Abstract
Folliculin Interacting Protein-1 (Fnip1) is a cytoplasmic protein originally discovered through its interaction with the master metabolic sensor 5’ AMP-activated protein kinase (AMPK) and Folliculin (FLCN), a protein mutated in individuals with Birt-Hogg-Dubé Syndrome. In response to low energy, AMPK stimulates catabolic pathways such as autophagy to enhance energy production, while inhibiting anabolic pathways regulated by mechanistic target of rapamycin complex 1 (mTORC1). We previously found that constitutive disruption of Fnip1 in mice resulted in a lack of peripheral B cells due to a block in B cell development at the pre-B cell stage. Both AMPK and mTORC1 were activated in Fnip1-deficient B cell progenitors. In this study we found inappropriate mTOR localization at the lysosome under nutrient-depleted conditions. Ex vivo lysine or arginine depletion resulted in increased apoptosis. Genetic inhibition of AMPK, inhibition of mTORC1, or restoration of cell viability with a Bcl-xL transgene failed to rescue B cell development in Fnip1-deficient mice. Fnip1-deficient B cell progenitors exhibited increased nuclear localization of Transcription Factor Binding to IgHM Enhancer 3 (TFE3) in developing B cells, which correlated with increased expression of TFE3-target genes, increased lysosome numbers and function, and increased autophagic flux. These results indicate that Fnip1 modulates autophagy and energy response pathways in part through regulation of AMPK, mTORC1, and TFE3 in B cell progenitors.
Introduction
Developing B cells must pass through well-defined checkpoints, which ensure they are capable of producing functional Ab molecules with minimal self-reactivity. Emerging evidence suggests that B cells may also be regulated by metabolic checkpoints which ensure that mature B cells are appropriately equipped with the molecular machinery to fuel cellular growth required for clonal expansion and Ab production. For example, B cell-specific disruption of Raptor, a positive regulator of mTORC1, inhibits pre-B cell metabolism and triggers a complete block in B cell development at the pre-B cell stage, suggesting pre-BCR-driven cell growth may act as a checkpoint for testing metabolic capacity(1). Similarly, mice deficient in PI3K and PLCγ1/PLCγ2, positive mediators of mTOR signaling, also exhibit blocks in B cell development at the pre-B cell stage(2-5). Conditional deletion of Raptor during peripheral B cell development inhibits the generation of plasma cells and germinal center B cells (see (6) for review), suggesting that metabolic checkpoints might also regulate peripheral B cell maturation.
Despite abundant information about the roles of mTORC1 in the development of immune cells, the roles of other metabolic pathways in B cell development remain unclear. Recently, the Fnip1/Folliculin/AMPK complex has emerged as a central mediator in maintaining metabolic homeostasis during B cell development (7, 8). Folliculin interacting protein-1 (Fnip1) is an evolutionarily conserved cytoplasmic protein originally discovered through its interaction with Folliculin (Flcn), a protein mutated in the rare autosomal dominant disorder Birt-Hogg-Dubé syndrome (BHDS)(9). Patients with BHDS develop benign hair follicle neoplasms, and are at high risk for developing lung cysts, pneumothorax, and renal tumors with a wide variety of histologies (reviewed in(10)). Fnip1 interacts in heteromultimeric complexes with Flcn, Fnip2, and AMPK, a master regulator of cellular metabolism (9). AMPK is phosphorylated during conditions of energy deprivation and responds by activating energy and nutrient producing processes such as mitochondrial biogenesis and autophagy, while simultaneously inhibiting energy and nutrient consuming pathways controlled by mTORC1. We previously generated Fnip1-deficient mice and showed that constitutive loss of Fnip1 resulted in a complete block in B cell development at the pre-B cell stage, due in part to increased apoptosis (7, 8). Enforced expression of IgH and IgL chain proteins in Fnip1-null pre-B cells failed to rescue mature B cell development despite surface IgM expression, suggesting an intrinsic defect in cellular growth and differentiation pathways following disruption of Fnip1.
In this study we took a genetic approach to investigate molecular mechanisms of how Fnip1 controls B cell development. Our results reveal that Fnip1 coordinates multiple metabolic pathways regulated by AMPK, mTORC1, and Transcription Factor Binding to IgHM Enhancer 3 (TFE3) in order to maintain metabolic homeostasis necessary for pre-B cell survival and differentiation during metabolic stress.
Materials and Methods
Mice
Fnip1−/−(7), Bcl-xLTg, Trp53tm1Tyj, Raptorfl/fl, Mb1-Cre, Tsc1fl/fl, Prkaa1tm1.1Sjm, Rag2tm1FwaIl2rgtm1Wjl, Eμ-Myc, and EGFP-LC3 mice were described previously (11-19). Bcl-xLTg mice were kindly provided by Tim Behrens, Mb1-Cre and Eμ-Myc mice were provided by Robert Eisenman, Tsc1fl/fl mice were provided by Raymond Yeung, and EGFP-LC3 were provided by Mike Bevan, and ROSA26lox-stop-lox tdTomato mice were provided by R. Palmiter (20). Trp53tm1Tyj, Prkaa1tm1.1Sjm, mice were purchased from Jackson Labs, and Rag2tm1FwaIl2rgtm1Wjl mice were purchased from Taconic Biosciences. Mice were maintained on a C57Bl/6J background or were backcrossed >10 generations, with the exception of Tsc1fl/fl and Raptorfl/fl crosses, which were on a mixed 129:C57Bl/6J background. Co-housed littermates of both sexes were used whenever possible. Animal studies were reviewed and approved by the University of Washington Institutional Animal Care and Use Committee.
Cell viability and proliferation assays
To assess apoptosis, cells were stained ex vivo with CellEvent Caspase 3/7 (Invitrogen, Carlsbad, CA) and Ghost dye live/dead viability stain (Tonbo Biosciences, San Diego, CA), and analyzed according to the manufacturers’ instructions. Analysis of cellular proliferation in vivo was performed by i.p. BrdU injection (1 mg, BD Biosciences, San Jose, CA) ~16 hrs prior to harvest. Intracellular (IC) staining were performed according to the manufacturer with α–BrdU PerCP/Cy5.5 (BD Pharmingen, San Jose, CA).
Antibodies and flow cytometry
Cells were stained using Abs specific for mouse Ags: CD45R (B220) (various fluorochromes) (BD Pharmingen, BioLegend, San Diego, CA and Tonbo Biosciences); IgM (various fluorochromes, Jackson ImmuoResearch Laboratories, West Grove, PA); CD19 eFlour450, CD25 APC, MHC II APC (Tonbo Biosciences); CD43 PE, BP-1 PE, CD117 PE, CD24 (HSA) PE-Cy7, IgD FITC (BD Pharmingen); CD21 PerCP/Cy5.5 (BioLegend). IC staining was performed with IC fixation and permeabilization buffer (eBiosciences). Methanol fixation was performed for IC phospho (p)-S6R detection. Abs used for IC staining were p-ribosomal S6 protein (S6R) S235/236 PE (eBiosciences); p-AMPKα T172, c-Myc AF488 and p-Ulk1 S555 (Cell Signaling Technology, Danvers, MA). Donkey anti-rabbit Alexa-Fluor 647 (Life Technologies, Carlsbad CA) secondary Ab was used to detect unlabeled primary Abs. Data was collected using FACS Canto II or LSR II flow cytometers (BD Biosciences) and analyses were performed using FlowJo software (TreeStar, Ashland, OR).
Immunoblotting
Immunoblotting was performed on whole cell extracts from cultured immortalized MEFs derived from Fnip1−/− mouse embryos (21). Proteins were detected using Abs against LC3B (D11, Cell Signaling Technology, Danvers, MA) and GAPDH (loading control;D16H11, Cell Signaling Technology).
Cell labeling using NBD-PS
The phospholipid incorporation assay was performed with Fnip1−/− or Fnip1+/− BM using the fluorescent analog of phosphatidylserine, NBD-PS (Avanti Polar Lipids, Inc.). Briefly, cells were labeled with 5 μM NBD-PS in HBSS (Gibco)+5.5mM D-glucose, 20 mM HEPES at 15°C for 5 min. Labeling was quenched in HBSS+5.5 mM D-glucose, 20 mM HEPES, 1% lipid-free BSA for 5 min on ice followed by two washes in HBSS +5.5 mM D-glucose, 20 mM HEPES and staining with fluorescent antibodies for flow cytometric analysis.
Cell labeling using DQ-BSA
BM cells were stained with Abs for flow cytometry as above, stimulated for 1 hr with 10 ng/mL IL-7 in complete media, then labeled with 20 μg/ml DQ-BSA Green (Invitrogen) in complete media at 37 °C for 1 hour, followed by two washes with DPBS+3% FBS before flow cytometric analysis.
Imagestream® imaging flow cytometry
Cells were cultured ex vivo for 2-3 hrs or overnight and fixed as described above for detection of IC Ags using rat α–LAMP2 (Abcam), rabbit α–mTOR (Cell Signaling Technology), or rabbit α–TFE3 (Sigma) Abs. Donkey α-rabbit Alexa Fluor 647 and donkey α-rat Alexa Fluor 488 (Life Technologies) secondary Abs were used to detect primary Abs. Data were collected using an Imagestream® imaging flow cytometry X (Millipore) and analyses were performed with IDEAS software (Millipore).
Mass spectrometry-based proteomics
B cells were enriched from total BM from Fnip1−/− and Fnip1+/+ littermates. Pre- and pro-B cells were enriched via positive selection with α-CD19 Ab-conjugated magnetic beads (EasySep, Stemcell Technologies) followed by FACS sorting for IgM negative cells, and were cultured overnight in rIL-7 (R&D systems). 2×106 cells from each replicate were then lysed using 8M Urea. Peptide enrichment and chromatography were performed as previously described. Following peptide quantification (Pierce quantitative fluorometric peptide assay), 1 ug of each sample was analyzed by mass spectrometry as described previously(22).
Statistical analyses
Statistical differences between two groups were analyzed by two-tailed unpaired Student’s t tests with equal variance. Multiple group comparisons were made using one-way ANOVA with multiple comparisons post-hoc t tests. GraphPad Prism software (version 6; GraphPad Software, San Diego, CA) was used for analyses, and data were considered significant at *p≤0.05, **p≤0.001, ***p≤0.0001, ****p<0.00001. Center values shown on graphs represent means and error bars represent standard deviations. For all graphs, each point plotted represents the analysis of primary cells derived from a single mouse.
Results
Disruption of Fnip1 results in increased apoptosis of B cell progenitors.
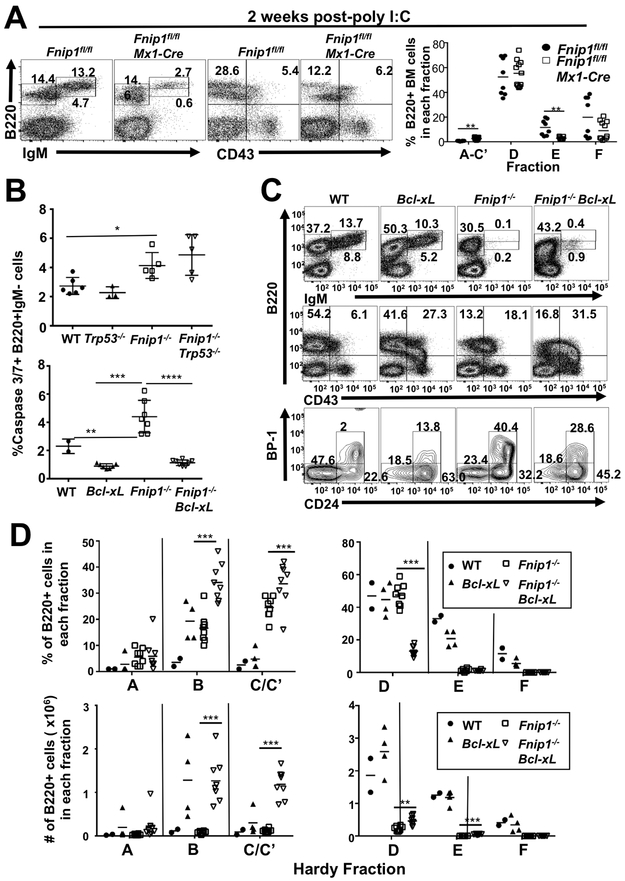
To assess whether acute disruption of Fnip1 affects B cell development in adult animals and not via deletion in embryonic/fetal/neonatal tissues, we crossed Fnip1fl/fl mice (8) with Mx1-Cre (21) mice, which express Cre recombinase from the IFN-responsive Mx1 gene promoter. Cre expression is transiently induced in response to injection of the dsRNA analog polyinosinic:polycytidylic acid (poly I:C). We found that the percentage of immature and mature B cells (Hardy Fractions E and F, (23)) in the bone marrow (BM) were markedly reduced two wks following poly I:C injection (Fig.1A), while the representation of pro-B/early pre-B cells (Hardy Fractions A-C’)) were slightly increased. This likely represents a developmental block, as Caspase 3/7+ apoptotic pro-B/pre-B cells increased 48 hrs after poly I:C administration (Supplemental Fig. 1A). To visualize Fnip1-deleted cells, Fnip1fl/flMx1Cre mice were crossed with tdTomato Cre-reporter mice, which allows Cre-expressing cells to be identified via the expression of red fluorescent protein. A single injection of poly I:C resulted in ~90% tomato fluorescent protein (FP)+ B220+IgM- proB/preB cells, ~80% tomatoFP+ B220loIgM+ immature B cells, and ~50% tomatoFP+ mature B220hiIgM+ B cells by 48 hrs post-injection (Supplemental Fig. 1B). The reduction in tomatoFP+ cells during the pre-B to immature B cell stage is consistent with increased apoptosis and selection against the deleted allele with maturation. Although poly I:C-induced α/β IFN might synergize with Fnip1-deficiency to mediate cell death, both experimental and control mice were treated with poly I:C. These findings indicate that acute disruption of Fnip1 leads to impaired development at the pre-B cell stage due in part to increased apoptosis.
FIGURE 1.
Disruption of Fnip1 results in increased apoptosis. (A) Representative flow cytometric analyses of bone marrow (BM) cells from Fnip1fl/fl or Fnip1fl/flMx1-Cre mice 13 days after deletion of Fnip1 via poly I:C (pI:C) injection. Shown are gated lymphocytes (FSC/SSC) following staining for B220 (CD45R), IgM and CD43 (Ly48). Numbers on the plots represent the frequencies of the gated populations. The scatter plot (right panel) shows frequencies of B220+ BM cells in various developmental fractions (Hardy fractions). **=p<0.005 t-test (B) p53 deletion and Bcl-xL overexpression fail to rescue B cell development in Fnip1-deficient mice. Scatter plots show frequencies of cleaved Caspase 3/7 substrate in B220+IgM− BM cells from wildtype (WT), Fnip1−/−, Trp53−/− and Fnip1−/−Trp53−/− mice (upper panel), or Bcl-xL and Fnip1−/−Bcl-xL mice (lower panel). p≤0.0001 one-way ANOVA (C) Representative flow cytometric analyses of BM cells from WT, Fnip1−/−, Bcl-xL and Fnip1−/−Bcl-xL mice are shown gated on lymphocytes and stained for B220, IgM, CD43, BP-1 (Ly-51, CD249) and CD24 (HSA). (D) Scatter plots show developmental (Hardy) fractions representing frequency (bottom left panel) and absolute number (bottom right panel) of cells from these mice. Gating for fractions: A=B220+CD43+CD24−BP-1−, B=B220+CD43+CD24+BP-1−, C/C’=B220+CD43+CD24+BP-1+, D=B220loCD43−IgM−, E=B220loCD43−IgM+, F=B220hiCD43−IgM+.
Increased apoptosis of Fnip1-deficient B cell progenitors occurs independently of p53.
We next investigated the molecular mechanisms leading to increased apoptosis of Fnip1-deficient B cell progenitors. V(D)J recombination activates a p53-dependent checkpoint during B cell development, which induces apoptosis if recombination is prolonged or defective(24). Whereas Fnip1−/− pre-B cells express intracellular μ (ICμ) indicative of effective recombination, levels of ICmu are lower than in WT cells(7) possibly indicating a deficiency in survival during V(D)J recombination. We therefore asked whether increased apoptosis of Fnip1−/− B cell progenitors proceeds through a p53-dependent mechanism. Fnip1−/− mice were crossed with p53−/− mice to generate Fnip1−/−p53−/− animals. p53 deficiency did not alter levels of apoptosis in Fnip1−/− B cell progenitors (Fig. 1B) and had no effect on B cell development (Supplementary Fig. 1C). These results suggest that the increase in pro-B/pre-B cell apoptosis following disruption of Fnip1 occurs independently of p53.
Bcl-xL overexpression restores cell viability but fails to rescue B cell development in Fnip1-deficient mice.
We next investigated whether overexpression of a pro-survival factor could restore normal B cell development. We crossed Fnip1−/− mice with transgenic (Tg) mice overexpressing Bcl-xL, an anti-apoptotic member of the Bcl-2 family, in B cells (18). Although expression of the Bcl-xL transgene markedly reduced apoptosis in Fnip1−/− pro- and pre-B cells (Fig. 1B), Bcl-xL only minimally (<10%) rescued the development of early and late pre-B (Fractions B, C/C’ and D) and immature B cells (Fraction E) in the BM, and completely failed to restore the development of mature B cells in the BM (Fig. 1C,D) and spleen (not shown). An increase in Hardy Fractions B and C is reflective of Bcl-xL Tg, which has been shown previously to increase the number of pro-B cells(18). Our results suggest that Fnip1 deficiency results in impaired B cell development that is only partially dependent on cell survival, and that additional mechanisms contribute to blocked B cell differentiation.
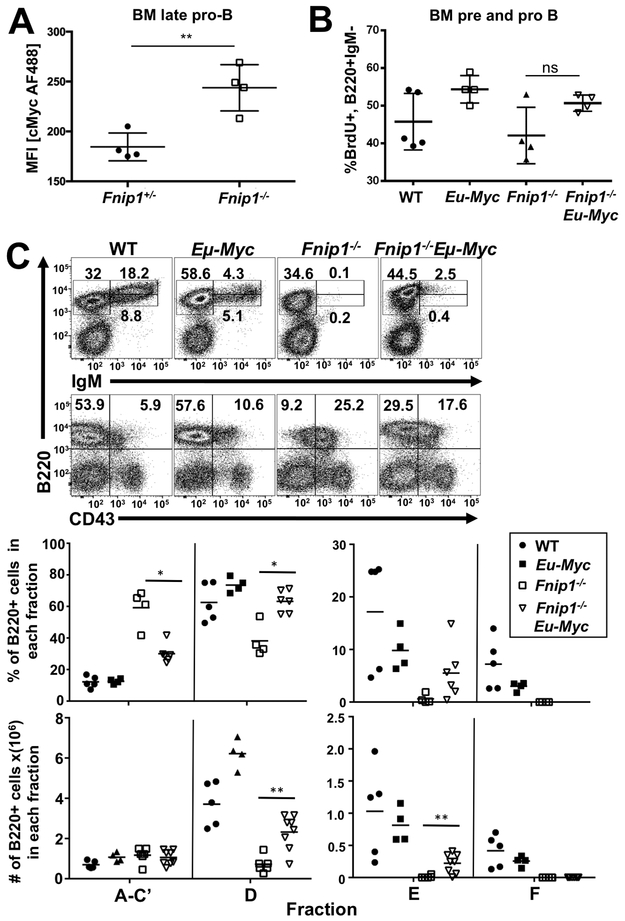
Increased c-Myc-mediated cell growth and division fails to restore B cell development in Fnip1-deficient mice.
We previously found that constitutive loss of Fnip1 resulted in enrichment for genes regulated by the Myc transcription factor (p=1.8 × 10−7) in microarray analysis of pre-B cells from Fnip1−/− compared to WT mice (7). c-Myc is important for pre-B cell development (21), and drives cell growth and proliferation while altering the apoptotic threshold of cells (reviewed in (25)). Analyses of c-Myc protein levels via IC staining revealed increased c-Myc in Fnip1−/− BM late pro-B cells (Fig. 2A, Supplemental Fig. 2D) indicating that decreased Myc levels were not responsible for blocked B cell development. Since increased c-Myc-driven metabolism might compensate for the absence of Fnip1, we tested whether B cell-specific overexpression of c-Myc could further stimulate B cell growth, metabolism, and drive maturation of Fnip1−/− pre-B cells by generating Fnip1−/− mice that overexpress c-Myc during B cell development (Eμ-c-Myc)(14). c-Myc overexpression failed to significantly increase proliferation of Fnip1−/− B cell progenitors (Fig. 2B), but efficiently increased cell growth, based on an increase in cell size (not shown). However, the c-Myc transgene only enabled development of a small population of pre-B and immature B cells in Fnip1−/− animals, and completely failed to rescue the development of circulating mature B cells in the BM (Fig. 2C) and periphery (not shown). These results indicate that stimulation of cellular growth, metabolism, and proliferation by overexpression of c-Myc is not sufficient to restore B cell development in the absence of Fnip1.
FIGURE 2.
Increased c-Myc-mediated cell growth fails to restore B cell development in Fnip1-deficient mice. Bar graphs show (A) mean fluorescence intensity (MFI) of IC c-Myc staining in B220+BP-1+CD25− late pro-B cells from Fnip1+/− or Fnip1−/− BM and (B) frequencies of BrdU+ BM pro- and pre-B cells (B220+IgM−), which represent dividing cells from WT, Eμ-Myc, Fnip1−/−, and Eμ-Myc Fnip1−/− mice. (C) Representative flow cytometric analyses of BM cells from WT, Eμ-Myc, Fnip1−/−, and Eμ-Myc Fnip1−/− mice are shown gated on lymphocytes and stained for B220, IgM and CD43. (D) Scatter plots show developmental (Hardy) fractions representing frequency (top panel) and absolute number (bottom panel) of cells from these mice. Fraction A-C’ is gated B220+CD43+.
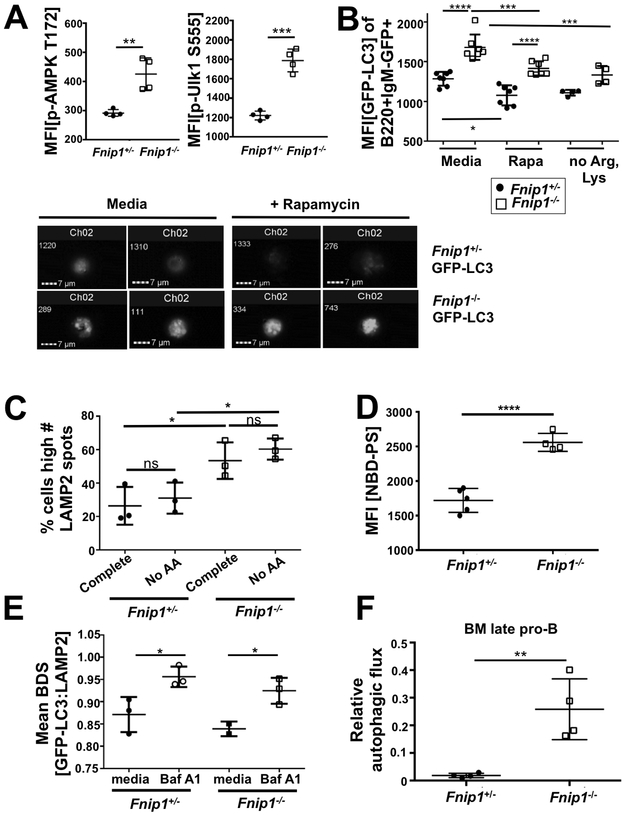
Disruption of Fnip1 increases AMPK activation, autophagy induction and flux.
Autophagy is a self-degradation process necessary to generate nutrients and energy during development and in response to energy stress. Because Fnip1 interacts with AMPK, a primary activator of autophagy, and mTORC1 inhibits autophagy (26), we examined whether Fnip1 deficiency impacts autophagy during B cell development. Consistent with our previous report, we found Fnip1−/− pro- and pre-B cells exhibited increased AMPKα phosphorylation at Thr172, as well as increased phosphorylation of a direct substrate of AMPK, Unc-51 Like Autophagy Activating Kinase 1 (Ulk1 phospho-Ser555), which is thought to initiate autophagy (Fig. 3A, Supplemental Fig. 2D).
FIGURE 3.
Disruption of Fnip1 increases autophagy induction and flux. Bar graphs show MFIs of (A) IC p-AMPK T172 staining (left panel) and IC p-Ulk1 S555 (right panel) on live-gated B220+IgM− cells. (B) MFIs of EGFP-LC3 and representative images (imaging cytometry) of B220+IgM−GFP+ BM cells from EGFP-LC3 Fnip1+/− or EGFP-LC3 Fnip1−/− mice without or with Rapamycin or arginine and lysine-free media for 24 hrs as measured by flow cytometry. p<0.0001 one-way ANOVA. (C) Frequency of B220+IgM− BM cells from Fnip1+/− or Fnip1−/− mice with a high number of LAMP2 (lysosomal marker) spots, as measured by Imagestream® imaging flow cytometry, without or with 2 hrs of incubation in media without AAs. p<0.0001 one-way ANOVA. (D) MFI of B220+IgM− BM cells from Fnip1+/− or Fnip1−/− mice incubated with fluorescent phospholipid analog NBD-phosphatidylserine for one minute. (E) Mean bright detail similarity (BDS) of GFP-LC3 and DyLight 650 LAMP2 from B220+IgM−GFP+ BM cells from EGFP-LC3 Fnip1+/− or EGFP-LC3 Fnip1−/− mice cultured 24 hrs ex vivo in complete media or media without AAs supplemented with 10 nM Bafilomycin A1 (Baf A1) (blocks autophagy through inhibition of lysosomal degradation), as measured by imaging cytometry. Mean BDS is a measure of colocalization. (F) Relative autophagic flux as measured by IC flow cytometric staining of LC3-II (autophagosome-bound LC3) in B220+IgM− BM cells from Fnip1+/− or Fnip1−/− mice. The ratio of LC3-II after 2 hrs 100 nM Baf A1 treatment compared to basal levels of LC3-II is shown.
Initiation of autophagy may not lead to increased flux through the pathway if autophagosome/lysosome fusion is blocked. Therefore, we visualized autophagosomes by generating Fnip1−/− microtubule-associated protein 1A/1B-light chain 3-GFP (LC3-GFP) fusion mice. During autophagy, cytosolic LC3-I is conjugated to phosphatidylethanolamine (PE) to form LC3-II, which subsequently becomes incorporated into autophagosomes. Fnip1−/− pro- and pre-B cells displayed increased levels of LC3-GFP overall (LC3-I&-II), which decreased in response to autophagy induction with rapamycin or amino acid (AA) restriction, but remained higher than Fnip1+/− cells (Fig. 3B). Imagestream® imaging flow cytometry revealed an increased number and size of LC3-GFP puncta (autophagosomes) in Fnip1−/− pro- and pre- B cells, which appeared largely unchanged during induction of autophagy with rapamycin, while puncta number decreased in control cells (Fig. 3B, lower panel). Since altered lysosome number or function can help differentiate between autophagy inhibition or maximally increased flux, we measured the number of lysosomes (number of IC spots of the lysosome marker LAMP2) using Imagestream® and examined lysosomal activity by evaluating IC fluorescence of proteolyzed DQ-BSA, which requires enzymatic cleavage to generate a fluorescent product. A higher percentage of Fnip1−/− pro- and pre-B cells had high numbers of lysosomes (Fig. 3C) as well as increased lysosomal activity during induction of autophagy with the Tat-D11 (Beclin 1) peptide compared to control cells (Supplementary Fig. S1D), which suggests increased autophagic flux. To investigate whether a defect in lipid turnover was responsible for increased autophagosome and lysosome numbers, we measured incorporation of the fluorescent phospholipid analog NBD-phosphatidylserine (NBD-PS) into Fnip1−/− pro- and pre-B cells. We found that incorporation of NBD-PS into IC membranes was increased in Fnip1−/− cells relative to Fnip1+/− controls (Fig. 3D), indicating lipid turnover is higher than usual in these cells. We next measured LC3-GFP co-localization with LAMP2 using Imagestream® to determine if disruption of Fnip1 results in defects in autophagosome-lysosome fusion, which could also result in increased organelle number and size. We found that autophagosome-lysosome co-localization was not different between Fnip1−/− and Fnip1+/− control cells, and both displayed increased co-localization following treatment with Bafilomycin A1, which blocks autophagy at the final step of the pathway by preventing digestion of autolysosomes (Fig. 3E).
To definitively assess the flux of autophagy in Fnip1−/− cells, we next performed IC staining of LC3-II in late pro-B cells after treatment with Bafilomycin A1 to block autophagy. Autophagic flux was then determined by comparing the ratio of LC3-II after Bafilomycin treatment to basal levels of LC3-II. Autophagy flux was increased in Fnip1−/− late pro-B cells relative to Fnip1+/− controls (Fig. 3F). We used immunoblotting to confirm this result, comparing LC3B-I and LC3B-II levels in response to Bafilomycin in Fnip1+/− and Fnip1−/− mouse embryonic fibroblasts (MEFs, Supplemental Fig. S1E). Fnip1−/− MEFs had a much higher basal ratio of membrane-bound:cytoplasmic LC3B (LC3B-II:LC3B-I, lower panel densitometry), and upon starvation converted LC3-I to LC3-II faster than control cells, confirming Fnip1 deficiency increases autophagic flux. Together, these results indicate that disruption of Fnip1 in B cell progenitors results in an increased number of autophagosomes and lysosomes, as well as increased autophagic flux compared to Fnip1+/− B cell progenitors.
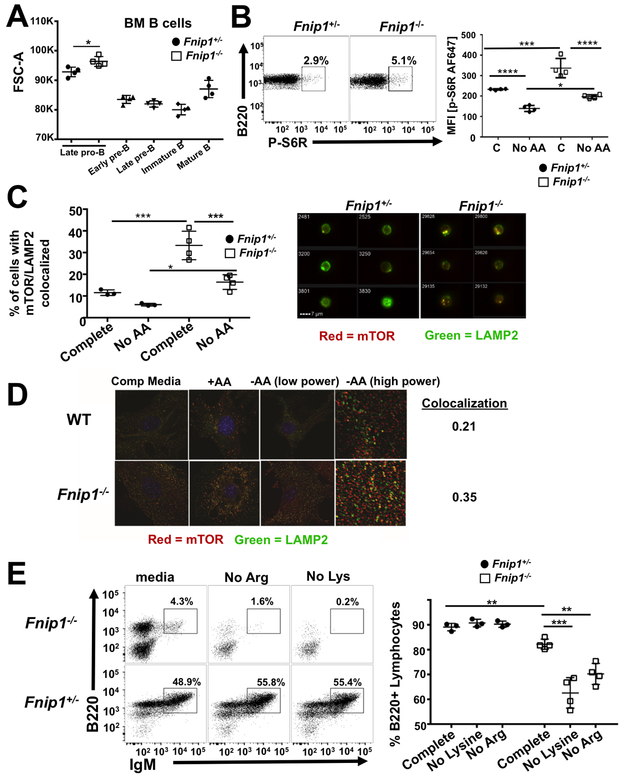
mTORC1 signaling is aberrantly increased in Fnip1-deficient B cell progenitors
mTORC1 is activated by growth factors and sufficient AAs, which are transported to the lysosome resulting in activation of vacuolar ATPases (v-ATPases). Activated v-ATPases then stimulate guanine nucleotide exchange activity of the Ragulator complex towards the RagA/B/C/D GTPases, which recruit mTORC1 to the lysosome where mTOR is activated (reviewed in(27)). While some studies have shown that constitutive Fnip1 or Flcn deficiency in mice is associated with mTORC1 hyperactivation (7, 28-30), other investigations in cell lines have shown that Fnip1 and Folliculin positively regulate mTORC1 activity by functioning as Rag C/D GTPase activating proteins (GAPs) or Rag A/B guanine nucleotide exchange factors (GEFs) at the lysosomal surface, thus helping recruit and retain mTORC1 at the lysosome (31, 32). To further assess the roles of Fnip1 in mTORC1 activation during B cell development, we measured the size of BM B cell progenitors (indicative of cell growth) in Fnip1−/− and Fnip1+/− mice using flow cytometry. Cell size was increased in Fnip1−/− late pro-B cells relative to Fnip1+/− control pro-B cells, which correlates with the stage at which pre-B cells normally decrease in size during early B cell development (Fig. 4A). IC phosphorylated S6 ribosomal subunit (p-S6R), a substrate downstream of mTORC1, was increased in Fnip1−/− versus Fnip1+/− pro-B cells (Fig. 4B). Although mTORC1 activity decreases in Fnip1−/− pro-B cells in response to AA restriction, it remains inappropriately high relative to Fnip1+/− controls, both in the presence or absence of AAs (Fig. 4B). To address if Fnip1 modulates the pre-B cell proteome, we performed whole cell mass spectroscopy-based proteomics on sorted pro- and pre-B cells from Fnip1−/− and control littermates. Consistent with increased mTORC1 activity, we found multiple ribosomal proteins were more abundant in Fnip1−/− relative to Fnip1+/− control cells (Supplementary Table 1).
FIGURE 4.
mTORC1 signaling is aberrantly increased in Fnip1-deficient B cell progenitors. (A) Forward scatter (FSC-A, a measure of cell size) of BM B cells as measured by flow cytometry. Summary data from Fnip1+/− or Fnip1−/− mice. Late pro-B: B220+BP-1+CD25−, early pre-B: B220+BP-1+CD25+, late pre-B: B220+BP-1−CD25+, immature B: B220loIgM+, mature B: B220hiIgM+. (B) Representative dot plots (left panel) or summary MFI flow cytometry data without and with incubation in AA free media (right panel) of IC phospho-S6 ribosomal (P-S6R) staining (mTORC1 downstream substrate) from B220+IgM− BM of Fnip1+/− or Fnip1−/− mice. p<0.0001 one-way ANOVA (C) Frequency of cells with colocalization (left panel) or representative images (right panels) of mTOR (red) and LAMP2 (green) IC staining in Fnip1+/− or Fnip1−/− BM gated B220+IgM−, as measured by imaging flow cytometry, without (left) or with (right) overnight incubation in AA free media. p<0.0001 one-way ANOVA. (D) Representative immunofluorescent images from Fnip1+/− or Fnip1−/− MEFs stained with anti-mTOR (red) and anti-LAMP2 (green) Abs. Cells were cultured ex vivo in AA-free media for 50 min +/− refeeding with AAs (1X concentration) for 10 min. Pearson’s coefficient representing colocalization is shown (0=random distribution, 1=100% colocalization). (E) Representative dot plots of B220+IgM+ cells (left panels) and frequency of B220+ lymphocytes (right panel) in Fnip1+/− or Fnip1−/− BM after 4 day ex vivo culture in complete media, media without lysine, or media without arginine.
Since mTORC1 activity is regulated by recruitment to the lysosomal surface in response to AA sufficiency, we examined whether the increase in mTORC1 activation in Fnip1−/− pre-B cells was associated with altered localization at the lysosome. We assessed mTOR co-localization with lysosomes in Fnip1−/− versus Fnip1+/− B cell progenitors using IC staining for mTOR and LAMP2, followed by visualization with Imagestream® imaging flow cytometry. As expected, mTOR/lysosomal co-localization decreased in response to AA restriction in control cells (Fig. 4C left panel). However, Fnip1−/− cells exhibited much higher basal levels of mTOR/lysosomal co-localization, which decreased upon AA starvation but remained aberrantly high (Fig. 4C). We also observed increased co-localization of mTOR with lysosomes in MEFs derived from Fnip1−/− mice using fluorescence microscopy (Fig. 4D). Relative to Fnip1+/− control MEFs under nutrient-rich conditions, which should show the most co-localization, mTOR remained associated with lysosomes during AA deprivation in Fnip1−/− MEFs, indicating that Fnip1 is necessary for optimal inactivation of mTOR.
We next determined whether increased (inappropriate) localization of mTOR to the lysosome in Fnip1−/− mice might impact cell survival in response to deprivation of arginine or lysine, which have been shown to be important for activating mTOR (33). We observed decreased numbers of B220+ lymphocytes in Fnip1−/− BM cultured for 24 hrs in the absence of lysine or arginine compared to control BM, which displayed no significant loss of B cells in response to AA deprivation (Fig. 4E). These results suggest that Fnip1 may help terminate mTORC1 activation and increase cell survival in part by enabling mTOR translocation from the lysosomal surface to the cytosol in response to AA deprivation.
In light of these findings, we wondered whether genetic inhibition of mTORC1 activation could restore B cell development in Fnip1−/− mice. Fnip1−/− mice were crossed with Raptorfl/flMb1-Cre mice (1), which disrupts mTORC1 activation during B cell development. Inhibition of mTORC1 did not restore B cell development in Fnip1- and Raptor- double deficient animals (Fnip1−/−Raptorfl/flMb1Cre+ mice) (Supplementary Fig. 2A-C) or Fnip1−/−Raptorfl/+Mb1Cre+ mice (data not shown), which express attenuated levels of Raptor. These results are consistent with our previous study showing that prolonged treatment of Fnip1−/− mice with the mTORC1 inhibitor rapamycin was also unable to rescue B cell development (7).
Increased mTORC1-mediated cell growth does not inhibit pre-B cell development and fails to restore B cell development in Fnip1-deficient mice.
Since mTORC1 activity is essential for pre-B cell development, we next addressed whether the high mTORC1 activity we observed in Fnip1−/− pre-B cells could be compensatory for Fnip1 deficiency. Thus, we tested whether further increasing mTORC1 activation would restore B cell development in Fnip1−/− mice. We disrupted Tsc1 (a negative regulator of mTORC1) by breeding B cell-specific Tsc1 null mice (Tsc1fl/flMb1-Cre) to Fnip1−/− mice to generate Tsc1 and Fnip1 double deficient mice. Fnip1−/−Tsc1fl/flMb1-Cre animals develop renal disease before 4 wks of age (29), so we transplanted total BM cells from Fnip1−/−Tsc1fl/flMb1-Cre animals into irradiated Rag2−/−Il2rg−/− or CD45.1 mismatched recipients. In accordance with high levels of mTORC1 signaling, both cell size and IC levels of p-S6R protein were increased in Tsc1fl/flMb-1Cre and Fnip1−/−Tsc1fl/flMb-1Cre animals relative to Fnip1−/− mice (Supplementary Fig. S3A and data not shown). However, Tsc1 deficiency failed to rescue B cell development, although a modest increase in B220loCD25+ late pre-B cells, B220loCD43-IgM- pro-and pre-B cells was noted in the BM (Supplementary Fig. S3B). Unlike Raptor deficiency, B cell-specific Tsc1 deficiency does not inhibit B cell development. This evidence combined with the observation that conditional deletion of Ampkα1 in B cells (Ampkα1fl/flMb1-Cre) does not rescue B cell development in Fnip1−/− mice (Supplementary Fig. S3C) indicate aberrant mTORC1 or AMPK activation alone are not responsible for the developmental block in Fnip1−/− B cell progenitors.
Fnip1 is necessary for TFE3 inactivation in pro-and pre- B cells.
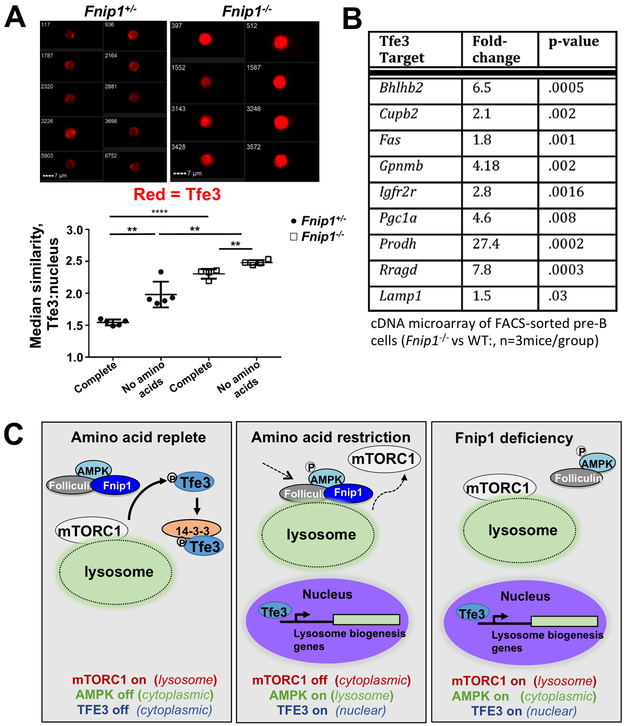
Both mTORC1 and the Folliculin/Fnip1/Fnip2 complex have been implicated in inhibiting lysosomal biogenesis through phosphorylation and cytoplasmic retention of TFE3 (34-37), which is considered a master regulator of lysosome gene expression. Since mTORC1 activity and lysosomal biogenesis were both increased in Fnip1−/− B cell progenitors, we hypothesized that disruption of Fnip1 leads to increased TFE3 nuclear localization. To investigate this hypothesis we utilized imaging flow cytometry to examine the IC localization of TFE3 in Fnip1−/− and Fnip1+/− B cell progenitors. TFE3 nuclear localization, based on co-localization of TFE3 and DAPI staining, increased in response to AA restriction in both control and Fnip1−/− pro/pre-B cells as expected (Fig. 5A). Notably, Fnip1−/− cells displayed significantly higher TFE3 nuclear localization than control cells both in the presence and absence of essential AAs (Fig. 5A), despite higher levels of lysosome-associated mTORC1 under the same conditions (Fig. 4C and 4D). Increased TFE3 nuclear localization also correlated with increased transcriptional activation of known TFE3 target genes including Gpnmb, Pgc1a, Rragd, and Lamp1 (Fig. 5B)(7). These results collectively suggest that in B cell progenitors, Fnip1 is required for retention of TFE3 in the cytosol under nutrient replete conditions, when mTORC1 is activated.
FIGURE 5.
Fnip1 regulates TFE3 nuclear localization in B cell progenitors. (A) Median similarity (a measure of image overlap, lower panel) of TFE3 IC staining and nuclear staining (DAPI), and representative images (upper panels) of B220+IgM− cells in Fnip1+/− or Fnip1−/− BM as measured by imaging flow cytometry. p<0.0001 one-way ANOVA. (B) Expression of selected TFE3 target genes as measured using Illumina cDNA microarray of FACs-sorted B220+IgM− pro- and pre-B cells, Fnip1−/− vs. WT, n=3 mice/group. (C) Schematic model of signaling inside Fnip1-deficient B cells compared to normal B cells under nutrient replete or AA restricted conditions, showing dysregulation of mTORC1 and TFE3 localization in the absence of Fnip1.
Discussion
Constitutive disruption of Fnip1 in mice results in a complete absence of mature B cells and Abs due to a block in B cell development at the pre-B cell stage (7, 8) and decreased survival of B cell progenitors in response to metabolic stressors such as nutrient deprivation and pre-BCR crosslinking. In this study, we found that increased apoptosis occurred independently of p53, and that rescuing pre-B cell survival with a Bcl-xL transgene failed to efficiently restore mature B cell development. These results suggest that Fnip1 regulates B cell development through mechanisms that are only partially dependent on pre-B cell survival. While our results differ from previously published results showing that Bcl-2 expression was able to rescue B cell development in Fnip1-deficient mice (8, 38), Bcl-2 and Bcl-XL are not completely functionally redundant; Bcl-xL is more stable than Bcl-2 (39) and inhibits both Bax and Bak, direct effectors of mitochondrial apoptosis, while Bcl-2 does not do so. Bcl-XL and Bcl-2 have different effects on hematopoietic cell fate when ectopically expressed in multipotent progenitor cells; Bcl-xL supports erythroid cell fate whereas Bcl-2 supports myeloid cell fates(40). Our studies suggest that the unique functions of Fnip1 in B cells include enabling efficient mTORC1 dissociation from the lysosome in response to AA deprivation, and limiting translocation of TFE3 to the nucleus. These results highlight multiple mechanisms whereby Fnip1 maintains metabolic homeostasis during B cell development.
Fnip1 was cloned based on physical interactions with Flcn and all three subunits of AMPK. AMPK phosphorylates both Fnip1 and Flcn, although the consequences of this modification remain unclear. In this study, we found that AMPK was activated in Fnip1-deficient pro-B/pre-B cells, which correlated with increased AMPK-mediated phosphorylation of ULK1 (a positive regulator of autophagy), increased induction of autophagy, and increased flux of autophagy relative to control cells. Elevated levels of autophagy in Fnip1−/− B cell progenitors did not increase further in response to AA deprivation, consistent with the notion that autophagy flux was near maximum. Previous studies have suggested that autophagy is also increased in flies lacking the Drosophila Flcn homologue DBHD (41), as well as in C. elegans and MEFs lacking Flcn (42). In addition, another group reported that basal autophagy was increased in Fnip1−/− B cell progenitors (38). These previous studies are consistent with our data showing increased autophagic flux in pro- and pre-B cells following disruption of Fnip1. However, Dunlop et al found that Flcn and Fnip1 positively regulate autophagy in human kidney cells via interaction with GABARAP, an autophagy component which is crucial for autophagosome formation and sequestration of cytosolic cargo into vesicles (42). Increased levels of SQSTM-1 and LC3 were observed in Flcn-deficient MEFs and renal tumor cells from a BHDS patient, but were interpreted as indicative of impaired autophagy, while autophagic flux was not examined. These studies highlight the complexity of the interactions between the Flcn/Fnip1/Fnip2 complex and autophagic pathways, suggesting that the impact of Flcn/Fnip deficiency on autophagic flux may be tissue dependent and/or influenced by the energetic state of the tissues examined.
Surprisingly, despite increased autophagy in Fnip1−/− B cell progenitors, we also found that mTORC1 activity and lysosomal localization were aberrantly increased, even under conditions of AMPK activation and AA restriction when mTORC1 should be cytosolic and inactive. These results suggest that at least in some cell types, Fnip1 may be necessary for AMPK to efficiently inactivate mTORC1, which is consistent with findings by others in human cell lines and MEFs. In particular, Nagashima et al found that ubiquitination and degradation of Fnip2 led to the dissociation of Flcn from the lysosome and constitutive association and activation of mTOR at the lysosomal surface regardless of nutrient status (43). Zhang et al demonstrated that the scaffold protein Axin tethers AMPK to the surface of endosomes and lysosomes where it is activated by the v-ATPase-Ragulator complex in response to glucose deprivation (44). These studies collectively suggest that the Fnip1/Fnip2/Flcn complex could serve as a molecular scaffold allowing AMPK to interact with the lysosomal v-ATPase at the surface of lysosomes, thus modulating the ability of AMPK to inactivate mTORC1 under nutrient poor conditions. With both catabolic and anabolic pathways active concurrently, our results suggest that a significant energy imbalance in Fnip1-deficient B cell progenitors precludes survival during cellular stresses such as AA restriction, since mTORC1-driven nutrient consumption remains inflexibly high.
Despite significant elevations in mTORC1 and AMPK activity following disruption of Fnip1, we also found that deletion of the AMPKα1 subunit, or attenuation of mTORC1 activity through disruption of Raptor or treatment with Rapamycin, were unable to rescue B cell development in Fnip1−/− mice. In addition, increasing mTORC1 activity through B cell specific disruption of Tsc1 did not recapitulate the block in early development of Fnip1−/− B cells. These results indicate that additional factors may contribute to impaired B cell development in Fnip1−/− mice. Indeed, we found increased nuclear localization of the TFE3 transcription factor in Fnip1-deficient pro- and pre-B cells relative to control cells. Nuclear localization of TFE3 correlated with increased lysosome numbers and activity, mitochondrial biogenesis, expression of TFE3 target genes (7), and autophagy. Accordingly, previous studies showed that inactivation of FLCN in human renal cancer cell lines induced TFE3 transcriptional activity by increasing its nuclear localization (36), and that inactivation of FLCN in human fetal lung fibroblasts inhibited canonical WNT signaling, which could be completely restored only by silencing TFE3 (45). Other studies have similarly reported that the Flcn/Fnip1/Fnip2 complex is required for mouse ES cell (35), hematopoietic stem cell (46), and osteoclast (37) differentiation by restricting nuclear localization and activity of TFE3.
Previous studies have shown that mTORC1 inhibits TFE3 transcriptional activity by phosphorylating and inhibiting TFE3 nuclear translocation (35). The apparent paradox of high levels of mTORC1 activation concurrent with nuclear TFE3 in Fnip1-deficient pre-B cells was similarly seen by Wada et al in Flcn-deficient murine adipose tissue (34). In that study, mTORC1-mediated phosphorylation and cytoplasmic retention of TFE3 was dependent on Flcn, whereas phosphorylation of other downstream substrates such as S6K appeared independent of Flcn function. This may be explained by the findings that the Flcn/Fnip complex acts as a GAP converting RagC/DGTP to RagC/DGDP(31), and TFE3 is recruited to the lysosome by binding inactive RagC/DGDP (47). Thus, despite increased mTORC1 activity and lysosomal localization in the absence of the Fnip1/Flcn complex, TFE3 may not be recruited to the lysosome to be phosphorylated by mTORC1. The resulting aberrantly high levels of TFE3 transcriptional activity may contribute to impaired B cell development, as has been observed with HSC and osteoclast differentiation. The ability of mTORC1 to phosphorylate and inhibit nuclear localization of TFE3/Tfeb in a Flcn/Fnip1/Fnip2-dependent manner may be a conserved mechanism enabling nutrient sensing pathways to regulate differentiation of immune and other cell types.
Supplementary Material
Key points:
Fnip1 is required for pre-B cell development/survival independent of p53 and BclXL
Fnip1 is required for optimal inhibition of mTORC1 in response to aa restriction
Fnip1 regulates TFE3 nuclear translocation and lysosome biogenesis in pre-B cells
Acknowledgements
The authors thank T. Chang for assistance with Imagestream® data collection and analysis, and R. Shaw for immortalization of Fnip1−/− MEFs. We thank L. Kang, J. Chan, and M. Torres for help with the mouse colony.
Support: This work was supported by NIH grants R21AI109020, R01AI092093, R56AI092093 (B.M.I.); K01OD010554 (J.A.R.); K01OD21421 (T.I). L.S.S. is funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, NIH under contract HHSN261200800001E.
Abbreviations:
- mTOR
mechanistic target of Rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- Fnip1
Folliculin Interacting Protein-1
- Flcn
Folliculin
- AMPK
5’AMP-activated protein kinase
- TSC1
tuberous sclerosis-1
- 4EBP1
eIF4E-binding protein 1
- BM
bone marrow
- IC
intracellular
- poly I:C
polyinosinic-polycytidylic acid
- LC3
Microtubule-associated protein 1A/1B-light chain
Footnotes
Disclosures
The authors declare no financial conflicts of interest.
References
- 1.Iwata TN, Ramirez JA, Tsang M, Park H, Margineantu DH, Hockenbery DM, and Iritani BM. 2016. Conditional Disruption of Raptor Reveals an Essential Role for mTORC1 in B Cell Development, Survival, and Metabolism. J Immunol 197: 2250–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, Corcoran AE, and Okkenhaug K. 2010. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci Signal 3: ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baracho GV, Cato MH, Zhu Z, Jaren OR, Hobeika E, Reth M, and Rickert RC. 2014. PDK1 regulates B cell differentiation and homeostasis. Proc Natl Acad Sci U S A 111: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu M, Chen Y, Zeng H, Zheng Y, Fu G, Zhu W, Broeckel U, Aggarwal P, Turner A, Neale G, Guy C, Zhu N, Chi H, Wen R, and Wang D. 2017. PLCgamma-dependent mTOR signalling controls IL-7-mediated early B cell development. Nat Commun 8: 1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng H, Yu M, Tan H, Li Y, Su W, Shi H, Dhungana Y, Guy C, Neale G, Cloer C, Peng J, Wang D, and Chi H. 2018. Discrete roles and bifurcation of PTEN signaling and mTORC1-mediated anabolic metabolism underlie IL-7-driven B lymphopoiesis. Sci Adv 4: eaar5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwata TN, Ramirez-Komo JA, Park H, and Iritani BM. 2017. Control of B lymphocyte development and functions by the mTOR signaling pathways. Cytokine Growth Factor Rev 35: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park H, Staehling K, Tsang M, Appleby MW, Brunkow ME, Margineantu D, Hockenbery DM, Habib T, Liggitt HD, Carlson G, and Iritani BM. 2012. Disruption of Fnip1 Reveals a Metabolic Checkpoint Controlling B Lymphocyte Development. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba M, Keller JR, Sun HW, Resch W, Kuchen S, Suh HC, Hasumi H, Hasumi Y, Kieffer-Kwon KR, Gonzalez CG, Hughes RM, Klein ME, Oh HF, Bible P, Southon E, Tessarollo L, Schmidt LS, Linehan WM, and Casellas R. 2012. The folliculin-FNIP1 pathway deleted in human Birt-Hogg-Dube syndrome is required for murine B-cell development. Blood 120: 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF 3rd, Hartley JL, Furihata M, Oishi S, Zhen W, Burke TR Jr., Linehan WM, Schmidt LS, and Zbar B. 2006. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A 103: 15552–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt LS, and Linehan WM. 2015. Clinical Features, Genetics and Potential Therapeutic Approaches for Birt-Hogg-Dube Syndrome. Expert Opin Orphan Drugs 3: 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, and Hall MN. 2008. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 8: 399–410. [DOI] [PubMed] [Google Scholar]
- 12.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, and Weinberg RA. 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7. [DOI] [PubMed] [Google Scholar]
- 13.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, and Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A 103: 13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, and Brinster RL. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538. [DOI] [PubMed] [Google Scholar]
- 15.Meikle L, McMullen JR, Sherwood MC, Lader AS, Walker V, Chan JA, and Kwiatkowski DJ. 2005. A mouse model of cardiac rhabdomyoma generated by loss of Tsc1 in ventricular myocytes. Hum Mol Genet 14: 429–435. [DOI] [PubMed] [Google Scholar]
- 16.Nakada D, Saunders TL, and Morrison SJ. 2010. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, and Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang W, Mueller DL, Pennell CA, Rivard JJ, Li YS, Hardy RR, Schlissel MS, and Behrens TW. 1996. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity 4: 291–299. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn R, Schwenk F, Aguet M, and Rajewsky K. 1995. Inducible gene targeting in mice. Science 269: 1427–1429. [DOI] [PubMed] [Google Scholar]
- 20.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, and Zeng H. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habib T, Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, Knoepfler PS, Andrews S, Rawlings DJ, Eisenman RN, and Iritani BM. 2007. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol 179: 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tampella G, Kerns HM, Niu D, Singh S, Khim S, Bosch KA, Garrett ME, Moguche A, Evans E, Browning B, Jahan TA, Nacht M, Wolf-Yadlin A, Plebani A, Hamerman JA, Rawlings DJ, and James RG. 2015. The Tec Kinase-Regulated Phosphoproteome Reveals a Mechanism for the Regulation of Inhibitory Signals in Murine Macrophages. J Immunol 195: 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy RR, Carmack CE, Shinton SA, Kemp JD, and Hayakawa K. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guidos CJ, Williams CJ, Grandal I, Knowles G, Huang MT, and Danska JS. 1996. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev 10: 2038–2054. [DOI] [PubMed] [Google Scholar]
- 25.McMahon SB 2014. MYC and the control of apoptosis. Cold Spring Harb Perspect Med 4: a014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, and Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxton RA, and Sabatini DM. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell 169: 361–371. [DOI] [PubMed] [Google Scholar]
- 28.Reyes NL, Banks GB, Tsang M, Margineantu D, Gu H, Djukovic D, Chan J, Torres M, Liggitt HD, Hirenallur SD, Hockenbery DM, Raftery D, and Iritani BM. 2015. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc Natl Acad Sci U S A 112: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centini R, Tsang M, Iwata T, Park H, Delrow J, Margineantu D, Iritani BM, Gu H, Liggitt HD, Kang J, Kang L, Hockenbery DM, Raftery D, and Iritani BM. 2018. Loss of Fnip1 alters kidney developmental transcriptional program and synergizes with TSC1 loss to promote mTORC1 activation and renal cyst formation. PLoS One 13: e0197973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh HB, Sciarretta S, Nagashima K, Haines DC, Schneider MD, Adelstein RS, Schmidt LS, Sadoshima J, and Marston Linehan W. 2014. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet 23: 5706–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, and Sabatini DM. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit CS, Roczniak-Ferguson A, and Ferguson SM. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202: 1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, and Sabatini DM. 2013. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wada S, Neinast M, Jang C, Ibrahim YH, Lee G, Babu A, Li J, Hoshino A, Rowe GC, Rhee J, Martina JA, Puertollano R, Blenis J, Morley M, Baur JA, Seale P, and Arany Z. 2016. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev 30: 2551–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, and Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor TFE3. Cell 153: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, and Linehan WM. 2010. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 5: e15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba M, Endoh M, Ma W, Toyama H, Hirayama A, Nishikawa K, Takubo K, Hano H, Hasumi H, Umemoto T, Hashimoto M, Irie N, Esumi C, Kataoka M, Nakagata N, Soga T, Yao M, Kamba T, Minami T, Ishii M, and Suda T. 2018. Folliculin Regulates Osteoclastogenesis Through Metabolic Regulation. J Bone Miner Res 33: 1785–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siggs OM, Stockenhuber A, Deobagkar-Lele M, Bull KR, Crockford TL, Kingston BL, Crawford G, Anzilotti C, Steeples V, Ghaffari S, Czibik G, Bellahcene M, Watkins H, Ashrafian H, Davies B, Woods A, Carling D, Yavari A, Beutler B, and Cornall RJ. 2016. Mutation of Fnip1 is associated with B-cell deficiency, cardiomyopathy, and elevated AMPK activity. Proc Natl Acad Sci U S A 113: E3706–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen HC, Kanai M, Inoue-Yamauchi A, Tu HC, Huang Y, Ren D, Kim H, Takeda S, Reyna DE, Chan PM, Ganesan YT, Liao CP, Gavathiotis E, Hsieh JJ, and Cheng EH. 2015. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat Cell Biol 17: 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haughn L, Hawley RG, Morrison DK, von Boehmer H, and Hockenbery DM. 2003. BCL-2 and BCL-XL restrict lineage choice during hematopoietic differentiation. J Biol Chem 278: 25158–25165. [DOI] [PubMed] [Google Scholar]
- 41.Liu W, Chen Z, Ma Y, Wu X, Jin Y, and Hou S. 2013. Genetic characterization of the Drosophila birt-hogg-dube syndrome gene. PLoS One 8: e65869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunlop EA, Seifan S, Claessens T, Behrends C, Kamps MA, Rozycka E, Kemp AJ, Nookala RK, Blenis J, Coull BJ, Murray JT, van Steensel MA, Wilkinson S, and Tee AR. 2014. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy 10: 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagashima K, Fukushima H, Shimizu K, Yamada A, Hidaka M, Hasumi H, Ikebe T, Fukumoto S, Okabe K, and Inuzuka H. 2017. Nutrient-induced FNIP degradation by SCFbeta-TRCP regulates FLCN complex localization and promotes renal cancer progression. Oncotarget 8: 9947–9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, Lian G, Liu Q, Guo H, Yin Z, Ye Z, Han J, Wu JW, Yin H, Lin SY, and Lin SC. 2014. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 20: 526–540. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy JC, Khabibullin D, Hougard T, Nijmeh J, Shi W, and Henske EP. 2019. Loss of FLCN inhibits canonical WNT signaling via TFE3. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baba M, Toyama H, Sun L, Takubo K, Suh HC, Hasumi H, Nakamura-Ishizu A, Hasumi Y, Klarmann KD, Nakagata N, Schmidt LS, Linehan WM, Suda T, and Keller JR. 2016. Loss of Folliculin Disrupts Hematopoietic Stem Cell Quiescence and Homeostasis Resulting in Bone Marrow Failure. Stem Cells 34: 1068–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villegas F, Lehalle D, Mayer D, Rittirsch M, Stadler MB, Zinner M, Olivieri D, Vabres P, Duplomb-Jego L, De Bont E, Duffourd Y, Duijkers F, Avila M, Genevieve D, Houcinat N, Jouan T, Kuentz P, Lichtenbelt KD, Thauvin-Robinet C, St-Onge J, Thevenon J, van Gassen KLI, van Haelst M, van Koningsbruggen S, Hess D, Smallwood SA, Riviere JB, Faivre L, and Betschinger J. 2019. Lysosomal Signaling Licenses Embryonic Stem Cell Differentiation via Inactivation of TFE3. Cell Stem Cell 24: 257–270 e258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.