range of imidazole derivatives to discover improved charge reducing reagents and to determine how their chemical properties influence charge reduction efficacy. We measured their effects on a soluble protein complex, a membrane protein complex in detergent, and lipoprotein nanodiscs with and without embedded peptides, and used computational chemistry to understand the observed charge-reduction behavior. Together, our data revealed that hydrophobic substituents at the 2 position on imidazole can significantly improve both charge reduction and gas-phase stability over existing reagents. These new imidazole derivatives will be immediately beneficial for a range of native MS applications and provide chemical principles to guide development of novel charge reducing reagents.
Graphical Abstract
Introduction
Native mass spectrometry (MS) has emerged as a powerful tool for characterizing biomolecular complexes. By preserving noncovalent interactions during electrospray ionization (ESI), native MS enables quantitation of protein complex stoichiometry, ligand binding, and lipid interactions.1–4 It has been applied to a range of different complexes, including soluble proteins, membrane proteins, nucleic acids,5,6 and lipid nanoparticles.7,8 However, even under nondenaturing ESI conditions, labile interactions can be disrupted. Because lowering the charge of ions reduces electrostatic repulsions that drive dissociation in the gas phase, charge reduction is widely used to mitigate gas-phase dissociation and unfolding.9–15 Charge reducing reagents may also improve stability of complexes by reducing the internal energy of ions through evaporative cooling as weakly-bound adducts are released in the gas phase.11,16 In addition to improving stability, charge reduction can also aid data analysis by reducing overlap between charge states and narrowing the charge state distribution, which improves the signal-to-noise ratio.
Several approaches to charge reduction have been employed for native MS. The most commonly used method is to add charge reducing reagents to solution prior to ESI. Imidazole11 and triethylamine (TEA)15 are commonly used, but recent work has shown that trimethylamine oxide (TMAO) can be a potent charge reducing reagent.17–19 However, TMAO tends to form adducts with protein complexes, which is an important component of its activity.19
Our prior studies have explored the effects of imidazole as a charge reducing reagent on lipoprotein nanodiscs with only lipids, with embedded membrane proteins,20 and with incorporated transmembrane peptides.21 TEA is less effective than imidazole for stabilizing nanodiscs, and TMAO forms adducts with nanodiscs that are not easily removed. Empty nanodiscs and nanodiscs with transmembrane peptides are significantly stabilized by imidazole, and charge reduction is essential for retaining labile lipids like cholesterol.22 However, cholesterol nanodiscs are not fully stabilized by imidazole, indicated by the fact that more cholesterol is retained in negative ionization mode.
To find more effective charge reducing reagents, we hypothesized that a systematic study of imidazole derivatives would provide not only more effective reagents but also inform on the chemical principles governing charge reduction. We discovered that the addition of hydrocarbon substituents to the 2 position on imidazole allowed for greater charge reduction, improved signal-to-noise ratios, and resulted in cleaner spectra across a range of different systems, including a soluble protein complex, a membrane protein complex, and lipoprotein nanodiscs. We also found that these derivatives could better stabilized noncovalent interactions during native MS. To explore the chemical factors contributing to the behavior of these new charge reducing reagents, we tested their influence on solution-phase thermal stability of proteins and calculated their gas-phase basicities, proton affinities, and interaction energies with protein side chain models. Together, these results reveal not only largely explain the efficacy of these new charge reduction reagents but also provide heuristics for development of future reagents.
Methods
Materials and Sample Preparation
Imidazole and trimethylamine oxide were purchased from Arcos Organics. Ammonium acetate, Amberlite XAD-2, 4(5)-hydroxymethylimidazole, 4(5)-methylimidazole, 2-methylimidazole, 1-methylimidazole, 2-ethylimidazole, and 5-ethylimidazole were purchased from Sigma Aldrich. 2-isopropylimidazole and 2-propylimdazole were purchased from Tokyo Chemical Industry. 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol (DMPG), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) lipids were purchased from Avanti Polar Lipids. Streptavidin was purchased from GBioscience. LL-37 was purchased from Bachem.
Stock solutions of streptavidin were prepared by dissolving it into 0.2 M ammonium acetate at 1 mg/ml. Samples were then buffer exchanged into ammonium acetate using Micro Bio-Spin Columns (Bio-Rad Laboratories Inc) to remove residual salts. From a single stock, replicate streptavidin samples were buffer exchanged and measured in triplicate.
As previously described,20,23 AmtB-TEV-MBP-HIS was expressed in E. coli and purified by immobilized metal affinity chromatography (IMAC) and size exclusion chromatography (SEC) on a Superdex 200 16/600 (GE Healthcare) with buffers containing 0.025% dodecyl-maltoside (DDM) from Anatrace. AmtB was prepared by incubating with TEV protease overnight to cleave the poly-histidine tags. The cleaved protein was then purified by reverse IMAC and detergent exchanged by SEC into 0.5% C8E4 detergent in 0.2 M ammonium acetate. Membrane protein detergent solutions were detergent exchanged and measured in triplicate. For lipid titration experiments, 2.5 mM POPE lipids were added to a solution of 50 μM AmtB containing 40 mM charge reducing reagent, diluting the charge reducing reagent to 36 mM.
MSP1D1(−) was expressed, purified, and assembled into nanodiscs as previously described.23–25 Briefly, nanodiscs were assembled by solubilizing dried DMPC or DMPG in sodium cholate. MSP1D1(−) was added to the lipids and the sodium cholate was removed by addition of Amberlite XAD-2 beads. Nanodiscs were purified by SEC using a Superose 6 Increase 10/300 column (GE Healthcare) in 0.2 M ammonium acetate and diluted to 2.5 μM. LL-37 was dissolved in methanol to a concentration of 0.1 mM. To create peptide-nanodisc complexes, the DMPG nanodiscs were mixed 19:3 v/v with 0.1 mM LL-37 for a final concentration of 0.01 mM. Nanodisc samples were assembled and measured in triplicate.
Charge Reduction
Stock solutions of imidazole, TMAO, and all imidazole derivatives were dissolved in water at a concentration of 400 mM. The pH was lowered to 7 using acetic acid. Preliminary experiments with streptavidin, AmtB in C8E4, and peptide-nanodiscs optimized the concentration of charge reducing agents in each sample. For streptavidin, protein was mixed 9:1 v/v for a final concentration of 40 mM. For charge reduction of AmtB, protein was mixed with charge reducing reagent 9:1 v/v for a final concentration of 40 mM. For AmtB lipid binding experiments, protein and lipid were mixed 10:1 v/v for a final concentration of 36 mM of charge reducing reagent. Empty nanodiscs were mixed 41:1 v/v for a final concentration of 10 mM. Peptide-nanodiscs were mixed 45:1 v/v with 400 mM charge reducing reagent for a final concentration of 9 mM.
Mass Spectrometry
Native MS was performed as previously described.21,23,26 Briefly, electrospray ionization (ESI) was performed with borosilicate needles pulled with a P-1000 micropipette puller (Sutter Instrument, Novato, CA). Mass spectrometry was performed with a Q-Exactive HF Orbitrap mass spectrometer equipped with the Ultra-High Mass Range (UHMR) research modifications (Thermo Fisher Scientific).27 Instrumental parameters applied to each of the samples tested included 200 °C capillary temperature, 1.1-1.5 kV capillary voltage, and resolution setting of 15,000 with 10 microscans summed into one scan. For AmtB, empty nanodiscs, and peptide-nanodiscs, an additional 50 V of source fragmentation was applied. Streptavidin scans were collected from 1,000-20,000 m/z with a trapping gas pressure of 3. The applied HCD voltage was increased from 0-200 V in 20 V increments of one minute for each voltage step. AmtB scans were collected from 2,000-30,000 m/z with a trapping gas pressure of 7. HCD voltage was increased from 0-200 V in 50 V increments for 1-minute acquisitions at each step. Peptide-nanodisc and empty nanodisc scans were collected from 2,000-25,000 m/z with a trapping gas pressure of 7. In-source trapping was increased from 0-200 V in 20 V increments of one minute for each step.
Mass Spectrometry Data Analysis
Native mass spectra were deconvolved using UniDec28 and MetaUniDec.26 The deconvolution settings for streptavidin included a mass range of 1–60 or 70 kDa, a charge range of 1–50, and a Gaussian peak FWHM of 1 m/z. A charge smooth width and point smooth width of 1.0 were also applied. The deconvolution settings for AmtB included a mass range of 5-500 kDa, a charge range of 1-30, and a Gaussian peak FWHM of 0.85 m/z. A charge smooth width and point smooth width of 1.0 were also applied. The settings for deconvolution of the nanodiscs included a mass range between 20-210 kDa and a charge range of 5-25. A Gaussian peak FWHM of 10.0 was also used. A charge smooth width and mass smooth width of 1.0 were applied with the mass of the lipid set as the mass difference. For the analysis of all spectra, a curved background subtraction of 100 was applied.
To determine the number of peptides incorporated into the nanodiscs, we used mass defect analysis29 in MetaUniDec. Mass defect analysis involves dividing the measured mass by a reference mass. Here, the reference mass was the mass of DMPG, 667 Da. The total intensities of the mass defects were summed across all masses to determine the number of peptides incorporated into the nanodisc as previously described.21 Any ambiguities in matching the mass defect peaks with the corresponding number of peptides were resolved using collisional dissociation experiments.
Nano-Differential Scanning Fluorimetry
Nano-differential scanning fluorimetry measurements were made using a NanoTemper Tycho NT.6 NanoDSF instrument as previously described.20 Streptavidin was mixed 9:1 v/v with 400 mM charge reducing reagent for a final concentration of charge reducing reagent of 40 mM. AmtB solubilized in C8E4 detergent was mixed 9:1 v/v with 400 mM charge reducing reagent for a final concentration of charge reducing reagent of 40 mM. 10 μL of each of the samples were added to Tycho NT.6 Capillaries (Nano Temper Technologies), and the temperature was raised from 35–95 °C while measuring the ratio of fluorescence at 320 and 350 nm. The change in the ratio of fluorescence was plotted against the change in temperature to determine the melting point of each sample. All the samples were tested in triplicate.
Calculations
Computational modelling was performed using the University of Oregon’s High-Performance Computing Cluster, Talapas. Initial structures for neutral and protonated conformers of charge stripping reagents were constructed in the molecular modelling program Avogadro v. 1.2.0 to determine the respective molecule’s proton affinity and gas basicity. Proton bound dimers of charge stripping reagents bonded to n-butylamine were also constructed to simulate the respective charge stripping reagent bound to a protonated lysine side chain of a protein ion. For each base, low-energy structures identified from a search of 5,000-conformers in Avogadro were optimized using the MMFF94 force field. The resulting structures were geometry optimized in Gaussian 09 (Gaussian, Inc.) first at the B3LYP/6-31G* level of theory. Output structures were then further geometry optimized and harmonic vibrational frequencies were computed using the B3LYP/6-31++G** level of theory. Enthalpies and Gibbs free energies at 298 K were calculated without rescaling of the vibrational frequencies. Proton affinities (PA) and gas-phase basicity (GB) values were computed as previously described.20 In short, the PA and GB of a given molecule were calculated by subtracting the 298 K enthalpy or Gibbs free energy, respectively, of the protonated structure from that of the neutral structure and then correcting for the standard 298 K enthalpy or Gibbs free energy30 of formation for a proton gas. When this method is applied to imidazole, the PA and GB are calculated to be 947.7 kJ/mol and 915.5 kJ/mol, respectively, at the B3LYP/6-31++G** level of theory. These computed values agree well with experimentally determined values reported by Hunter and Lias31 (942.8 and 909.2 kJ/mol, respectively). 298 K enthalpies and Gibbs free energies of binding for proton-bound dimers of charge reducing reagents with n-butylamine were computed by adding the 298 K enthalpy or Gibbs free energy, respectively, of the protonated structure of the more basic substituent to that of the neutral structure of the other substituent. The 298 K enthalpy or Gibbs free energy of the proton bound dimer complex was then subtracted from the sum to yield the dimer enthalpy or Gibbs free energy of binding.
Results and Discussion
Charge Reduction and Stabilization of Streptavidin
Based on prior results with trialkylamines,32 we predicted that imidazole derivatives with added nonpolar substituents and higher gas-phase basicities would provide greater charge reduction and enhance the stability of biomolecular complexes during native MS. We first tested our hypothesis with streptavidin, a soluble tetrameric protein. Control spectra were collected for streptavidin with no additive, with imidazole, and with TMAO (Figure 1A–D). Addition of imidazole significantly reduced the charge states of streptavidin compared to spectra without additives. We found that TMAO resulted in an even greater charge reduction, but only when collision voltage was applied (Figure 1D). TMAO spectra were poorly resolved without collision voltage, likely due to formation of TMAO adducts that require collisional activation to remove (Figure 1C).
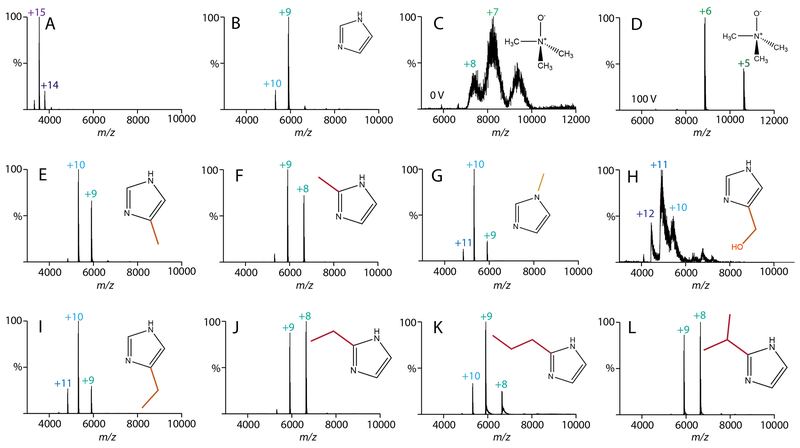
Figure 1.
Native mass spectra of streptavidin with 40 mM of charge reducing agent added. All spectra were collected at 0 V in-source trapping collision voltage except (D), which was 100 V. Additives were (A) water, (B) imidazole, (C, D) TMAO, (E) 4(5)-methylimidazole, (F) 2-methylimidazole, (G) 1-methylimidazole, (H) 4(5)-hydroxy methylimidazole, (I) 4(5)-ethylimidazole, (J) 2-ethylimidazole, (K) 2-propylimidazole, and (L) 2-isopropylimidazole.
We then monitored the effects of adding a methyl to imidazole at three different locations: 4(5)-methylimidazole, 2-methylimidazole, and 1-methylimdazole (Figure 1E–G). Interestingly, 4(5)-methyl and 1-methyl derivatives were less charge reducing than imidazole, but the 2-methyl derivative was more charge reducing. Switching to a hydrophilic substituent, such as 4(5)-hydroxymethylimidazole, resulted in spectra that were poorly resolved, likely due to adduction to the protein complex (Figure 1H). Thus, we concluded that non-polar derivatives are superior to polar derivatives.
Returning to non-polar derivatives, we next tested 5-ethylimidazole and 2-ethylimidazole (Figure 1I and 1J). Like the methyl derivatives, ethyl derivatives were more effective at the 2 position. Furthermore, 2-ethylimidazole was slightly more charge reducing than 2-methylimidazole, which suggested that longer carbon chains might be more effective. Thus, we tested 2-propylimidazole and 2-isopropylimidazole (Figure 1K and 1L). Here, the 2-propyl derivative was less effective whereas the 2-isopropyl derivative was slightly more charge reducing than the 2-ethyl derivative. Butyl and tert-butyl derivatives were not sufficiently soluble in ammonium acetate solution and were thus not explored further.
To test whether these imidazole derivatives also improved the overall stability of streptavidin, we performed collision induced dissociation (CID) experiments by increasing the collision voltage from 0-200 V (Figure 2 and S1). Without charge reducing reagents, we observed significant dissociation from tetramer to trimer by 40 V and mostly monomer and peptide fragments by 200 V. With added imidazole, streptavidin dissociation did not begin until around 150 V. Across the imidazole derivatives, gas-phase stability correlated with charge reduction. Interestingly, the 2-ethyl, 2-propyl, and 2-isopropyl derivatives were more stabilizing than TMAO despite having overall higher charges. This suggests that additional factors such as evaporative cooling11,16 contribute to the improved stability. With all the charge reducing reagents tested, we observed fragmentation into peptides at 200 V, which is consistent with prior results with low charge states of TTR.33 Comparing the laboratory collision energy34—defined as the applied collision voltage times the average charge state—revealed that the stability enhancements were not simply the result of lower effective collision energy. Overall, the addition of 2 or 3 carbon alkyl substituents at the 2 position of imidazole reduced the charge and improved the stability of streptavidin for native MS.
Figure 2.
The mass spectra (A, B, C) and deconvolved mass distributions (D, E, F) of streptavidin in the presence of no additive (A, D), 40 mM imidazole (B, E), and 40 mM 2-isopropylimidazole (C, F). The in-source trapping collision voltage was increased from 0-180 V in increments of 20 V as shown by various colors.
Charge Reduction and Lipid Retention with AmtB
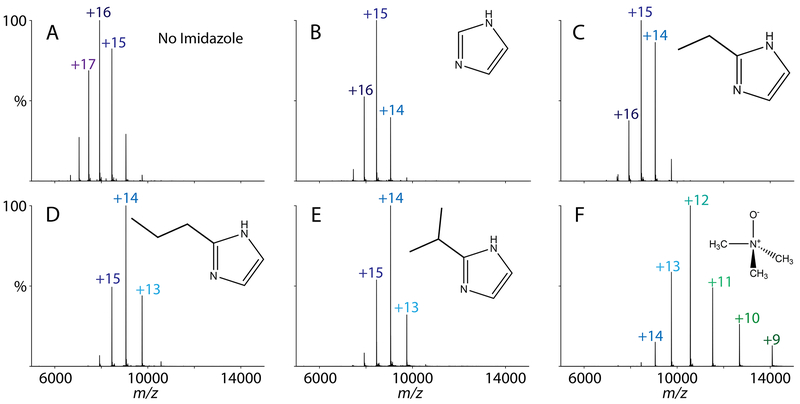
Using the imidazole derivatives that were effective with streptavidin, we next tested their effects on AmtB, a membrane protein trimer, in C8E4 detergent. AmtB with no additive, with imidazole, and with TMAO were compared as controls. Because C8E4 is known to be charge reducing,35 addition of imidazole had less of a charge reduction effect for AmtB than for streptavidin (Figure 3A and 3B).
Figure 3.
Charge states of AmtB with 40 mM charge reducing agent added. The charge reducing agents tested were (A) with water added as a control, (B) imidazole, (C) 2-ethylimidazole, (D) 2-propylimidazole, (E) 2-isopropylimidazole, and (F) TMAO.
Like streptavidin, we discovered that imidazole derivatives with the longer hydrocarbon substituents were more charge reducing for AmtB (Figure 3C–E). For example, the predominant charge state was +15 with imidazole, but the predominant charge state was +14 for both 2-propylimidazole and 2-isopropylimidazole. Adding TMAO produced the lowest charge state but created a wider charge state distribution (Figure 3F), which has the potential to reduce the signal-to-noise ratio. Because collisional activation is needed to remove detergents from AmtB, we did not observe any TMAO adducts on AmtB. Overall, the most charge reducing imidazole derivatives for AmtB were 2-propylimidazole and 2-isopropylimidazole.
To evaluate how imidazole derivatives impacted stability of lipid bound AmtB, we added POPE at a final concentration of 0.21 mM. We then performed CID from 0-200 V in 50 V steps. There were only minor differences in initial lipid binding, but we found that the presence of imidazole and imidazole derivatives stabilized bound lipids at higher collision voltages, as shown in Figure S2 and S3. Although TMAO was more stabilizing in this case, imidazole derivatives were as effective as imidazole at stabilizing membrane protein-lipid complexes.
Charge Reduction on Nanodisc-Peptide Complexes
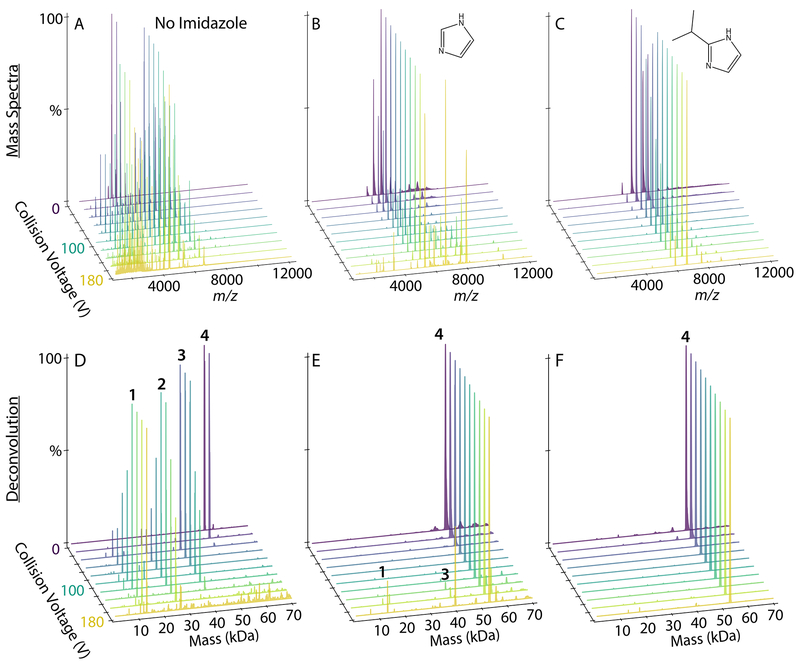
Finally, we tested the most promising imidazole derivatives on lipoprotein nanodiscs. Prior research has shown that imidazole is charge reducing and stabilizing for “empty” nanodiscs with both zwitterionic and anionic embedded lipids.20 We first tested DMPC nanodiscs, which eject lipids under increasing collisional activation.36 Although imidazole has a clear charge reduction and stabilizing effect on DMPC nanodiscs, 2-methyl, 2-ethyl, 2-propyl, and 2-isopropyl derivatives showed only minor differences from imidazole (Figure S4 and S5). Similar results were observed for empty DMPG nanodiscs (Figure S6 and S7). Thus, although these imidazole derivatives are useful for stabilizing empty nanodiscs for ESI, they are not significantly better than conventional imidazole.
However, significant differences were observed upon addition of an antimicrobial peptide to nanodiscs. We previously found that imidazole stabilizes peptide-nanodisc complexes and allows for the detection of higher stoichiometries of peptides in nanodiscs.21 To test whether imidazole derivatives would further stabilize peptide-nanodisc complexes, we added LL-37, an antimicrobial peptide, at a 6/1 molar ratio with DMPG nanodiscs. As a control, we collected spectra with no additive, imidazole, and TMAO. There was poor signal intensity and unresolvable spectra when TMAO was added to nanodisc samples.
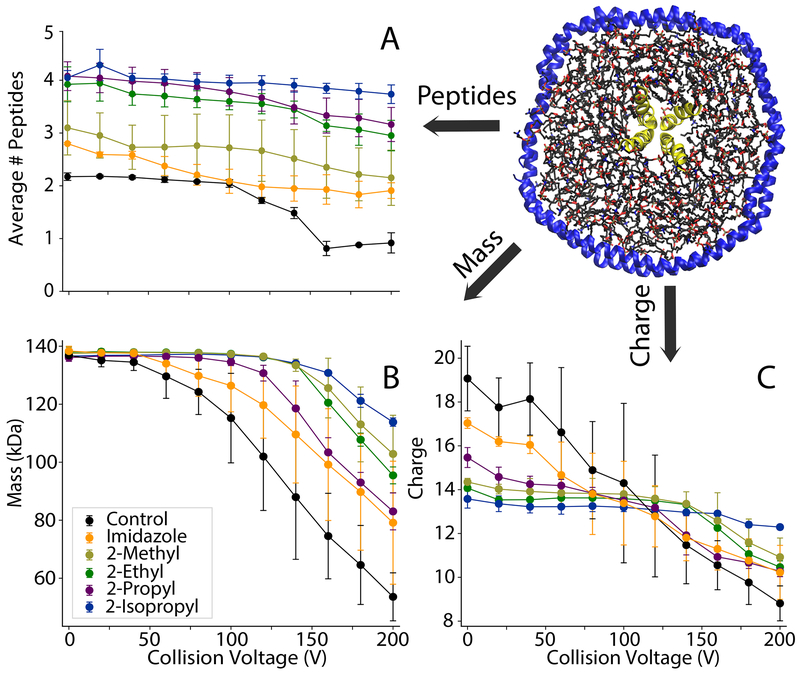
Adding imidazole increased the average number of incorporated peptides from 2.17±0.07 with controls to 2.80±0.08 (Figure 4A). 2-methylimidazole did not significantly increase the average number of peptides and caused a wider standard deviation between replicate samples. In contrast, 2-ethyl, 2-propyl, and 2-isopropyl all significantly increased the average number of peptides incorporated to around 4. Thus, imidazole derivatives improve retention of labile peptides and provide more accurate quantitation of the stoichiometry of peptides associated with nanodiscs.
Figure 4.
(A) The average number of peptides incorporated, (B) average mass, and (C) average charge of DMPG nanodiscs with 6/1 LL-37 and different charge reducing agents added. The collision voltage was increased from 0-200 V in 20 V increments.
To investigate the effects of imidazole derivatives on the stability of peptide-nanodisc complexes, we performed CID and monitored the average number of peptides incorporated, the average mass of the entire nanodisc complex, and the average charge (Figure 4). Deconvolved mass distributions are also shown in Figure S8. Without charge reducing reagents, there was a substantial loss of peptides, mass, and charge at increasing CID. Some degree of stabilization was observed with imidazole, but imidazole derivatives were more stabilizing towards peptide and average mass loss and showed lower charge states. Interestingly, stability of mass is not always correlated with stability of peptide incorporation. For example, the 2-propyl derivative was significantly more stabilizing towards peptide loss than the 2-methyl derivative but less stabilizing towards overall mass loss. This could be due to different reagents shifting the competition between dissociation of lipids and dissociation of peptides. It could also be due to interactions of the more hydrophobic derivatives with the lipid bilayer that alter dissociation pathways. In any case, the 2-isopropyl derivative was the most stabilizing across all three measures of dissociation. Thus, 2-isopropylimidazole provides an improved reagent for native MS of nanodisc complexes with embedded antimicrobial peptides.
Mechanisms of Improved Charge Reduction
To understand why the imidazole derivatives were superior charge reducing reagents, we first sought to rule out solution-phase effects. Thus, we tested the thermal stability of streptavidin, AmtB, and empty nanodiscs by nano-differential scanning fluorimetry (DSF).20 Addition of imidazole, TMAO, and imidazole derivatives generally did not significantly influence the melting temperature (Figure S9 and Table S1). Thus, we do not expect that solution-phase stability is affected by these reagents. Imidazole is often used in protein purification and likely has only minor influences on protein activity and interactions in most cases. However, we cannot rule out that imidazole derivatives might interact with the protein in some cases, particularly with the more hydrophobic reagents.
We next considered the gas-phase interactions of the different derivatives with protons and with model side chains. Our hypothesis was that more effective charge reducing reagents would show stronger interactions with protons but weaker interactions with proteins. Gas-phase basicity (as measured by PA, negative ΔH of protonation, or GB, negative ΔG of protonation) has been shown to correlate with the degree of charge reduction for other reagents and to be an important factor in gas-phase ion structure.13,15,32,37–39
Experimental GB values are not available for many of the reagents studied here, so we used ab initio computations to determine them, as well as the PA and GB of models of lysine and arginine side chains (n-butylamine and methylguanidine, respectively). Computational results for PA and GB affinity are summarized in Figure S10 and Table S2. Because GB showed nearly identical trends to PA, we focus the discussion on PA. All charge reducing reagents were found to be less basic in the gas phase than methylguanidine but more basic than n-butylamine, indicating that these reagents likely will not deprotonate arginine side chains upon dissociation but should be able to deprotonate many lysines, histidines, and other less basic groups on the protein, thereby reducing the charge.
Imidazole derivatives showed higher PA values with larger alkyl substituents. Thus, the trend of increased charge reduction with increasing alkyl chain length can be partially explained by increasing PA. Derivatives in the 4 position had lower PA than those in the 1 or 2 position, in line with the observed lower efficacy of 4-subsituted reagents at charge reduction. Derivatives in the 2 position had similar GB values to derivatives in the 1 position, indicating that factors not well-described by these computations, such as sterics, likely play a role in the observed differences between 1- and 2-substituted isomers in the experiments. A notable exception to the strong correlation between PA and ability reduce charge was unsubstituted imidazole, which had a much lower calculated PA than the other imidazole derivatives (though still higher than n-butylamine) despite its excellent charge reduction capabilities. TEA and TMAO have similar PA values to one another and to propyl and isopropyl imidazole derivatives but have very different charge reducing effects, indicating that factors beyond basicity contribute to charge reduction efficacy.
To understand why some charge reducing reagents are more prone to adduction during ESI, we computed dissociation energies of proton-bound heterodimers formed between each charge reducing reagent and n-butylamine, a model of the lysine side chain. For imidazole derivatives, heterodimer dissociation energies were generally found to decrease with increasing alkyl chain length due to an increasing difference between the PA of the charge reducing reagent and that of n-butylamine. Thus, higher PA values led to derivatives that are easier to dissociate and hence less prone to adduction to lysines and other basic protein side chains. Interestingly, despite having a high PA, TMAO has an especially high dimerization energy with n-butylamine. This can be explained by the large charge rearrangement in the polar N-O bond that occurs upon dimerization with lysine and which does not occur for the other bases studied, including TEA. This result is consistent with TMAO’s tendency to remain adducted to positively charge biomolecular ions under low-activation conditions (Figure 1C).18,19 In contrast, TEA has a similar PA to TMAO, but TEA undergoes little change in C-N bond polarity upon dimerization with n-butylamine, resulting in a relatively low dimerization energy as compared to TEA or the imidazole derivatives studied. This result is consistent with TEA’s low tendency toward adduction. As for the calculated PA/GB results discussed above, unsubstituted imidazole is an exception, with a relatively high computed proton-bound dimerization energy (ΔG of 59 kJ/mol at 298 K), even higher than TMAO (57 kJ/mol), but it does not show adduction like TMAO. 4-hydroxymethylimidazole shows much higher adduction but similar dimerization energies to other methyl derivatives, likely because our calculations do not fully reflect the contributions of the hydroxyl interacting with other hydrogen-bonding sites on the protein.
Comparing computations with experimental results, imidazole derivatives with higher computed PA/GB values and lower shared-proton dimer binding energies with n-butylamine are generally more effective at charge reduction while avoiding adduction. Based on the experimental data presented above and these computational data, we conclude that the following factors are helpful in optimizing ESI charge-reducing reagents for protonated biomolecular ions: 1) the reagent should have a high PA/GB; 2) it should have a low proton-bound dimer dissociation energy with n-butylamine; and 3) other chemical properties, such as solubility in the electrospray solvent, volatility, and strength of interactions with the analyte, should be taken into consideration as appropriate.
Conclusions
We have shown that the imidazole derivatives with alkyl substituents, particularly isopropyl, in the 2 position can provide superior charge reduction and stability for a range of different biomolecular complexes, including a soluble protein, a membrane protein in detergent, and lipoprotein nanodiscs with embedded transmembrane peptides. Given their superior performance and ease of use, we expect that these reagents will have wide-ranging impacts in native MS analysis. These improvements are largely explained by increases in computed gas-phase basicity/proton affinity and decreases in proton-shared heterodimer energies between imidazole derivatives and lysine side chains, which reveal opportunities for future development of charge reduction reagents with even more potent effects.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Alexander Makarov, Maria Reinhardt-Szyba, and Kyle Fort at Thermo Fisher Scientific for support on the UHMR Q-Exactive HF. The pMSP1D1 plasmid was a gift from Stephen Sligar (Addgene plasmid No. 20061). This work was funded by the Bisgrove Scholar Award from Science Foundation Arizona, the American Society for Mass Spectrometry Research Award, the National Science Foundation (CHE-1845230), and the National Institute of General Medical Sciences and National Institutes of Health (NIH)/National Institute of General Medical Sciences (R35 GM128624) to M.T.M. Z.M.M. and J.S.P. are funded by the NIH/National Institute of Allergy and Infectious Diseases (R21AI125804). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors thank Larry Walker for helpful discussions.
Footnotes
Supporting Information. The Supporting Information is available free of charge via the Internet at http://pubs.acs.org and contains supplemental figures and tables.
REFERENCES
- 1.Hopper JT; Robinson CV Mass spectrometry quantifies protein interactions--from molecular chaperones to membrane porins. Angew. Chem. Int. Ed 2014, 53, 14002–14015. [DOI] [PubMed] [Google Scholar]
- 2.Frick M; Schmidt C Mass spectrometry-A versatile tool for characterising the lipid environment of membrane protein assemblies. Chem. Phys. Lipids 2019, 221, 145–157. [DOI] [PubMed] [Google Scholar]
- 3.Jovcevski B; Pukala TL: Mass Spectrometry and Its Applications. In Biomolecular and Bioanalytical Techniques, 2019; pp 219–253. [Google Scholar]
- 4.Bolla JR; Agasid MT; Mehmood S; Robinson CV Membrane Protein-Lipid Interactions Probed Using Mass Spectrometry. Annu. Rev. Biochem 2019, 88, 85–111. [DOI] [PubMed] [Google Scholar]
- 5.Fabris D A role for the MS analysis of nucleic acids in the post-genomics age. J. Am. Soc. Mass Spectrom 2010, 21, 1–13. [DOI] [PubMed] [Google Scholar]
- 6.Hofstadler SA; Griffey RH Analysis of Noncovalent Complexes of DNA and RNA by Mass Spectrometry. Chem. Rev 2001, 101, 377–390. [DOI] [PubMed] [Google Scholar]
- 7.Marty MT; Zhang H; Cui W; Blankenship RE; Gross ML; Sligar SG Native mass spectrometry characterization of intact nanodisc lipoprotein complexes. Anal. Chem 2012, 84, 8957–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marty MT; Hoi KK; Robinson CV Interfacing Membrane Mimetics with Mass Spectrometry. Acc. Chem. Res 2016, 49, 2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacholarz KJ; Barran PE Use of a charge reducing agent to enable intact mass analysis of cysteine-linked antibody-drug-conjugates by native mass spectrometry. EuPA Open Proteomics 2016, 11, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehmood S; Marcoux J; Hopper JTS; Allison TM; Liko I; Borysik AJ; Robinson CV Charge Reduction Stabilizes Intact Membrane Protein Complexes for Mass Spectrometry. J. Am. Chem. Soc 2014, 136, 17010–17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J; Kitova EN; Klassen JS Method for Stabilizing Protein-Ligand Complexes in Nanoelectrospray Ionization Mass Spectrometry. Anal. Chem 2007, 79, 416–425. [DOI] [PubMed] [Google Scholar]
- 12.Sun N; Soya N; Kitova EN; Klassen JS Nonspecific interactions between proteins and charged biomolecules in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom 2010, 21, 472–481. [DOI] [PubMed] [Google Scholar]
- 13.Hall Z; Politis A; Bush MF; Smith LJ; Robinson CV Charge-State Dependent Compaction and Dissociation of Protein Complexes: Insights from Ion Mobility and Molecular Dynamics. J. Am. Chem. Soc 2012, 134, 3429–3438. [DOI] [PubMed] [Google Scholar]
- 14.Bornschein RE; Hyung S-J; Ruotolo BT Ion Mobility-Mass Spectrometry Reveals Conformational Changes in Charge Reduced Multiprotein Complexes. J. Am. Soc. Mass. Spectrom 2011, 22, 1690. [DOI] [PubMed] [Google Scholar]
- 15.Lemaire D; Marie G; Serani L; Laprévote O Stabilization of Gas-Phase Noncovalent Macromolecular Complexes in Electrospray Mass Spectrometry Using Aqueous Triethylammonium Bicarbonate Buffer. Anal. Chem 2001, 73, 1699–1706. [DOI] [PubMed] [Google Scholar]
- 16.Bagal D; Kitova EN; Liu L; El-Hawiet A; Schnier PD; Klassen JS Gas Phase Stabilization of Noncovalent Protein Complexes Formed by Electrospray Ionization. Anal. Chem 2009, 81, 7801–7806. [DOI] [PubMed] [Google Scholar]
- 17.Patrick JW; Laganowsky A Generation of Charge-Reduced Ions of Membrane Protein Complexes for Native Ion Mobility Mass Spectrometry Studies. J. Am. Soc. Mass Spectrom 2019, 30, 886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gault J; Lianoudaki D; Kaldmae M; Kronqvist N; Rising A; Johansson J; Lohkamp B; Lain S; Allison TM; Lane DP; Marklund EG; Landreh M Mass Spectrometry Reveals the Direct Action of a Chemical Chaperone. J. Phys. Chem. Lett 2018, 9, 4082–4086. [DOI] [PubMed] [Google Scholar]
- 19.Kaldmae M; Osterlund N; Lianoudaki D; Sahin C; Bergman P; Nyman T; Kronqvist N; Ilag LL; Allison TM; Marklund EG; Landreh M Gas-Phase Collisions with Trimethylamine-N-Oxide Enable Activation-Controlled Protein Ion Charge Reduction. J. Am. Soc. Mass Spectrom 2019, 30, 1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keener JE; Zambrano DE; Zhang G; Zak CK; Reid DJ; Deodhar BS; Pemberton JE; Prell JS; Marty MT Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J. Am. Chem. Soc 2019, 141, 1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker LR; Marzluff EM; Townsend JA; Resager WC; Marty MT Native Mass Spectrometry of Antimicrobial Peptides in Lipid Nanodiscs Elucidates Complex Assembly. Anal. Chem 2019, 91, 9284–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostelic MM; Ryan AM; Reid DJ; Noun JM; Marty MT Expanding the Types of Lipids Amenable to Native Mass Spectrometry of Lipoprotein Complexes. J. Am. Soc. Mass Spectrom 2019, 30, 1416–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid DJ; Keener JE; Wheeler AP; Zambrano DE; Diesing JM; Reinhardt-Szyba M; Makarov A; Marty MT Engineering Nanodisc Scaffold Proteins for Native Mass Spectrometry. Anal. Chem 2017, 89, 11189–11192. [DOI] [PubMed] [Google Scholar]
- 24.Bayburt TH; Grinkova YV; Sligar SG Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar]
- 25.Ritchie TK; Grinkova YV; Bayburt TH; Denisov IG; Zolnerciks JK; Atkins WM; Sligar SG: Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs In Methods Enzymol.; Nejat D, Ed.; Academic Press: San Diego, CA, 2009; Vol. 464; pp 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid DJ; Diesing JM; Miller MA; Perry SM; Wales JA; Montfort WR; Marty MT MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J. Am. Soc. Mass Spectrom 2019, 30, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Waterbeemd M; Fort KL; Boll D; Reinhardt-Szyba M; Routh A; Makarov A; Heck AJ High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 2017, 14, 283–286. [DOI] [PubMed] [Google Scholar]
- 28.Marty MT; Baldwin AJ; Marklund EG; Hochberg GK; Benesch JL; Robinson CV Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem 2015, 87, 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marty MT; Hoi KK; Gault J; Robinson CV Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew. Chem. Int. Ed. Engl 2016, 55, 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fifen JJ; Dhaouadi Z; Nsangou M Revision of the thermodynamics of the proton in gas phase. J. Phys. Chem. A 2014, 118, 11090–11097. [DOI] [PubMed] [Google Scholar]
- 31.Hunter EPL; Lias SG Evaluated gas phase basicities and proton affinities of molecules: An update. J. Phys. Chem. Ref. Data 1998, 27, 413–656. [Google Scholar]
- 32.Quintyn RS Applying Tandem Mass Spectrometry coupled with Ion Mobility to probe the Structure of Non-Covalent Protein Complexes and their Interactions with Ligands, Peptides and other Proteins PhD Dissertation, The Ohio State University, 2015. [Google Scholar]
- 33.Pagel K; Hyung SJ; Ruotolo BT; Robinson CV Alternate dissociation pathways identified in charge-reduced protein complex ions. Anal. Chem 2010, 82, 5363–5372. [DOI] [PubMed] [Google Scholar]
- 34.Dongré AR; Jones JL; Somogyi Á; Wysocki VH Influence of Peptide Composition, Gas-Phase Basicity, and Chemical Modification on Fragmentation Efficiency: Evidence for the Mobile Proton Model. J. Am. Chem. Soc 1996, 118, 8365–8374. [Google Scholar]
- 35.Reading E; Liko I; Allison TM; Benesch JL; Laganowsky A; Robinson CV The role of the detergent micelle in preserving the structure of membrane proteins in the gas phase. Angew. Chem. Int. Ed 2015, 54, 4577–4581. [DOI] [PubMed] [Google Scholar]
- 36.Marty MT; Zhang H; Cui W; Gross ML; Sligar SG Interpretation and deconvolution of nanodisc native mass spectra. J. Am. Soc. Mass Spectrom 2014, 25, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catalina MI; van den Heuvel RH; van Duijn E; Heck AJ Decharging of globular proteins and protein complexes in electrospray. Chemistry 2005, 11, 960–968. [DOI] [PubMed] [Google Scholar]
- 38.Prell JS; O’Brien JT; Steill JD; Oomens J; Williams ER Structures of Protonated Dipeptides: The Role of Arginine in Stabilizing Salt Bridges. J. Am. Chem. Soc 2009, 131, 11442–11449. [DOI] [PubMed] [Google Scholar]
- 39.Roscioli JR; McCunn LR; Johnson MA Quantum Structure of the Intermolecular Proton Bond. Science 2007, 316, 249–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.