Abstract
PARP1 inhibitors (PARPi) are known to kill tumor cells via two mechanisms (i.e., PARP1 catalytic inhibition vs. PARP1 trapping). The relative contribution of these two pathways in mediating the cytotoxicity of PARPi, however, is incompletely understood. Here we designed a series of small molecule PARP degraders. Treatment with one such compound iRucaparib results in highly efficient and specific PARP1 degradation. iRucaparib blocks the enzymatic activity of PARP1 in vitro, and PARP1-mediated PARylation signaling in intact cells. This strategy mimics PARP1 genetic depletion, which enables the pharmacological decoupling of PARP1 inhibition from PARP1 trapping. Finally, by depleting PARP1, iRucaparib protects muscle cells and primary cardiomyocytes from DNA damage-induced energy crisis and cell death. In summary, these compounds represent “non-trapping” PARP1 degraders that block both the catalytic activity and scaffolding effects of PARP1, providing an ideal approach for the amelioration of the various pathological conditions caused by PARP1 hyperactivation.
Introduction
PARP1 is an abundant nuclear protein that is critically involved in a number of biological processes linked to cellular stress responses1–3. The enzymatic function of PARP1 is to catalyze a protein posttranslational modification called Poly-ADP-ribosylation (PARylation)4. PARylation is tightly connected to DNA damage response (DDR)5, 6. PARP1 becomes activated upon sensing DNA strand breaks, leading to the generation of a large number of PARylated proteins, including itself. PAR polymers then recruit various DNA repair factors that contain PAR-binding motifs (e.g., OB-fold, WWE, PBZ, BRCT and macrodomain). These PBMs (PAR-binding motifs) bind to different topological units within PAR (e.g., ADP-ribose and iso-ADP-ribose), and thereby trigger downstream DDR signaling6.
BRCA1/2-mutated cancers are deficient for homologous recombination. These tumors rely on PARP1 for genome integrity, and become selectively vulnerable to PARP1 inhibitors (PARPi) (i.e., “synthetic lethality”)7, 8. A number of recent studies revealed that besides simply blocking PAR synthesis, PARPi may also kill tumor cells via a “trapping” mechanism9. Upon auto-PARylation, PARP1 is dissociated from DNA, due to steric hindrance and charge repulsion. PARPi treatment prevents PARP1 auto-PARylation and causes it to be trapped at DNA lesions. This PARP1-PARPi-DNA complex interferes with subsequent DNA replication and therefore is highly cytotoxic10. Although this model suggests that catalytic inhibition of PARP1 is functionally inequivalent to PARP1 deletion, there lacks a pharmacological approach to dissecting these two pathways (i.e., PARP1 catalytic inhibition vs PARP1 trapping), in order to assess their relative contribution to the cytotoxicity of PARPi.
Besides the role as an oncology target, PARP1 is hyperactivated upon sensing genotoxic stresses associated with various pathological conditions (e.g., ischemia-reperfusion (IR) injury11 and neurodegeneration12). Over-stimulation of the catalytic activity of PARP1 represents the primary mechanism for NAD+ catabolism13. NAD+ depletion during these genotoxic conditions leads to forced NAD+ replenishment through the salvage pathway. This, in turn, lowers the cellular ATP level, resulting in energy crisis, and eventually cell death (i.e., necrosis)1. Furthermore, recent studies have proposed PAR itself as a death signal, the accumulation of which could cause apoptosis-inducing factor (AIF) to translocate from mitochondria to the nucleus, leading to another form of cell death known as parthanatos14, 15. Based on these observations, PARPi are also being evaluated for the treatment of IR injury and neurodegeneration. However, current PARPi may not be useful in these circumstances, because of the significant cytotoxicity associated with PARP1 trapping. Towards this, the development of compounds that mimic PARP1 genetic deletion (i.e., inhibit PARP1 without causing PARP1 trapping) will likely have substantial therapeutic potential.
Recently, a number of groups reported a strategy where a small molecule ligand of a protein of interest is linked to a binder of a ubiquitin E3 ligase. In doing so, this bivalent agent brings the E3 ligase and the target protein into proximity where this protein is ubiquitinated and subsequently degraded through the proteasome pathway. This strategy, termed proteolysis targeting chimera (PROTAC), has been designed utilizing small molecule binders of various E3 ligases (e.g., cereblon/CRBN, VHL, MDM2, etc.,)16. Compared to regular catalytic inhibitors, PROTACs offer a range of unique advantages. In particular, by pharmacological depletion of a protein of interest (e.g., PARP1), this strategy is able to eliminate the scaffolding (i.e., non-enzyme-dependent) function of this protein16.
Results
Development of PARP1 and PARP2 degraders
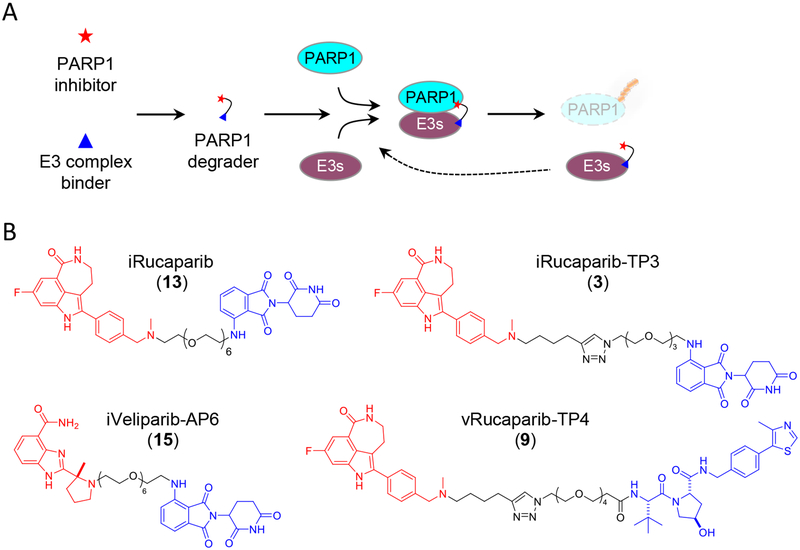
Using the PROTAC approach, we initially designed PARP1 degraders based on an FDA-approved PARP1 inhibitor, Rucaparib (1) (Figure 1A, Supplementary Figure 1 and Supplementary Table 1). Rucaparib and the other NAD+ competitive PARP1 inhibitors all contain a pharmacophore of an amide17, a moiety that is essential for its inhibitory activity against PARP1 (Supplementary Figure 1A). Examination of the co-crystal structure of PARP1 in complex with Rucaparib18 also reveals that a tether attached at the amino group distal to the indolyl lactam moiety in Rucaparib would be an ideal position for derivatization (Supplementary Figure 1A). For example, pomalidomide (2) (a CRBN ligand, and a known immunomodulatory imide drug, IMiD) is conjugated to Rucaparib through a tri(ethylene glycol) (PEG3) linker to afford a compound that we designate as iRucaparib-TP3 (3)—IMiD-Rucaparib with a triazole-PEG3 linker (Figure 1B).
Figure 1.
Targeted degradation of PARP1. (A) The overall strategy for PARP1 degradation. A PARP1 inhibitor is conjugated to an E3 binder (e.g., a CRBN ligand or VHL ligand). A PARP1 degrader recruits the E3 complex to PARP1 to induce its ubiquitination and subsequent proteasomal degradation. (B) Chemical structure of the PARP degraders, including iRucaparib, iRucaparib-TP3, iVeliparib-AP6 and vRucaparib-TP4. The PARP1 inhibitor and the E3 binder are highlighted in red and blue, respectively.
Even though our initial results with iRucaparib-TP3 were very encouraging (Supplementary Figure 1B) (considering PARP1 is one of the most abundant nuclear proteins19 with a long half-life (>60 hrs)20), we sought to further improve its potency. Building upon this prototype compound, we synthesized a large panel of additional PARP1 degraders where we systematically evaluated the warhead, linker and E3 binder (Supplementary Figures 1–3 and Supplementary Table 1). We found that the linker lengths have a profound impact on the activity of these compounds. In a series of iRucaparib-TP compounds, only iRucaparib-TP3 and iRucaparib-TP4 (4) (IMiD-Rucaparib with a triazole-PEG4 linker) were able to induce PARP1 degradation (Supplementary Figure 1B–1G). We found that switching the linker conjugation sites to the indole-N position of Rucaparib (iRucaparib-ITP3 (5) and iRucaparib-ITP4 (6)), completely abolished their PARP1 degradation activities (Supplementary Figure 2A and 2B). We also synthesized compounds wherein we conjugated a VHL ligand21 (7) to Rucaparib via a PEG3 or PEG4 linker (vRucaparib-TP3 (8) and vRucaparib-TP4 (9)). Even though PARP1 was successfully downregulated by these two compounds, they were, however, less potent compared to iRucaparib-TP3 (Supplementary Figure 2C–2H).
Next we fine-tuned the linker chemistries in the iRucaparib compounds. We found that replacing the triazole group in iRucaparib-TP3 with an amide rendered them completely inactive (iRucaparib-AMP3–1 (10) and iRucaparib-AMP3–2 (11)) (Supplementary Figure 2I and 2J). We also tested iRucaparib compounds with different lengths of all PEG linkers (Supplementary Figure 2K). Intriguingly, two of these compounds—iRucaparib-AP5 (12) and iRucaparib-AP6 (13)—emerged as highly potent PARP1 degraders with a DC50 value of 36 nM (Dmax = 90 %) and 82 nM (Dmax = 92 %), respectively (Figure 1B, Figure 2 and Supplementary Table 1).
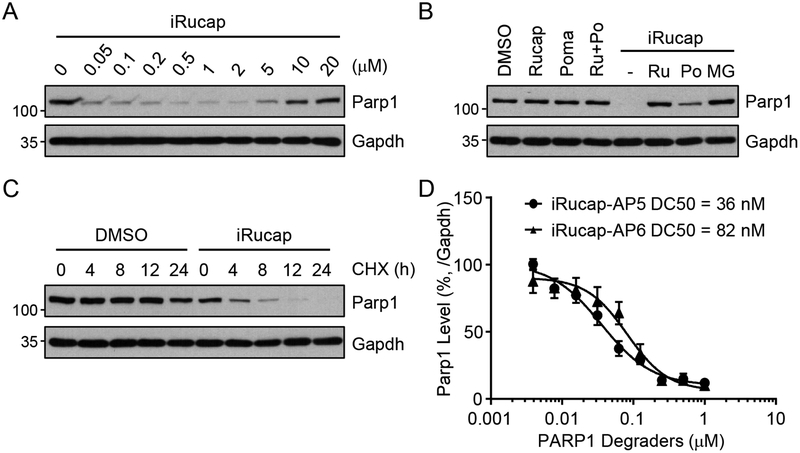
Figure 2.
iRucaparib induces PARP1 degradation. (A-D) Degradation of PARP1 by iRucaparib in primary rat neonatal cardiomyocytes. (A) Cells were treated with increasing concentrations of iRucaparib for 24 hrs. iRucap, iRucaparib-AP6. The result is a representative of two biologically independent experiments. (B) Cells were treated with Rucaparib (1 μM), pomalidomide (1 μM), Rucaparib (1 μM) plus pomalidomide (1 μM), iRucaparib (1 μM), or iRucaparib (1 μM) plus Rucaparib (1 μM) or Pomalidomide (10 μM) or MG132 (1 μM) for 24 hrs. Rucap/Ru, Rucaparib; Poma/Po, Pomalidomide; MG, MG-132. The result is a representative of three biologically independent experiments. (C) Cells were pre-treated with iRucaparib (1 μM) for 3 hrs followed by cycloheximide (10 μg/ml) treatment for the indicated times. CHX, Cycloheximide. The result is a representative of two biologically independent experiments. (D) Degradation curves of PARP1 for iRucaparib-AP5 and iRucaparib-AP6 (iRucaparib). Values represent mean ± SEM (n = 3 biological independent samples). In these experiments, whole cell lysates were analyzed by immunoblotting assays using the indicated antibodies, with the molecular weight standards shown (kDa). Uncropped blots are shown in Supplementary Figure 14.
Finally, we replaced the warhead in iRucaparib-AP compounds with different clinically relevant PARP1 inhibitors, including Niraparib, Olaparib and Veliparib (Supplementary Figure 3 and Supplementary Table 1). Among these compounds, only iOlaparib-AP6 (14) displayed a weak activity in inducing PARP1 degradation (Supplementary Figure 3H). Intriguingly, one of the iVeliparib compounds (iVeliparib-AP6 (15)) was able to potently induce the degradation of PARP2 (DC50 = 63 nM) (Figure 1B, Supplementary Figure 3K and Supplementary Table 1).
We treated primary rat neonatal cardiomyocytes with increasing concentrations of iRucaparib-AP6 (designated herein as iRucaparib), and found that this compound induced robust PARP1 degradation at a concentration of as low as 50 nM (Figure 2A). We found that iRucaparib-induced PARP1 degradation was blocked by MG132, Rucaparib or pomalidomide treatment (Figure 2B). PARP1 degraders post-translationally regulate the stability of PARP1 (Figure 2C and Supplementary Figure 4A–4D). We also performed washout experiments and showed that the effect of PARP1 degraders is reversible (Supplementary Figure 4E–4H). Consistent with the essential role of CRBN for PARP degraders, these compounds were completely inactive in CRBN-knockout cells (Supplementary Figure 5A, 5B and 5C). Finally, we showed that iRucaparib was able to induce the interaction between PARP1 and CRBN (Supplementary Figure 5D), leading to the robust ubiquitination of PARP1 (Supplementary Figure 5E).
iRucaparib selectively targets PARP1 for degradation
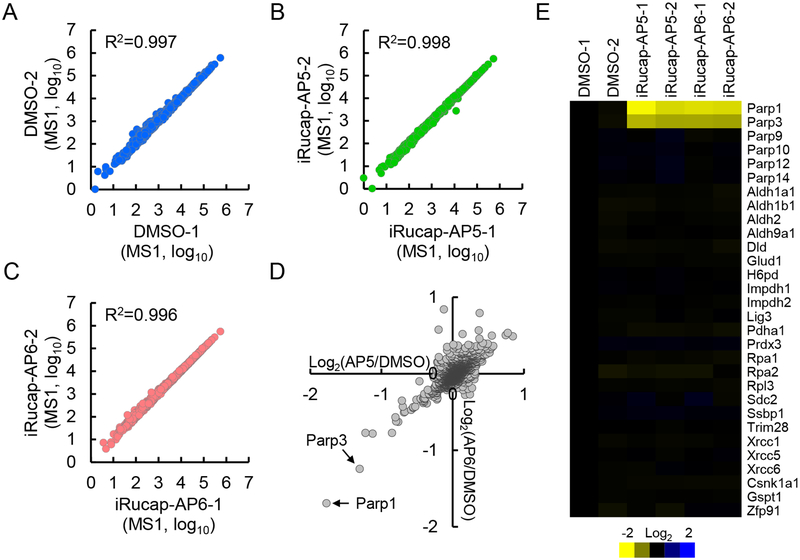
To characterize the effects of iRucaparib in an unbiased manner, we performed global protein expression analysis using a TMT (tandem mass tag)-based, multiplexed quantitative mass spectrometric approach. Proteome-wide expression profiling was performed on primary rat neonatal cardiomyocytes where we included two independent biological replicate samples for the following three conditions: (1) DMSO treatment; (2) iRucaparib-AP5 treatment; and (3) iRucaparib-AP6 (iRucaparib) treatment, with the goal of comparing the specificity of these two potent PARP1 degraders (Figure 3 and Supplementary Figure 6A). From the above-mentioned 6-plex sample, we were able to identify and quantify a total of 195,382 peptides from 6,097 proteins (peptide false discovery rate = 0.11% and protein false discovery rate = 1%). A representative MS2 spectrum leading to the identification and quantification of a PARP1 peptide (TLGDFAAEYAK) is shown in Supplementary Figure 6B. Correlation analysis revealed that an excellent reproducibility was achieved in our quantification analysis between the biological replicate analyses (Figure 3A–3C).
Figure 3.
iRucaparib selectively targets PARP1 for degradation. (A-C) Reproducibility of the TMT experiments in primary cardiomyocytes. The S/N values (signal-to-noise ratios) of the corresponding TMT channels for each protein were extracted and were Log10-transformed for the (A) control group (DMSO), (B) iRucaparib-AP5 treatment group and (C) iRucaparib treatment group (n = 2 biological independent samples). (D) Log-Log plot comparing protein expression in iRucaparib-AP5 (AP5) treatment vs. iRucaparib (AP6) treatment in primary cardiomyocytes (n = 2 biological independent samples). The corresponding ratio (compared to DMSO) was log2-transformed. PARP1 and PARP3 are indicated by the corresponding arrows. (E) Heatmap presentation of the protein expression changes in the primary cardiomyocyte TMT experiment. All data were normalized to the first control sample, which were then log2-transformed.
We first extracted the protein expression data of the DMSO and iRucaparib-AP5 group and performed a binary comparison (Supplementary Figure 6C). iRucaparib-AP5 caused rather limited perturbation of the overall proteome, with the abundance of only four proteins decreased by more than 2-fold after the treatment. Among the quantified proteins, PARP1 and PARP3 ranked as the most and second most downregulated protein, with a Log2(iRucaparib-AP5/DMSO) ratio of −1.77 and −1.31, respectively (Figure 3D, 3E and Supplementary Figure 6C). We then extracted the protein expression data of the DMSO and iRucaparib group. Compared with iRucaparib-AP5, iRucaparib treatment resulted in even smaller perturbation of the overall proteome, with PARP1 and PARP3 being the only two proteins that decreased by more than 2-fold after the treatment (Log2(iRucaparib/DMSO) ratio of −1.69 and −1.24, respectively) (Supplementary Figure 6D).
In this dataset, we were also able to identify a number of other PARPs, including PARP9, PARP10, PARP12 and PARP14 (Figure 3E). Even though the parent compound Rucaparib is a quite promiscuous PARP inhibitor (e.g., it binds to PARP10)22, none of these additional PARPs were degraded by either iRucaparib-AP5 or iRucaparib (Figure 3E). In addition to PARPs, a recent chemoproteomic study demonstrated that Rucaparib is also able to interact with a number of other NAD+/NADP+-utilizing enzymes (e.g., hexose-6-phosphate dehydrogenase (H6PD) and aldehyde dehydrogenase, mitochondria (ALDH2))23. We found that these proteins and several DDR-related proteins (e.g., XRCC5) also showed no downregulation upon the treatment of iRucaparib-AP5 or iRucaparib (Figure 3E). Finally, we also identified ZFP91, CK1α and GSPT1, which are recently reported lenalidomide/pomalidomide (IMiD)-dependent substrates of the ubiquitin E3 ligase CRBN24–26. These proteins also showed no degradation upon the treatment of iRucaparib-AP5 or iRucaparib (Figure 3E).
We used the similar quantitative proteomic approach, and characterized the specificity of additional PARP1 degraders, including iRucaparib-TP3 and vRucaparib-TP4 (Supplementary Figure 6E–6O). Despite a slight difference in their degradation profiles (e.g., besides PARP1, iRucaparib-TP3 also degrades ZFP91), all of these PARP1 degraders (in particular, iRucaparib) offer an exceptionally specific strategy for PARP1 degradation.
iRucaparib blocks ADP-ribosylation-dependent signaling
To study how the PARP1 downstream signaling network is affected by the treatment of PARP1 degraders, we first examined the biochemical activity of the various CRBN-based PARP1 degraders in vitro (Supplementary Figure 7). We found that the PARP1 IC50 for the parent compound Rucaparib is 2.5 nM (Supplementary Figure 7A), which is similar to that reported in a recent study23. The iRucaparib series compounds maintained nanomolar biochemical inhibition of PARP1 (e.g., iRucaparib-TP3 IC50 = 18 nM and iRucaparib IC50 = 29 nM) (Supplementary Figure 7A and 7B). The slight decrease in the inhibitory activity of these iRucaparib compounds is not surprising, due to the potential steric hindrance introduced by the linker creating unfavorable interactions with the helical subdomain (HD) of PARP1 (Supplementary Figure 1A). We also measured the IC50 values of some these compounds against PARP2 (Supplementary Figure 7C). In particular, although iVeliparib-AP6 degrades PARP2, it is able to inhibit both PARP1 and PARP2 with an IC50 value of 69 nM and 21 nM, respectively (Supplementary Figure 7B and 7C). These data indicate that the degradative activities of the PARP degraders are not simple reflection of their in vitro IC50 values. Besides in vitro PARP1/PARP2 IC50 values, we also probed how the in vivo PARP1 inhibitory activities might be affected by the different structural features of these compounds. PARP1 was strongly inhibited by iRucaparib-AP5 in intact cells (Supplementary Figure 7D). However, the in vivo inhibitory activity of iRucaparib-AMP3–1 and iRucaparib-AMP3–2 was much weaker. These three compounds differed in their linker chemistries, but had otherwise similar linker lengths. This difference likely resulted from a change in cell membrane-permeability introduced by the amide group in iRucaparib-AMP3–1 and iRucaparib-AMP3–2.
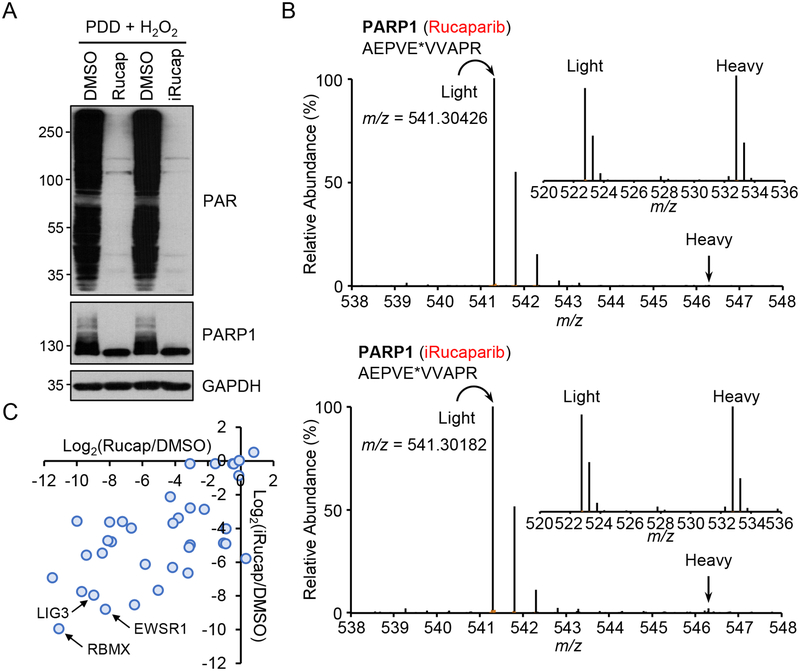
To characterize how individual PARylation events respond to Rucaparib vs. iRucaparib, we performed global analysis of the Asp/Glu-ADP-ribosylated proteome that is regulated by these two compounds (Supplementary Figure 8A). Specifically, we generated SILAC (stable isotope labeling by amino acids in cell culture)-labeled HeLa cells, and treated the light and heavy cells with PDD00017273 (1 hr). The light and heavy cells were then treated with DMSO and Rucaparib (1 hr), respectively. Both cells were then treated with H2O2 and the lysates were combined at a 1:1 ratio. PARylated peptides were enriched with boronate affinity chromatography, and were eluted with NH2OH27, 28. In a second experiment, we also performed a similar analysis using iRucaparib (1 hr treatment) (Figure 4A and Supplementary Figure 8A). We were able to identify and quantify a total of 326 and 353 PARylated peptides from the Rucaparib and iRucaparib group, respectively.
Figure 4.
iRucaparib inhibits Poly-ADP-ribosylation (PARylation)-mediated signaling events downstream of PARP1. (A) Immunoblotting analysis of PARylation levels in HeLa cells treated with either Rucaparib or iRucaparib. HeLa cells were pretreated with a PARG inhibitor (PDD 00017273, 2 μM) for one hour and then treated with Rucaparib or iRucaparib (10 μM) for another hour. Cells were then challenged with H2O2 (2 mM) for 5 min, and whole cell lysates were analyzed by immunoblotting assays using the indicated antibodies, with the molecular weight standards shown (kDa). The result is a representative of two biologically independent experiments. Uncropped blots are shown in Supplementary Figure 14. (B) Identification of a Rucaparib-sensitive (upper panel) and iRucaparib-sensitive (lower panel) PARP1 auto-modified peptide (AEPVE*VVAPR). The site of modification is indicated by an asterisk. The inset shows a ~1:1 ratio (heavy/light) of a non-PARylated peptide (RGVACTQPR) from DHX15. The upper panel shows the peptides extracted from the Rucaparib SILAC experiment, and the lower panel shows the peptides extracted from the iRucaparib SILAC experiment. (C) Correlation analysis for the ADP-ribosylated peptides identified in the Rucaparib and iRucaparib SILAC experiments. Log2(compound/control) values are shown (median values if identified multiple times).
As expected, we observed a dramatic change in the overall Asp-/Glu-ADP-ribosylated proteome after Rucaparib treatment, with many Asp/Glu-ADP-ribosylation sites showing exquisite sensitivity to this compound. For example, the abundance of a PARP1 auto-modified peptide AEPVE*VVAPR (* indicates the site of ADP-ribosylation) decreased by more than 99% after Rucaparib treatment (Figure 4B). As a control, a non PARylated peptide (RGVACTQPR) from DHX15 showed a 1:1 ratio between the light and heavy cells. The PARP1 automodified peptide AEPVE*VVAPR was also detected in the iRucaparib experiment, which showed a very similar response to iRucaparib (Figure 4B). We then extracted the SILAC ratio for all the PARylated peptides identified in the Rucaparib vs. iRucaparib experiments, and found that the change in protein ADP-ribosylation correlated well between the two compounds (Figure 4C). These results indicate that despite the addition of the degradation moiety, iRucaparib retains the capability to potently inhibit the catalytic activity of PARP1 and its downstream signaling in intact cells. We also repeated these SILAC experiments comparing Rucaparib vs. iRucaparib-TP3 and obtained very similar results (Supplementary Figure 8B–8D).
Decoupling of PARP1 catalytic inhibition and trapping
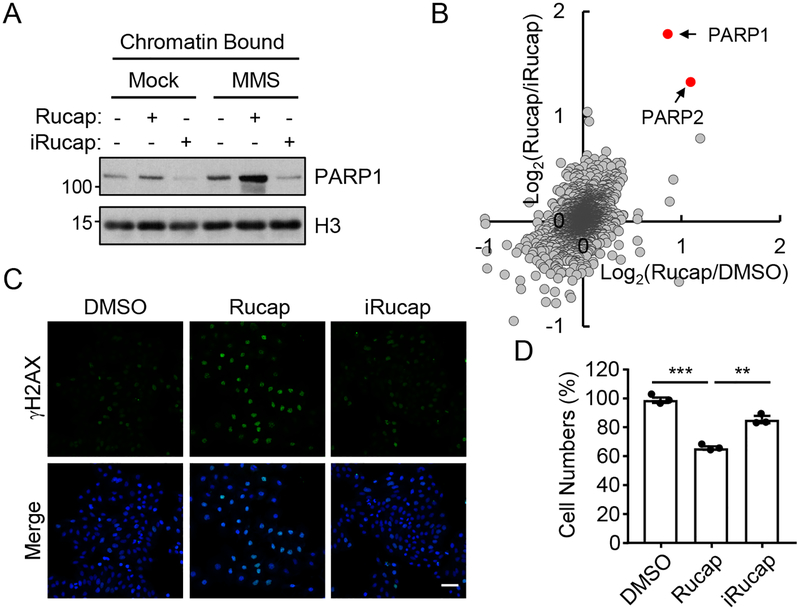
To dissect PARP1 catalytic inhibition vs. trapping, we first treated HeLa cells with Rucaparib or iRucaparib. The cells were then challenged with MMS (an agent that induces alkylating DNA damage), and subject to subcellular fractionation. Under the control conditions (without the treatment of MMS), most PARP1 was in the nuclear soluble fraction (Supplementary Figure 9A). We found that MMS treatment alone led to a slight enrichment of PARP1 in the chromatin-bound fraction (Figure 5A and Supplementary Figure 9A). Consistent with the previous studies, PARP1 trapping on chromatin was dramatically increased after the cells were treated with a combination of MMS and Rucaparib. In contrast, PARP1 was degraded upon the treatment of iRucaparib in cells (Figure 2 and Supplementary Figure 5). Because the dramatic decrease in the total amount of PARP1, PARP1 was barely detectable in the chromatin-bound fraction from the cells treated with MMS and iRucaparib (Figure 5A). As an independent assay, we also studied the kinetics of PARP1 recruitment at DNA lesions (Supplementary Figure 9B). We stably expressed GFP-PARP1 in HeLa cells, and treated them with laser microirradiation. Consistent with previous results6, DNA damage induced robust PARP1 recruitment. Furthermore, Rucaparib pre-treatment greatly stabilized PARP1, which led to a profound accumulation of PARP1 at the DNA damage site (trapped PARP1). In contrast, iRucaparib pretreatment led to PARP1 degradation, and there was very little PARP1 signal at the DNA lesion in these samples (Supplementary Figure 9B).
Figure 5.
iRucaparib is a non-trapping PARP1 degrader. (A) PARP1 trapping in HeLa cells treated with Rucaparib or iRucaparib. Cells were pretreated with Rucaparib or iRucaparib (1 μM) for 24 hrs followed by a 2-hour MMS (0.01%) treatment. Chromatin-bound proteins were extracted and analyzed using the indicated antibodies, with the molecular weight standards shown (kDa). The result is a representative of two biologically independent experiments. Uncropped blots are shown in Supplementary Figure 14. (B) Log-Log plot comparing proteins abundances (by TMT experiments) quantified from chromatin-bound fractions in Rucaparib treatment vs. control treatment or iRucaparib treatment in HeLa cells (n = 2 biologically independent samples). The corresponding ratio was log2-transformed. PARP1 and PARP2 are indicated by the corresponding arrows. (C) DNA damage response as a result of the treatment of Rucaparib or iRucaparib in HeLa cells. Cells were treated with Rucaparib or iRucaparib (10 μM) for 48 hrs, and γH2A.X signals were detected by immunofluorescence experiments. The result is a representative of two biologically independent experiments. Scale bar = 50 μm. (D) Cell proliferation analyses of HeLa cells after the treatment of Rucaparib or iRucaparib. Cells were treated with Rucaparib or iRucaparib (both at 10 μM) for 72 hrs, and values represent mean ± SEM (n = 3 biological independent samples). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing Rucaparib treatment to DMSO or iRucaparib treatment, ** p < 0.01, *** p < 0.001. p values were 1.0×10−4 and 0.0022.
To characterize the “trapped proteome” in an unbiased manner, we performed multiplexed quantitative mass spectrometric analyses of the chromatin-bound fractions isolated from HeLa cells treated with (1) MMS+DMSO; (2) MMS+Rucaparib; and (3) MMS+iRucaparib (Figure 5B). We included two biological replicates for each condition, and from this TMT6-plex sample, we were able to identify and quantify a total of 3,392 proteins (protein FDR = 1%). Consistent with our biochemical results, we found that PARP1 and PARP2 were two of the proteins that are most enriched in the chromatin-bound fraction, consistent with the robust trapping activity of Rucaparib. These two proteins were, however, depleted (compared to MMS treatment) from the chromatin after iRucaparib treatment (Figure 5B and Supplementary Figure 9C).
The trapped PARP1/DNA protein complex is known to impair replication fork progression and subsequently, induce a DNA damage response9, 10. To determine the contribution of trapped PARP1 in mediating the cytotoxicity of PARP1 inhibitors under the basal conditions, we treated HeLa cells with DMSO, Rucaparib or iRucaparib for 72 hrs. Cell cycle analyses revealed that Rucaparib, but not iRucaparib, induced G2 accumulation (Supplementary Figure 9D and 9E). In addition, treatment of Rucaparib but not iRucaparib caused DNA damage (as shown by the accumulation of γH2AX) (Figure 5C) and impaired cell proliferation (Figure 5D). These data are consistent with a model where spontaneously generated base lesions are recognized by PARP1, which, upon Rucaparib treatment, leads to the formation of trapped PARP1 and causes cell death. These toxic PARP1/DNA complexes, however, are abrogated by PARP1 degraders, resulting reduced suppression of cell survival (Figure 5D). We also repeated these experiments using iRucaparib-TP3 and obtained very similar results (Supplementary Figure 9A and 9F–9K).
Protection against genotoxic stress-induced cell death
We sought to test whether iRucaparib treatment mimics PARP1 genetic deletion, and therefore protects cells against genotoxic stress-induced cell death. Similar to our previous observation in primary cardiomyocytes, we found that iRucaparib treatment also resulted in efficient PARP1 degradation in mouse C2C12 myoblasts, and fully differentiated C2C12 myotubes (Supplementary Figure 10A and 10B). Besides iRucaparib, we found that the treatment of C2C12 myotubes and primary cardiomyocytes with iRucaparib-AP5 also led to robust and specific PARP1 degradation (Supplementary Figure 10C–10F).
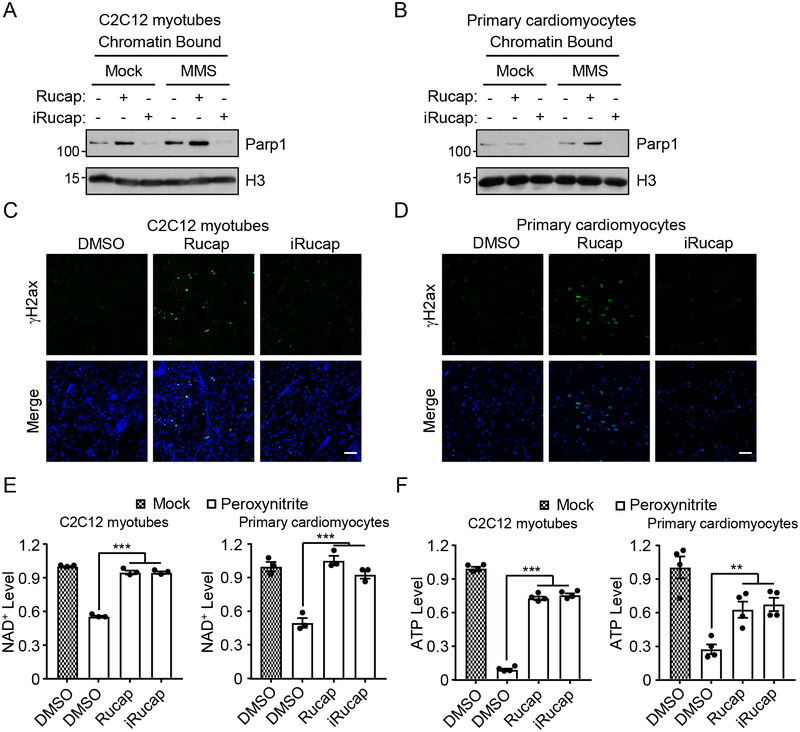
Importantly, MMS- and H2O2-induced PARP1 activation was completely blocked by pretreating C2C12 myotubes with either Rucaparib or iRucaparib (Supplementary Figure 10G). Furthermore, we also tested peroxynitrite, which is a reactive nitrogen species generated during ischemia-reperfusion, and a critical contributor to IR-dependent tissue injury29. We showed that peroxynitrite treatment robustly activated PARP1, leading to profound accumulation of PAR. Peroxynitrite-induced PARP1 activation was completely inhibited by either Rucaparib or iRucaparib (Supplementary Figure 10G). Next we examined PARP1 trapping in C2C12 myotubes and primary cardiomyocytes, and the results showed that the amount of PARP1-DNA complexes was dramatically increased upon the treatment of Rucaparib and MMS (Figure 6A and 6B). However, upon the treatment of iRucaparib, PARP1 was degraded, and became undetectable in the chromatin-bound fraction, in both the control and the MMS treatment group. Accordingly, Rucaparib, but not iRucaparib treatment resulted in robust accumulation of γH2AX in C2C12 myotubes and primary cardiomyocytes (Figure 6C and 6D). Similar results were also obtained in C2C12 myoblasts (Supplementary Figure 11A and 11B).
Figure 6.
iRucaparib protects cells from genotoxicity-induced cell death. (A-B) PARP1 trapping as a result of the treatment of Rucaparib and iRucaparib in (A) C2C12 myotubes and (B) primary rat neonatal cardiomyocytes. Cells were pretreated with Rucaparib or iRucaparib (1 μM) for 24 hrs, which was followed by a 2-hour treatment of MMS (0.01%). Chromatin-bound proteins were extracted and analyzed by immunoblotting assays using the indicated antibodies, with the molecular weight standards shown (kDa). The results are representatives of two biologically independent experiments. Uncropped blots are shown in Supplementary Figure 14. (C-D) DNA damage response as a result of the treatment of Rucaparib or iRucaparib in (C) C2C12 myotubes and (D) primary rat cardiomyocytes. Cells were treated with Rucaparib or iRucaparib (10 μM) for 48 hrs, and γH2A.X signals were detected by immunofluorescence. The results are representatives of two biologically independent experiments. Scale bar = 50 μm. (E) iRucaparib protects cells from genotoxicity-induced NAD+ depletion. Cells were pretreated with Rucaparib or iRucaparib (1 μM) for 24 hrs, and then challenged with peroxynitrite (500 μM) for 2 hrs (C2C12 myotubes, left panel) or 30 min (primary cardiomyocytes, right panel). NAD+ levels were then determined, and values represent mean ± SEM (n = 3 biological independent samples). Statistical significance was calculated with one-way ANOVA comparing peroxynitrite treated DMSO group to Rucaparib and iRucaparib groups, *** p < 0.001. p values were 6.9×10−7 and 1.6×10−4. (F) iRucaparib protects cells from genotoxicity-induced ATP depletion. Cells were pretreated with Rucaparib or iRucaparib (1 μM) for 24 hrs, and then challenged with peroxynitrite for 8 hrs (500 μM for C2C12 myotubes, left panel) or 30 min (100 μM for primary cardiomyocytes, right panel). ATP levels were measured by CellTiter-Glo assays, and values represent mean ± SEM (n = 4 biological independent samples). Statistical significance was calculated with one-way ANOVA comparing peroxynitrite treated DMSO group to Rucaparib and iRucaparib groups, **p < 0.01, ***p < 0.001. p values were 1.5×10−10 and 0.002.
We then examined whether iRucaparib protects cells against genotoxic stress-induced cell death. First we studied how the bioenergetic status of a cell is modulated following DNA damage with or without the treatment of PARP1 inhibitors. We pretreated C2C12 myotubes with Rucaparib or iRucaparib, challenged the cells with MMS, H2O2 or peroxynitrite. Consistent with PARP1 activation being a major mechanism for NAD+ catabolism, DNA alkylation and oxidative damage caused a dramatic decrease in the NAD+ levels in these cells. In contrast, NAD+ decrease was potently blocked in cells that were pre-treated with either Rucaparib or iRucaparib (Figure 6E and Supplementary Figure 12A–12B). Similar results were also obtained in primary cardiomyocytes (Figure 6E and Supplementary Figure 12A–12B). Depletion of the cellular NAD+ pool leads to forced NAD+ synthesis through the salvage pathway, which dramatically lowers the cellular ATP level. Indeed, we found that ATP abundances in C2C12 cells and primary cardiomyocytes were dramatically lowered upon the treatment of MMS, H2O2 or peroxynitrite, which could be rescued by treating the cells with either Rucaparib or iRucaparib (Figure 6F and Supplementary Figure 12C–12D). Finally, Rucaparib, but not iRucaparib decreased in the proliferation of C2C12 myoblasts (Supplementary Figure 11C–11E). We also repeated these abovementioned experiments using iRucaparib-TP3 and obtained very similar results (Supplementary Figure 12E, 12F and 13A–13J).
Discussion
We systematically evaluated the three modules in a PARP degrader, i.e., the warhead, linker and E3 binder. It has been shown that in order to achieve efficient target degradation, bivalent degraders have to recruit their respective targets into productive proximity. Furthermore, this is a dynamic process that involves various fine-tuning mechanisms (e.g., back-folding of the linker) to generate additional contact points/surfaces that are critical for the degradative activity of these compounds30. Consistent with this model, we found that all of these three factors are critical for the efficacy of a PARP degrader. Besides these abovementioned parameters, the efficiency of protein knockdown could also be affected by several other factors, including (1) the expression level of the relevant E3 ligase in a cell (i.e., CRBN for iRucaparib, and VHL for VHL-Rucaparib) (Supplementary Figure 2C). (2) the efficiency of ubiquitin transfer between the E3 ligase and the target protein (Supplementary Figure 5E); (3) the rate at which the ubiquitinated protein is degraded; and (4) the rate of de-ubiquitination and protein re-synthesis. Future studies are warranted to examine how each of these factors contributes to the efficiency of a PARP1 degrader in a specific cellular context.
All of the current PARP1 inhibitors block this enzyme by competitively occupying the NAD+-binding pocket, a domain that is highly conserved among all PARPs. As a result, it is likely that these PARP1 inhibitors will have additional targets, including other PARPs and/or NAD+-binding proteins (similar to many promiscuous ATP-competitive kinase inhibitors). Indeed, in addition to its primary target (PARP1), Rucaparib has been shown to bind to many other PARP proteins, including PARP2, PARP3, PARP4, TNKS1, TNKS2, PARP10, PARP15 and PARP1622. Similar to degradation efficiency, the specificity of degraders could also be affected by the linker chemistry. Although a degrader might bind to multiple targets, only a subset of these proteins might be conducive to degradation because of the presence of favorable protein-protein interfaces (i.e., without opposing charges), and efficient ubiquitination sites (i.e., Lys residues accessible to ubiquitin transfer). In this regard, the selectivity of a promiscuous ligand might be improved by an optimized linker chemistry, when this compound is incorporated into a degrader molecule. Indeed, we showed that despite a slight difference in their degradation profiles (e.g., degradation of ZFP91), all of the PARP1 degraders (in particular, iRucaparib) offer an exceptionally specific strategy for PARP1 degradation. Intriguingly, we found that a Veliparib-based compound (iVeliparib-AP6) showed promise as a PARP2 degrader. Further development is warranted to fully define its mechanism of selectivity and evaluate its potential for PARP2 degradation.
It has been proposed that PARP1 inhibitors kill tumors via two distinct but interconnected mechanisms. First, by catalytically inhibiting PARP1, these compounds block PARylation and PARylation-dependent DDR. Second, PARPi may also kill tumor cells by inducing PARP1 trapping. Although several PARP1 inhibitors have similar catalytic inhibition potency, they demonstrate markedly different cytotoxicity, an effect that could be ascribed to their unequal PARP1 trapping abilities31. We reason that by depleting PARP1, PARP1 degraders offer a pharmacological approach to dissecting PARP1 inhibition vs. trapping in mediating the cytotoxic effects. We first showed that despite the presence of the bulky linker and the pomalidomide moiety in the molecule, iRucaparib retains the capability to block PARP1 enzymatic activity both in vitro and in vivo. Next we showed that Rucaparib caused PARP1 trapping during genotoxic stress. In contrast, PARP1 was degraded by iRucaparib during these conditions, and hence was not detected in the chromatin-bound fractions. Consistent with these findings, iRucaparib treatment did not induce DDR, and had no effect on cell cycle progression. These results are consistent with a model where PARP1 trapping is likely to be a main contributor of the anticancer activity of PARP1 inhibitors.
Besides the role in mediating cell stress responses in cancer cells, PARP1 is hyperactivated upon sensing DNA damage caused by other pathophysiological triggers, including those produced during ischemia-reperfusion (IR) in cardiac myocytes, NMDA receptor activation in neurons, endotoxin stimulation in macrophages, and elevated extracellular glucose in endothelial cells11. In these circumstances, DNA damage-induced PARP1 activation is known to cause the collapse of the bioenergetic cycle, and eventually cell death (e.g., necrosis and parthanatos32). PARP1 knockout in animals has been shown to provide profound protection against IR injury (e.g., stroke, cardiac infarction and ischemia renal injury)33–35, cerebellar ataxia36, streptozocin-induced diabetes37 and neurodegeneration12. Thus, it has been proposed that PARP1 inhibitors, by preventing NAD+ depletion and PAR formation, have the potential to sustain cell viability. We showed that iRucaparib treatment mimics PARP1 genetic deletion in muscle cells and primary cardiomyocytes. Specifically, iRucaparib was able to inhibit the enzymatic activity of PARP1, and protect these cells from genotoxic stress-induced NAD+/ATP depletion. However, unlike Rucaparib, iRucaparib treatment does not cause PARP1 trapping, DNA damage or growth suppression.
In summary, we have designed, synthesized and evaluated a number of PARP1 degraders. By fine-tuning the warhead, the linker, as well as the specific E3 ligase system, we identified iRucaparib as a promising PARP1 degrader that achieves highly potent and specific degradation of PARP1. Using this compound, we were able to dissect the contribution of PARP1 catalytic inhibition vs. PARP1 trapping in mediating the cytotoxicity of PARP1 inhibitors in cancer cells. Treatment of this compound in non-cancerous cells mimics PARP1 genetic deletion, which protects cells against DNA damage-induced PARP1 activation and energy depletion, without eliciting the deleterious PARP1 trapping. Besides offering mechanistic insights into PARP1 trapping, our results suggest that PARP1 degraders offer a promising approach for the amelioration of pathological conditions caused by PARP1 hyperactivation. Future studies are warranted to define the full therapeutic potential of this class of compounds.
Materials and Methods
Chemicals and Reagents
Olaparib was purchased from LC laboratory. BMN-673, Veliparib, Niraparib, Rucaparib, MG-132 and Pomalidomide were obtained from Selleck. PDD 00017273 was purchased from Tocris. Peroxynitrite was purchased from Cayman Chemical. Antibodies against GAPDH and Poly-(ADP-Ribose) were purchased from Thermo and Trevigen, respectively. Antibodies against PARP1, SP1, H3, γH2A.X, CRBN, HA-Tag and VHL were purchased from Cell Signaling Technology. The PARP1-GFP plasmid was a gift from Dr. Xiaochun Yu (City of Hope). HA-Ub, lentiCRISPR-CRBN and control plasmids were constructed by standard molecular methods. All other chemicals and reagents were obtained from Sigma, unless stated otherwise.
Compound synthesis and characterization
Details of compound synthesis and characterization are placed in Supplementary Note.
Cell Culture
All cell lines were purchased from ATCC. HeLa cells and murine C2C12 myoblasts were maintained in high glucose DMEM medium supplemented with 10% fetal bovine serum. C2C12 myoblasts were differentiated into myotubes using high glucose DMEM supplemented with 2% horse serum38. 786-O cells and PC-3 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum. BT-549 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1 μg/mL insulin. Neonatal rat cardiomyocytes were isolated and cultured as described previously39. The protocol for isolation of neonatal rat cardiomyocytes was approved by the Institutional Animal Care and Use Committee (IACUC) of UT Southwestern and the performance was in adherence to the relevant ethical regulations.
Immunoblotting
Cells were washed once with cold PBS and lysed with the SDS lysis buffer (1% SDS, 10 mM HEPES pH 7.0, 2 mM MgCl2, 500U universal nuclease). Protein concentrations were determined by the BCA assay kit (Thermo Fisher Scientific). A total of 20 μg proteins (40 μg proteins for PAR immunoblotting experiments) were loaded onto the SDS-PAGE gel and were then transferred to a nitrocellulose membrane. Nitrocellulose membranes were then blocked with the TBST buffer containing 5% milk (Bio-Rad). Membranes were incubated with the primary antibodies overnight at 4℃ and the secondary antibodies for 1 hour at room temperature (RT). The blots were developed using enhanced chemiluminescence and were exposed on autoradiograph films.
Generating of CRBN deficient HeLa cells
HeLa cells were transiently transfected with lentiCRISPR plasmids (Addgene #52961) expressing the CRBN sgRNA or control sgRNA for 24 hrs, and were selected by puromycin (2 μg/mL) for two generations. Cells were then cultured in 96 well plates to generate single colonies. CRBN expression levels were confirmed by immunoblotting assays. The sgRNA sequence for CRBN is 5’-ACCAATGTTCATATAAATGG-3’, and the sequence for control sgRNA is 5’-GTATTACTGATATTGGTGGG-3’.
Cycloheximide treatment
Cells (1.5×106) were plated in a 60 mm dish, which were allowed to adhere, and were cultured overnight. Cells were pre-treated with iRucaparib or vehicle for 12 hrs (HeLa cells) or 3 hrs (primary cardiomyocytes), then treated with cycloheximide (10 μg/ml). At the indicated time points, cells were washed with cold PBS, and were lysed with the SDS lysis buffer. PARP1 expression was determined by immunoblotting assays.
Calculation of DC50
Cells (~90% confluency) were treated with increasing concentrations of PARP degraders for 24 hrs. The expression of PARPs and GAPDH were determined by immunoblotting experiments and were quantified by the ImageJ software. PARP expression was normalized by GAPDH expression. The expression level of PARP in untreated cells (DMSO) was defined as 100%. The PARP expression of the treated cells was defined as the percentage of the PARP level in treated cells, compared to that in the untreated cells. Values represent mean ± SEM (n = 3).
Real-Time Quantitative PCR
Total RNA was isolated from HeLa cells using the Trizol reagent (Invitrogen) and cDNA was synthesized using the SuperScript III first-strand synthesis kit (Invitrogen). Quantitative PCR reactions were performed on a CFX real-time system using the SYBR Green PCR Supermix according to manufacturer’s instructions (ABI). GAPDH levels were used for normalization. Primer sequences (all human genes) are as following: GAPDH forward: 5’-GAGTCAACGGATTTGGTCGT-3’, GAPDH reverse: 5’-GACAAGCTTCCCGTTCTCAG-3’; PARP1 forward: 5’- TGGAAAAGTCCCACACTGGTA-3’, PAPR1 reverse: 5’-AAGCTCAGAGAACCCATCCAC-3’; PAPR2 forward: 5’-GGCACAAATCAAGGCAGGTTA-3’, PAPR2 reverse: 5’-AAGTCATGCGGAATCCTGGTG-3’; PARP3 forward: 5’-GACCAACATCGAGAACAACAACA-3’, PAPR3 reverse: 5’-GCCTTGTGAAGT GGTTGATCT-3’.
Immunoprecipitation and Ubiquitination assays
For immunoprecipitation, HeLa cells (8×106) were left untreated (DMSO) or treated with PARP degraders for 24 hrs and incubated with 10 μM MG132 for last 12 hrs. Cells were then lysed with 1 mL Triton X-100 lysis buffer (50 mM Tris, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, 1 mM N-ethylmaleimide, 2 mM Na3VO4, 20 mM NaF, 1 mM PMSF and 1× protease inhibitor cocktail). Cell lysates were sonicated and clarified by centrifuging at 14,000 g for 15 min at 4°C. The supernatants (1 mg) was pre-incubated with protein G agarose beads for pre-clearing and further incubated with 1 μg of the corresponding antibodies and new protein G agarose beads overnight at 4°C. The beads were washed three times with 1 ml Triton X-100 lysis buffer. The immunocomplexes were eluted from the beads by boiling at 95°C for 10 min, and were analyzed by immunoblotting assays. For ubiquitination analysis, HeLa cells were transfected with an HA-Ub plasmid using lipofectamine 2000 for 24 hrs, and were left untreated (DMSO) or treated with PARP degraders as described above. Standard immunoprecipitation procedures were then performed.
Quantitative Mass Spectrometry
Cells were left untreated (DMSO) or treated with the indicated conditions for 24 hrs. Two biological replicate samples were prepared for each treatment condition. Cells were washed with cold PBS and lysed using the SDS lysis buffer, or washed with PBS and trypsinized, and were fractionated to extract chromatin bound proteins. Protein concentrations were determined by the BCA assay (Thermo Fisher Scientific). For each sample, 500 μg of protein was used for the subsequent TMT experiments40. The labeling scheme was as following: (1) HeLa cell TMT: 126 for DMSO-1, 127 for DMSO-2, 128 for iRucaparib-TP3–1, 129 for iRucaparib-TP3–2, 130 for iRucaparib-TP3+Rucaparib-1 and 131 for iRucaparib-TP3+Rucaparib-2; (2) BT-549 cell TMT: 126 for DMSO-1, 127 for DMSO-2, 128 for iRucaparib-TP3–1, 129 for iRucaparib-TP3–2, 130 for vRucaparib-TP4–1 and 131 for vRucaparib-TP4–2; (3) Primary cardiomyocyte TMT: 126 for DMSO-1, 127 for DMSO-2, 128 for iRucaparib-AP5–1, 129 for iRucaparib-AP5–2, 130 for iRucaparib-AP6–1 and 131 for iRucaparib-AP6–2; (4) HeLa cell chromatin TMT: 126 for DMSO-1, 127 for DMSO-2, 128 for Rucaparib-1, 129 for Rucaparib-2, 130 for iRucaparib-AP6–1 and 131 for iRucaparib-AP6–2.
For the TMT experiments, proteins were reduced with 2 mM DTT for 10 min and alkylated with 50 mM iodoacetamide for 30 min in dark. Proteins were then extracted using methanol-chloroform precipitation. Protein pellets were dissolved in 400 μL 8 M Urea buffer (8 M urea, 50 mM Tris-HCl, 10 mM EDTA, pH 7.5) and were digested by Lys-C (Wako, at a 1:100 enzyme/protein ratio) for 2 hrs at RT. The urea concentration was then reduced to 2 M using freshly made 100 mM ammonium bicarbonate solution. Proteins were subsequently digested with trypsin (Thermo Fisher Scientific, at 1:100 enzyme/protein ratio) overnight at RT. Peptides were desalted with Oasis HLB cartridges (Waters) and resuspended in 200 mM HEPES (pH 8.5) to a final concentration of 1 μg/μL. For each sample, 100 μg of peptides were reacted with the corresponding amine-based TMT six-plex reagents (Thermo Fisher Scientific) for 1 hour. The reactions were quenched with 5% hydroxylamine solution and were combined.
Samples were desalted and fractioned by bRPLC (basic pH reversed phase HPLC) on a ZORBAX 300 Extend-C18 column (Narrow Bore RR 2.1 mm × 100 mm, 3.5 μm particle size, 300 Ǻ pore size). Buffer A is (10 mM Ammonium Formate in H2O, pH 10.0). Gradient was developed at a flow rate of 0.2 mL/min from 0% to 70% buffer B (1 mM Ammonium Formate, pH 10.0, 90% ACN). Seventeen fractions were collected, which were lyophilized, desalted and analyzed by LC-MS/MS as described previously40. Briefly, peptides were separated on a hand-pulled fused silica microcapillary column (75 μm × 15 cm, packed with Magic C18AQ, Michrom Bioresources). A 75 min linear gradient was developed ranging from 7% to 32% acetonitrile in 0.1% formic acid at 300 nL/min to elute the peptides (Thermo EASY-nLC system). Samples were then analyzed on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) using a top 15 HCD (higher-energy collisional dissociation) method.
MS/MS spectra were searched against a composite database of the human or Rat UniProt protein database (02-04-2014) and its reversed complement using the Sequest (Rev28) algorithm. Search parameters allowed for a static modification of 57.02146 Da on cystine (Carbamidomethyl), a variable modification of 15.994915 Da on methionine (oxidation), and a static modification of TMT labels (229.16293 Da) on peptide N-terminus and lysine. For TMT quantification, a 0.03 Th window was scanned around the theoretical m/z of each reporter ion (126:126.127725; 127:127.124760; 128:128.134433; 129:129.131468; 130:130.141141; 131:131.138176). The maximum intensity of each reporter ion was extracted. For each reporter ion channel, the observed signal-to-noise ratio was summed across all quantified proteins and was normalized.
In Vitro PARP1 and PARP2 Activity Assays
PARP1 and PARP2 IC50 for Rucaparib, Veliparib and the various CRBN-based PARP degraders were measured by using a PARP universal chemiluminescent assay kit (Trevigen, #4676–096-K) and PARP2 Chemiluminescent Assay Kit (BPS Bioscience, #80552), respectively. Briefly, the histone-coated strip wells (PARP1) were hydrated with the assay buffer for 30 min. The blank strip wells (PARP2) were coated with histone proteins overnight at 4℃ and blocked with blocking buffer for 90 min. The various compounds were added and were incubated with the PARP enzyme for 10 min. The reaction cocktail containing biotinylated NAD+ was then added into the system, which was incubated for 1 hour to activate PARP enzyme. After the reaction, the wells were washed twice with 0.1% Triton X-100/PBS and twice with PBS (PARP1) or were washed three times with 0.05% Tween-20/PBS (PARP2). The wells were incubated with Strep-HRP for another hour, and were washed again as described above. Peroxy-Glow reagents were mixed and added into the strip wells, and the biotin signal on coated histone was immediately measured with chemiluminescence on a Synergy microplate reader (Bio-Tek). The well without the PARP enzyme, and the well with the PARP enzyme only was used as the negative (0%) and positive (100%) control, respectively.
Quantitative analysis of the Asp- and Glu-ADP-ribosylated proteome
The D/E-ADP-ribosylation analyses were performed as described previously27, 28. Briefly, SILAC-labeled HeLa cells ([12C614N2]lysine and [12C614N4]arginine, “Light”, or [13C615N2]lysine and [13C615N4]arginine, “Heavy”) were pretreated with 2 μM of the PARG inhibitor41 PDD 00017273 for 1 hour, and then were left untreated (DMSO) or treated with 10 μM Rucaparib or iRucaparib for another hour. The cells were challenged with 2 mM H2O2 for 5 min, and were harvested in the SDS lysis buffer. Protein concentrations were determined by the BCA assay kit. The same amounts of proteins (20 mg) were combined between the light and heavy samples. Lysates were reduced with DTT (3 mM for 20 min at RT), alkylated with iodoacetamide (50 mM for 30 min at RT in dark), and digested overnight with Lys-C and then trypsin (both at a 1:100 enzyme/protein ratio). PARylated peptides were enriched using m-Aminophenyl-boronic acid-agarose beads and were eluted using 2 M NH2OH.
Samples were analyzed by LC-MS/MS on an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) using a top 20 CID (Collision-induced dissociation) method. The isolation window and the minimal signal threshold for MS/MS experiments were set to be 2 Th and 500 counts, respectively. MS/MS spectra were searched against a composite database of the human UniProt database (02-04-2014) and its reversed complement using the Sequest (Rev28) algorithm. Search parameters allowed for a static modification of 57.02146 Da on cysteine, a dynamic modification of addition of 15.0109 Da to aspartic acid and glutamic acid, a variable modification of 15.994915 Da on methionine and a variable modification of 8.01420 Da on lysing and 10.00827 Da on arginine (SILAC label), respectively. Search results were filtered to include < 1% matches to the reverse database by the linear discriminator function.
Subcellular Protein Extraction
Cells (1×106) were left untreated (DMSO) or pretreated with PARP inhibitors or PARP degraders for 24 hrs and were challenged with 0.01% MMS for 2 hrs. Subcellular proteins were extracted from the cells using a subcellular fractionation kit (Thermo Fisher Scientific, #78840). Briefly, cells were digested with a 0.25% trypsin-EDTA solution, harvested by centrifugation at 500 g for 5 min, and were washed once with cold PBS. The cytosolic proteins were isolated by incubating the cells with the CEB buffer for 10 min at 4℃, and centrifuged at 500 g for 5 min. The membrane proteins were isolated by incubating the pellet from the previous step with the MEB buffer for 10 min at 4℃, and centrifuged at 3,000 g for 5 min. The soluble nuclear proteins were isolated by incubating the pellet from the previous step with the NEB buffer for 30 min at 4℃, and centrifuged at 5,000 g for 5 min. The chromatin bound proteins were isolated by incubating the pellet from the previous step with the NEB buffer supplemented with nuclease for 15 min at RT, and centrifuged at 16,000 g for 5 min. Protein concentrations were determined by the BCA assay kit.
Laser microirradiation and microscope image acquisition
Cells were grown on 35-mm glass bottomed culture dishes (Mattek) and were transfected with a PARP1-GFP plasmid for 24 hrs. Cells were then left untreated (DMSO) or treated with 1 μM Rucaparib or iRucaparib for another 24 hrs. Laser micro-irradiation was performed on a Zeiss LSM 780 inverted confocal microscope coupled with the MicroPoint Laser Illumination and Ablation System (Photonic Instruments, Inc.). The GPF fluorescence strips were recorded at the indicated time points and then analyzed with the Image J software.
Cell Viability Assays
HeLa cells were seeded in 96 well plates with a density of 2,000 cells/well. Sixteen hours later, cells were treated with MMS alone or in combination with 10 μM Rucaparib or PARP degraders for 72 hrs. Cell viability was determined using a CellTiter-Glo luminescence kit (Promega). Briefly, cells were balanced for 30 min at RT. One hundred microliters of the CellTiter-Glo reagent was added into each well and the cells were incubated for 10 min. The luminescence was measured using a Synergy microplate reader (Bio-Tek). The ATP level in untreated cells was defined as 100%. The viability of the treated cells was defined as the percentage of the ATP level in treated cells, compared to that in the untreated cells.
Cell Cycle Analysis
Cells (1×105) were left untreated (DMSO) or treated with 10 μM Rucaparib or iRucaparib for 48 hrs. Cells were then trypsinized and harvested by centrifuge at 300 g for 5 min, fixed with 70 % pre-chilled methanol for 2 hrs at −20℃. Fixed cells were washed once with PBS and incubated with RNase A (250 μg/mL) in PBS for another 2 hrs at 37℃ and then stained with PI (2 μg/mL). Stained cells were analyzed using BD FACS lyric and analyses were performed using the Flowjo software.
ATP and NAD+ measurement
ATP levels were measured by a luminescence assay using the CellTiter-Glo luminescent kit (Promega). Cells were left untreated (DMSO) or pretreated with Rucaparib or iRucaparib for 24 hrs, and then were challenged with 0.01% MMS, 2 mM H2O2 (50 μM for primary cardiomyocytes) or 500 μM Peroxynitrite (100 μM for primary cardiomyocytes). ATP levels were determined according the manufacturer’s instruction. NAD+ levels were assayed using an NAD/NADH quantification kit (Sigma). Cells were pretreated with Rucaparib or iRucaparib for 24 hrs, and then were challenged with 0.01% MMS, 2 mM H2O2 or 500 μM Peroxynitrite. Approximately 2×105 cells were washed with 1 mL cold PBS and lysed in 400 μL of the extraction buffer. The total NAD/NADH levels were extracted following two cycles of freezing on dry ice for 20 min followed by 10 min at RT. The samples were vortexed for 15 sec and centrifuged at 13,000 g for 10 min to remove the insoluble materials. NAD+ was quantified according to the manufacturer’s instruction. Fifty microliters of the extracts were used in the NAD+ assay, and the values were normalized by the amount of protein in each sample.
Immunofluorescence Microscopy
Cells were plated in 35-mm glass bottomed culture dishes (Mattek), and were cultured overnight. The cells were then left untreated (DMSO) or treated with 10 μM Rucaparib or iRucaparib for 48 hrs before washing with PBS. Cells were fixed with 4% Paraformaldehyde for 15 min at RT and were washed three times with PBS. The cells were then permeabilized with 0.1% Triton X-100 in PBS for 5 min and were blocked with 1% BSA in PBS for 30 min. Fixed cells were incubated with a γH2AX antibody (1:200, Cell Signaling Technology, #9718) at 4℃ for overnight, washed three times with PBS for 5 min, and incubated with an Alexa Fluor 488 conjugated anti-rabbit antibody (1:500, Thermo Fisher Scientific, A-11008) for one hour at RT. Cells were washed three times with PBS for 5 min and stained with DAPI (1:1000, Thermo, #62248) for 2 min. Cells were washed with PBS and mounted with the FluorSave reagent (Millipore, #345789). Images were collected on a Zeiss LSM 880 Airyscan inverted confocal microscope.
Statistical Analysis
Statistical analyses (t-tests or one-way ANOVA) were performed using GraphPad Prism software (v7). Data were derived from three biological replicate experiments and presented as the mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001.
Data Availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD014838 (Supplementary Dataset 1), PXD014836 and PXD014837 (Supplementary Datasets 2–3), PXD014840 (Supplementary Datasets 4–5), and PXD014839 (Supplementary Dataset 6). The source data underlying Figure 2d, 5d, 6e, 6f and Supplementary Figure 12a–12f are provided in Source Data files and Supplementary Dataset 7. Fully uncropped blots are shown in Supplementary Figure 14.
Supplementary Material
Acknowledgements
We thank J.A. Hill for sharing primary rat neonatal cardiomyocytes (UT Southwestern Medical Center), and X. Zhong and C. Kim for the help with the immunofluorescence microscopy experiments and PARP1 immunoprecipitation experiments, respectively. We thank X. Yu for proving the GFP-PARP1 construct (City of Hope). We also thank X.D. Wang, L. Yuan and the other members of the Yu laboratory for helpful discussions. This work was supported by grants from NIH (GM122932 to Y.Y.) and Welch foundation (I-1800 to Y.Y.).
Competing interests
Y.Y. receives research support from Pfizer. A provisional patent application on the PARP degraders and technologies described herein has been filed (Y.Y., C. C., S. W., and L. H.). The remaining authors declare no competing interests.
References
- 1.D’Amours D, Desnoyers S, D’Silva I & Poirier GG Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J 342, 249–268 (1999). [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson BA & Kraus WL New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Hottiger MO Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem 84, 227–263 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Chambon P, Weill JD & Mandel P Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun 11, 39–43 (1963). [DOI] [PubMed] [Google Scholar]
- 5.Gupte R, Liu Z & Kraus WL PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31, 101–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Vyas A, Kassab MA, Singh AK & Yu X The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res 45, 8129–8141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant HE et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Farmer H et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Pommier Y, O’Connor MJ & de Bono J Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med 8, 362ps317 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Lord CJ & Ashworth A PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger NA et al. Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br J Pharmacol 175, 192–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kam TI et al. Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson’s disease. Science 362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shieh WM et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem 273, 30069–30072 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Yu SW et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Dawson VL & Dawson TM Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp. Neurol 218, 193–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burslem GM & Crews CM Small-Molecule Modulation of Protein Homeostasis. Chem. Rev 117, 11269–11301 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Ferraris DV Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J. Med. Chem 53, 4561–4584 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Thorsell AG et al. Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J. Med. Chem 60, 1262–1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamanaka H, Penning CA, Willis EH, Wasson DB & Carson DA Characterization of human poly(ADP-ribose) polymerase with autoantibodies. J. Biol. Chem 263, 3879–3883 (1988). [PubMed] [Google Scholar]
- 20.Schwanhausser B et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Galdeano C et al. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel-Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) alpha subunit with in vitro nanomolar affinities. J. Med. Chem 57, 8657–8663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahlberg E et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol 30, 283–288 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Knezevic CE et al. Proteome-wide Profiling of Clinical PARP Inhibitors Reveals Compound-Specific Secondary Targets. Cell Chem Biol 23, 1490–1503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kronke J et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 523, 183–188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An J et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat Commun 8, 15398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishoey M et al. Translation Termination Factor GSPT1 Is a Phenotypically Relevant Off-Target of Heterobifunctional Phthalimide Degraders. ACS Chem Biol 13, 553–560 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang J, Ding M & Yu Y Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods 10, 981–984 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Zhen Y, Zhang Y & Yu Y A Cell-Line-Specific Atlas of PARP-Mediated Protein Asp/Glu-ADP-Ribosylation in Breast Cancer. Cell Rep 21, 2326–2337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacher P, Beckman JS & Liaudet L Nitric oxide and peroxynitrite in health and disease. Physiol. Rev 87, 315–424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadd MS et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 13, 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murai J & Pommier Y PARP Trapping Beyond Homologous Recombination and Platinum Sensitivity in Cancers. Annual Review of Cancer Biology 3, 131–150 (2019). [Google Scholar]
- 32.Dawson TM & Dawson VL Mitochondrial Mechanisms of Neuronal Cell Death: Potential Therapeutics. Annu. Rev. Pharmacol. Toxicol 57, 437–454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eliasson MJ et al. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat. Med 3, 1089–1095 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Zingarelli B & Szabo C Effect of genetic disruption of poly (ADP-ribose) synthetase on delayed production of inflammatory mediators and delayed necrosis during myocardial ischemia-reperfusion injury. Shock 13, 60–66 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Zheng J, Devalaraja-Narashimha K, Singaravelu K & Padanilam BJ Poly(ADP-ribose) polymerase-1 gene ablation protects mice from ischemic renal injury. Am J Physiol Renal Physiol 288, F387–398 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Hoch NC et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541, 87–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Luo C, Chowdhury S, Gao ZH & Liu JL Parp1 deficient mice are protected from streptozotocin-induced diabetes but not caerulein-induced pancreatitis, independent of the induction of Reg family genes. Regul. Pept 186, 83–91 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Pirinen E et al. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab 19, 1034–1041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang QJ et al. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J. Clin. Invest 121, 2447–2456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu R et al. Quantitative Secretomic Analysis Identifies Extracellular Protein Factors That Modulate the Metastatic Phenotype of Non-Small Cell Lung Cancer. J Proteome Res 15, 477–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James DI et al. First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib. ACS Chem Biol 11, 3179–3190 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD014838 (Supplementary Dataset 1), PXD014836 and PXD014837 (Supplementary Datasets 2–3), PXD014840 (Supplementary Datasets 4–5), and PXD014839 (Supplementary Dataset 6). The source data underlying Figure 2d, 5d, 6e, 6f and Supplementary Figure 12a–12f are provided in Source Data files and Supplementary Dataset 7. Fully uncropped blots are shown in Supplementary Figure 14.