Abstract
The wingless and integration site growth factor-5a (Wnt5a) is a ligand of the receptor tyrosine kinase-like orphan receptor-1 (ROR1). Since both Wnt5a and ROR1 are expressed in circulating chronic lymphocytic leukemia (CLL) cells and because in other cell types STAT3, which is constitutively activated in CLL, induces Wnt-5a signaling, we wondered whether STAT3 induces the expression of Wnt5a in CLL cells. Sequence analysis detected four putative STAT3-binding sites in close proximity to the Wnt5a gene promoter’s start codon. Chromatin immunoprecipitation and electrophoretic mobility shift assay revealed that STAT3 binds to the Wnt5a gene promoter, and a luciferase assay showed that STAT3 activates the Wnt5a gene. Additionally, transfection of peripheral blood CLL cells with STAT3-short hairpin-RNA downregulated Wnt5a mRNA and protein levels, suggesting that STAT3 binds to the Wnt5a gene promoter and induces the expression of Wnt5a in CLL cells. Flow cytometry and confocal microscopy determined that both Wnt5a and its receptor ROR1 are co-expressed on the surface of CLL cells and Western immunoblotting showed an inverse correlation between Wnt5a and ROR1 protein levels, implying that, regardless of CLL cells’ ROR1 levels, blocking the interaction between Wnt5a and ROR1 might be beneficial to patients with CLL. Indeed, transfection of CLL cells with Wnt5a small interfering RNA reduced wnt5a mRNA and protein levels and significantly increased the spontaneous apoptotic rate of CLL cells. Taken together our data unravel an autonomous STAT3-driven pro-survival circuit that provides circulating CLL cells with a microenvironment-independent survival advantage.
Keywords: CLL, Wnt5a, ROR1, STAT3
Introduction
Receptor tyrosine kinase-like orphan receptor-1 (ROR1) is an evolutionary conserved type I surface membrane protein that is commonly expressed during embryogenesis but repressed in most adult tissues (1). Like most adult cells normal B cells do not express ROR1. Conversely, B-cell chronic lymphocytic leukemia (CLL) cells express high levels of ROR1 (2) whose activation provides the cells with survival advantage (3) and induces cellular proliferation (4).
The ligand that binds to and activates ROR1 is Wnt5a. Wnt5a is a member of the wingless and integration site growth factor (WNT) family of secreted glycoproteins known to modulate several biological processes including embryogenesis, organogenesis and tumorigenesis (5). Wnt5a binds to ROR1 and induces activation of β-catenin-dependent and -independent pathways (4). Bone marrow mesenchymal stromal cells (MSC) are the only cells known to produce Wnt5a in human bone marrow where Wnt5a also binds ROR2 and participates in the induction of osteogenesis (6, 7). Recent studies demonstrated that in CLL cells Wnt5a induced signaling depends on the interaction between ROR1 and ROR2 (8), and that Wnt5a induces ROR1 to recruit DOCK2 and activate Rac1/2 to enhance CLL cell proliferation (4). Like ROR1 transcripts (3), Wnt5a transcripts have been detected in CLL cells of nearly all patients (9). However, what factor(s) activate the transcription of Wnt5a in CLL cells is currently unknown.
Previously, we found that signal transducer and activator of transcription (STAT)-3 induces the expression of ROR1 in CLL cells (3). STAT3 is constitutively phosphorylated and activated in CLL cells (10). Phosphorylated STAT3 forms dimers, shuttles to the nucleus, and induces the expression of several STAT3-target genes (10–13). Because Wnt5a is constitutively expressed in CLL cells (9), and phosphorylated STAT3 was found to induce Wnt5a signaling in various cell types (14), we sought to determine whether, in addition to inducing the expression of ROR1 (3), STAT3 also induces the expression Wnt5a in CLL cells.
Materials and Methods
Fractionation of CLL cells and normal B cells
Peripheral blood (PB) cells were obtained from previously untreated CLL patients who were followed at The University of Texas MD Anderson Cancer Center leukemia clinic, after obtaining an Institutional Review Board approved informed consent. The patients’ clinical characteristics are depicted in Supplemental Table 1. To isolate low-density cells, peripheral blood cells were fractionated using Ficoll-Hypaque 1077 (Sigma-Aldrich, St. Louis, MO). More than 90% of the PB lymphocytes obtained from the low-density fraction of the studied CLL patients co-expressed CD19 and CD5 antigens, as assessed by flow cytometry using an upgraded FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). To isolate normal CD19+ B cells, normal donors’ PB low-density cells were fractionated using micro-immunomagnetic beads and Miltenyi columns (Miltenyi Biotec, San Diego, CA) in accordance with the manufacturer’s instructions. More than 97% of the fractionated cells were CD19+ cells as assessed by flow cytometry.
Western immunoblotting
Western blot analysis was performed as previously described (10). Briefly, cell lysates were assayed for their protein concentrations using the BCA protein assay reagent (Pierce Chemical Co., Rockford, IL). Each set of paired lyasates were adjusted for the same protein concentration. A lysate of CLL cell extract was mixed with 4x Laemmli sample buffer and was then denatured by boiling for 5 minutes. Forty micrograms of each lysate were separated using 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane. The transfer was done overnight at 30 V in a cooled (4oC) reservoir. The nitrocellulose membrane was then placed in Ponceaus S stain to verify equal loading of protein. The membranes were blocked with 5% dried milk dissolved in 50 ml of phosphate buffered saline (PBS) with Tween-20. After blocking, the membrane was incubated with the following primary antibodies: monoclonal mouse anti-human Wnt5a (Invitrogen, Waltham, MA), monoclonal mouse anti-human ROR1 (Invitrogen), monoclonal mouse anti-human STAT3 antibodies (BD Biosciences, San Jose, CA), and mouse anti-human β-actin (Sigma-Aldrich, St. Louis, MO). After incubation with horseradish peroxidase–conjugated secondary antibodies (GE Healthcare, Buckinghamshire, UK) for 1 hour, blots were visualized with an enhanced chemiluminescence detection system (GE Healthcare).
Immunoprecipitation studies
Immunoprecipitation studies were done as previously described (15). Briefly, CLL cell lysates were incubated with rabbit anti–Wnt5a antibodies (Invitrogen) for 16 hours at 4°C. Protein A agarose beads (Cell Signaling Technologies, Danvers, MA) were added for 2 hours at 4°C. For negative controls, the cytoplasmic lysates were incubated either with rabbit serum plus protein A agarose beads or with protein A agarose beads alone. After three washes with radioimmunoprecipitation assay buffer (Ripa), the beads were suspended in sodium dodecyl sulfate (SDS) sample buffer, boiled for 5 minutes, the beads removed by centrifugation, and the supernatant proteins were separated by SDS–polyacrylamide gel electrophoresis (PAGE). Normal B cells from 2 healthy volunteers and HeLa cells were used as controls.
Flow cytometry
Live unpermeabilized cells were used for flow cytometry. Before staining cells were washed three times in PBS with 2% fetal bovine serum (FBS; Invitrogen). Cells were then stained with Wnt5a (Invitrogen) and ROR1 (Invitrogen) antibodies, and their corresponding isotypic antibody. Cells were analyzed using an upgraded FACSCalibur flow cytometer (Becton Dickinson) and data analysis was performed using CellQuest software (BD Biosciences). Graphics were created with CellQuest (BD Biosciences) and WinList (Verity Software House, Topsham, ME) software.
Confocal microscopy
CLL cells were fixed in 2% paraformaldehyde for 10 minutes at 37°C and permeabilized overnight at −20°C. Before staining, cells were washed three times in PBS with 2% fetal bovine serum (FBS; Invitrogen). Then, the cells were incubated in micro tubes in PBS supplemented with 5% FBS. After 1 hour of incubation, the cells were washed three times with PBS and then incubated with rabbit anti-Wnt5a antibodies (Invitrogen) and mouse anti-ROR1 antibodies (Invitrogen) for 1 hour. After being washed three times with phosphate-buffered saline, the cells were incubated with Alexa flour 488 labeled anti-rabbit and Alexaflour 647 labeled anti-mouse antibodies. After being washed three times with PBS, the cells were suspended in 5 mg/ml solution of 4’,6-diamidino-2-phenylindole (DAPI) dye (Invitrogen) for 15 minutes and then washed in PBS to remove the unbound dye. The cells were then placed into μ-slide VI chamber slides (ibidi, LLC) for microscopic analysis. The slides were viewed using an Olympus FluoView 500 confocal laser scanning microscope (Olympus America, Waltham, MA), and images were analyzed using the FluoView software (Olympus America).
Transfection of MM-1 cells with Wnt5a promoter fragments and luciferase assay
Four different Wnt5a promoter fragments were transfected into MM-1 cells via electroporation using the Gene Pulser Xcell Electroporation System (Bio-Rad Laboratories, Hercules, CA). Each construct included a luciferase reporter gene and a Wnt5a promoter fragment that included between 1 to 4 gamma interferon activation site (GAS)-like elements. The luciferase activity of unstimulated or IL-6-stimulated MM-1 cells was assessed 24 hours after transfection using a Dual-Luciferase Reporter Assay System (Promega) and a BD MonolightTM 3010 luminometer (BD Biosciences, San Jose, CA, USA). The luciferase activity of each of the human Wnt5a promoter constructs was determined by calculating the constructs’ luciferase activity relative to the activity of the Renilla luciferase produced by the pRL-SV40 control vector.
Chromatin immunoprecipitation (ChIP) assay
A chromatin immunoprecipitation (ChIP) assay was performed using a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, Boston, MA) according to the manufacturer’s instructions. Briefly, cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature and then harvested and incubated on ice for 10 minutes in lysis buffer. Nuclei were pelleted and digested with micrococcal nuclease. Following sonication and centrifugation, sheared chromatin was incubated with anti-STAT3 or rabbit serum (negative control) overnight at 4oC. Then, protein G beads were added, and the chromatin was incubated for 2 hours in rotation. Antibody-bound protein-DNA complexes were eluted and subjected to polymerase chain reaction (PCR) analysis. The primers to amplify the human c-Myc promoter were F: 5-TGA GTA TAA AAG CCG GTT TTC-3 and R: 5-AGT AAT TCC AGC GAG AGG CAG-3, which generated a 63 bp product; to amplify the Wnt5a promoter were F: 5-CAG AGA GGA GGA GCT GGA GAT-3 and R:5-CCC AGT TCA TTC ACA CCA CAG-3, which generated a 74bp product; to amplify the ROR1 promoter were F: 5-TTT GAG GAG TGT GGG GGA GGG-3 and R:5-GTT GAG AGG CTG CAG CAG AGG−3 which generated a 110 bp product; to amplify the STAT3 promoter: F: 5-CCG AAC GAG CTG GCC TTT CAT-3 and R: 5-GGA TTG GCT GAA GGG GCT GTA-3 which generated a 86 bp product; to amplify the VEGF promoter were F: 5-CTT CTC CAG GCT CAC AGC TT-3 and R: 5- CCT GGA AAT AGC CAG GTC AG-3 which generated a 181 bp product; and primers to amplify the human RPL30 gene were provided by Cell Signaling Technologies. The primers used to amplify fragments of the human Wnt5a gene promoter were F’: +56 and R’ +130 encompassing a 74-bp product that covers the γ-interferon activation sequence (GAS) binding sites from 100 bp to 108 bp upstream of the Wnt5a start codon; F’ +110 and R’+254, a 144-bp product that covers the upstream GAS binding sites from 208 bp to 216 bp; F’+234 and R’ +294, a 60-bp product that covers the upstream GAS-binding sites from 234 bp to 243 bp; F’ +244 and R’ +348, a 104-bp product that covers the upstream GAS-binding sites from 249 bp to 257 bp.
Electrophoretic mobility shift assay (EMSA)
Non-denatured cellular nuclear extracts were prepared using a NE-PER extraction kit (Thermo Scientific Pierce, Rockford, IL). Nuclear protein extracts were incubated with 4’ biotin-labeled DNA probes derived from the Wnt5a promoter sequence. Each probe was synthesized by Sigma-Genosys (The Woodlands, TX) and designed to include one of the above described 4 GAS-like elements in the Wnt5a promoter. The probes were incubated for 30 minutes on ice. Following incubation, the samples were separated on a 5% polyacrylamide gel, transferred onto a nylon membrane, and fixed on the membrane via ultraviolet cross-linking. The biotin-labeled probe was detected with streptavidin-horseradish peroxidase (Gel-Shift Kit; Panomics, Fremont, CA). The control consisted of a 7-fold excess of unlabeled cold probe. To test the effect of STAT3, anti-STAT3 antibodies (BD Biosciences) or mouse IgG1 (BD Biosciences) were added to the nuclear extracts, as previously described (10).
Generation of GFP-conjugated lentiviral STAT3 short hairpin RNA (shRNA) and infection of CLL cells
293T cells were co-transfected with GFP-conjugated lentiviral STAT3 shRNA or a GFP-conjugated empty lentiviral vector and with packaging vectors (pCMV delta R8.2 and pMDG generously provided by Dr. Giorgio Inghirami, (Department of Pathology, University of Torino, Italia) using the Superfect transfection reagent (QIAGEN Inc.) as previously described (10). The 293T cell culture medium was replaced after 16 hours and collected after 48 hours. Then, the culture medium was filtered through a 45-μm syringe filter to remove floating cells, the lentivirus was concentrated by filtration through an Amicon Ultra centrifugal filter device (EMD; Millipore, Burlington, MA), and the concentrated supernatant was used to infect CLL cells. CLL cells (5 × 106/ml) were incubated in six-well plates (Becton Dickinson, Franklin Lakes, NJ) in 2 ml of Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, Grand Island, NY) supplemented with 10% FBS and were transfected with 100 μl of viral supernatant. Polybrene (10 ng/ml) was added to the viral supernatant at a ratio of 1:1000 (v/v) and allowed to complex for 10 minutes before infection. Transfection efficiency was measured after 48 hours and ranged between 40% and 50% (calculated based on the ratio of propidium iodide–negative and green fluorescet protein-positive cells). These experiments were conducted using an upgraded FACSCalibur flow cytometer (Becton Dickinson).
Quantitative reverse-transcription polymerase chain reaction analysis
We used 500 ng of total RNA in one-step quantitative reverse-transcription polymerase chain reaction (qRT-PCR; Applied Biosystems, Foster City, CA) analysis with an ABI Prism 7700 sequence detection system (Applied Biosystems) using a TaqMan gene expression assay for STAT3, Bcl2, Cyclin D1, c-Myc, p21, caspase 3, colony stimulating factor-2 receptor (CSF2R)-α, lipoprotein lipase (LPL), ROR1, and Wnt5a genes according to the manufacturers’ instructions. Samples were run in triplicate, and relative quantification was performed by using the comparative CT method.
Transfection of CLL cells with Wnt5a small interfering RNA (siRNA)
Twenty μM human Wnt5a-siRNA or scrambled-siRNA, 20 μM 6-carboxyfluorescein (FAM)-labeled human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or reagent control (Applied Biosystems, Foster City, CA) were added to 10 μl siPORT NeoFX transfection reagent diluted in 50 μl Opti-MEMI reduced serum medium (Thermo Fisher Scientific), and incubated at room temperature for 10 minutes. Then, the reagents were incubated at room temperature with 1 × 107 CLL cells suspended in 0.2 ml Opti-MEM I medium. After 1 hour of incubation, electroporation was performed using the Gene Pulser Xcell Electroporation System (Bio-Rad Laboratories) and the cells were incubated in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% FBS for 24 hours. Transfection efficiency of the FAM-conjugated siRNA was assessed by Flow cytometry using a FACSCalibur flow cytometer (Becton Dickinson Biosciences).
Annexin V/propidium iodide assay
The rate of cellular apoptosis was analyzed using double staining with a Cy5-conjugated annexin V kit and propidium iodide (PI; BD Biosciences) according to the manufacturer’s instructions. Briefly, 1×106 cells were washed once with phosphate-buffered saline and re-suspended in 200 μL binding buffer with 0.5 μg/mL annexin V-Cy5 and 2 μg/ml PI. After incubation for 10 minutes in the dark at room temperature, the samples were analyzed on a FACSCalibur flow cytometer (Becton Dickinson). Cell viability was calculated as the percentage of annexin V positive cells.
Results
CLL cells co-express Wnt5a and ROR1
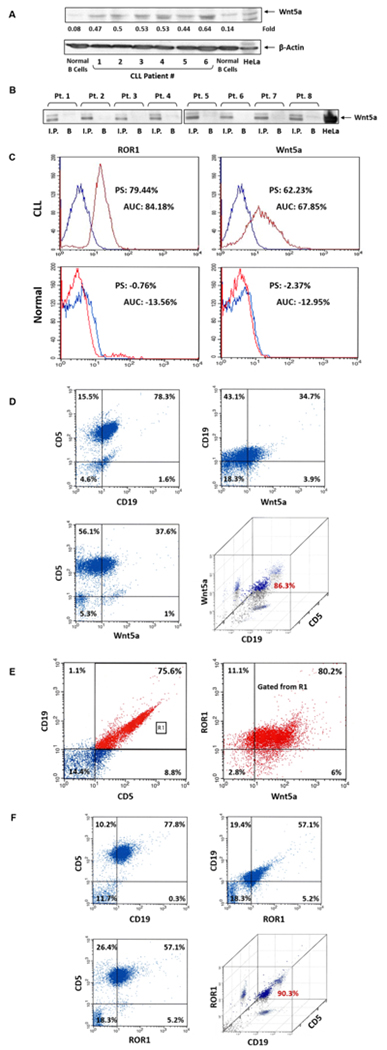
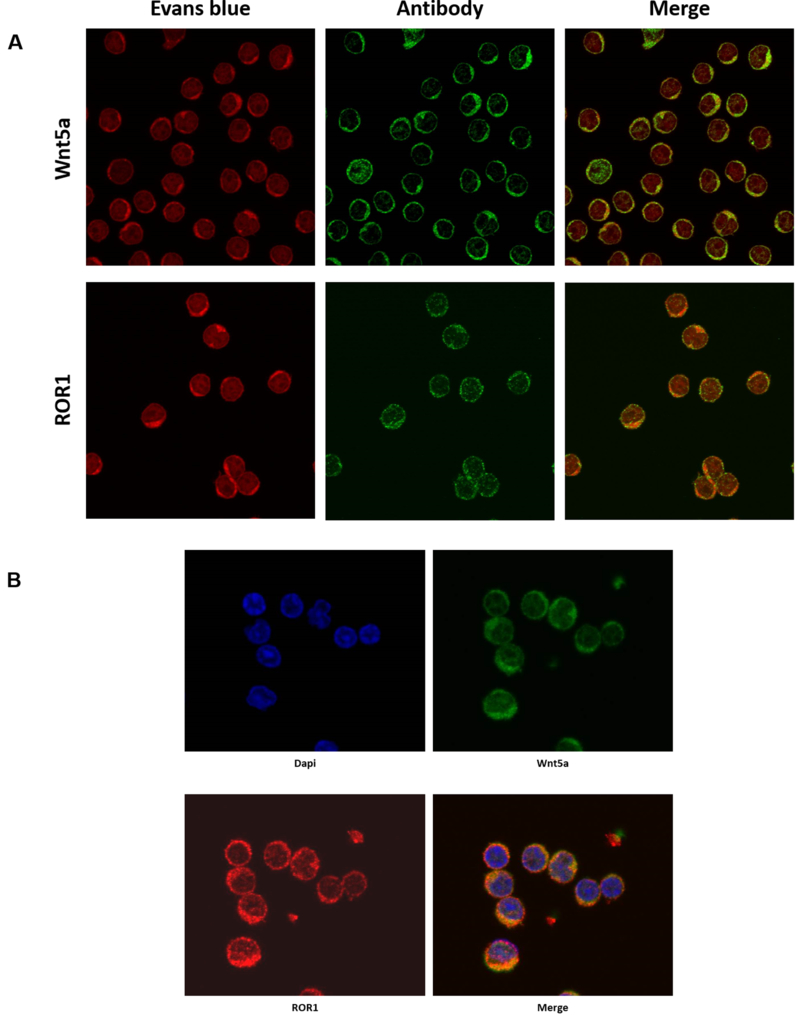
Previous studies demonstrated that CLL cells express ROR1 (2, 3). To assess whether CLL cells also express the ROR1 ligand Wnt5a, we performed Western Immunoblotting of PB samples of 6 randomly selected patients. As shown in Figure 1A, we detected Wnt5a protein in CLL cell extracts of all patient samples. To confirm these findings, we obtained CLL cells from 8 additional patients, immunoprecipitated the cell lysates with anti-Wnt5a antibodies and by using Western immunoblotting, detected Wnt5a protein in all samples (Figure 1B). To further confirm these data we used flow cytometry and detected cell surface Wnt5a and ROR1 on the majority of CLL cells but not on normal CD19+ B lymphocytes (Figure 1C – E). To determine whether Wnt5a and ROR1 are present in CLL cells we used confocal microscopy. We found that Wnt5a and ROR1 are expressed in all CLL cells (Figure 2A). Then, to assess whether Wnt5a and ROR1 are expressed in the same cell we used dual staining and confirmed that CLL cells co-express Wnt5a and ROR1 (Figure 2B).
Figure 1.
CLL cells co-express cell surface Wnt5A and ROR1 protein. (A) Western blot analysis of CLL lysates of PB low-density cells obtained from 6 patients. As shown, Wnt5A was detected in all patients but not in normal B cells. Beta actin was used for loading control. (B) Immunoprecipitation of CLL cell lysates with Wnt5a antibodies. CLL cells from 6 additional CLL patients were lysed and immunoprecipitated with Wnt5a antibodies. The immune complexes were separated using SDS-PAGE and, as shown, Wnt5a was detected in all patients’ samples. B, beads coated with the Wnt5a antibodies’ isotype control. (C) Flow cytometry analysis of live unfixed, CLL and normal CD19+ cells using anti-ROR1 (left panel) and anti-Wnt5a (right panel) antibodies is depicted. The Kolmogorov–Smirnov test was used to assess the difference in expression between ROR1, Wnt5a and their corresponding isotopyic controls. As shown, ROR1 and Wnt5a were detected on the surface of CLL but not normal B cells. The percentages of differences in peak shifts (PS) and the areas under the curve (AUC) are depicted. Blue: isotype, red: specific antibody. In CLL cells the difference of the area under the curve was 84.18% (P < 0.001) for ROR1 and 67.85% (P < 0.001) for Wnt5a. (D) Flow cytometry analysis of PB cells from another patient with CLL shows that 86.3% of the cells that co-express CD5 and CD19 express Wnt5a. (E) Flow cytometery analysis of PB low density cells from another patient with CLL. In this sample only 76.5% of the cells co-expressed CD5 and CD19 (left panel). Of the gated cells (R1) 80.2% co-expressed ROR1 and Wnt5a (right panel). As shown in the right lower quadrant panel 86% CD19+CD5+ cells express Wnt5a.
Figure 2.
ROR1 and Wnt5a are detected in CLL cells. (A) Confocal microscopy images of fixed and permiabilized CLL cells co-stained with either anti-Wnt5A or ROR1 antibodies show that Wnt5A and ROR1 are detected in CLL cells. (B) Confocal microscopy analysis of fixed and permiabilized CLL cells from another patient show that both Wnt5a and ROR1 are present in the same CLL cells. DAPI staining (blue) was used to detect the cells’ nuclei.
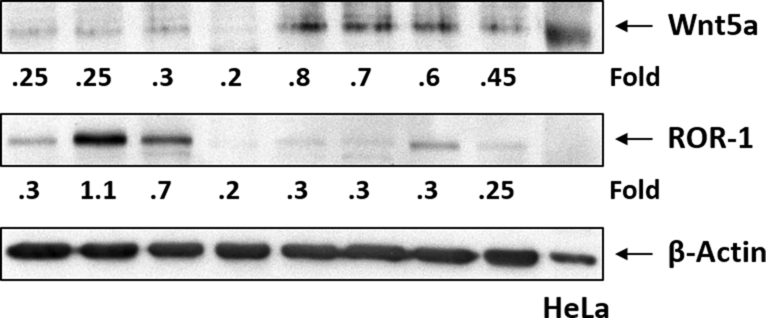
A recent study suggested that high levels of ROR1 are associated with an accelerated disease progression (16). Because the expression of both ROR1 and Wnt5a is induced by STAT3, we wondered whether in patients with high ROR1 levels Wnt5a levels would be high as well. Using Western immunoblotting we assessed the levels of ROR1 and Wnt5a on the same CLL CLL patients’ cell samples. We found that CLL cells with high ROR1 levels expressed low Wnt5a levels whereas most samples with low ROR1 levels expressed high levels of Wnt5a (Figure 3), excluding the possibility that we detected receptor-bound ligand and suggesting that low levels of the ROR1 receptor are ‘compensated’ by increased levels of the Wnt5a ligand.
Figure 3.
Both Wnt5a and ROR1 protein are detected in cell extracts obtained from the same CLL patients. As shown, an inverse correlation between ROR1 and Wnt5a levels was detected in most patient samples.
STAT3 binds to the Wnt5a promoter and activates the Wnt5a gene
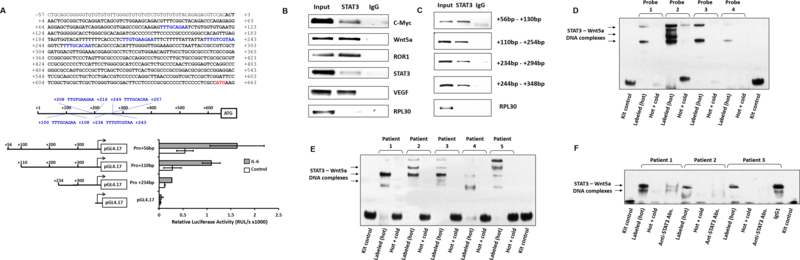
Because we previously found that ROR1 expression in CLL cells is driven by STAT3 (3) and STAT3 was found to induce Wnt5a signaling in various cell types (14), we sought to assess the effect of STAT3 on the expression of Wnt5a. Using the TFsearch database (http://diyhpl.us/~bryan/irc/protocol-online/protocol-cache/TFSEARCH.html) we found 4 GAS-like elements, known as putative STAT3 binding sites (17) within the Wnt5a promoter, upstream and in close proximity to the Wnt5a start codon (Figure 4A, upper panel). Then, to test whether STAT3 binds to any of those GAS-like elements, we constructed 4 DNA fragments derived from the Wnt5a promoter, containing 1, 2, 3, and 4 STAT3 putative binding sites attached to a luciferase reporter gene (Figure 4A, middle panel). These constructs were transfected into MM1 cells to which IL-6 was added in order to phosphorylate and activate STAT3. As shown in the lower panel of Figure 4A, each of the putative STAT3-binding sites induced luciferase activity and with each added STAT3 binding site an increased luciferase activity was observed, suggesting that STAT3 binds to each of these GAS-like elements and activates the Wnt5a promoter. To validate these data and test whether STAT3 binds to the Wnt5a promoter in CLL cells we used ChIP. Using the 104-bp product, that covers the upstream GAS-binding sites from 249 bp to 257 bp, we found that like the STAT3-target genes c-Myc, ROR1, STAT3, and VEGF, Wnt5a DNA co-immunoprecipitated with STAT3 (Figure 4B). Then, using the same assay, we confirmed that all 4 GAS-like elements bind STAT3 in CLL cells (Figure 4C). To further confirm these we used an EMSA and found that, like in MM1 cells, all 4 GAS-like elements bind to the Wnt5a promoter in CLL cells (Figure 4D). Furthermore, we found that in nuclear extracts from 5 randomly chosen patients, probe 1, previously found to bind to GAS-like elements at +100 bp - +108 bp, formed STAT3 – Wnt5a DNA complexes (Figure 4E) and that the binding of nuclear extract to the Wnt5a DNA probe was significantly attenuated by anti-STAT3 antibodies but not by their isotype (IgG1) (Figure 4F).
Figure 4.
STAT3 binds to and activates the Wnt5a gene promoter. (A) Sequence analysis of the Wnt5a gene promoter, performed by using the NCBI the website https://www.ncbi.nlm.nih.gov/nucleotide/NG_031992.1?report=genbank&log$=nuclalign&blast_rank=1&RID=RJ0BMXJ8014, detected 4 putative STAT3-binding sites (shown in blue) located in close proximity to the Wnt5a gene start codon outlined (shown in red in the upper panel). Three truncated forms of those putative STAT3-binding sites (middle panel) were attached to the luciferase reporter gene (pGL4.17) and transfected into MM1 cells (lower panel). Each horizontal bar in the lower panel shows the mean luciferase activity ± standard error in MM1 transfected cells incubated without (control) or with 20 ng/ml IL-6 which activates STAT3. As shown, the luciferase activity was increased with the addition of another STAT3 binding site in IL-6-stimulated but not in –unstimulated MM1 cells (P =.02, 0.22 and .001, respectively). ATG (shown in red) is the start codon and +1 denotes the first nucleotide of the first exon as well as the first nucleotide of the Wnt5a promoter. Arrowheads indicate start codon. (B) ChIP shows that Wnt5a DNA co-immunoprecipitates with STAT3 protein. Protein extracts from CLL cells were incubated with or without anti-STAT3 antibodies, and DNA was extracted from the immunoprecipitated chromatin fragments. As shown, anti-STAT3 antibodies co-immunoprecipitated with DNA of Wnt5a promoter region and of several other known STAT3-target genes including C-myc, ROR1, STAT3 and VEGF. RPL30 was used as negative control. “Input” denotes DNA extracted obtained from chromatin fragments of non-immunoprecipitated CLL cell (negative control). (C) Four primers for each of the above described STAT3 putative binding sites were prepared and their binding to STAT3 protein was assessed using ChIP. Protein extract of CLL cells was incubated with anti-STAT3 antibodies and each of these 4 primers was used to amplify the co-immunoprecipitated DNA. As shown, DNA was amplified with all 4 primers. RPL30 was used as negative control and IgG as the isotype of the anti-STAT3 antibodies. (D) EMSA confirmed that CLL nuclear protein extract binds to the putative STAT3 binding sites in the Wnt5a gene promoter. CLL-cell nuclear protein extract was incubated with each of the 4 labeled Wnt5a promoter fragments harboring one putative STAT3-binding site. As shown, STAT3-Wnt5a promoter complexes were detected with each labeled fragment and the binding was attenuated with the addition of unlabeled (cold) fragments. (E) A labeled Wnt5a promoter fragment harboring a single putative STAT3 binding site (probe 1) was added to nuclear protein extracts of CLL cells from 5 different patients. As shown, STAT3-Wnt5a complexes were detected in samples of all patients and STAT3-Wnt5a binding was significantly attenuated with the addition of unlabeled (cold) Wnt5a promoter fragment. (F) Anti-STAT3 antibodies, but not their isotype (IgG1), significantly attenuated STAT3-Wnt5a binding in nuclear extract from 3 CLL patient cells, further confirming that STAT3 binds to and activates the Wnt5a promoter.
STAT3 shRNA downregulates Wnt5a mRNA and protein levels
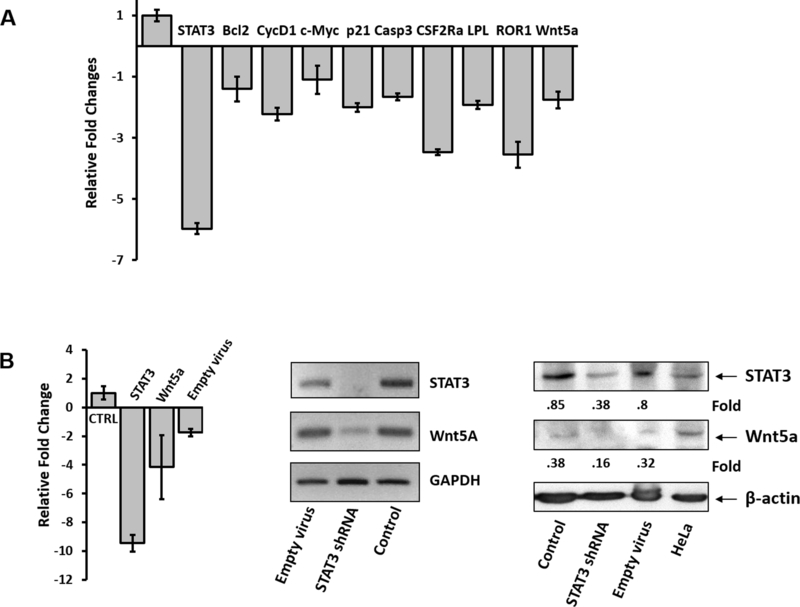
To confirm that STAT3 induces the expression of Wnt5a, we infected CLL cells with a lentiviral STAT3-shRNA or with an empty vector. As shown in Figure 5A, infection of CLL cells with STAT3-sh RNA downregulated mRNA levels of STAT3, Wnt5a and several STAT3-regulated genes including STAT3, Bcl2, Cyclin D1, c-Myc, p21, caspase 3, CSF2Rα, LPL, and ROR1. Furthermore, STAT3-shRNA downregulated STAT3 and Wnt5a mRNA (Figure 5B, left and middle panels) and protein levels (Figure 5B, right panel), confirming that STAT3 induces the expression of Wnt5a in CLL cells.
Figure 5.
STAT3 induces the expression of Wnt5a in CLL cells. (A) CLL cells were infected with STAT3-shRNA or with an empty vector. As shown, infection of CLL cells with STAT3-sh-RNA significantly downregulated mRNA levels of STAT3, Wnt5a and several STAT3-regulated genes including Bcl2, Cyclin D1, c-Myc, p21, caspase 3, CSF2Rα, LPL, and ROR1 (P <.0001; Student’s t-test; left panel). Ctrl., transfection with empty virus (control). (B) Infection of CLL cells with STAT3-shRNA, but not the empty lentiviral vector, significantly downregulated STAT3 and Wnt5a mRNA (assessed by qRT-PCR, P <.0001; Student’s t-test; left panel) and conventional PCR (middle panel)) and protein levels (right panel).
Wnt5a siRNA induces apoptosis of CLL cells
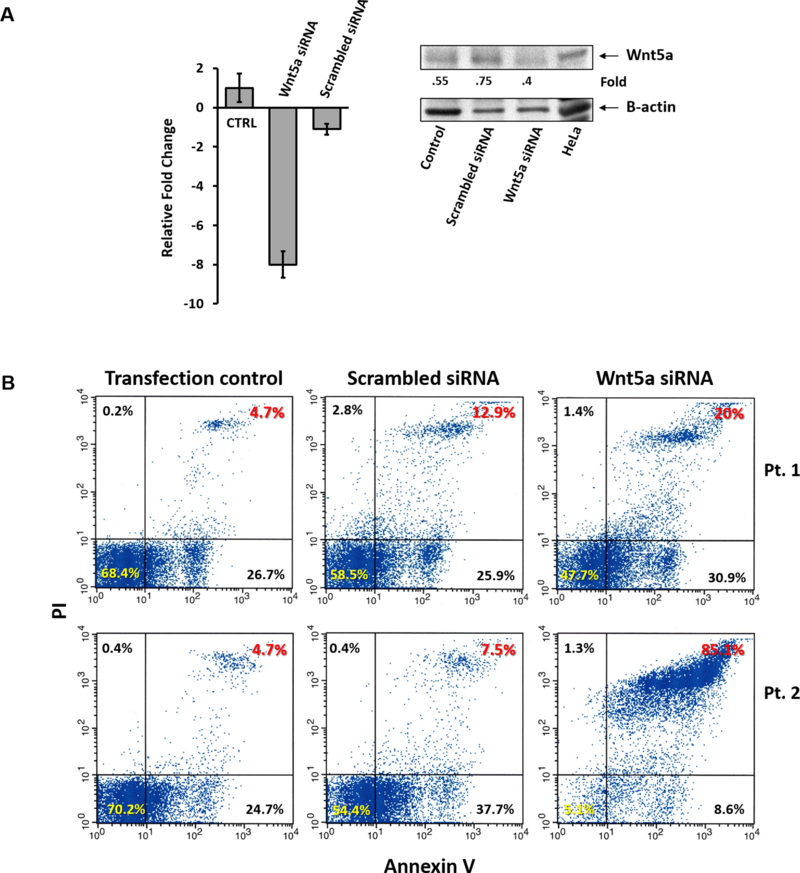
Because ROR1 protects CLL cells from apoptosis (3, 18) and CLL cells express both ROR1 and its ligand Wnt5a, we wondered how knockdown of Wnt5a would affect the survival of CLL cells. To answer this question we transfected CLL cells with Wnt5a siRNA. We found that Wnt5a siRNA, but not scrambled siRNA, downregulated Wnt5a mRNA and protein levels (Figure 6A) and significantly increased the apoptosis rate of CLL cells (Figure 6B), suggesting that endogenously produced Wnt5a provides CLL cells with a survival advantage.
Figure 6.
Wnt5a provides CLL cells with a survival advantage. (A) Transfection of CLL cells with Wnt5a-siRNA, but not with scrambled siRNA (transfection efficiency 73%), induced a 8-fold reduction in Wnt5a RNA levels (left panel) (P <.0001; Student’s t-test) and significantly downregulated Wnt5a protein levels (right panel). (B) Flow cytometry of Annexin/PI-stained cells was used to assess the effect of Wnt5a expression on CLL cell spontaneous apoptosis rate. As shown, CLL cells transfected with Wnt5a-siRNA had a significantly higher spontaneous apoptosis rate than that of CLL cells transfected with scrambled-siRNA or untransfected cells incubated in the transfection reagents alone (transfection control). In one patient (Pt. 1; upper panel) the transfection efficiency was 73% wereas in Pt. 2 (lower panel) it was 88%. The rate of cells in active early and late apoptosis (right lower and upper corner) was 38.8% in CLL cells transfected with scrambled-siRNAand 50.9% in CLL cells transfected with Wnt5a-siRNA in Pt. 1, and 45.2% in CLL cells transfected with scrambled-siRNA and 93.7% in CLL cells transfected with scrambled-siRNA in Pt.2.
Discussion
Here, we show that both ROR1 and its ligand Wnt5a are co-expressed on the surface of CLL cells, that the interaction between Wnt5a and ROR1 provides CLL cells with survival advantage, and that the expression of Wnt5a is driven by the transcriptional activity of STAT3.
Several types of neoplastic cells secrete soluble ligands that bind to their corresponding cell surface receptor and activate signaling in an autocrine fashion (19). For example, in breast cancer cells autocrine activation of the canonical WNT signaling pathway by secreted Frizzled-related protein-1 induces β-catenin-driven transcriptional activity of genes that activate cell proliferation (20). Similarly, in glioblastoma enodgenous secreted platelet-derived growth factors (PDGF) bind to and activate PDGF receptors-α and -β in an autocrine manner (21). Our data suggest that in CLL cells endogenously produced Wnt5a activates endogenous ROR1.
We found that in CLL cells STAT3 induces the expression of both ROR1 (3) and its ligand Wnt5a and that the ROR1 ligand Wnt5a provides the cells with survival advantage. Because ROR1 and Wnt5a are co-expressed on the surface of CLL cells, Wnt5a might stimulate ROR1 in an autocrine, paracrine or acrine manner through attachment to adjacent CLL cells. Indeed, overexpression of Wnt5a was found to increase cell motility (9) and high Wnt5a levels were shown to correlate with unfavorable prognostic indicators such as mutated TP53, SF3B1 or unmutated IgHV, and with short time to first treatment (9).
We have recently found that CLL cells harbor a unique protein complex that enables serine creatine kinase 2 to phosphorylate STAT3 on serine 727 residues (22). As a result, constitutively phosphorylated STAT3 induces STAT3-driven transcriptional activity in unstimulated CLL cells (10). STAT3 activates a metabolic program which provides the cells with adequate means to meet their metabolic requirements (22), regulate translational activity (12, 23) and induce STAT3-target gene expression (3, 24). We have previously shown that STAT3 induces aberrant expression of ROR1 in CLL cells (3). Here we show that STAT3 binds to the Wnt5a gene promoter and transcription. By driving the expression of both the ROR1 receptor and its ligand Wnt5a, STAT3 activates a potent autonomous pro-survival signaling circuit. Inhibition of the expression of either ROR1 or Wnt5a induced marked increase in the spontaneous apoptosis rate of CLL cells, suggesting that this circuit provides CLL cells with survival advantage.
Remarkably, we found an inverse correlation between ROR1 and Wnt5a protein levels in CLL cells. CLL cells with high ROR1 levels expressed low levels of Wnt5a and cells with low ROR1 levels express high levels of Wnt5a. Although ROR1 levels correlate with poor clinical outcome (16) and inhibition of ROR1 might be most beneficial in these patients, our data suggest that inhibiting the interaction between ROR1 and its ligand Wnt5a might be therapeutically beneficial even in CLL patients in whom the neoplastic cells express low levels of ROR1.
Whereas circulating CLL cells are quiescent, CLL cell proliferate and progressively expand in lymph nodes, bone marrow and spleen (25). We demonstrated that endogenously produced Wnt5a protects CLL from apoptosis, an effect that is further enhanced by CLL cell-to-cell interaction and/or direct attachment with mesenchymal stromal cells (26), endothelial cells (27, 28) and monocyte derived cells (29). Yet, circulating CLL cells detached from their microenvironment, benefit from both endogenously produced Wnt5a and ROR1 that enact autonomous pro-survival pathways that enable CLL cells to survive even in the presence of unfavorable environmental pressures. Therefore, inhibiting the interaction between Wnt5a and ROR1 might prove to be beneficial in patients with CLL.
Supplementary Material
Key Points.
STAT3 induces the expression of Wnt5a in CLL cells
Wnt5a provides CLL cells with survival advantage
Acknowledgments
Financial support
This study was supported by a grant from the CLL Global Research Foundation and by the Cancer Center Support Grant from the NIH/NCI, P30 CA016672.
Footnotes
Conflicts of interest
All authors report no conflict of interest.
References
- 1.Borcherding N, Kusner D, Liu GH, and Zhang W 2014. ROR1, an embryonic protein with an emerging role in cancer biology. Protein Cell 5: 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daneshmanesh AH, Mikaelsson E, Jeddi-Tehrani M, Bayat AA, Ghods R, Ostadkarampour M, Akhondi M, Lagercrantz S, Larsson C, Osterborg A, Shokri F, Mellstedt H, and Rabbani H 2008. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer 123: 1190–1195. [DOI] [PubMed] [Google Scholar]
- 3.Li P, Harris D, Liu Z, Liu J, Keating M, and Estrov Z 2010. Stat3 activates the receptor tyrosine kinase like orphan receptor-1 gene in chronic lymphocytic leukemia cells. PLoS One 5: e11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasan MK, Yu J, Widhopf GF 2nd, Rassenti LZ, Chen L, Shen Z, Briggs SP, Neuberg DS, and Kipps TJ 2018. Wnt5a induces ROR1 to recruit DOCK2 to activate Rac1/2 in chronic lymphocytic leukemia. Blood 132: 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi A, Yamamoto H, Sato A, and Matsumoto S 2012. Wnt5a: its signalling, functions and implication in diseases. Acta Physiol (Oxf) 204: 17–33. [DOI] [PubMed] [Google Scholar]
- 6.Baksh D, Boland GM, and Tuan RS 2007. Cross-talk between Wnt signaling pathways in human mesenchymal stem cells leads to functional antagonism during osteogenic differentiation. Journal of cellular biochemistry 101: 1109–1124. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Rubin B, Bodine PV, and Billiard J 2008. Wnt5a induces homodimerization and activation of Ror2 receptor tyrosine kinase. Journal of cellular biochemistry 105: 497–502. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Chen L, Cui B, Widhopf GF 2nd, Shen Z, Wu R, Zhang L, Zhang S, Briggs SP, and Kipps TJ 2016. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. The Journal of clinical investigation 126: 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janovska P, Poppova L, Plevova K, Plesingerova H, Behal M, Kaucka M, Ovesna P, Hlozkova M, Borsky M, Stehlikova O, Brychtova Y, Doubek M, Machalova M, Baskar S, Kozubik A, Pospisilova S, Pavlova S, and Bryja V 2016. Autocrine Signaling by Wnt-5a Deregulates Chemotaxis of Leukemic Cells and Predicts Clinical Outcome in Chronic Lymphocytic Leukemia. Clin Cancer Res 22: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, Shanker S, Ferrajoli A, Keating MJ, and Estrov Z 2010. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood 115: 2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozovski U, Grgurevic S, Bueso-Ramos C, Harris DM, Li P, Liu Z, Wu JY, Jain P, Wierda W, Burger J, O’Brien S, Jain N, Ferrajoli A, Keating MJ, and Estrov Z 2015. Aberrant LPL Expression, Driven by STAT3, Mediates Free Fatty Acid Metabolism in CLL Cells. Mol Cancer Res 13: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozovski U, Calin GA, Setoyama T, D’Abundo L, Harris DM, Li P, Liu Z, Grgurevic S, Ferrajoli A, Faderl S, Burger JA, O’Brien S, Wierda WG, Keating MJ, and Estrov Z 2013. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol Cancer 12: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Hazan-Halevy I, Harris DM, Li P, Ferrajoli A, Faderl S, Keating MJ, and Estrov Z 2011. STAT-3 activates NF-kappaB in chronic lymphocytic leukemia cells. Mol Cancer Res 9: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh M, and Katoh M 2007. STAT3-induced WNT5A signaling loop in embryonic stem cells, adult normal tissues, chronic persistent inflammation, rheumatoid arthritis and cancer (Review). International journal of molecular medicine 19: 273–278. [PubMed] [Google Scholar]
- 15.Liu D, Huang Y, Zeng J, Chen B, Huang N, Guo N, Liu L, Xu H, Mo X, and Li W 2011. Down-regulation of JAK1 by RNA interference inhibits growth of the lung cancer cell line A549 and interferes with the PI3K/mTOR pathway. Journal of cancer research and clinical oncology 137: 1629–1640. [DOI] [PubMed] [Google Scholar]
- 16.Cui B, Ghia EM, Chen L, Rassenti LZ, DeBoever C, Widhopf GF 2nd, Yu J, Neuberg DS, Wierda WG, Rai KR, Kay NE, Brown JR, Jones JA, Gribben JG, Frazer KA, and Kipps TJ 2016. High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia. Blood 128: 2931–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker T, Kovarik P, and Meinke A 1997. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res 17: 121–134. [DOI] [PubMed] [Google Scholar]
- 18.Hojjat-Farsangi M, Khan AS, Daneshmanesh AH, Moshfegh A, Sandin A, Mansouri L, Palma M, Lundin J, Osterborg A, and Mellstedt H 2013. The tyrosine kinase receptor ROR1 is constitutively phosphorylated in chronic lymphocytic leukemia (CLL) cells. PLoS One 8: e78339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sporn MB, and Roberts AB 1985. Autocrine growth factors and cancer. Nature 313: 745–747. [DOI] [PubMed] [Google Scholar]
- 20.Schlange T, Matsuda Y, Lienhard S, Huber A, and Hynes NE 2007. Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res 9: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarenko I, Hede SM, He X, Hedren A, Thompson J, Lindstrom MS, and Nister M 2012. PDGF and PDGF receptors in glioma. Upsala journal of medical sciences 117: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozovski U, Harris DM, Li P, Liu Z, Jain P, Veletic I, Ferrajoli A, Burger J, O’Brien S, Bose P, Thompson P, Jain N, Wierda W, Keating MJ, and Estrov Z 2017. Constitutive Phosphorylation of STAT3 by the CK2-BLNK-CD5 Complex. Mol Cancer Res 15: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, Ikuo M, Ghosh AK, Lerner S, Rassenti LZ, Xiao L, Hu J, Reuben JM, Calin S, You MJ, Manning JT, Wierda WG, Estrov Z, O’Brien S, Kipps TJ, Keating MJ, Kay NE, and Calin GA 2013. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 122: 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozovski U, Harris DM, Li P, Liu Z, Jain P, Ferrajoli A, Burger J, Thompson P, Jain N, Wierda W, Keating MJ, and Estrov Z 2018. STAT3-activated CD36 facilitates fatty acid uptake in chronic lymphocytic leukemia cells. Oncotarget 9: 21268–21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koczula KM, Ludwig C, Hayden R, Cronin L, Pratt G, Parry H, Tennant D, Drayson M, Bunce CM, Khanim FL, and Gunther UL 2016. Metabolic plasticity in CLL: adaptation to the hypoxic niche. Leukemia 30: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trimarco V, Ave E, Facco M, Chiodin G, Frezzato F, Martini V, Gattazzo C, Lessi F, Giorgi CA, Visentin A, Castelli M, Severin F, Zambello R, Piazza F, Semenzato G, and Trentin L 2015. Cross-talk between chronic lymphocytic leukemia (CLL) tumor B cells and mesenchymal stromal cells (MSCs): implications for neoplastic cell survival. Oncotarget 6: 42130–42149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng CW, Smith SK, and Charnock-Jones DS 2003. Wnt-1 signaling inhibits human umbilical vein endothelial cell proliferation and alters cell morphology. Exp Cell Res 291: 415–425. [DOI] [PubMed] [Google Scholar]
- 28.Wright M, Aikawa M, Szeto W, and Papkoff J 1999. Identification of a Wnt-responsive signal transduction pathway in primary endothelial cells. Biochem Biophys Res Commun 263: 384–388. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Wu T, Shao S, Shi B, and Zhao Y 2016. Phenotype, development, and biological function of myeloid-derived suppressor cells. Oncoimmunology 5: e1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.