Abstract
Efforts to construct synthetic biological circuits with more complex functions have often been hindered by the idiosyncratic behavior, limited dynamic range, and crosstalk of commonly utilized parts. Here, we employ de novo RNA design to develop two high-performance translational repressors with sensing and logic capabilities. These synthetic riboregulators, termed toehold repressors and three-way junction (3WJ) repressors, detect transcripts with nearly arbitrary sequences, repress gene expression by up to 300-fold, and yield orthogonal sets of up to 15 devices. Automated forward engineering is used to improve toehold repressor dynamic range and SHAPE-Seq is applied to confirm the designed switching mechanism of 3WJ repressors in living cells. We integrate the modular repressors into biological circuits that execute universal NAND and NOR logic and evaluate the four-input expression NOT ((A1 AND A2) OR (B1 AND B2)) in Escherichia coli. These capabilities make toehold and 3WJ repressors valuable new tools for biotechnological applications.
INTRODUCTION
Synthetic biology aims to provide an engineering-driven approach towards building biological entities with complex and novel functionality using well-characterized modular parts. Precise and programmable control of gene expression is becoming increasingly important for many synthetic biology applications as well as biotechnology in general. The ability to control the expression of multiple genes, for instance, will aid in the optimization of biosynthetic pathways for industrial chemical production while maximizing productivity and minimizing host toxicity1. Over the years, synthetic biology approaches have yielded increasingly sophisticated means of controlling gene expression including synchronized oscillators2,3, logic gates4–7, memory devices8,9, analog signal processors10,11, and state machines12. However, many of the regulatory elements in previous work overlap or are incompatible with each other, thereby limiting integration of such diverse components towards more complex circuits.
A basic requirement for engineering complex systems is a large repertoire of regulators that are modular, programmable, homogeneous, predictable, and easy to compose. The idiosyncratic nature of many protein regulators presents challenges for their use in circuits, but is being addressed using insulation, computer-aided design strategies, and orthogonal proteins and enzymes13–16. RNA molecules provide an alternative to proteins for constructing genetic circuits with excellent programmability and composability due to their predictable base-pairing rules and well-characterized thermodynamics. An assortment of RNA-based regulators have been developed using interaction strategies from nature with the hok/sok, pT181, and IS10 antisense systems being used for translational activation13,14, transcriptional attenuation17,18, and translational repression19, respectively.
To expand the potential of RNA-based regulation, we have recently harnessed de novo RNA design to develop toehold switch riboregulators20 that detect trigger RNAs with virtually arbitrary sequences to activate translation. Sequence design coupled with toehold-mediated strand-displacement reactions21 enabled the toehold switches to provide substantially improved, protein-like performance20. Moreover, the modular and programmable toehold switch design facilitated their integration into multi-input logic-processing ribocomputing devices22 and deployment in diagnostic systems23–26. At the same time, similar RNA design strategies have also generated high-performance transcriptional riboregulators27–29. Despite these developments, riboregulators that provide translational repression with protein-like dynamic range and low crosstalk are currently lacking. Recent work has highlighted the importance of robust universal logic gates such as NAND and NOR as building blocks for constructing complex genetic circuitry13,14,30. A crucial factor in the development of these systems has been the generation of large libraries of repressors that carry out NOT logic by repressing gene expression31,32. Accordingly, the development of libraries of high-performance RNA-based repressors could enable more efficient and complex forms of biomolecular logic.
Here we describe two de-novo-designed RNA-based repressors termed toehold and three-way junction (3WJ) repressors that employ toehold-mediated interactions to achieve effective translational inhibition. Both repressors strongly repress translation in response to trigger RNAs with nearly arbitrary sequences and can decrease gene expression in excess of 100-fold, a substantial improvement over previous RNA-based translational repressors19 and comparable to protein repressors31. Thermodynamics-based forward engineering is used to enhance the performance of the toehold repressors. In-cell SHAPE-Seq is used to directly confirm the formation of three-way junction structures in the 3WJ repressors. Validated repressors are integrated into ribocomputing devices to achieve NOR and NAND logic with up to four sequence-independent input RNAs, providing universal building blocks for logical computation.
RESULTS
Design of Synthetic Translational Repressors
Previously, we developed toehold switches that inhibit translation using a hairpin secondary structure that sequesters the RBS and start codon within a hairpin loop and stem, respectively (Figure 1a). A single-stranded toehold domain a* at the 5′ end of the switch RNA hairpin provides the initial binding site for a single-stranded trigger RNA strand, which has a complementary domain a. Upon binding of the cognate trigger molecule to the switch hairpin and completion of a toehold-mediated branch migration process, the RBS and start codon are available for ribosome binding and translation of the downstream gene. The lack of sequence constraints in designing trigger RNA molecules greatly expanded the orthogonality of toehold switches and the use of thermodynamically and kinetically favorable toehold-mediated interactions provided wide dynamic range20.
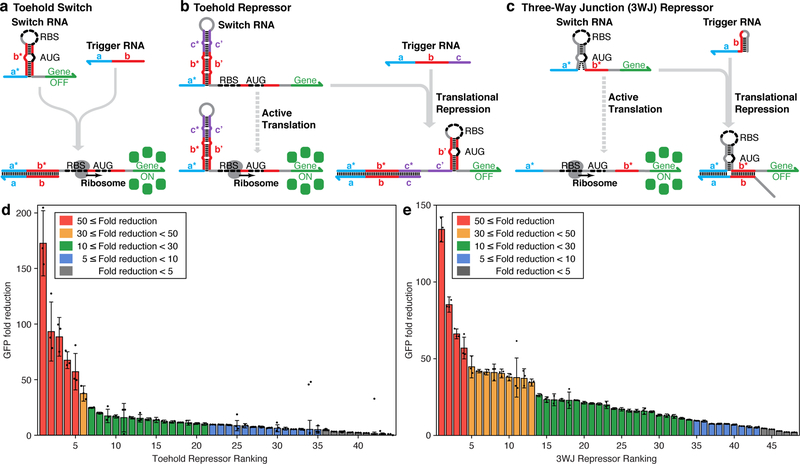
Figure 1 |. Operating mechanisms of de-novo-designed repressors and in vivo characterization.
a, Toehold switches repress translation through base pairs before and after the start codon (AUG). Interactions are initiated via a single-stranded toehold domain a* in the switch RNA that binds to a complementary a domain on the trigger RNA. A branch migration through domain b frees the ribosomal binding site (RBS) and start codon to allow translation. b, Toehold repressors harbor a strong hairpin structure upstream of an exposed RBS and start codon for translation of a downstream gene. The toehold domain a* of the switch binds to the trigger, initiating a branch migration that opens the strong hairpin stem. Newly freed domains form a hairpin structure that represses translation. c, The switch RNA of a three-way junction (3WJ) repressor contains an unstable hairpin structure that allows ribosomal access to the RBS and start codon and two single-stranded domains a* and b* for trigger binding. The trigger RNA employs a toehold to bind to the switch RNA and establish a three-way junction structure that prevents ribosomal access to the RBS and start codon. d, Fold reduction of GFP fluorescence levels obtained 3 hr after induction for 44 first-generation toehold repressors. e, Fold reduction of GFP fluorescence levels obtained 3 hr after induction for 48 3WJ repressors. Fold reduction is the ratio of the arithmetic mean of the GFP fluorescence level for the ON and OFF translation states. The relative errors for the ON and OFF states are from the SD of n=3 biologically independent samples. Relative errors for GFP fold reduction were obtained by adding the relative errors of the repressor ON- and OFF-state fluorescence measurements in quadrature. Individual points show the fold reduction from n=3 pairs of biologically independent samples.
We sought to obtain a library of programmable, wide dynamic range translational repressors analogous to the toehold switches and devised two types of repressors inspired by the design principles of these earlier riboregulators. The first repressor employs a switch RNA with a 5’ toehold domain and is referred to as a toehold repressor (Figure 1b, see Online Methods and Supplementary Figure 1a for details). The 15-nt toehold domain of the switch RNA is followed by a hairpin structure and a single-stranded expression region containing an RBS, start codon, and the coding sequence of the output gene. Without the trigger RNA, the exposed RBS and start codon enable active translation of the output gene. The trigger RNA of the toehold repressors is a 45-nt single-stranded RNA sequence that is complementary to the toehold and stem of the switch RNA. After binding of the trigger to the switch RNA toehold, the ensuing branch migration process unwinds the hairpin stem and releases the domains b′ and c′. Domain b′ is complementary to the sequences upstream and downstream of the start codon, and thus forms a hairpin structure with these domains. This newly formed hairpin recapitulates the repressed structure of the toehold switch and thus represses translation upon trigger binding. The toehold repressor trigger sequence does not possess bases complementary to the RBS or the start codon, which allows arbitrary choice of potential trigger sequences. If a trigger RNA sequence leads to in-frame stop codons in the expression region, bulges can be introduced or shifted in the b′ domain of the switch RNA hairpin to compensate.
The second repressor adopts a three-way junction structure to suppress translation and is referred to as a three-way junction (3WJ) repressor (Figure 1c, see Online Methods and Supplementary Figure 1b for details). Here, the switch RNA employs an unstable hairpin secondary structure that contains an RBS in the loop region and a start codon in the stem region. Despite its high secondary structure, this unstable hairpin was previously demonstrated to be translationally active in toehold switch mRNA sensors24. On either side of the unstable hairpin are single-stranded domains a* and b*. We hypothesized that transient formation of the bottom stem domain of the hairpin would co-localize these two domains to provide an effective binding site for a complementary trigger RNA. To take advantage of this design feature and improve repressor orthogonality, we designed cognate triggers where domain b is mostly contained in a hairpin secondary structure and a toehold composed of domain a and part of domain b is located at the 3’ end. When the trigger RNA is expressed, the toehold binds to the a* domain and part of the b* domain of the switch RNA. The switch RNA b* domain then completes a branch migration to unwind the trigger RNA stem. The resulting trigger-switch complex has a stable three-way junction structure that effectively sequesters the RBS and start codon within the loop and stem of the switch RNA, respectively, and strongly represses translation. Despite the use of a trigger with a hairpin structure to improve device orthogonality, the 3WJ repressors can also detect nearly arbitrary trigger RNAs provided that the trigger RNA sequence does not lead to an in-frame stop codon in domain b*.
In Silico Design of Repressors and In Vivo Validation
We generated libraries of both translational repressors de novo using the NUPACK sequence design package33 (see Online Methods for details). A total of 44 toehold repressors and 48 3WJ repressors were designed and validated in vivo (see Supplementary Tables 1–3 for sequence information). Members of the toehold repressor library were selected to reduce the potential for a non-cognate trigger RNA to disrupt the switch RNA stem. Members of the 3WJ repressor library were selected to minimize the potential for the non-cognate trigger RNAs to interact with the switch RNA. The E. coli BL21 Star DE3 strain with an IPTG-inducible genomic T7 RNA polymerase and decreased RNase activity was used for repressor characterization. A medium-copy plasmid containing the switch RNA regulating GFP and a high-copy plasmid encoding the trigger RNA were transformed into E. coli. For measurements in the absence of a cognate trigger, a non-cognate RNA strand with high secondary structure was transcribed from the high-copy plasmid.
Figure 1d shows the fold reduction of GFP fluorescence observed for the toehold repressor library. The GFP fold reduction was measured from the geometric mean fluorescence of GFP obtained from flow cytometry for cells in the ON state expressing the non-cognate trigger RNA and in the repressed OFF state expressing a cognate trigger RNA (see Supplementary Figure 2a for ON- and OFF-state GFP expression levels). Cell autofluorescence was not subtracted from either the ON- or OFF-state fluorescence for determination of the GFP fold reduction. Although the toehold repressor devices show wide variations in performance, 48% or 21 out of 44 provide at least a 10-fold change in gene expression upon detection of the trigger RNA. Five devices or 11% exhibit GFP fold reduction of at least 50-fold, corresponding to over 98% repression of GFP signal. The 3WJ repressors overall provided improved performance compared to the toehold repressors (Figure 1e). A substantially higher fraction of these devices at 71% or 34 out of 48 provided at least 10-fold reduction of GFP expression, while a smaller fraction (8% or 4 out of 48) yielded exceptionally high 50-fold or more reduction in GFP (see Supplementary Figure 2b for ON- and OFF-state GFP expression levels).
We also tested the repressors in a variety of different conditions to determine their effects on performance (see Online Methods for details). Although library screening was conducted in BL21 Star DE3, an RNase-deficient strain, we found that both types of repressors provided greater than 20-fold reduction of GFP in E. coli BL21 DE3 cells with wild-type RNase levels (Supplementary Figure 3). We observed that significant repression occurs within an hour of induction and that the fold reduction of both repressors increased over time (Supplementary Figure 4). The repressors also functioned well when transcribed using the endogenous E. coli RNA polymerase with inducible promoters in E. coli MG1655/Marionette-Wild34 and provided stronger repression as the trigger RNA concentration increased (see Supplementary Figure 5 and Supplementary Table 4 for sequence information). In cell-free in vitro translation reactions, the systems achieved 10-fold reductions in expression when supplied with as low as a two-fold excess of the trigger RNA over the switch RNA (Supplementary Figure 6). Variant 3WJ repressors designed with different stem sequences also operated successfully in E. coli when their secondary structures were sufficiently weak to allow translation to occur (see Supplementary Figure 7 and Supplementary Table 5 for sequence information). Finally, we found that the dynamic range of the 3WJ repressors could be increased by roughly an order of magnitude using a faster-degrading GFP variant or decreased 1.2- to 1.7-fold using a more stable GFP (Supplementary Figure 8).
Automated Forward Engineering of Toehold Repressors
To generate higher performance toehold repressors, we implemented an automated strategy for ranking putative riboregulator devices. We first compiled a set of 114 thermodynamic parameters that could be computed rapidly from the sequence information of the trigger and switch RNAs (see Supplementary Note and Supplementary Figures 9 and 10 for details). This set of thermodynamic parameters and experimental GFP fluorescence data from the toehold repressor library were then used in linear regressions to generate a scoring function for the devices (Supplementary Figure 10).
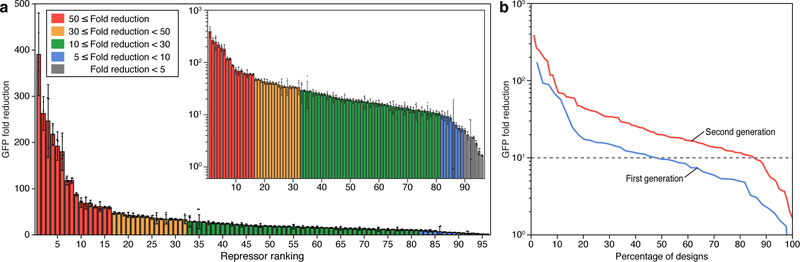
NUPACK was used to generate an additional set of 265 toehold repressor sequences using identical secondary structures and design parameters as the first-generation library. The scoring function was applied to rank each of the devices and select the top 96 for a second-generation library (see Supplementary Table 6 for sequence information). Figure 2a presents the fold reduction of GFP fluorescence for the second-generation toehold repressors (see Supplementary Figure 2c for ON- and OFF-state GFP expression levels). There is a dramatic increase in GFP fold reduction for the devices in general, with eight switches exhibiting a dynamic range greater than 100 and 81 switches exhibiting a dynamic range greater than 10. The second-generation systems exhibit an average GFP fold reduction of 40 compared to 20 for the first-generation library. High-performing toehold repressors exhibit fold changes rivaling the dynamic range of protein-based regulators31 without requiring any in vitro evolution or large-scale screening experiments. We quantified the effectiveness of our selection criteria by calculating the percentage of toehold repressors with GFP fold reductions exceeding a given minimal level (Figure 2b). The yield of high-performance devices is higher for the second-generation devices across all fold reductions.
Figure 2 |. Characterization of forward-engineered toehold repressors.
a, Fold reduction of GFP fluorescence levels obtained 3 hr after induction for 96 second-generation toehold repressors. Inset, GFP fold reduction on a logarithmic scale. Fold reduction is the ratio of the arithmetic mean of the GFP fluorescence level for the ON and OFF translation states. The relative errors for the ON and OFF states are from the SD of n=3 biologically independent samples. Relative errors for GFP fold reduction were obtained by adding the relative errors of the repressor ON- and OFF-state fluorescence measurements in quadrature. Individual points show the fold reduction from n=3 pairs of biologically independent samples. b, Percentage of first-generation and second-generation library components that had GFP fold reduction that exceeded the value defined on y-axis. The GFP fold reduction of 10 is marked by a gray dashed line.
SHAPE-Seq Measurements of 3WJ Repressor Structure
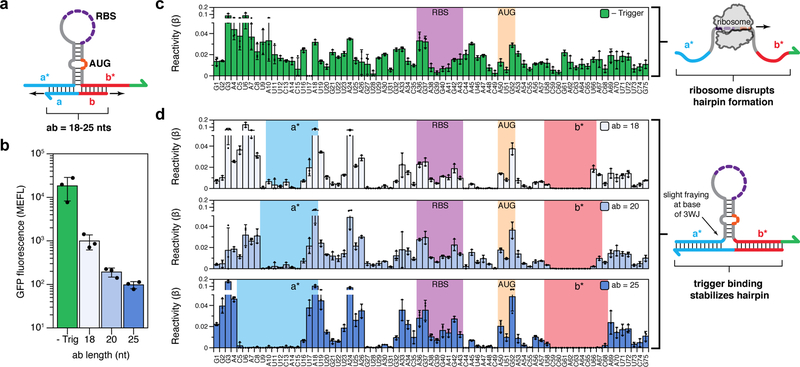
To better understand the operating mechanism of the synthetic repressors, we performed in-cell SHAPE-Seq35 on devices with varying repression efficiencies. The strong secondary structure of the toehold repressors, however, prevented interrogation by SHAPE-Seq. Fortunately, the weaker secondary structures of the 3WJ repressors enabled SHAPE-Seq studies for multiple trigger-switch interaction lengths. In the SHAPE-Seq experiment, 1-methyl-7-nitroisatoic anhydride (1M7) is introduced into the cell culture and used to covalently modify cellular RNAs at unstructured or unconstrained positions. These modifications can be detected by reverse transcription stops coupled to high-throughput sequencing, and the mapped modification positions can then be used to calculate a reactivity value (β) at each nucleotide. Higher reactivities correspond to flexible or unstructured nucleotides, and lower reactivities indicate constrained interactions such as base-pairing or stacking effects. Simultaneous measurements of GFP expression using the same cell cultures allow direct links to be drawn between the performance of repressor variants and their structures.
We studied a 3WJ repressor switch RNA and three trigger variants, with interaction lengths ranging from 18 to 25 nts (Figure 3a, see Supplementary Table 7 for sequence information). Functional characterization demonstrated active translation from the switch RNA and strong translational repression upon trigger expression (Figure 3b). SHAPE-Seq reactivity measurements of these variants showed remarkable agreement with the proposed in silico design strategy. Without the trigger RNA, we observed a trend of high reactivities across the switch RNA sequence (Figure 3c). This reactivity signature supports the design hypothesis that the switch hairpin is sufficiently weak to facilitate structural disruption by ribosomes, leading to active translation. A striking difference is seen when a trigger RNA is expressed (Figure 3d). Sharp drops in reactivity are observed precisely at the predicted binding sites of each trigger (a-a* in blue and b-b* in red), providing structural evidence of trigger binding across the junction. Moreover, drops in reactivity also occur within the stem of the switch RNA hairpin at regions predicted to form the hairpin structure, providing direct evidence that trigger binding leads to the formation of a stable, translationally inaccessible 3WJ structure. Interestingly, higher reactivities are observed at several positions around the base of the hairpin when the triggers are present (specifically U16-U19), suggesting slight fraying or flexibility at the base of the trigger-switch three-way junction. We also studied a second 3WJ repressor with different triggers, which also showed formation of the three-way junction structure upon repression (Supplementary Figure 11). To the best of our knowledge, these results represent the first structural confirmation of the regulatory mechanism of a completely de-novo-designed riboregulator.
Figure 3 |. In-cell SHAPE-Seq confirmation of the 3WJ repressor mechanism.
a, Design schematic for testing 3WJ repressor variants. A 3WJ repressor switch was characterized using in-cell SHAPE-Seq, either expressed alone or co-expressed with a trigger RNA. Several triggers were tested, varying in their designed binding length (ab) to either side of the switch hairpin. b, Functional characterization of switch plasmid expressed without trigger (green) and with triggers of increasing interaction length (blue). Strong repression is observed upon trigger binding, with longer triggers showing increased repression efficiency. c, In-cell SHAPE-Seq reactivity profile of the switch RNA expressed alone. A trend of high reactivities is observed across the molecule, consistent with the design hypothesis that the switch hairpin can be disrupted by ribosome binding, leading to active translation. d, In-cell SHAPE-Seq reactivity profiles of the switch co-expressed with trigger RNAs. Sharp drops in reactivity are observed at the predicted trigger binding sites (a-a* and b-b*) and within the switch hairpin, suggesting formation of a stable 3WJ structure when the trigger is bound. The RBS and start codon (AUG) positions are indicated. Fluorescence (b) and reactivity (c,d) values are the arithmetic mean of n=3 biologically independent samples. Error bars represent the SD from n=3 biologically independent samples and individual points are shown for the fluorescence and reactivity of each sample.
Evaluation of Repressor Orthogonality
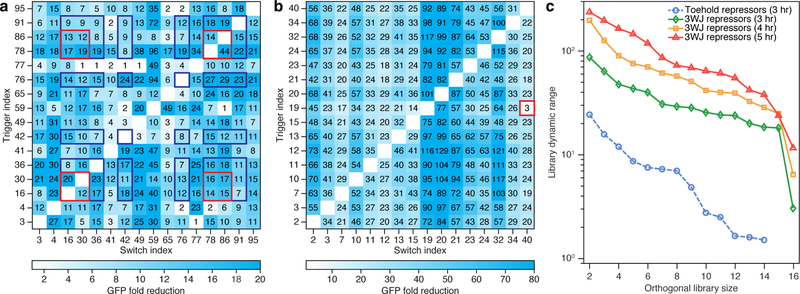
One of the prerequisites for higher-order logic processing is the orthogonality of regulatory components with respect to one another. We thus measured in vivo the interactions between pairwise combinations of different repressor trigger and switch RNAs. For the second-generation toehold repressors, we performed in silico screening to identify 16 devices that provided more than 10-fold GFP reduction and also displayed low levels of predicted crosstalk with non-cognate triggers. Flow cytometry was used to quantify GFP output in E. coli for all 256 trigger-switch interactions (Figure 4a). Crosstalk was calculated by dividing the GFP fluorescence obtained from a non-cognate trigger and a given switch RNA by the fluorescence of the switch in its triggered state. Typically, non-cognate trigger-switch pairs showed higher GFP output compared to cognate trigger-switch pairs as shown in Figure 4a. However, the crosstalk level was high in many instances such that the set of orthogonal devices that maintained at least 12-fold dynamic range was reduced to four devices (red boxes in Figure 4a). For the less stringent orthogonality condition of at least 7-fold dynamic range, the toehold repressor library yielded a set of eight independent riboregulators (blue boxes in Figure 4a).
Figure 4 |. Assessment of toehold and 3WJ repressor orthogonality.
a, Toehold repressor crosstalk measured by flow cytometry for 256 trigger-switch combinations 3 hr after induction. Red boxes designate a subset of four repressors that exhibit sufficiently low crosstalk to provide at least 12-fold GFP reduction. Blue boxes designate a subset of eight repressors that provide at least 7-fold GFP reduction. b, Three-way junction repressor crosstalk measured by flow cytometry for 256 trigger-switch combinations 3 hr after induction. All but one of the 240 non-cognate combinations provide at least 14-fold higher GFP expression than cognate pairs. The red box marks the trigger-switch combination with the most substantial crosstalk. c, Comparison of overall library dynamic range and orthogonal library size for the toehold repressors and 3WJ repressors. Three-way junction repressor orthogonal library size and dynamic range increase over the 3- to 5-hr induction time. Crosstalk was determined by dividing the arithmetic mean of the GFP fluorescence from a given trigger-switch pair by the arithmetic mean of the GFP fluorescence for the cognate trigger-switch interaction. GFP fluorescence was measured from n=3 biologically independent samples.
Based on the shorter exposed single-stranded regions of the 3WJ repressor trigger RNAs, we anticipated that these repressors would show improved orthogonality compared to the toehold repressors. We measured the pairwise trigger-switch interactions for 16 of the top devices using the same methods (Figure 4b). The 3WJ repressors showed substantially reduced crosstalk while maintaining strong repression of cognate trigger-switch pairs (see Supplementary Figure 12 for GFP expression levels). In fact, we found that a set of 15 3WJ repressors provided at least 17-fold reductions in GFP expression in the presence of the cognate trigger compared to any of the other 14 non-cognate triggers. Moreover, we only observed substantial crosstalk in a single pairwise interaction (red box in Figure 4b).
To quantify device orthogonality, we determined the maximum number of repressors that could be used to provide a given minimum level of overall dynamic range (Figure 4c, see Supplementary Table 8 for the orthogonal repressor sets). This analysis showed large improvements in orthogonal library size and dynamic range for the 3WJ repressors compared to the toehold repressors. For example, the most orthogonal eight-device toehold repressor set provided an overall dynamic range of at least 7-fold, while the corresponding eight-device 3WJ repressor set yielded an overall dynamic range of 29-fold. As the induction time increased, we also observed steady increases in the fold reduction of GFP in the cells, leading to parallel increases in device orthogonality for the 3WJ repressors. For instance, a set of six 3WJ repressors provided a remarkable library dynamic range of 118 at the five-hour time point. Based on the orthogonality of the repressors, we also investigated their ability to respond to intracellular mRNAs. These mRNA-sensing toehold and 3WJ repressors, which were designed to bind to regions of low secondary structure in the trigger mRNAs, successfully detected multiple antibiotic resistance genes using GFP and mCherry reporter proteins (Supplementary Figure 13, see Supplementary Table 9 for sequence information).
Two-Input Repressor-Based Logic Circuitry
The modular and programmable nature of the toehold and 3WJ repressors makes them ideal candidates for integration into ribocomputing devices for implementing sophisticated genetic programs. We have previously demonstrated that toehold switch riboregulators can be incorporated into such RNA-based computing systems for multi-input intracellular computation using RNA input signals and protein output signals22,36. We thus applied the ribocomputing strategy to the repressors to enable efficient computation of NAND and NOR logic functions in living cells (see Online Methods and Supplementary Figures 14 and 15 for circuit design details and Supplementary Table 10 for circuit sequence information).
We studied two-input NAND gates based on toehold repressors optimized for these logic operations. The trigger RNA sequence was divided into two input RNAs A1 and A2 and complementary bridging domains u and u* were appended to each input (Figure 5a and Supplementary Figure 14a). Only when both inputs A1 and A2 are present do they hybridize to one another through the u-u* interaction and bring both trigger halves into close proximity for binding to the gate RNA, which consists of a single switch RNA hairpin upstream of an output gene. Similar associative toehold mechanisms have been demonstrated in vitro37,38 and have been used for AND logic in ribocomputing devices22. Figure 5b shows the mean GFP measured from the two-input NAND circuit (see Supplementary Figure 16 for GFP population histograms of all ribocomputing circuits). We found that the dynamic range of the repressor-based ribocomputing devices increased with IPTG induction time up to ~6 hours (see Online Methods and Supplementary Figure 17). For the logical TRUE input conditions with at least one input missing, GFP output from the gate RNA remained high. When the logical FALSE condition occurred with two input RNAs expressed, the NAND gate provided strongly reduced GFP expression. Mean GFP fluorescence for the null input condition with no cognate input RNAs expressed was divided by the mean GFP fluorescence obtained from each of the input conditions to compute the fold reductions for the circuit (Figure 5c). GFP was reduced by 40-fold in the logical FALSE state. A noticeable decrease of 2.5-fold GFP reduction was observed upon expression of input A2 alone, potentially due to A2 binding causing a partial disruption of the gate RNA stem or cross-interactions between the u* bridging domain with exposed single-stranded regions of the gate.
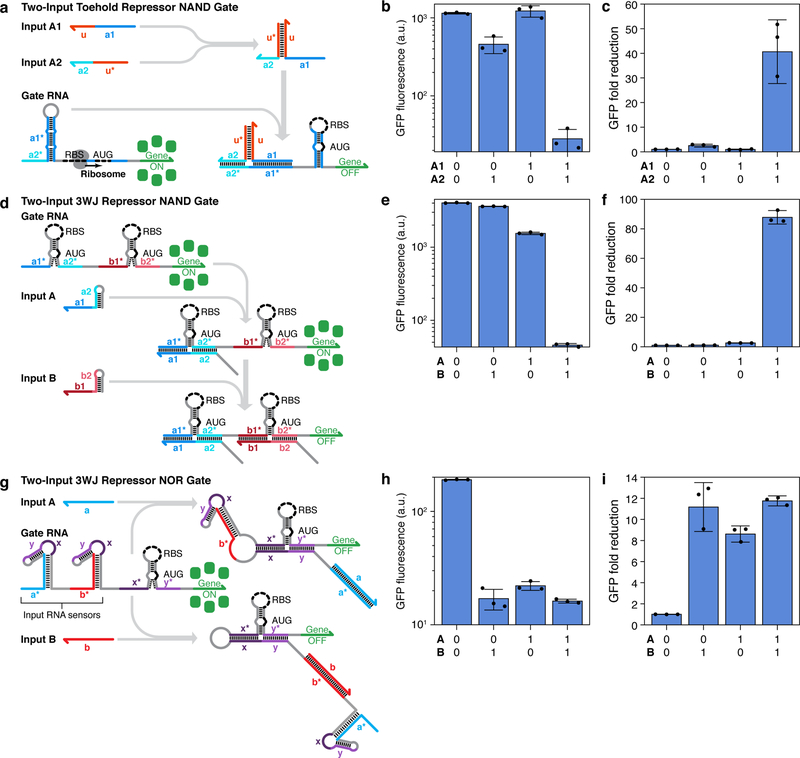
Figure 5 |. Two-input logic operations using repressor-based devices.
a, Design of a toehold repressor NAND gate where the input RNAs hybridize to form a complete trigger for repression. b, c, GFP fluorescence (b) and fold reduction (c) for the toehold repressor NAND device. d, Design of a 3WJ repressor NAND gate. In the gate RNA, two switch modules are inserted in-frame and upstream of the reporter gene and both input RNAs must bind to the gate to prevent gene expression. e, f, GFP fluorescence (e) and fold reduction (f) for the 3WJ repressor NAND device. g, Design of a 3WJ repressor NOR gate. The gate RNA contains a repressor module to regulate the output gene and two trigger modules sequestered within the loops of strong hairpin secondary structures that sense the input RNAs. Binding of either input RNA causes the corresponding hairpin to unwind, which releases the trigger to bind to and inhibit the 3WJ repressor module. h, i, GFP fluorescence (h) and fold reduction (i) for the 3WJ repressor NOR device. Devices were measured 6 hr after induction. GFP fluorescence values and their error bars are the arithmetic mean and SD, respectively, of n=3 biologically independent samples. Fold reductions for each device were calculated by dividing the GFP fluorescence value from the gate RNA obtained for the null input case by the GFP fluorescence value for each input combination. Relative errors for the fold reductions were obtained by adding the relative fluorescence errors in quadrature. Individual points show the fluorescence measured from n=3 biologically independent samples (b,e,h) or the fold reduction (c,f,i) from n=3 pairs of biologically independent samples.
We also implemented repressor-based gate RNAs integrating multiple repressor hairpin modules upstream of the output gene. Attempts using toehold repressor hairpins proved unsuccessful since their strong hairpin secondary structure and long target RNA binding sites both prevented efficient translation of the output gene by impeding translation from upstream RBS regions. However, the comparatively weak secondary structure of the 3WJ repressors and their short trigger RNA binding sites were ideal for incorporation into gate RNAs. We implemented a two-input NAND gate RNA composed of two orthogonal 3WJ repressor hairpins separated by a 17-nt single-stranded spacer domain (Figure 5d). With only one input RNA present, translation of the output gene will continue from the unrepressed hairpin module, since the ribosome can translate through weak hairpin secondary structures and duplexes formed by the input and gate RNAs. As a result, only simultaneous binding of both input RNAs to the gate RNA will fully inhibit gene expression. We evaluated the two-input NAND gate and found that GFP expression remained strong except for the logical FALSE case with both inputs expressed (Figure 5e). Small decreases in GFP expression were observed when only one of the input RNAs was present, likely as a result of inhibition of one of the two translation initiation sites from the gate RNA. The GFP fold reductions of the circuit show a large 88-fold decrease in expression in response to the two input RNAs compared to null input case (Figure 5f). This reduction was at least a factor of 33 higher than any of the changes in expression observed for single-input cases.
To implement NOR ribocomputing devices responsive to sequence-independent input RNAs, we developed a gate RNA architecture that exploited co-localized intramolecular interactions (Figure 5g and Supplementary Figure 15a). These NOR gate RNAs contain multiple sequestered trigger RNA sequences upstream of a 3WJ repressor module regulating the output gene. The trigger RNA domains x and y, which are complementary to the downstream repressor hairpin, are confined within the loops of strong hairpin secondary structures. These hairpins function as input RNA sensors that provide toehold domains for binding to complementary input RNA sequences. When an input RNA is expressed, binding to the input sensor leads to a branch migration that unwinds the sensor stem. This interaction releases the trigger RNA domain and enables the trigger to repress the downstream 3WJ repressor domain through an efficient gate RNA intramolecular interaction. We constructed the two-input NOR gate RNA using a validated 3WJ repressor and two input sensor hairpins, resulting in a gate RNA regulatory region of 312 nts. Measurements of GFP fluorescence from this gate RNA showed a substantial reduction in fluorescence upon expression of any of the cognate input RNAs (Figure 5h). Analysis of the GFP fold reductions from the circuit show between 8- to 12-fold decrease in GFP output in response to one or two input RNAs (Figure 5i).
Three- and Four-Input Repressor-Based Logic Circuitry
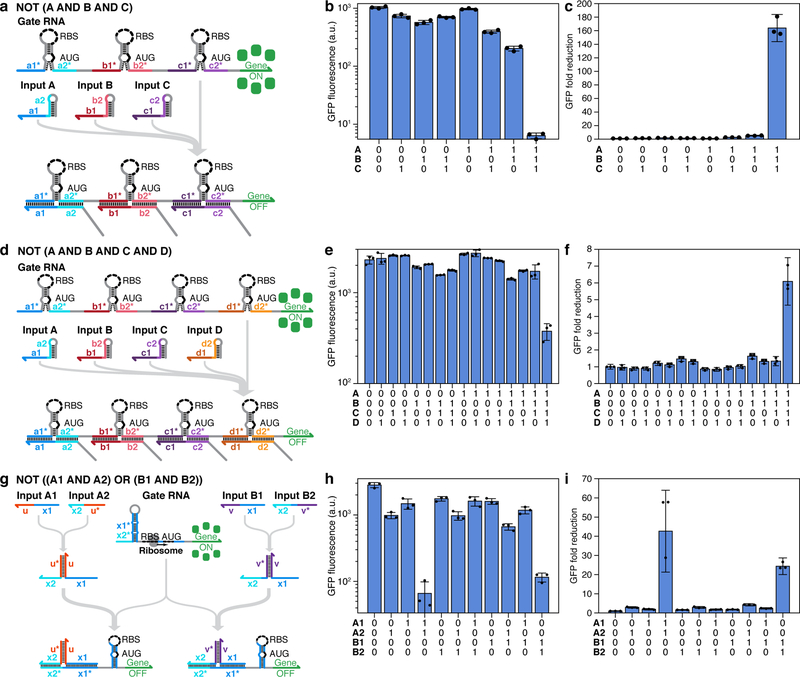
NAND gate RNAs based on 3WJ repressors were extended to three- and four-input operation by adding additional repressor modules upstream of the output gene. A three-input gate RNA for NOT (A AND B AND C) computations was constructed using three orthogonal 3WJ repressor hairpins separated by 11-nt single-stranded spacer domains (Figure 6a). This device showed high GFP expression for all logical TRUE conditions lacking at least one of the input RNAs (Figure 6b), while providing low expression for the logical FALSE condition with all inputs. The GFP fold reduction for this NAND circuit was 163-fold over the null input case and provided at least 33-fold lower GFP expression than the other input RNA combinations (Figure 6c). We constructed a four-input device for the expression NOT (A AND B AND C AND D) using four orthogonal 3WJ repressor modules and a regulatory region of 365 nts (Figure 6d and Supplementary Figure 15b). The GFP fluorescence for this device remained high for all logical TRUE conditions and decreased substantially when all input RNAs were expressed (Figure 6e). Calculation of the GFP fold changes for this circuit showed a 6-fold reduction in GFP expression in the sole logical FALSE state and provided at least a 3.7-fold reduction in GFP compared to all logical TRUE states (Figure 6f). We also tested multiple additional NAND gates with two to four inputs using combinations of six orthogonal 3WJ repressor modules. Overall, we found that 23 out of 25 gates operated successfully in E. coli (Supplementary Figures 18 and 19). Further studies also indicated that the performance of the NAND gates can be affected by the copy number of the plasmids used for the input RNAs, with a higher-copy plasmid in one case causing lower expression when targeting a 3WJ repressor downstream of another translation start site (see Supplementary Figure 20 and Online Methods for details on additional NAND gates).
Figure 6 |. Multi-input ribocomputing devices employing toehold and 3WJ repressors.
a, Design of a three-input 3WJ repressor NAND gate. The gate RNA contains three orthogonal 3WJ repressor hairpins in-frame and upstream of the output gene. b, c, GFP fluorescence (b) and fold reduction (c) for the three-input 3WJ repressor NAND device. d, Design of a four-input 3WJ repressor NAND gate. The gate RNA contains four orthogonal 3WJ repressor hairpins in-frame and upstream of the output gene. e, f, GFP fluorescence (e) and fold reduction (f) for the four-input 3WJ repressor NAND device. g, Design of a NOT ((A1 AND A2) OR (B1 AND B2)) logic circuit based on toehold repressors. Independent two-input NAND gate behavior is enabled by two partial-trigger domains x1 and x2 coupled with bridging domains u and u* for inputs A1 and A2 or with orthogonal bridging domains v and v* for inputs B1 and B2. The two partial-triggers present the full-length trigger for the toehold repressor. h, i, GFP fluorescence (h) and fold reduction (i) for the four-input ribocomputing device based on toehold repressors. Devices were measured 6 hr after induction. GFP fluorescence values and their error bars are the arithmetic mean and SD, respectively, of n=3 biologically independent samples. Fold reductions for each device were calculated by dividing the GFP fluorescence value from the gate RNA obtained for the null input case by the GFP fluorescence value for each input combination. Relative errors for the fold reductions were obtained by adding the relative fluorescence errors in quadrature. Individual points show the fluorescence measured from n=3 biologically independent samples (b,e,h) or the fold reduction (c,f,i) from n=3 pairs of biologically independent samples.
An additional four-input logic system was implemented using the toehold repressors. Starting from the A1-A2 NAND gate in Figure 5a–c, we designed a second pair of bridge domains v and v* and shifted the trigger splitting point 4 nts to generate new NAND inputs B1 and B2 (Figure 6g and Supplementary Figure 14b). The resulting ribocomputing device performed the computation NOT ((A1 AND A2) OR (B1 AND B2)) (Figure 6h–i). As expected, we observed substantial reductions in GFP expression only when A1 and A2 or B1 and B2 were expressed simultaneously. Further, only weak crosstalk was observed when the non-interacting A triggers and B triggers were tested in pairs. The crosstalk observed for the trigger A2 and B1 combination was at least 5-fold less than the cognate pair of triggers.
DISCUSSION
We have developed two types of high-performance translation-repressing riboregulators using de novo RNA sequence design. Toehold repressors exploit strong RNA secondary structures to provide very wide dynamic range of gene expression and are best suited for applications requiring tight translational control. Three-way junction repressors exhibit very low device crosstalk while using weaker RNA secondary structures and are optimal for multiplexed sensing and multi-input logic. While this work was being performed, an independent study uncovered similar designs for RNA-based translational repressors39. These designs exhibited weaker repression efficiency than the systems presented here, but they enabled targeting of endogenous mRNA transcripts.
The synthetic RNA-based repressors provide wide dynamic range comparable to protein-based transcriptional repressors. A previous 20-component protein repressor library yielded an average 51.3-fold reduction in reporter expression31. In comparison, the top 20 second-generation toehold and 3WJ repressors provided average GFP reductions of 122- and 43-fold, respectively. The 3WJ repressors also exhibited good orthogonality with 15 devices providing 17-fold dynamic range, while the toehold repressors had eight devices with 7-fold GFP reduction.
The toehold and 3WJ repressors were also incorporated into genetically compact ribocomputing devices that effectively computed NOT-related logic expressions with up to four different input RNAs. 3WJ repressor designs, in particular, were amenable to integration into long NAND gate RNAs to simultaneously detect multiple sequence-independent trigger RNAs and displayed excellent modularity with 92% of the 25 devices tested operating correctly. NOR gates based on 3WJ repressors that exploited intramolecular RNA interactions enabled two-input regulation without requiring translation through downstream hairpins, in contrast to previously reported OR gate systems22.
We also successfully applied in-cell SHAPE-Seq35 to simultaneously characterize RNA structure and function for the 3WJ repressors. Analysis of 3WJ repressors yielded the first direct structural evidence to support the mechanistic model of a de-novo-designed riboregulator and also revealed potential pitfalls in our design strategies. These results highlight how SHAPE-Seq can be used to confirm design principles and understand potential failure modes of synthetic riboregulators, which can be used to guide future design improvements.
Overall, the toehold and 3WJ repressors represent versatile new components for the rapidly expanding RNA synthetic biology toolkit. The development of these NOT, NAND, and NOR logic devices coupled with advances in RNA-guided CRISPR/Cas systems40,41, RNA-based transcriptional regulators17,27,28, and systems that merge these capabilities29,42–44 point to increasingly sophisticated forms of RNA-enabled genetic circuits that exploit regulation at the transcriptional, translational, and post-transcriptional levels to achieve more dynamic and programmable cellular functions.
ONLINE METHODS
Strains and growth conditions.
These E. coli strains were used in this study: BL21 Star DE3 (F− ompT hsdSB (rB−mB−) gal dcm rne131 (DE3); Invitrogen), BL21 DE3 (F− ompT hsdSB (rB−mB−) gal dcm (DE3); Invitrogen), E. coli MG1655/Marionette-Wild34, and DH5α (endA1 recA1 gyrA96 thi-1 glnV44 relA1 hsdR17 (rK−mK+) λ−). All strains were grown in LB medium at 37 °C with appropriate antibiotics: ampicillin (50 μg mL−1), spectinomycin (25 μg mL−1), and kanamycin (30 μg mL−1). E. coli MG1655/Marionette-Wild34 (sAJM.1506) was a gift from Christopher Voigt (Addgene bacterial strain # 108254).
Synthetic Repressor Computational Design.
Toehold repressors were designed to provide a 15-nt toehold region for trigger binding and refold into a repressing hairpin structure identical to that used in toehold switches (Supplementary Figure 1a). The repressed hairpin had a 12-nt loop and the top 3-bp of the stem was specified to contain only A-U base pairing, which was previously associated with high-performance toehold switches20. An additional 12-bp stem domain c* was used to ensure that the repressing hairpin structure would only form upon binding to the trigger RNA. A 4-nt single-stranded region (AAAC) was used upstream of the main ribosomal binding site (RBS) sequence (AGAGGAGA) to allow efficient translation of the output gene in the absence of the trigger. These design considerations resulted in a 30-nt long hairpin stem region for the switch RNA in its active translation state. Three bulges were included in this hairpin structure at 8-nt increments to discourage transcriptional termination through the strong secondary structure. As part of the RNA design, the switch RNA sequence was considered up to the 30 nts following the repressed hairpin structure, which included a 21-nt linker previously used for toehold switches and the first 9 nts of GFPmut3b (see Supplementary Table 1). Trigger RNAs for the toehold repressors were designed with a 5’ hairpin region to increase RNA stability followed by the c, b, and a domains responsible for binding to the switch RNA. The trigger RNA was also designed with the 47-nt T7 terminator sequence (see Supplementary Table 1). Three-nucleotide spacers were added between the interaction domains and the outer hairpins as part of the trigger design.
The 3WJ repressor switch RNAs were designed using the core sequence of first-generation toehold switch number 1.20 This core region is indicated by the gray and black bases within the hairpin structure shown in Supplementary Figure 1b and has the sequence:
5′-UUGUUAUAGUUAUGAACAGAGGAGACAUAACAUGAACAA-3′
where the RBS and start codon are shown in bold. This hairpin sequence provided very high translational output despite its secondary structure in previous studies20. The core translational element was integrated into the 3WJ repressor by appending binding domains a* and b* with lengths of 15 nts and 12 nts, respectively, to either side. A 7-nt single-stranded domain was added downstream of b* to preserve the correct reading frame followed by the 21-nt linker sequence and the first 9 nts of the GFPmut3b coding sequence (see Supplementary Table 1). The 3WJ repressor trigger RNAs featured a 17-nt toehold domain comprising the a domain and the last 2 nts of the b domain. The remaining 10 nts of the b domain were contained within an 8-bp stem structure and part of the 6-nt loop. A 3-nt single-stranded spacer region was used to separate the binding domains of the trigger RNA from the T7 terminator at the 3’ end of the transcript.
Designs for the repressor libraries were generated using the NUPACK complex design package33, which enabled the trigger, switch, and trigger-switch complex to be optimized simultaneously. The toehold repressors were designed using a specified temperature of 37 °C; Serra and Turner, 1995 energy parameters45; and the prevented sequences AAAA, CCCC, GGGG, UUUU, KKKKKKKKKK, MMMMMMMMMM, RRRRRRRRRR, SSSSSSSSSS, WWWWWWWWWW, YYYYYYYYYY. The 3WJ repressors used a specified temperature of 37 °C; Mathews et al., 1999 energy parameters46; and the prevented sequences AAAA, CCCC, GGGG, UUUU, KKKKKKKK, MMMMMM, RRRRRR, SSSSSS, WWWWWW, YYYYYY. The test tube capability of NUPACK was not used in the designs, nor was the pseudoknot setting used in any of the designs.
Toehold and 3WJ repressor library construction.
Plasmids were constructed using PCR and Gibson assembly. DNA templates for repressor switch and trigger RNA expression were assembled from single-stranded DNAs purchased from Integrated DNA Technologies. The synthetic DNA strands were amplified via PCR and then inserted into plasmid backbones using 30-bp homology domains via Gibson assembly47. All plasmids were cloned in the E. coli DH5α strain and validated through DNA sequencing. Backbones for the plasmids were taken from the commercial vectors pET15b (ampicillin resistance, ColE1 origin), pCOLADuet (kanamycin resistance, ColA origin), and pCDFDuet (spectinomycin resistance, CDF origin) from EMD Millipore, and the repressor DNA was inserted upstream of the T7 terminator sequence to replace their respective multiple cloning sites. GFPmut3b-ASV, GFPmut3b with an ASV degradation tag48, was used as the reporter for the repressor switch plasmids, except for experiments studying GFPs with different degradation tags. In addition, the kanR mRNA toehold repressor used an mCherry reporter without a degradation tag. Sequences of elements commonly used in the plasmids are provided in Supplementary Table 1.
Toehold and 3WJ repressor expression.
Toehold and 3WJ repressor switch and trigger RNAs were expressed using T7 RNA polymerase in BL21 Star DE3, an RNase-deficient strain, with the T7 RNA polymerase induced with the addition of isopropyl β–D-1-thiogalactopyranoside (IPTG). Selected sets of toehold and 3WJ repressor switch and trigger RNAs were also tested in BL21 DE3 strain with the T7 RNA polymerase induced with the addition of IPTG. For both strains, cells were grown overnight in 96-well plates with shaking at 900 rpm and 37 °C. Overnight cultures were then diluted by 100-fold into fresh LB media with antibiotics and returned to the shaker (900 rpm, 37 °C). After 80 minutes, both strains were induced with 0.1 mM IPTG and cells were returned to the shaker (900 rpm, 37 °C) until the flow cytometry measurements at specified times post-induction.
Flow cytometry measurements and analysis.
Flow cytometry measurements of toehold repressor libraries and their ribocomputing devices were performed using a BD LSRFortessa cell analyzer with a high-throughput sampler. Prior to loading to flow cytometer, cells were diluted by a factor of ~65 into phosphate-buffered saline. Cells were detected using a forward scatter (FSC) trigger and at least 30,000 cells were recorded for each measurement. Flow cytometry measurements of 3WJ repressor libraries and their ribocomputing devices were performed using a Stratedigm S1300EXi cell analyzer equipped with a A600 high-throughput autosampler. Cells with the 3WJ repressor systems were diluted by a factor of ~17 into phosphate-buffered saline and detected as described above with 40,000 cells recorded for each measurement. Cell populations were gated according to their FSC and side scatter (SSC) distributions as described previously20 (Supplementary Figure 21), and the GFP or mCherry fluorescence levels of these gated cells were used to measure circuit output via the geometric mean from at least three biological replicates. Fold reductions of GFP or mCherry fluorescence levels were then evaluated by taking the geometric mean fluorescence output of the toehold or 3WJ repressor switch with a non-cognate trigger and dividing it by the fluorescence output with a cognate trigger. Cellular autofluorescence was not subtracted prior to determining the fold reduction.
Evaluation of Repressors with Inducible Promoters.
Inducible expression was implemented using the 3OC6-HSL-inducible promoter PluxB to regulate trigger RNA transcription and the anhydrotetracycline (aTc)-inducible promoter Ptet* for the switch RNA34. Trigger and switch RNAs were expressed using separate plasmids. The trigger was on a high-copy plasmid with a ColE1 origin and ampicillin resistance. The switch was expressed using the 5’ insulating riboJ ribozyme49 from a medium-copy plasmid with a ColA origin and kanamycin resistance (see Supplementary Table 4 for sequence information). Previous experiments have shown that co-transformation of E. coli with both plasmids results in a 6- to 8-fold higher copy number for the trigger plasmids compared to the switch plasmid20. The two plasmids were transformed into E. coli MG1655/Marionette-Wild34. Overnight cultures of the transformed cells were then diluted 100-fold into fresh LB with antibiotics and incubated with shaking for 80 minutes at 37 °C. The cultures were then induced with varying concentrations of 3OC6-HSL and aTc and returned to shaking at 37 °C. Aliquots of the cells were then harvested at one-hour time increments to measure the expression of GFPmut3b-ASV via flow cytometry (Supplementary Figure 5).
Evaluation of Repressors in Cell-Free Systems.
Trigger and switch RNAs for toehold and 3WJ repressors were separately transcribed, quantitated, and added to transcription-translation reactions (PURExpress, NEB) at different concentrations. Time-course measurements were then conducted on the reactions using a plate reader (Biotek H1MF) to determine GFP expression (Supplementary Figure 6).
3WJ Repressors with Stem Sequence Variations.
Alternative 3WJ repressors with the same secondary structure but different RNA sequences were studied to determine the effect of sequence changes on 3WJ repressor performance. The variants all used the trigger RNA of 3WJ repressor index 20. The “NN” variants were generated by allowing any base (i.e. N bases) to be present within the stem region of the switch RNA (white bases in Supplementary Figure 7a). To generate weaker stems, “SW” variants were designed with a combination of Strong (i.e. G-C) base pairs and Weak (i.e. A-U) base pairs that matched those of the original hairpin sequence. Both types of designs were screened to ensure that they did not have any in-frame stop codons following the start codon in the stem.
Two versions of the NN design and three versions of the SW design were tested in E. coli BL21 Star DE3 (see Supplementary Table 5 and Supplementary Figure 7b for sequence information). Both NN designs were unable to modulate GFP expression and provided near-background GFP levels even in the absence of the trigger (Supplementary Figure 7c–d). Two out of the three SW designs (devices B and C) provided substantial GFP expression without the trigger, while expression from the third (device A) was nearly undetectable (Supplementary Figure 7c–d).
Repressors Using GFP with Different Degradation Tags.
To examine the effect of the degradation tag on riboregulator performance, 3WJ repressor systems were tested in E. coli BL21 Star DE3 using three different types of output GFPmut3b proteins: ASV-tagged (~110-minute half-life)48, LVA-tagged (~40-minute half-life)48, and untagged. The resulting repressor systems were tested using the same conditions and plasmid combinations as the repressor libraries and characterized using flow cytometry (Supplementary Figure 8).
SHAPE-Seq measurements and analysis.
In-cell SHAPE-Seq measurements were carried out as described by Watters et al.35 Briefly, 3WJ repressor variants were transformed into BL21 Star DE3 as in the functional characterization experiments. Overnight cultures were diluted 100-fold into 1.2 mL of fresh LB with antibiotics. Following IPTG induction and 5 hr additional subculture, 100 µL of culture was removed and diluted by ~100-fold for functional characterization using a BD Accuri cell analyzer with a high-throughput sampler. 500 µL of the remaining culture was then added to 13.3 µL of 250 mM 1M7 or 13.3 µL of DMSO (control solvent). Cells were returned to shaking for 3 minutes to allow 1M7 to react, then cellular RNAs were Trizol extracted and reverse transcribed using a custom reverse transcription primer specific for GFPmut3b (5’-CAACAAGAATTGGGACAACTCCAGTG-3’). Additional 5’ and 3’ sequencing adapters were then added. Following 2 × 35 bp paired-end Illumina sequencing, ß reactivities were calculated as described by Aviran et al.50 Error bars represent the standard deviation of three samples, each probed from a separate transformation on a separate day. Replicate samples were only processed in parallel during final sequencing.
Design and Testing of mRNA-Sensing Repressors.
mRNA-sensing toehold repressors were implemented by extending their toehold domains to 30 nts to compensate for the increased secondary structure of target mRNAs. In silico screening was then used to identify fragments along the target mRNA that provided the lowest secondary structure to facilitate repressor binding (see Supplementary Table 9 for sequence information). For the 3WJ repressors, the riboregulator design was left unchanged and target mRNA binding sites were selected by determining 27-nt regions having low secondary structure.
The mRNA-sensing repressors were validated against mRNAs encoding antibiotic resistance genes: the kanamycin resistance protein (kanR), beta-lactamase (bla) conferring resistance to the antibiotic ampicillin, and aadA conferring resistance to spectinomycin (Supplementary Figure 13). For the toehold repressors, a sensor was constructed to repress the translation of GFP output after binding to bla transcripts, while another sensor repressed mCherry expression after binding to kanR. For the 3WJ repressors, kanR and aadA sensors were developed to repress GFP output. The mRNA-sensing repressors were then tested using procedures employed for library validation and expression induced for 5 hr using IPTG. Flow cytometry was then used to assess the output based on fluorescent protein (FP) output with and without the trigger mRNAs expressed.
Design of Toehold Repressor Ribocomputing Devices.
A modified toehold repressor design was used in two-input NAND devices (Supplementary Figure 14). The toehold domain length was increased to 16 nts and the stem of the gate RNA was reduced to 24 nts, which enabled the trigger RNA sequence to be divided into two segments of similar length. Two different segment lengths were used in devices. Inputs A1 and A2 used a1 and a2 domains with lengths of 24 and 16 nts, respectively (Supplementary Figure 14a). Inputs B1 and B2 used b1 and b2 domains that were both 20 nts in length (Supplementary Figure 14b). The repressed fold of the gate RNA retained the same stem secondary structure but had a loop domain of 18 nts rather than 12 nts. The additional loop bases comprise the first 6 nts released upon toehold-mediated disruption of the gate RNA stem. The input RNAs were designed to hybridize through a u or v domain of 23 nts and form an RNA duplex with a single-nucleotide bulge at the midpoint.
Design of 3WJ Repressor Two-Input NOR Gate RNA.
The two-input NOR gate RNA was designed using two hairpin sensor modules upstream of a 3WJ repressor hairpin (Supplementary Figure 15a). Each sensor module consisted of a 15-nt toehold domain followed by a hairpin with an 18-bp stem. The loop region of this hairpin contained a sequestered internal trigger for the downstream repressor hairpin. The trigger sequence length was reduced by 5 nts within the x domain compared to that used for the original library characterization to reduce the probability of leakage from the internal trigger. The two sensor modules and the repressor hairpin were separated by 18-nt spacer sequences having small hairpin secondary structures. These spacers were used to ligate the three modules together through Gibson assembly during plasmid construction and their hairpin structures were used to reduce the effective distance between the intramolecular triggers and the downstream 3WJ repressor hairpin. Input RNAs complementary to the toehold and stem domain of the two sensor modules were also designed. These input RNAs had 33-nt binding domains to the gate RNA and were flanked by a 5’ hairpin structure and the T7 terminator sequence.
Design of 3WJ Repressor NAND Gate RNAs.
The NAND gate RNAs based on 3WJ repressors were generated by taking the core regulatory sequence of the repressors running from the 5’ end of the a* domain through to the nucleotide immediately before the 21-nt linker sequence. Since this core regulatory sequence had a length of 73 nts, spacers of 3n+2, where n is a non-negative integer, were used to connect different 3WJ repressors together. Spacers of this length enabled successive repressor modules to remain in-frame through the full length of the gate RNA. For testing purposes, 11- and 17-nt spacers were inserted between different 3WJ repressor hairpins. The spacers were designed using NUPACK to have single-stranded secondary structures when flanked by the two repressor hairpins. These spacers were then used to connect different 3WJ repressors together to form multi-input NAND gates (see Supplementary Figure 15b for the designed structure of a four-input NAND gate RNA).
Experimental testing of ribocomputing devices.
For device testing, the RNA inputs and the gate RNA were expressed from separate plasmids through the T7 promoter in BL21 Star DE3 cells. In cases where an input RNA was not present, a non-cognate input RNA was expressed in its place. Culturing and induction of the cells was performed in the same way as the repressor libraries.
Study of 3WJ Repressor NAND Gate Modularity.
Multiple versions of the two-, three-, and four-input 3WJ NAND gates were tested to assess their modularity (see Supplementary Table 10 for sequence information). NAND gates were designed from a parent library of six 3WJ repressor modules selected based on their fold reduction levels and their low crosstalk. The NAND gates were selected for testing based on their relatively low expected ensemble defect in NUPACK. GFP fluorescence and GFP fold reduction were obtained in E. coli BL21 Star DE3 for 16 different two-input NAND systems: eight with 11-nt spacers (Supplementary Figure 18a) and eight with 17-nt spacers (Supplementary Figure 18b). Six different three-input NAND gates and three different four-input NAND gates were generated from a smaller subset of four 3WJ repressor modules after elimination of index 10 and index 21 hairpins based on their lower performance in two-input NAND computations (Supplementary Figure 19a).
Supplementary Material
Acknowledgements
This work was supported by an NIH Director’s New Innovator Award (1DP2GM126892), an Alfred P. Sloan Research Fellowship (FG-2017-9108), Gates Foundation funds (OPP1160667), an Arizona Biomedical Research Commission New Investigator Award (ADHS16-162400), a DARPA Young Faculty Award (D17AP00026), Gordon and Betty Moore Foundation funds (#6984), NIH funds (1R21AI136571), and Arizona State University funds to A.A.G; an NIH Director’s Pioneer Award (1DP1GM133052-01), Office of Naval Research funds (N00014-16-1-2410), and NSF funds (CCF-1317291, MCB-1540214) to P.Y.; an NSF CAREER award to J. B.L. (1452441); Defense Threat Reduction Agency funds (HDTRA1-14-1-0006), Air Force Office of Scientific Research funds (FA9550-14-1-0060), and Paul G. Allen Frontiers Group funds to J.J.C; and BMBF funds (Erasynbio project UNACS - 031L0011) and DFG funds (SFB 10327TPA2) to F.C.S. J.K. acknowledges a Wyss Institute Director’s Cross-Platform Fellowship. The views, opinions, and/or findings contained in this article are those of the authors and should not be interpreted as representing the official views or policies, either expressed or implied, of DARPA or the Department of Defense.
Footnotes
Competing financial interests
US provisional patents have been filed by J.K., A.A.G., J.J.C., & P.Y. and Y.Z. & A.A.G. based on this work. P.Y. is the co-founder of Ultivue Inc. and NuProbe Global.
Data Availability Statements
The datasets generated during and/or analysed during the current study are available from the corresponding authors on reasonable request. Supplementary Tables are available from A.A.G. in spreadsheet format upon request. The following plasmids from the study are available from Addgene: pYZ_3WJrep_N20_switch 132722; pYZ_3WJrep_N19_switch 132723; pYZ_3WJrep_N10_switch 132724; pYZ_3WJrep_N24_switch 132725; pYZ_3WJrep_N20_trigger 132726; pYZ_3WJrep_N19_trigger 132727; pYZ_3WJrep_N10_trigger 132728; pYZ_3WJrep_N24_trigger 132729; pYZ_NAND2_L17_S19_S11 132730; pYZ_NAND3_L11_S11_S13_S19 132731; pYZ_NAND4_L17_S24_S11_S19_S13 132732; pYZ_3WJrep_N11_trigger 132733; pYZ_3WJrep_N12_trigger 132734; pYZ_3WJrep_N13_trigger 132735; pAG_PluxB_ToeRep_N01_trigger 132736; pAG_Ptet*_ToeRep_N01_switch 132737; pYZ_PluxB_3WJrep_N19_trigger 132738; pYZ_Ptet*_3WJrep_N19_switch 132739; pAG_ToeRep_N09_trigger 132740; pAG_ToeRep_N09_switch 132741; pJK_ToeRepG2_N02_switch 132742; pJK_ToeRepG2_N64_switch 132743; pJK_ToeRepG2_N19_switch 132744; pJK_ToeRepG2_N02_trigger 132745; pJK_ToeRepG2_N64_trigger 132746; pJK_ToeRepG2_N19_trigger 132747.
REFERENCES
- 1.Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA & Voigt CA, “Synthetic biology to access and expand nature’s chemical diversity,” Nature Reviews Microbiology 14, 135–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danino T, Mondragon-Palomino O, Tsimring L & Hasty J, “A synchronized quorum of genetic clocks,” Nature 463, 326–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS & Hasty J, “A sensing array of radically coupled genetic ‘biopixels’,” Nature 481, 39–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausländer S, Ausländer D, Müller M, Wieland M & Fussenegger M, “Programmable single-cell mammalian biocomputers,” Nature 487, 123–127 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Moon TS, Lou C, Tamsir A, Stanton BC & Voigt CA, “Genetic programs constructed from layered logic gates in single cells,” Nature 491, 249–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Win MN & Smolke CD, “Higher-order cellular information processing with synthetic RNA devices,” Science 322, 456–460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siuti P, Yazbek J & Lu TK, “Synthetic circuits integrating logic and memory in living cells,” Nature Biotechnology 31, 448 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Nielsen AA, Fernandez-Rodriguez J, McClune CJ, Laub MT, Lu TK & Voigt CA, “Permanent genetic memory with >1-byte capacity,” Nature methods 11, 1261–1266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet J, Subsoontorn P & Endy D, “Rewritable digital data storage in live cells via engineered control of recombination directionality,” Proceedings of the National Academy of Sciences 109, 8884–8889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel R, Rubens JR, Sarpeshkar R & Lu TK, “Synthetic analog computation in living cells,” Nature 497, 619–623 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Farzadfard F & Lu TK, “Genomically encoded analog memory with precise in vivo DNA writing in living cell populations,” Science 346, 1256272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roquet N, Soleimany AP, Ferris AC, Aaronson S & Lu TK, “Synthetic recombinase-based state machines in living cells,” Science 353, aad8559 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Nielsen AA, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D & Voigt CA, “Genetic circuit design automation,” Science 352, aac7341 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Andrews LB, Nielsen AAK & Voigt CA, “Cellular checkpoint control using programmable sequential logic,” Science 361, aap8987 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Weinberg BH, Pham NTH, Caraballo LD, Lozanoski T, Engel A, Bhatia S & Wong WW, “Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells,” Nature Biotechnology 35, 453–462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao XJ, Chong LS, Kim MS & Elowitz MB, “Programmable protein circuits in living cells,” Science 361, 1252–1258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucks JB, Qi L, Mutalik VK, Wang D & Arkin AP, “Versatile RNA-sensing transcriptional regulators for engineering genetic networks,” Proceedings of the National Academy of Sciences of the United States of America 108, 8617–8622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi MK & Lucks JB, “A modular strategy for engineering orthogonal chimeric RNA transcription regulators,” Nucleic Acids Research 41, 7577–7588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutalik VK, Qi L, Guimaraes JC, Lucks JB & Arkin AP, “Rationally designed families of orthogonal RNA regulators of translation,” Nature Chemical Biology 8, 447–454 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Green AA, Silver PA, Collins JJ & Yin P, “Toehold switches: de-novo-designed regulators of gene expression,” Cell 159, 925–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yurke B, Turberfield AJ, Mills AP Jr., Simmel FC & Neumann JL, “A DNA-fuelled molecular machine made of DNA,” Nature 406, 605–608 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Green AA, Kim J, Ma D, Silver PA, Collins JJ & Yin P, “Complex cellular logic computation using ribocomputing devices,” Nature 548, 117–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P & Collins JJ, “Paper-based synthetic gene networks,” Cell 159, 940–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O’Connor DH, Gehrke L & Collins JJ, “Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components,” Cell 165, 1255–1266 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Ma D, Shen L, Wu K, Diehnelt CW & Green AA, “Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment,” Synthetic Biology 3, ysy018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi MK, Tan X, Dy AJ, Braff D, Akana RT, Furuta Y, Donghia N, Ananthakrishnan A & Collins JJ, “A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers,” Nature Communications 9, 3347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chappell J, Takahashi MK & Lucks JB, “Creating small transcription activating RNAs,” Nature Chemical Biology 11, 214–220 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Meyer S, Chappell J, Sankar S, Chew R & Lucks JB, “Improving fold activation of small transcription activating RNAs (STARs) with rational RNA engineering strategies,” Biotechnology and Bioengineering 113, 216–225 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Chappell J, Westbrook A, Verosloff M & Lucks JB, “Computational design of small transcription activating RNAs for versatile and dynamic gene regulation,” Nature Communications 8, 1051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gander MW, Vrana JD, Voje WE, Carothers JM & Klavins E, “Digital logic circuits in yeast with CRISPR-dCas9 NOR gates,” Nature Communications 8, 15459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanton BC, Nielsen AAK, Tamsir A, Clancy K, Peterson T & Voigt CA, “Genomic mining of prokaryotic repressors for orthogonal logic gates,” Nature Chemical Biology 10, 99–105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS & Qi LS, “CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes,” Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM & Pierce NA, “NUPACK: Analysis and design of nucleic acid systems,” Journal of computational chemistry 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J & Voigt CA, “Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors,” Nature Chemical Biology 15, 196–204 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Watters KE, Abbott TR & Lucks JB, “Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq,” Nucleic Acids Research 44, e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Yin P & Green AA, “Ribocomputing: Cellular Logic Computation Using RNA Devices,” Biochemistry 57, 883–885 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, “Expanding the Rule Set of DNA Circuitry with Associative Toehold Activation,” Journal of the American Chemical Society 134, 263–271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genot AJ, Bath J & Turberfield AJ, “Combinatorial Displacement of DNA Strands: Application to Matrix Multiplication and Weighted Sums,” Angewandte Chemie International Edition 52, 1189–1192 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Carlson PD, Glasscock CJ & Lucks JB, “De novo Design of Translational RNA Repressors,” bioRxiv, 501767 (2018).
- 40.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP & Lim WA, “Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression,” Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F & Marraffini LA, “Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system,” Nucleic Acids Res 41, 7429–7437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siu K-H & Chen W, “Riboregulated toehold-gated gRNA for programmable CRISPR–Cas9 function,” Nature Chemical Biology 15, 217–220 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Oesinghaus L & Simmel FC, “Switching the activity of Cas12a using guide RNA strand displacement circuits,” Nature Communications 10, 2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanewich-Hollatz MH, Chen Z, Hochrein LM, Huang J & Pierce NA, “Conditional Guide RNAs: Programmable Conditional Regulation of CRISPR/Cas Function in Bacterial and Mammalian Cells via Dynamic RNA Nanotechnology,” ACS Central Science 5, 1241–1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serra MJ & Turner DH, “Predicting thermodynamic properties of RNA,” Methods in enzymology 259, 242–261 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Mathews DH, Sabina J, Zuker M & Turner DH, “Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure,” Journal of molecular biology 288, 911–940 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA & Smith HO, “Enzymatic assembly of DNA molecules up to several hundred kilobases,” Nature Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Andersen JB, Sternberg C, Poulsen LK, Bjørn SP, Givskov M & Molin S, “New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria,” Applied and environmental microbiology 64, 2240–2246 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lou C, Stanton B, Chen Y-J, Munsky B & Voigt CA, “Ribozyme-based insulator parts buffer synthetic circuits from genetic context,” Nature Biotechnology 30, 1137–1142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aviran S, Lucks JB & Pachter L, “RNA structure characterization from chemical mapping experiments,” 49th Annual Allerton Conference on Communication, Control, and Computing 1743–1750 (2011). 10.1109/Allerton.2011.6120379 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.