Highlights
-
•
Osteoporosis is a disease that impacts over 200 million people worldwide.
-
•
The probiotic bacterium Lactobacillus reuteri (L. reuteri) has been shown to prevent bone loss during estrogen deficiency.
-
•
Lactobacillic acid is important for L. reuteri-induced suppression of in vitro osteoclastogenesis.
-
•
Osteoclastogenesis was inhibited by L. reuteri and lactobacillic acid via GPR120 signaling.
Keywords: Bone, Probiotic, Osteoclast, Lactobacillic acid, Osteoporosis
Abstract
Osteoporosis is a disease that impacts over 200 million people worldwide. Taking into consideration the side effects stemming from medications used to treat this illness, investigators have increased their efforts to develop novel therapeutics for osteoporosis. In a previous study, we demonstrated that ovariectomy-induced bone loss in mice was prevented by treatment with the probiotic bacterium Lactobacillus reuteri 6475 (L. reuteri), an effect that correlated with reduced osteoclastogenesis in the bone marrow of L. reuteri treated mice. We also demonstrated that L. reuteri directly inhibited osteoclastogenesis in vitro. To better understand how L. reuteri impacts osteoclast formation, we used additional in vitro analyses to identify that conditioned supernatant from L. reuteri inhibited osteoclastogenesis at the intermediate stage of fused polykaryons. To elucidate the effect of L. reuteri treatment on host cell physiology, we performed RNAseq at multiple time points during in vitro osteoclastogenesis and established that L. reuteri downregulated several KEGG pathways including osteoclast differentiation as well as TNF-α, NF-κB, and MAP kinase signaling. These results were consistent with Western Blot data demonstrating that NF-κB and p38 activation were decreased by L. reuteri treatment. We further identified that lactobacillic acid (LA), a cyclopropane fatty acid produced by L. reuteri, contributed significantly to the suppression of osteoclastogenesis. Additionally, we demonstrated that L. reuteri is signaling through the long chain fatty acid receptor, GPR120, to impact osteoclastogenesis. Overall, these studies provide both bacterial and host mechanisms by which L. reuteri impacts osteoclastogenesis and suggest that long chain fatty acid receptors could be targets for preventing osteoclastogenesis.
1. Introduction
Osteoporosis remains a growing problem worldwide. According to the International Osteoporosis Foundation, over 200 million people have low bone mass and are at increased risk for sustaining bone fractures. Unfortunately, several medications used to treat osteoporosis have undesirable side effects. For example, the first line use of bisphosphonates to suppress osteoclast activity can result in the disruption of normal bone remodeling, which can result in atypical femur fractures or osteonecrosis of the jaw in a small number of patients (Rasmusson and Abtahi, 2014; Khan et al., 2015; Crandall et al., 2014). Hormone replacement therapy, while highly effective for preventing bone loss, can increase risk of invasive breast cancer and venous thromboembolism (Grossman et al., 2017). While these responses are rare, the fear of such side effects has reduced the number of patients taking osteoporosis medications. Thus, there is an impetus for the development of novel therapeutics with few side effects to combat osteoporosis.
One potential option is to utilize our understanding of the role that bacteria, especially in the intestine, play in shaping bone health. The intestinal microbiota is a collection of microbes, including bacteria, archaea, viruses, helminths, fungi, and their genetic material, that inhabit our gut. Investigating host microbe interactions at this interface has led to recognizing microbiota as a critical determinant of health and disease. In particular, discoveries demonstrating the impact of the microbiota on bone health have led to the emergence of the term osteomicrobiology, which refers to the study of the role of microbes in bone health and disease (Jones et al., 2017; Yan and Charles, 2017). Furthermore, increased attention has been given to the use of probiotics, defined as live microorganisms that confer health benefits when consumed according to the Food and Agriculture Organization (FAO) and World Health Organization (WHO), for the treatment of bone diseases (Morelli and Capurso, 2012). The efficacy of several probiotic strains in improving bone health has been demonstrated in multiple animal models (Narva et al., 2004; Narva et al., 2007; Mutuş et al., 2006; Chiang and Pan, 2011; Tomofuji et al., 2012; McCabe et al., 2013; Britton et al., 2014; Collins et al., 2016; Ohlsson et al., 2014; Schwarzer et al., 2016).
In our previous studies, we showed that treating OVX mice with the immunomodulatory probiotic strain Lactobacillus reuteri PTA 6475 (L. reuteri) resulted in decreased osteoclast formation from primary bone marrow outgrowth studies (Britton et al., 2014). Moreover, using the in vitro RAW264.7 cell line, we demonstrated that the differentiation of this monocyte/macrophage cell line into osteoclasts was arrested by the addition of a <3 kDa cell culture supernatant (CCS) fraction from L. reuteri 6475. Other studies involving probiotics and bone health have highlighted the efficacy of different bioactive compounds in preventing bone loss and this prevention has often times been attributed to suppression of osteoclastogenesis (Narva et al., 2007; Rahman et al., 2006; Ewaschuk et al., 2006; Li et al., 2016; Tyagi et al., 2018). Together, these studies strongly suggest that the identification of bioactive molecule(s), produced by bacteria that target osteoclastogenesis, may lead to understanding how bacteria contribute to bone health and optimize probiotic strain selection for treating bone disease.
In this study, we expanded the initial findings on L. reuteri suppression of osteoclastogenesis and characterized this interaction by describing the host response following L. reuteri stimulation. Through a guided RNA sequencing experiment, we identified that L. reuteri modulates genes involved in osteoclastogenesis as well as TNFα and NFκB pathways. We further demonstrate that L. reuteri suppression of osteoclastogenesis is in part mediated by lactobacillic acid (LA) interaction with the GPR120 receptor. Together, our studies identify specific host mechanisms as well as bacterial mechanisms by which L. reuteri modulates osteoclastogenesis. These findings have important implications in future drug development for bone disease.
2. Materials and methods
2.1. Chemicals and reagents used
The GPR40 and GPR120 antagonists, DC260126 (Cat. No. 5357) and AH7614 (Cat. No. 5256), respectively, were purchased from Tocris Biosciences. Lactobacillic acid (LA, also known as phytomonic acid) was purchased from Cayman chemical. The antibodies, p38 (Cat. No. 8610), phosphorylated p38 (Cat. No. 4511), ERK (Cat. No. 3552), phosphorylated ERK (Cat. No. 4377), JNK (Cat. No. 9258), phosphorylated JNK (Cat. No 9251), p65 (Cat. No. 8242), phosphorylated p65 (Cat. No. 3033), and beta actin (Cat. No. 4970) used for this study were purchased from Cell Signaling Technology.
2.2. Bacterial strains used in this study and growth conditions
L. reuteri ATCC PTA 6475 was provided to the Britton laboratory by Biogaia inc. (Sweden) and was cultured anaerobically in deMan, Rogosa, Sharpe media (MRS, Difco) for 18 h at 37 °C. To generate L. reuteri cell-free conditioned supernatant (CCS), the overnight culture was subcultured into fresh MRS and grown until log phase (OD600 = 0.4) and cells were pelleted by centrifugation at 4000 rpm for 10 min. The pellet was washed twice with sterile PBS to remove any residual MRS. The L. reuteri CCS was generated by resuspending the bacterial cell pellet in Minimum Essential Medium (MEM-α, Invitrogen) to an OD600 = 3.0 (5 ml in 50 ml BD conical tube) and incubated for 3 h at 37 °C with gentle orbital shaking (60 rpm). The bacterial cells were pelleted and the supernatant collected, filter-sterilized using a PVDF membrane filter (0.22 μm pore size, Millipore), and fractionated (Amicon filter, Millipore) to include only the <3 kDa fraction. The L. reuteri CCS were pipetted into 96-well plates (deep well, 3 ml volume) in 250 μL aliquots, lyophilized and stored at −80 °C. Sterile MEM-α also underwent processing in parallel to serve as the vehicle control for each experiment. This was also performed with the bacterial strains: E.coli DH5-α.
2.3. Culture conditions and osteoclastogenesis differentiation assay
The murine macrophage cell line, RAW264.7, was obtained from ATCC and maintained in phenol red-free medium (MEM-α, Invitrogen) supplemented with charcoal stripped fetal bovine serum (Invitrogen) at 37 °C with 5% CO2. In 24-well tissue culture grade plates (Costar), 2*104 cells were seeded. Following one day of incubation, cells were stimulated for differentiation with the addition of receptor activator of NF-kappa B ligand (RANKL, 100 ng/ml, R&D systems). Lyophilized L. reuteri CCS or CCS from other bacterial strains were resuspended in culture medium and used to treat the cells. For all experiments, each well contained 20% (0.2x) of the OD600 = 3.0 CCS (50 μL) unless otherwise noted. For dose response experiments, a CCS of 100% is equivalent to containing 250 μL of CCS (OD600 = 3.0) in each well. Fresh medium is replenished after every 2 days for a week. On day 7, cells were fixed and stained for tartrate-resistant acid phosphatase (TRAP) using a commercially available staining kit (Sigma Cat. No. 387A). Giant cells with ≥ 3 nuclei that stained positive were considered osteoclasts.
2.4. Pharmacological inhibition studies
For the antagonist studies, the osteoclastogenesis assay was performed as previously described (Britton et al., 2014) with an additional preincubation step with the inhibitor. Prior to the addition of L. reuteri or LA, 1 μM of GPR40 or GPR120 antagonist was added for 1 h. The fresh medium that was replenished every 2 days contained 1 μM of GPR40 or GPR120 antagonist. Following 7 days of culture, the cells were fixed, stained for TRAP, and the enumeration of osteoclasts was performed as previously described (Britton et al., 2014).
2.5. Cell viability or metabolic activity assay
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was used to assess cell viability as a function of metabolic activity. This is signified by the NAD(P)H-dependent oxidoreductase enzymatic activity of a cell. In the presence of these enzymes, MTT becomes reduced to formazan, which is an insoluble product possessing chromogenic properties. Briefly, 1 × 10^4 RAW264.7 cells are seeded on a 96-well culture plate (Cat. No. 07-200-760, Fisher Scientific) and incubated at 37C with 5% CO2. Following 24 h, the media is exchanged with fresh media and the cells are stimulated with the vehicle control or cell culture supernatant from L. reuteri. The cells are co-incubated with the different treatment conditions for 4 and 24 h. Upon completion of each treatment, 0.02 ml of MTT solution (5 mg/ml) dissolved in MEMα + 10% FBS was added to each well and the plate is incubated at 37 °C with 5% CO2 for 4 h. The plate is washed twice with PBS and then DMSO is added to each well to dissolve the formazan formed. The plate is shaken for 5 min and absorbance readings at 570 nm are taken to measure the amount of metabolic activity for each condition. The data is expressed as a ratio between the specific treatment groups to that of the control group containing untreated cells.
2.6. RNA extraction from RAW264.7 cells and sequencing
RNA extraction was performed using TRIzol Reagent (Cat. No. 15596018, Thermo Fisher Scientific) according to the manufacturer’s instructions. Library preparation and sequencing of Mus musculus transcriptome was performed by the Baylor Human Genome Sequencing Center. Libraries were prepared from 500 ng of total RNA with the TruSeq Stranded Total RNA Library Prep Kit according to the manufacturer’s directions (Illumina). Sequencing was performed on an Illumina HiSeq 2000 platform. Sequencing data were mapped onto the Mus musculus 10 genome available from UCSC Genome Bioinformatics.
2.7. Analysis of RNA sequencing data
On average, there were 31,060,000 high quality bases sequenced per sample with a mean fragment length of 225 bases per read. There was an average of 51,905 transcripts detected per sample. The count table signifying the amount of reads that fell into each gene was generated with using Bioconductor with Rsamtools and GenomicAlignments in R. Differential expression analysis was performed by the DESeq package in R (Anders et al., 2013). Pathway analysis was performed by GAGE in R (Luo et al., 2009).
2.8. Quantitative reverse transcriptase (RT) polymerase chain reaction (qPCR)
Following RNA extraction, cDNA synthesis was performed with Superscript III Reverse Transcriptase (Cat. No. 8080093, Thermo Fisher Scientific). A total of 1 μg of RNA was reverse transcribed. Briefly, an Eppendorf Mastercycler EP S was preheated to 65 °C. The mixture containing RNA, 100 ng of random hexamers, (Cat. No.C1181, Promega) and 1 μl of 10 nM dNTPs (Cat. No. 18427088) was placed into the thermocycler for 5 min. Following 1 min on ice, a mixture containing the reverse transcriptase and RNaseOUT was added to complete the cDNA synthesis. A cycle of 10 min at 25 °C, 50 min at 50 °C, and then 85 °C for 5 min was used. The cDNA was used immediately or stored at −20 °C. The qPCR reactions reactions contained 1 μl of cDNA, 1 μl of each forward and reverse primer (10 μM), 7 μl of nuclease free water, and 10 μl of Power SYBR Green PCR Master Mix (Cat. No. 4367659, Thermo Fisher Scientific). A 2-step PCR amplification protocol was used with acquisition at the annealing and melting curve steps. The protocol included an initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturing at 95 °C for 10 s, annealing at 51 °C for 20 s. A melting curve was performed at the end at 95 °C for 15 s and ramping up from 60 °C to 95 °C at a rate of +0.2 °C/sec. Data analysis was performed according to the method described by Pfaffl (2001).
2.9. Primer sequences for qPCR
The following primer pairs were used for the RNA sequencing validation studies:
2.10. Western blot analysis
The osteoclastogenesis assay was performed as previously described and proteins were extracted using the commercially available Cell Extraction Buffer (Cat. No. FNN0011, Thermo Fisher Scientific). It was used according to the manufacturer’s protocol. Timepoints were taken at 15, 30, 45, and 60 min. Chemiluminescent imaging was performed with the ProteinSimple FluorChem E system. Analysis was performed by comparing densitometry measurements obtained using the ImageJ software package (Girish and Vijayalakshmi, 2004). The following antibodies were used from Cell Signaling Technology: 1) Phospho-NF-κB p65 (Ser536) (93H1) Rabbit mAb (Cat No. 3033), 2) Phospho-p38 MAPK (Thr180/Tyr182) (D3F9) XP® Rabbit mAb (Cat. No. 4511), 3) p38 MAPK (D13E1) XP® Rabbit mAb (Cat. No.8690), 4) Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® Rabbit mAb (HRP Conjugate) (Cat. No. 8544), 5) Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® Rabbit mAb (HRP Conjugate) (Cat. No. 8544), 6) Phospho-SAPK/JNK (Thr183/Tyr185) (81E11) Rabbit mAb (Cat. No. 4668), 7) SAPK/JNK Antibody (Cat. No. 9252), 8) β-Actin (13E5) Rabbit mAb (Cat. No. 4970).
2.11. Bone resorption assay
Osteoassay (Cat. No. 3988) plates coasted with inorganic crystalline calcium phosphate were purchased from Corning Inc. The osteoclastogenesis assay was performed as previously described. Resorption pits were measured using the NIH imaging software, ImageJ (Girish and Vijayalakshmi, 2004).
2.12. Statistical analysis
The results presented are as means +/- SEM. An unpaired Student’s t test was used to assess differences between groups. One-way ANOVA analysis was applied when more than 2 groups were compared. The cutoff for significance was p ≤ 0.05. The post-hoc Tukey HSD test was used as a follow-up to experiments with statistically significant ANOVA results.
3. Results
3.1. L. reuteri suppressed osteoclastogenesis in a concentration-dependent manner
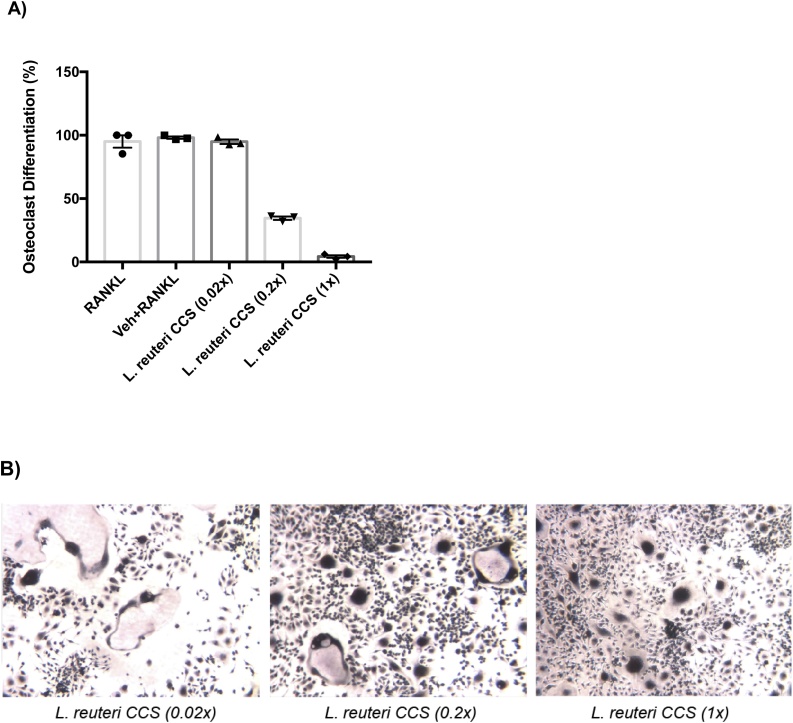
In a previous study we showed that concentrated conditioned supernatant (CCS) from L. reuteri inhibits osteoclastogenesis in vitro from RAW264.7 cells (Britton et al., 2014). This was also demonstrated in primary murine bone marrow macrophages, as osteoclast differentiation could be suppressed in cells treated with conditioned media from L. reuteri (Supplementary Fig. 1). Further characterization of this response was performed using the RAW264.7 cell line. We confirm here that L. reuteri CSS inhibited osteoclastogenesis in a concentration-dependent manner (Fig. 1). We further show specificity of this response by demonstrating that Escherichia coli CSS did not affect osteoclast formation in the same cell culture model (Supplementary Fig. 2).
Fig. 1.
Dose-dependent inhibition of osteoclast formation byL.reuteri. Osteoclast differentiation was induced with the addition of 100 ng/mL RANKL. Giant multinucleated cells that stained positive for TRAP and with ≥ 3 nuclei were considered osteoclasts (A). Light microscopy images (B) were taken and demonstrate an increase in osteoclast formation with decreasing levels of L. reuteri CCS used. This was performed three times in total and the reported result is a representative experiment with associated standard deviation, *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions as determined by one-way ANOVA.
3.2. L. reuteri halts osteoclastogenesis at the polykaryon stage
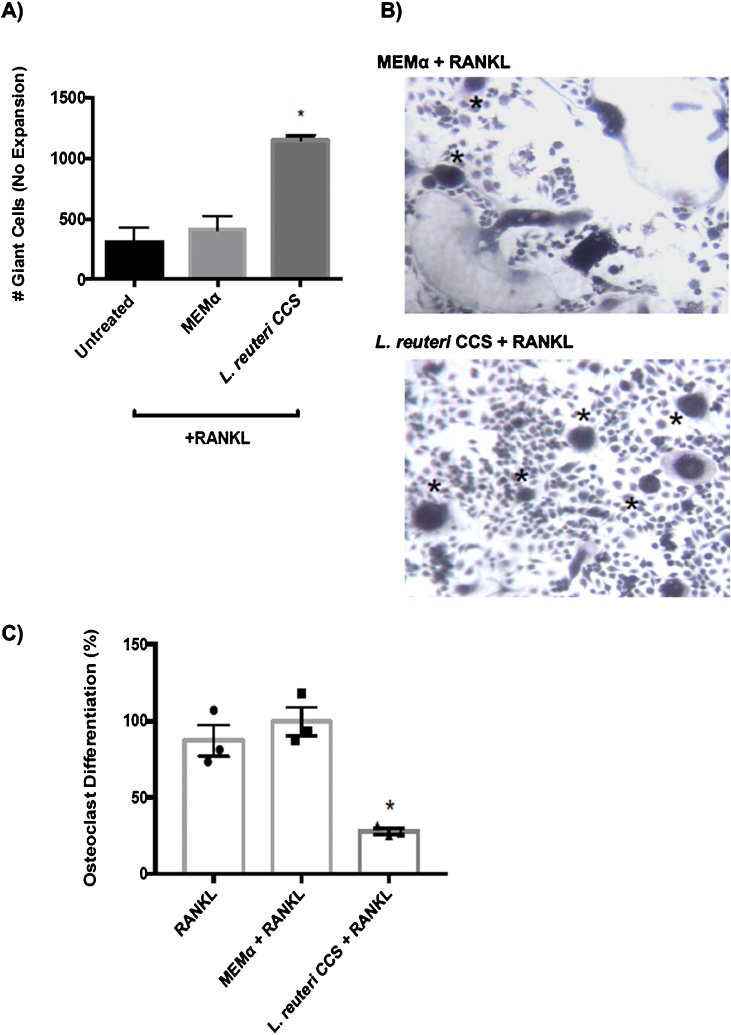
Osteoclast differentiation is a multi-step process consisting of osteoclast precursor activation, fusion, and differentiation before fully mature osteoclasts are developed (Yang et al., 2008a; Yagi et al., 2005; Cuetara et al., 2006; Asagiri and Takayanagi, 2007; Ikeda and Takeshita, 2015). To gain a better understanding at which stage(s) L. reuteri CSS inhibits osteoclastogenesis, the phenotypic progression was observed for the duration of the assay by microscopy (Supplementary Fig. 3). During an intermediate stage of osteoclastogenesis, mononuclear preosteoclasts fuse to form giant multinucleated cells known as polykaryons (Teitelbaum et al., 1997; Teitelbaum et al., 1997; Zhang et al., 2000; Weinberg et al., 1984); these cells display cytoskeletal actin rearrangement and increased cytosolic space (Kwon et al., 2015; Aharon and Bar-Shavit, 2006). Interestingly L. reuteri treatment resulted in an accumulation of fused polykaryons by days 5 and 7 (Fig. 2a, b) suggesting that differentiation was halted at this stage (Boyle et al., 2003; Brodbeck and Anderson, 2009; Miyamoto et al., 2012).
Fig. 2.
Progression of osteoclastogenesis over time and accumulation of polykaryons. RAW264.7 cells were stimulated for osteoclast differentiation with RANKL (100 ng/ml) and treated with a vehicle (MEM-α) or L. reuteri CCS. (A, B) Fused polykaryons (asterisks) were present in both conditions but L. reuteri treatment led to an accumulation of them by day 7. (C) After a 10 day washout period, osteoclast differentiation was still inhibited in cells treated by L. reuteri CCS at day 17. *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions as determined by one-way ANOVA. n = 3.
To test whether the osteoclastogenesis process was delayed rather than inhibited, additional experiments were performed where the duration of the experiment was extended to 17 days. Following the final treatment on day 5, there was a washout period where only fresh medium containing RANKL was replenished without L reuteri CCS treatment. At the end of day 17, cells treated with L. reuteri still had suppressed osteoclastogenesis with an accumulation of polykaryons (Fig. 2c). Taken together, these results indicated that L. reuteri halted osteoclastogenesis at a fused polykaryon stage and that the effects were long lasting.
3.3. L. reuteri suppresses mineral resorption in RANKL-stimulated RAW264.7 cells
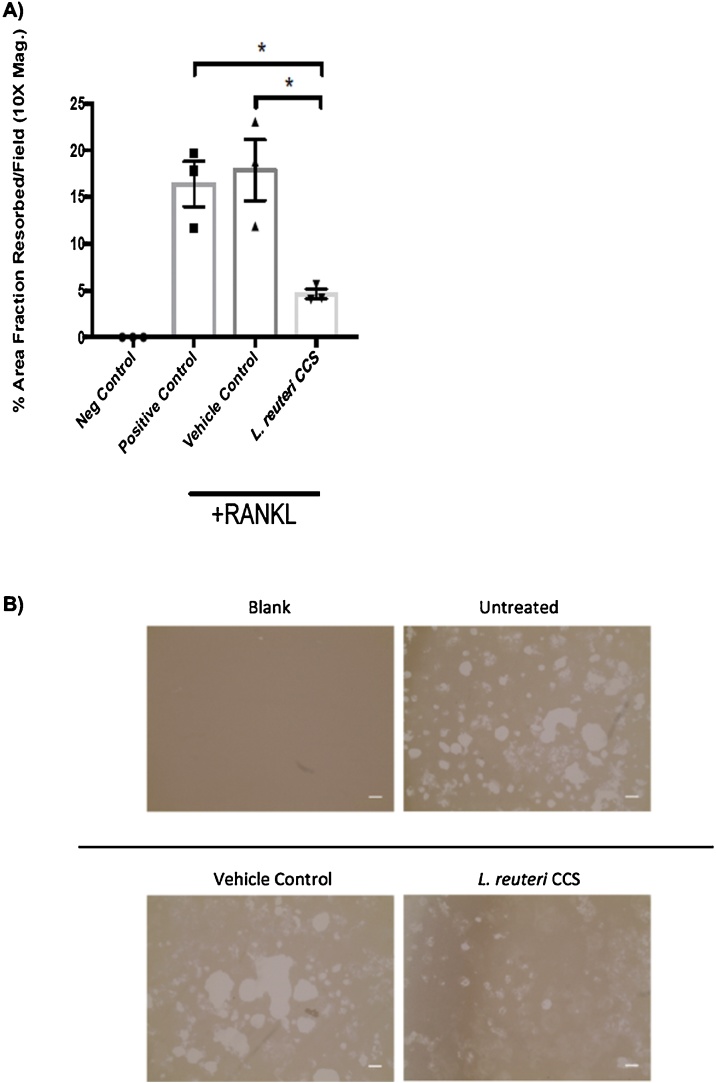
To address if inhibition of osteoclast formation by L. reuteri at the polykaryon stage impacted osteoclast function, we performed bone resorption assays using Osteoassay plates coated with calcium phosphate to mimic bone resorption. These plates were incubated with RAW264.7 cells for 24 h and they stimulated with RANKL. To assess the impact of L. reuteri on calcium phosphate resorption, we further added 0.2X CSS or equivalent vehicle control. Consistent with the ability to suppress osteoclastogenesis, L. reuteri CCS significantly decreased the amount of mineral resorption compared to the controls by ∼75 % (Fig. 3A, B).
Fig. 3.
Suppression of mineral resorption byL.reuteri. RAW264.7 cells were plated for on Osteoassay plates for 1 day and then stimulated concurrently with RANKL (100 ng/ml) and either the vehicle control (MEM-α) or L. reuteri CCS. Fresh medium was replenished every 2 days. The analysis was performed using the Image J software package to measure densitometry. (A) L. reuteri CCS significantly decreased the amount of absorption from calcium and phosphate coated plates. (B) The microscopy images also support that less absorption had taken place in the presence of L. reuteri CCS. Three biological replicates are depicted with associated standard error of mean, *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions as determined by one-way ANOVA. Scale bars signify 100 μm increments. n = 3.
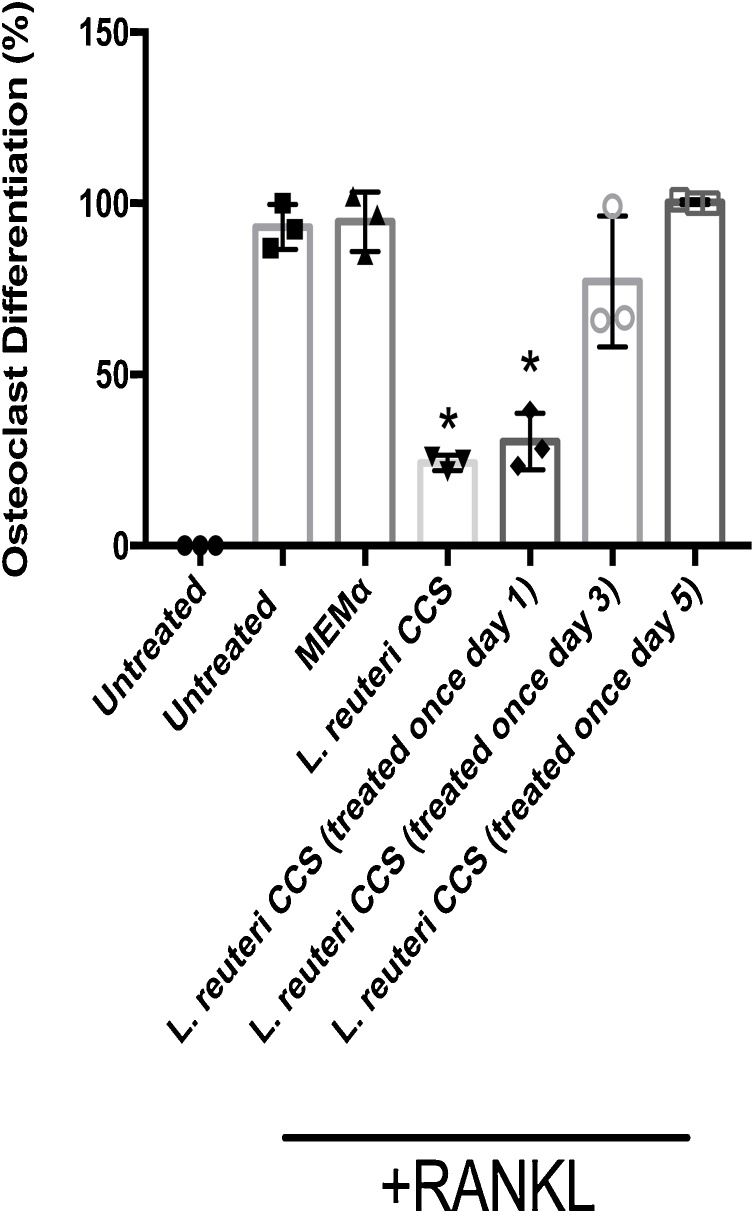
3.4. Treatment with L. reuteri CCS early during osteoclastogenesis is required for inhibition of osteoclast formation
Because osteoclastogenesis was inhibited at a late stage, we were interested in identifying when CCS treatment was required for inhibition. To test this idea, we performed a time course experiment where RAW264.7 cells were stimulated by RANKL for osteoclast differentiation and treated with L. reuteri 0.2X CCS throughout the duration of the assay (Fig. 1A) or added just once on day 1, 3, or 5. The addition of L. reuteri CCS on day 1 alone inhibited osteoclast differentiation while treatment at day 3 (p = 0.07) or day 5 had only a modest to no impact on osteoclastogenesis (Fig. 4). These results, coupled with the phenotypic studies (Fig. 2b, c), suggested that L. reuteri targeted an early signaling event during osteoclastogenesis that resulted in halting development at the late polykaryon stage.
Fig. 4.
Impact ofL.reuteri CCS on osteoclast differentiation at different time points. RAW264.7 cells were plated and osteoclast differentiation was induced with the addition of 100 ng/mL RANKL. L. reuteri CCS was added normally when the media was replenished (days 1, 3, and 5) or just once on day 1, 3, or 5. After 7 days, the number of giant multinucleated (≥ 3 nuclei) cells staining positive for TRAP were quantified. Treatment by L. reuteri CCS on day 1 was sufficient to suppress osteoclastogenesis. Three biological replicates are depicted with associated standard error of mean, *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions as determined by one-way ANOVA.
3.5. Lactobacillic acid is involved in L. reuteri suppression of osteoclastogenesis
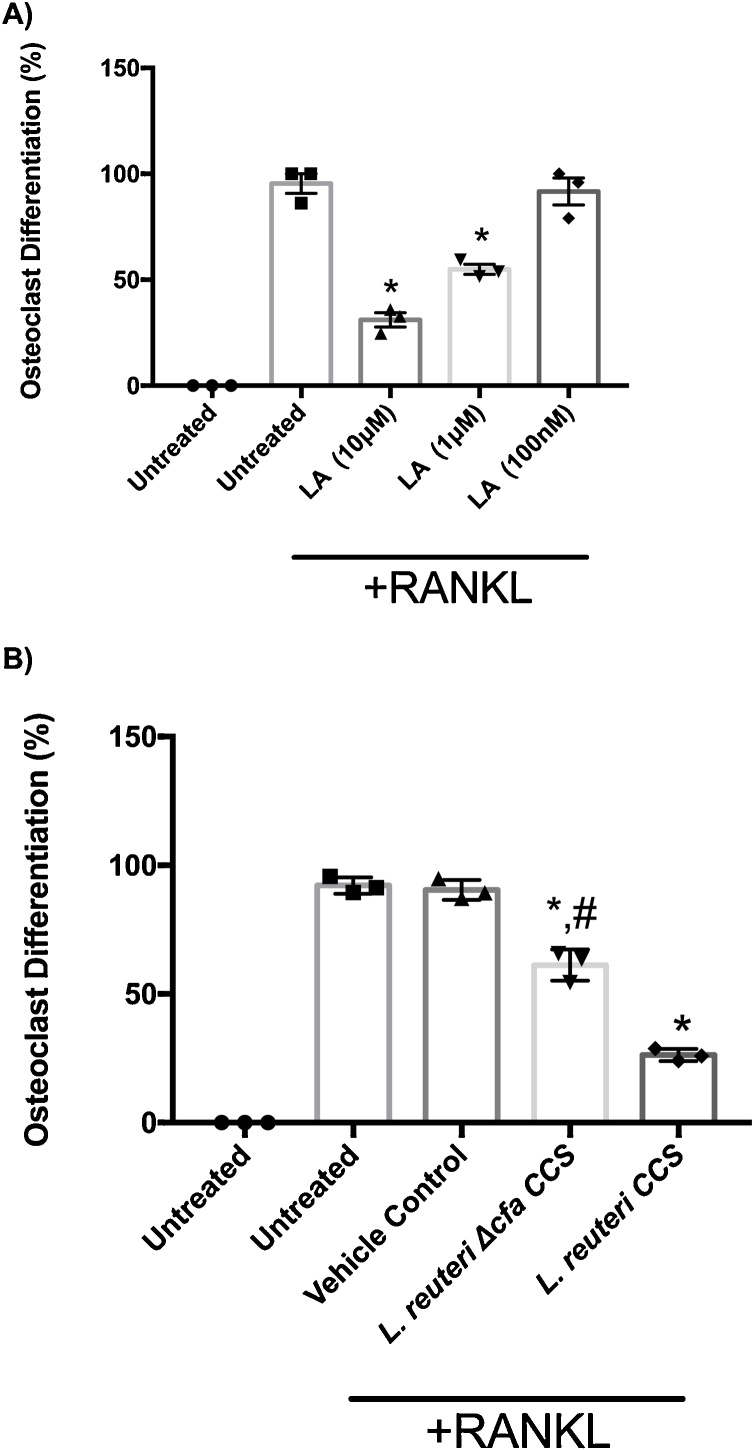
It was previously shown that the ability of L. reuteri to suppress TNF-α production from a human monocyte line was related to the presence of lactobacillic acid (LA), a long chain cyclopropyl fatty acid (Jones et al., 2011). Other long chain fatty acids have been shown to suppress in vitro osteoclastogenesis (Kwon et al., 2015; Drosatos-Tampakaki et al., 2014). Therefore, to test if LA is able to directly inhibit osteoclastogenesis, RAW264.7 macrophages were incubated without or with varying concentrations of LA, during osteoclastogenesis. Our results demonstrate that LA caused a concentration-dependent inhibition of osteoclast formation (Fig. 5a). A concentration of 1μM LA was sufficient to suppress osteoclast formation by 50% whereas 10 μM suppressed 80%. These result show that LA is important for the inhibition of osteoclast formation.
Fig. 5.
Lactobacillic acid (LA) involved with the suppression of osteoclastogenesis. RAW264.7 cells were plated and osteoclast differentiation was induced with the addition of 100 ng/mL RANKL. (A) Dose-dependent suppression of osteoclastogenesis was observed with LA at the concentrations of 10 μM, 1 μM, and 100 nM. (B) A mutant unable to produce LA in the L. reuteri genetic background was not as effective as the WT strain in suppressing osteoclastogenesis. Three biological replicates are depicted with associated standard error of mean, *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions, #p < 0.05 compared to L. reuteri as determined by one-way ANOVA.
LA is formed from vaccenic acid by the action of the enzyme cyclopropyl fatty acid synthase that is encoded by the cfa gene in L. reuteri 6475. To directly test whether L. reuteri CCS inhibits osteoclastogenesis via LA, we used a L. reuteri Δcfa mutant, in which the cyclopropyl fatty acid synthase (cfa) gene in disrupted and is unable to produce LA (Jones et al., 2011). Interestingly, we found that CCS from the Δcfa mutant was less effective (by more than 50%) in suppressing osteoclastogenesis in comparison with WT L. reuteri (Fig. 5b). Though, the L. reuteri Δcfa mutant retained some ability to suppress osteoclastogenesis.
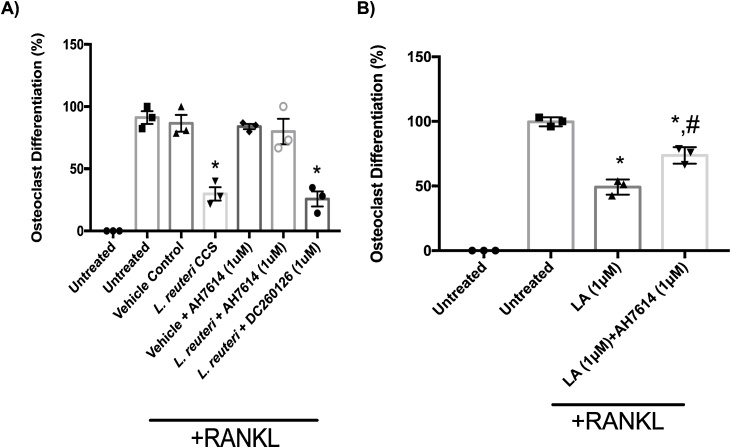
To identify possible mechanisms by which LA produced by L. reuteri mediates anti-osteoclastogenic effects, we focused on the free fatty acid receptor 4 (FFAR4; aka GPR120), a G protein-coupled receptor that has been shown to be a receptor for long chain fatty acids (Oh et al., 2014; Oh et al., 2010). Interestingly, activation of GPR120 in RAW264.7 cells and bone marrow macrophages suppresses osteoclastogenesis (Kim et al., 2015). Therefore, we hypothesized that LA from L. reuteri CCS inhibits osteoclastogenesis potentially via GPR120 activation. To test this, we induced osteoclast differentiation of RAW264.7 cells and treated them with L. reuteri CCS in the absence or presence of a GPR120 antagonist (AH7614). Inhibition of GPR120 with 1 μM AH7614 completely abolished the ability of L. reuteri CCS to inhibit osteoclast formation (Fig. 6a). GPR40 is another receptor for long chain fatty acids and has been shown to have a positive impact on bone health (Covington et al., 2006; Wauquier et al., 2013). Blocking GPR40 signaling with an antagonist (DC260126) however, did not impact the ability of L. reuteri to suppress osteoclastogenesis.
Fig. 6.
Suppression of osteoclastogenesis byL.reuteri was mediated through GPR120 signaling. RAW264.7 cells were plated. After 24 h, the cells were pretreated with 1 μM of AH7614 for 1 h. Then, osteoclast differentiation was induced with the addition of 100 ng/mL RANKL. Cell differentiation medium (containing RANKL and the inhibitor) was replenished every 2 days. (A) The presence of the inhibitor attenuated the suppression of osteoclastogenesis by L. reuteri. (B) Pharmacological inhibition of GPR120 decreased the ability of LA to suppress osteoclast formation. Three biological replicates are depicted with associated standard error of mean, *p < 0.05 compared to untreated and MEM-α (vehicle control), #p <0.05 compared to LA conditions as determined by one-way ANOVA.
To directly demonstrate LA inhibition of osteoclast formation occurs via GPR120, we induced osteoclastogenesis of RAW 264.7 cells in the absence or presence of AH7614 (GPR120) antagonist as well as LA. Again, we observed that 1 μM LA reduced osteoclast formation by 50% and that addition of AH7614 was able to partially block this suppression (Fig. 6b). However, it is worth noting that attempts to increase the concentration of the antagonist past 2.5 μM resulted in cellular toxicity. Nevertheless, these results support the hypothesis that L. reuteri inhibits osteoclastogenesis via LA stimulation of GPR120.
3.6. Transcriptomic profiling of RAW264.7 cells during L. reuteri treatment supports the suppression of osteoclastogenesis
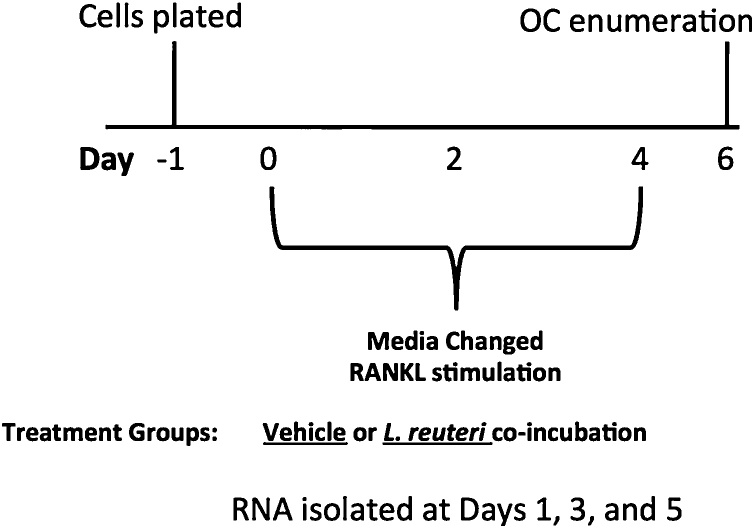
To gain a better understanding of how L. reuteri impacts osteoclastogenesis, we performed an RNA sequencing experiment to measure the levels of gene expression from the entire transcriptome at days 1, 3 and 5 after induction of osteoclast formation by RANKL. Cells were also treated with the vehicle control or L. reuteri CCS, with biological replicates for each condition at each timepoint (Fig. 7). Differential gene expression analysis was performed using the Bioconductor software package DESeq2 (Love et al., 2014). Using a fold change cutoff of 3, we observed 2,993 genes were differentially regulated by L. reuteri in comparison to the vehicle control when combining all time points (Table 1). However, there were substantially more genes differentially regulated at the day 3 and 5 timepoints.
Fig. 7.
Experimental layout for gene expression analysis. RAW264.7 cells were plated for 1 day and then stimulated concurrently with RANKL (100 ng/ml) and either the vehicle control (MEM-α) or L. reuteri CCS. RNA was extracted at days 1, 3, and 5.
Table 1.
Genes with significantly changed expression between vehicle and L. reuteri treatment (Fold change > 3). V denotes Vehicle (MEM-α), T denotes treatment, and the numbers signify time points day 1, 3, and 5.
| V1T1 | V3T3 | V5T5 | |
|---|---|---|---|
| Up-regulated | 24 | 1334 | 1635 |
| Down-regulated | 21 | 3097 | 2611 |
The gage software package was used to gain a better understanding of pathways that were differentially regulated by L. reuteri (Luo et al., 2009). The cutoff for statistical significance was determined using a q value < 0.05 (p value that takes into account the false discovery rate based off of multiple comparisons). Similar to the gene expression data, more KEGG pathways were differentially regulated by L. reuteri at days 3 and 5 with 90 out of the total 98 pathways being significant at those 2 time points (Table 2).
Table 2.
Pathways differentially regulated between vehicle and L. reuteri treatment. V denotes Vehicle (MEM-α), T denotes treatment, and the numbers signify time points day 1, 3, and 5.
| V1T1 | V3T3 | V5T5 | |
|---|---|---|---|
| Up-regulated | 4 | 2 | 4 |
| Down-regulated | 4 | 43 | 41 |
At day 1, few changes were observed. There was an activation of 4 pathways and down-regulation of 4 pathways, but none that were involved with osteoclast differentiation (Supplementary Table 1). Out of the up-regulated pathways, one was associated with innate immunity (Complement and coagulation cascades, mmu04610) and another associated with the detection of fatty acids (PPAR signaling pathway, mmu 03320). Notably, the down-regulated pathways suggested that cell cycle and/or proliferation were affected by L. reuteri treatment (Supplementary Table 1). However, light microscopy and trypan blue exclusion staining did not reveal any discernible phenotypic or viability differences (Fig. 5). Additionally, an MTT assay was utilized to detect metabolic activity and no differences were observed up to 3 days following L. reuteri treatment (data not shown).
By day 3, there were 2 pathways that were up-regulated and 43 that were down-regulated (Supplementary Table 2). Notable pathways that were down-regulated included osteoclast differentiation (mmu04380), NF-kappa B signaling (mmu04064), MAPK signaling (mmu04010), and TNF signaling (mmu04668). Pathway analysis from the day 5 timepoint suggested that the majority of the processes being impacted involved cellular metabolism and immune signaling (Supplementary Table 3). Consistent with day 3, osteoclast differentiation, NF-kappa B signaling, MAPK signaling, and TNF signaling were down-regulated.
To validate the data set, we performed quantitative PCR analysis of genes that were well characterized in the osteoclast differentiation pathway. The genes measured were NFatC1, TRAP, MMP9, CtsK, DC-STAMP, and ATP6v0d2 (Zhang et al., 2008; Yang et al., 2008b). For all the genes tested, the RNA sequencing results closely paralleled the quantitative PCR results (Supplementary Fig. 4). Taken together with previous studies showing that L. reuteri has immunomodulatory activity, the transcriptional analysis suggests that immune signaling may be targeted to impact osteoclastogenesis (Jones et al., 2011; Iyer et al., 2008; Thomas et al., 2012).
3.7. Activation of NF-κB and p38 in RAW264.7 cells was inhibited by L. reuteri
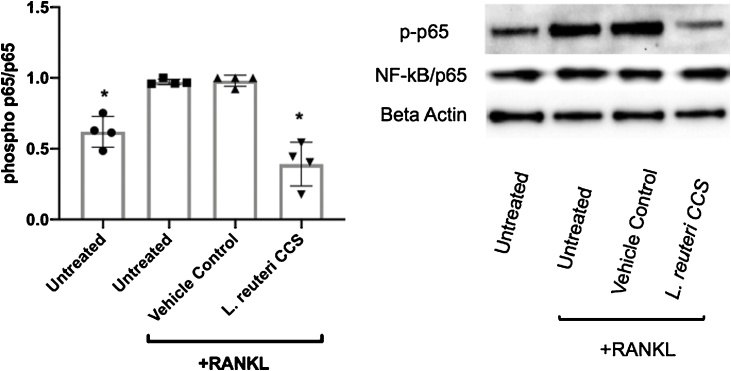
Based on the RNAseq results, we tested the impact of L. reuteri CCS treatment during osteoclastogenesis on NF-kB activation. RANKL initiates a cascade of intracellular signaling that culminates in the activation of NF-κB that is essential for osteoclastogenesis (Strait et al., 2008; Boyce et al., 2015). We observed that RANKL treatment of RAW264.7 cells resulted in the induction of NF-κB/p65 phosphorylation and treatment with L. reuteri CCS significantly inhibited this stimulation (Fig. 8). This was consistent with the transcriptional analyses that indicated the NF-κB signaling pathway being down-regulated by L. reuteri.
Fig. 8.
Effect ofL.reuteri on RANKL-induced NF-κB/p65 phosphorylation. RAW264.7 cells were concurrently treated with RANKL (100 ng/ml) and MEM-α or L. reuteri CCS for 60 min. Total intracellular contents were prepared and an equal amount of protein was analyzed. Western blot analysis using antibodies to the phosphorylated NF-κB/p65 subunit was performed. Band intensity was measured by densitometry using the ImageJ software package. Results were normalized to levels of total p65. Four biological replicates are depicted with associated standard error of mean, *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions as determined by one-way ANOVA.
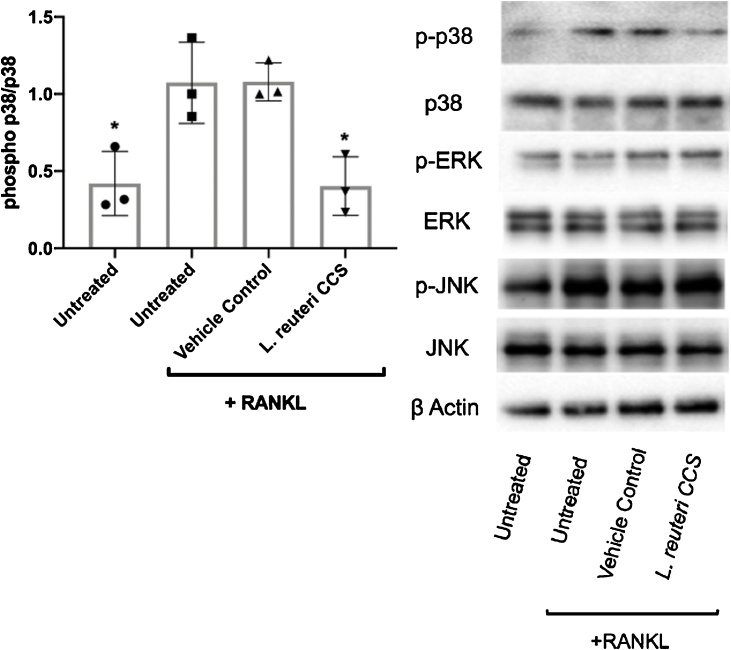
Mitogen-activated protein kinases (MAPK) signaling pathways have also been shown to be important for various facets of host physiology such as the immune response and cellular metabolism (Eriksson et al., 2006; Li et al., 2015; Arthur and Ley, 2013; Alam et al., 2015). Additionally, their role in osteoclast differentiation has also been heavily studied (Sharma et al., 2007; Huang et al., 2006; Ihn et al., 2015; Nie et al., 2016; Chen et al., 2016). To corroborate our transcriptomics findings, we performed Western blot analysis to assess the impact of L. reuteri on MAPK signaling. The MAPKs p38, ERK, and JNK were activated with RANKL and treated with the vehicle control or L. reuteri CCS for 15 min. Interestingly, our results suggested that while ERK and JNK were unaffected by L. reuteri treatment, p38 activation is inhibited (Fig. 9). Taken together, our data suggests that L. reuteri impacts NF-κB and p38 to suppress RANKL-induced differentiation of RAW264.7 cells.
Fig. 9.
Impact ofL.reuteri on MAPK signaling. RAW264.7 cells were concurrently treated with RANKL (100 ng/ml) and MEM-α or L. reuteri CCS for 60 min. Total intracellular contents were prepared and an equal amount of protein was analyzed. Western blot analysis using antibodies to the p38, phosphorylated p38, ERK, phosphorylated ERK, JNK, phosphorylated JNK. Band intensity was measured by densitometry using the ImageJ software package. Results were normalized to levels of total p38. Three biological replicates are depicted with associated standard error of mean, *p < 0.05 compared to untreated and MEM-α (vehicle control) conditions as determined by one-way ANOVA.
4. Discussion
Recent work has indicated that probiotics and the microbiota have a key role in regulating bone health. Not only do probiotics inhibit bone loss in localized inflammatory reactions such as bone infections or periodontitis, but they have also been shown to regulate bone mass from a distal site such as the gut (Britton et al., 2014; Ohlsson et al., 2014; Maekawa and Hajishengallis, 2014). Despite establishing this link between gut bacteria and bone health, our mechanistic understanding of how gut bacteria regulates bone health is only beginning to emerge. In previous studies, our group has shown that L. reuteri can protect against bone loss or accelerate bone formation under different scenarios using a mouse model of estrogen deficiency, an inflammatory model mediated by dorsal surgical incision, and under healthy conditions in a male murine model (McCabe et al., 2013; Britton et al., 2014; Collins et al., 2016). Under estrogen deficiency, the data suggests that L. reuteri can protect against bone loss by inhibiting osteoclast activity and bone resorption, and altering the gut microbiota (Britton et al., 2014). In healthy male mice, L. reuteri treatment lead to decreased intestinal inflammation and increased osteoblast activity and bone formation (McCabe et al., 2013). Lastly, under inflammatory conditions mediated by dorsal surgical incision in female mice, L. reuteri treatment also lead to decreased intestinal inflammation and increased levels of bone formation in the distal femur metaphyseal trabecular region (Collins et al., 2016). Given that L. reuteri has been shown to improve bone health in several different ways, it is likely that targeting the process of osteoclast differentiation is just one of several probiotic effects mediated by L. reuteri supplementation. This study aimed at further characterizing this response and identifying the factors involved.
Our results indicated that L. reuteri conditioned medium contains factor(s) that targeted an early stage of osteoclastogenesis since a single treatment with L. reuteri at the beginning of RAW264.7 cell culture was able to suppress RANKL-induced osteoclastogenesis by 75%. However, phenotypic changes were not apparent until days 5 and 7 where L. reuteri treatment resulted in an accumulation of fused polykaryons. A majority of the changes in the transcriptional profile also were apparent later in the assay (days 3 and 5) as very few genes were differentially regulated by L. reuteri at the day 1 time point. Although, it is worth noting that the transcriptional differences evident in later time points would be more pronounced as cells that were not treated by L. reuteri CCS would be farther down the path of osteoclastogenesis as compared to the treatment group. Together, the results indicated that while early stimulation by L. reuteri did not impact initial phenotypic changes, the early transcriptional changes that were present likely play a significant role in the suppression of osteoclastogenesis and arrest in a state of fused polykaryons. As a result, we focused our attention on early signaling pathways involved in osteoclastogenesis.
Pathway analysis revealed 3 pathways up-regulated by L. reuteri that may account for its activity in suppressing osteoclastogenesis. First, it has been previously shown that the complement cascade plays a role in bone metabolism. For example, C3a has been demonstrated to stimulate osteoblast formation (Matsuoka et al., 2014), while other studies have shown the importance of C3 in osteoclast formation (Ignatius et al., 2011; Tu et al., 2010). It is possible that the early promotion of monocyte fusion ultimately leads to the suspension in the polykaryon stage. The Jak-STAT pathway was another early pathway that was up-regulated by L. reuteri CCS treatment. It was previously shown that STAT3 activation was important for RANKL-induced osteoclastogenesis (hong Li et al., 2013; Li, 2013), while the phosphorylation and activation of STAT6 resulted in the inhibition of NFATc1 (Yamada et al., 2007). Additionally, the PPAR signaling pathway was also up-regulated by L. reuteri treatment. While PPAR-γ signaling has been shown to promote osteoclastogenesis and decreased bone mass under certain circumstances (Zou et al., 2016; Duque et al., 2013), activation of PPAR signaling has also been shown to inhibit osteoclastogenesis (Kasonga et al., 2019; Hounoki et al., 2008; Cho et al., 2012). These results, coupled with the analysis of decreased p65 and p38 signaling activation after L. reuteri treatment, suggested that these pathways are involved with the mediated suppression of osteoclastogenesis by L. reuteri.
The next issue we addressed was whether this inhibition of osteoclastogenesis by L. reuteri was a direct or indirect effect. It has been shown that osteoclast differentiation can be impacted indirectly by preventing the interaction between RANKL and its receptor, RANK (Zhao et al., 2015). However, the time course study suggested that L. reuteri does not interact with RANKL as an early pulse of RAW264.7 cells with CCS was sufficient to prevent osteoclast formation (Fig. 3). One potential candidate is lactobacillic acid (LA, also known as phytomonic acid), which is a long chain fatty acid containing a cyclopropane ring. LA is produced by L. reuteri and a mutant defective in the production of LA is deficient in the suppression of TNF- α production from a human monocyte cell line (Jones et al., 2011). The immunomodulatory activity of this strain contributed towards strain selection in the original bone health studies in the OVX osteoporosis model since TNF-α plays such an important role in promoting osteoclastogenesis and subsequent bone loss during estrogen deficiency (Kimble et al., 1997; Cenci et al., 2000; Srivastava et al., 1999; Weitzmann and Pacifici, 2006). LA is a 19-carbon long chain cyclopropane fatty acid derived from the conversion of vaccenic acid by cyclopropane fatty acid synthase (Jones et al., 2011). Dihydrosterculic acid, another CFA, is also present in L. reuteri and is derived from oleic acid. Interestingly, oleic acid has been shown to suppress osteoclastogenesis (Drosatos-Tampakaki et al., 2014; Cornish et al., 2008). Other fatty acids have also been shown to inhibit osteoclastogenesis through the inhibition of NF-κB activation, which is a crucial signaling event in osteoclast differentiation (Rahman et al., 2006; Rahman et al., 2011; Zwart et al., 2010). However, LA has yet to be studied in this context and warranted further investigation. We demonstrated that LA impacted osteoclastogenesis in two ways (Fig. 5). Using purified LA, we obtained a concentration-dependent inhibition of osteoclastogenesis. Additionally, CCS isolated from a mutant strain of L. reuteri incapable of producing LA was not as effective in suppressing osteoclastogenesis. Taken together, these results indicate that exogenous LA directly impacts osteoclastogenesis and its presence in L. reuteri contributes to its suppression of osteoclast differentiation. However, L. reuteri still maintained partial inhibition of osteoclastogenesis despite the absence of LA, suggesting that other bacterial product(s) that have not been identified also contribute to this process.
While LA production is not exclusive to L. reuteri, its presence in bacteria and potential for bone health suggests an evolutionary relationship that may elucidate the purpose of receptors that detect fatty acids of different carbon chain lengths. G-protein-coupled receptors (GPCR) signaling systems have been evolutionarily conserved over time (Krishnan and Schiöth, 2015; Corrêa-Oliveira et al., 2016; Mushegian et al., 2012) and their importance in regulating diseases such as cancer, obesity, and development has been the topic of much research (Oh et al., 2014; Oh et al., 2010; Marivin et al., 2016; Yu and Brown, 2015). More recently, the roles of the long chain fatty acid receptors, GPR40 and GPR120, have been shown to be important in bone health and regulating osteoclastogenesis (Kim et al., 2015; Wauquier et al., 2013; Philippe et al., 2016; Ahn et al., 2016). Although studies have demonstrated the potential impact of long chain fatty acids on bone health, LA has yet to be investigated in the same context and remains a promising candidate as a novel therapeutic (Bonnet and Ferrari, 2011; Chen et al., 2013). In our studies, we established that GPR120, but not GPR40, was important for the suppression of osteoclastogenesis by L. reuteri or LA. Future studies directed at-testing the impact of LA on in vivo osteoclastogenesis and bone density will be crucial for determining whether it can potentially be a novel therapeutic for bone health.
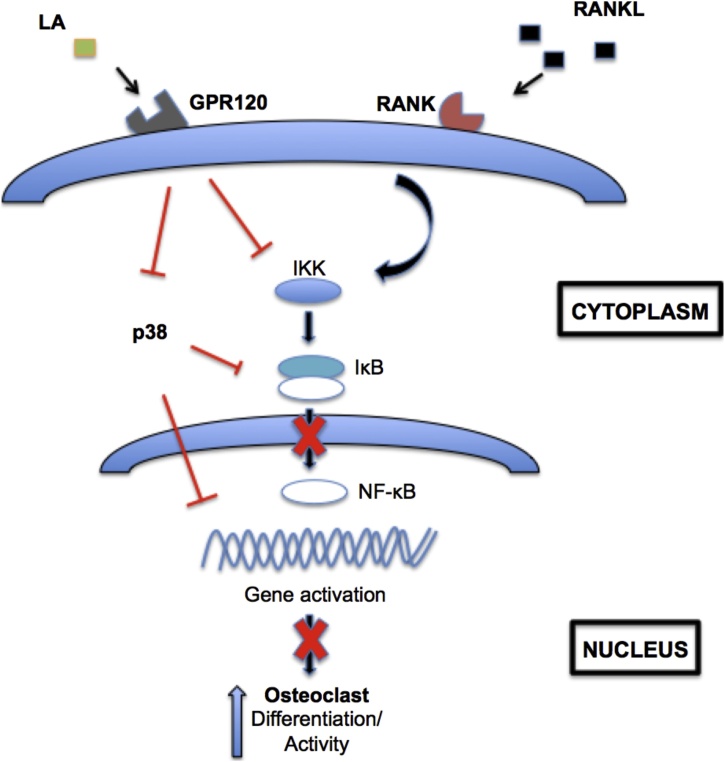
In the present study, we have established that early signaling events in osteoclastogenesis are targeted by L. reuteri. Additionally, we have uncovered a role for the fatty acid LA in osteoclastogenesis. In addition to suppressing osteoclastogenesis, we also demonstrated that L. reuteri inhibited osteoclast function, as indicated by reduced mineral resorption of calcium and phosphate coated surfaces. Together, our studies suggest that L. reuteri impacts osteoclast differentiation by targeting p38 and NF-κB activation. LA produced by L. reuteri is important for this inhibition and GPR120 signaling also plays an important role in the suppression of osteoclastogenesis (Fig. 10). However, there are many interesting observations that are made. Since osteoclast differentiation is not completely suppressed using a mutant deficient in LA production, another mechanism mediated by L. reuteri is suggested to be involved. Moreover, the fact that inhibition of GPR120 was unable to completely block the ability of LA to suppress osteoclast formation either suggests that LA has another target receptor or the concentration of LA used could overcome inhibition by AH7614. As previously mentioned, increasing the concentration of AH7614 past 2.5 μM lead to cellular toxicity. Thus, the concentration of 1 μM was used for the studies possibly rendering it not 100% effective.
Fig. 10.
Working model of osteoclastogenesis suppression byL.reuteri and LA. RAW264.7 cells are stimulated for osteoclastogenesis by RANKL. L. reuteri and LA activated GPR120 or a receptor yet to be identified to suppress osteoclastogenesis. GPR120 receptor antagonism partially inhibited the ability of L. reuteri and LA to suppress osteoclastogenesis. MAPK and NF-κB signaling has been shown to be downstream of GPR120 signaling and we demonstrated that L. reuteri impacts several arms of these pathways.
5. Conclusion
With the emergence of studies demonstrating the beneficial impact that gastrointestinal microbes can have on bone health, additional studies will be required to validate its true therapeutic potential. The pathogenesis of bone diseases is multifactorial, spanning influences from several organ systems, and a better understanding of the interactions between the different systems will be crucial for this purpose. Specifically, when it comes to osteomicrobiology, the increasing use of genetically tractable organisms in murine models of disease will aid in uncovering potential probiotic mechanisms of action. Moreover, prospective clinical trials establishing the efficacy of probiotics to promote bone health in humans are also warranted. Novel treatment options that take advantage of microbes that co-evolved and co-existed with humans may develop as we continue filling in these gaps of knowledge.
Data
The RNAseq data reported in this manuscript has been deposited in NCBI’s Gene Expression Omnibus (GEO) database. Accession number: GSE135755.
Funding information
This work was supported by NIH grant NCCIH R01AT007695-05 to NP, LM, and RB as well as seed funding from Baylor College of Medicine to RB.
Declaration of Competing Interest
The authors report that they have no competing interests.
Acknowledgements
We thank Fraser Collins and Zach Criss for technical support. Fraser Collins and Zach Criss provided valuable input and expertise in tissue culture experiments.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.bonr.2019.100227.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Aharon R., Bar-Shavit Z. Involvement of aquaporin 9 in osteoclast differentiation. J. Biol. Chem. 2006;281:19305–19309. doi: 10.1074/jbc.M601728200. [DOI] [PubMed] [Google Scholar]
- Ahn S.H., Park S.-Y., Baek J.-E., Lee S.-Y., Baek W.-Y., Lee S.-Y., Lee Y.-S., Ju Yoo H., Kim H., Lee S.H., Im D.-S., Lee S.-K., Kim B.-J., Koh J.-M. Free fatty acid receptor 4 (GPR120) stimulates bone formation and suppresses bone resorption in the presence of elevated n -3 fatty acid levels. Endocrinology. 2016;4 doi: 10.1210/en.2015-1855. [DOI] [PubMed] [Google Scholar]
- Alam H., Gu B., Lee M.G. Histone methylation modifiers in cellular signaling pathways. Cell. Mol. Life Sci. 2015;72:4577–4592. doi: 10.1007/s00018-015-2023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., Robinson M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. doi:nprot.2013.099[pii]\r10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Arthur J.S., Ley S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- Asagiri M., Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Bonnet N., Ferrari S.L. Effects of long-term supplementation with omega-3 fatty acids on longitudinal changes in bone mass and microstructure in mice. J. Nutr. Biochem. 2011;22:665–672. doi: 10.1016/j.jnutbio.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Boyce B.F., Xiu Y., Li J., Xing L., Yao Z. NF-κB-Mediated Regulation of Osteoclastogenesis. Endocrinol. Metab. (Seoul, Korea) 2015;30:35–44. doi: 10.3803/EnM.2015.30.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Britton R.A., Irwin R., Quach D., Schaefer L., Zhang J., Lee T., Parameswaran N., Mccabe L.R. Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J. Cell. Physiol. 2014;229:1822–1830. doi: 10.1002/jcp.24636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck W.G., Anderson J.M. Giant cell formation and function. Curr. Opin. Hematol. 2009;16:53–57. doi: 10.1097/MOH.0b013e32831ac52e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci S., Weitzmann M.N., Roggia C., Namba N., Novack D., Woodring J., Pacifici R. Estrogen deficiency induces bone loss by enhancing T-cell production of TNF-alpha. J. Clin. Invest. 2000;106:1229–1237. doi: 10.1172/JCI11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Gao R.-F., Yuan F.-L., Zhao M.-D. Recombinant human endostatin suppresses mouse osteoclast formation by inhibiting the NF-κB and MAPKs signaling pathways. Front. Pharmacol. 2016;7:1–10. doi: 10.3389/fphar.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.Y., Zhang Z.M., Zheng X.C., Wang L., Huang M.J., Qin S., Chen J., Lai P.L., Yang C.L., Liu J., Dai Y.F., Di Jin D., Bai X.C. Endogenous n-3 polyunsaturated fatty acids (PUFAs) mitigate ovariectomy-induced bone loss by attenuating bone marrow adipogenesis in FAT1 transgenic mice, Drug Des. Devel. Ther. 2013;7:545–552. doi: 10.2147/DDDT.S45263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S.S., Pan T.M. Antiosteoporotic effects of lactobacillus-fermented soy skim milk on bone mineral density and the microstructure of femoral bone in ovariectomized mice. J. Agric. Food Chem. 2011;59:7734–7742. doi: 10.1021/jf2013716. [DOI] [PubMed] [Google Scholar]
- Cho E.S., Kim M.K., Son Y.O., Lee K.S., Park S.M., Lee J.C. The effects of Rosiglitazone on osteoblastic differentiation, osteoclast formation and bone resorption. Mol. Cells. 2012;33:173–181. doi: 10.1007/s10059-012-2240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.L., Irwin R., Bierhalter H., Schepper J., Britton R.A., Parameswaran N., McCabe L.R. Lactobacillus reuteri 6475 increases bone density in intact females only under an inflammatory setting. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish J., MacGibbon A., Lin J.M., Watson M., Callon K.E., Tong P.C., Dunford J.E., Van Der Does Y., Williams G.A., Grey A.B., Naot D., Reid I.R. Modulation of osteoclastogenesis by fatty acids. Endocrinology. 2008;149:5688–5695. doi: 10.1210/en.2008-0111. [DOI] [PubMed] [Google Scholar]
- Corrêa-Oliveira R., Fachi J.L., Vieira A., Sato F.T., Vinolo M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016;5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington D.K., Briscoe C.A., Brown A.J., Jayawickreme C.K. The G-protein-coupled receptor 40 family (GPR40-GPR43) and its role in nutrient sensing. Biochem. Soc. Trans. 2006;34:770–773. doi: 10.1042/BST0340770. [DOI] [PubMed] [Google Scholar]
- Crandall C.J., Newberry S.J., Diamant A., Lim Y.W., Gellad W.F., Booth M.J., Motala A., Shekelle P.G. Comparative effectiveness of pharmacologic treatments to prevent fractures: an updated systematic review. Ann. Intern. Med. 2014;161:711–723. doi: 10.7326/M14-0317. [DOI] [PubMed] [Google Scholar]
- Cuetara B.L.V., Crotti T.N., O’Donoghue A.J., McHugh K.P. Cloning and characterization of osteoclast precursors from the RAW264.7 cell line. In Vitro Cell. Dev. Biol. Anim. 2006;42:182–188. doi: 10.1290/0510075.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosatos-Tampakaki Z., Drosatos K., Siegelin Y., Gong S., Khan S., Van Dyke T., Goldberg I.J., Schulze P.C., Schulze-Späte U. Palmitic acid and DGAT1 deficiency enhance osteoclastogenesis, while oleic acid-induced triglyceride formation prevents it. J. Bone Miner. Res. 2014;29:1183–1195. doi: 10.1002/jbmr.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque G., Li W., Vidal C., Bermeo S., Rivas D., Henderson J. Pharmacological inhibition of PPARγ increases osteoblastogenesis and bone mass in male C57BL/6 mice. J. Bone Miner. Res. 2013;28:639–648. doi: 10.1002/jbmr.1782. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Taskinen M., Leppä S. Mitogen activated protein kinase-dependent activation of c-Jun and c-Fos is required for neuronal differentiation but not for growth and stress reposne in PC12 cells. J. Cell. Physiol. 2006;207:12–22. doi: 10.1002/jcp.20907. [DOI] [PubMed] [Google Scholar]
- Ewaschuk J.B., Walker J.W., Diaz H., Madsen K.L. Bioproduction of conjugated linoleic acid by probiotic Bacteria Occurs in vitro and in vivo in Mice1,2. J. Nutr. 2006;136:1483–1487. doi: 10.1093/jn/136.6.1483. https://login.proxy.library.emory.edu/login?url=http://search.proquest.com/docview/197427286?accountid=10747%5Cnhttp://sfxhosted.exlibrisgroup.com/emu?url_ver=Z39.88-2004&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&genre=article&sid=ProQ:ProQ:pqrl&atitle=Biopr [DOI] [PubMed] [Google Scholar]
- Girish a., Vijayalakshmi V. Affordable image analysis using NIH Image/ ImageJ. Indian J. Cancer. 2004;41:47. [PubMed] [Google Scholar]
- Grossman D.C., Curry S.J., Owens D.K., Barry M.J., Davidson K.W., Doubeni C.A., Epling J.W., Kemper A.R., Krist A.H., Kurth A.E., Landefeld C.S., Mangione C.M., Phipps M.G., Silverstein M., Simon M.A., Tseng C.-W. Hormone therapy for the primary prevention of chronic conditions in postmenopausal women. JAMA. 2017;318:2224. doi: 10.1001/jama.2017.18261. [DOI] [PubMed] [Google Scholar]
- hong Li C., xia Zhao J., Sun L., qiang Yao Z., li Deng X., Liu R., yuan Liu X. AG490 inhibits NFATc1 expression and STAT3 activation during RANKL induced osteoclastogenesis. Biochem. Biophys. Res. Commun. 2013;435:533–539. doi: 10.1016/j.bbrc.2013.04.084. [DOI] [PubMed] [Google Scholar]
- Hounoki H., Sugiyama E., Mohamed S.G.K., Shinoda K., Taki H., Abdel-Aziz H.O., Maruyama M., Kobayashi M., Miyahara T. Activation of peroxisome proliferator-activated receptor γ inhibits TNF-α-mediated osteoclast differentiation in human peripheral monocytes in part via suppression of monocyte chemoattractant protein-1 expression. Bone. 2008;42:765–774. doi: 10.1016/j.bone.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Huang H., Ryu J., Ha J., Chang E.-J., Kim H., Kim H.-M., Kitamura T., Lee Z., Kim H.-H. Osteoclast differentiation requires TAK1 and MKK6 for NFATc1 induction and NF-kappaB transactivation by RANKL. Cell Death Differ. 2006;13:1879–1891. doi: 10.1038/sj.cdd.4401882. [DOI] [PubMed] [Google Scholar]
- Ignatius A., Schoengraf P., Kreja L., Liedert A., Recknagel S., Kandert S., Brenner R.E., Schneider M., Lambris J.D., Huber-Lang M. Complement C3a and C5a modulate osteoclast formation and inflammatory response of osteoblasts in synergism with IL-1β. J. Cell. Biochem. 2011;112:2594–2605. doi: 10.1002/jcb.23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihn H.J., Lee D., Lee T., Kim S.H., Shin H.I., Bae Y.C., Hong J.M., Park E.K. Inhibitory effects of KP-A159, a thiazolopyridine derivative, on osteoclast differentiation, function, and inflammatory bone loss via suppression of RANKL-Induced MAP Kinase signaling pathway. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0142201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Takeshita S. The role of osteoclast differentiation and function in skeletal homeostasis. J. Biochem. 2015;159 doi: 10.1093/jb/mvv112. mvv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer C., Kosters A., Sethi G., Kunnumakkara A.B., Aggarwal B.B., Versalovic J. Probiotic Lactobacillus reuteri promotes TNF-induced apoptosis in human myeloid leukemia-derived cells by modulation of NF-κB and MAPK signalling. Cell. Microbiol. 2008;10:1442–1452. doi: 10.1111/j.1462-5822.2008.01137.x. [DOI] [PubMed] [Google Scholar]
- Jones R.M., Mulle J.G., Pacifici R. Osteomicrobiology: The influence of gut microbiota on bone in health and disease. Bone. 2017 doi: 10.1016/j.bone.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Jones S.E., Whitehead K., Saulnier D., Thomas C.M., Versalovic J., Britton R.A. Cyclopropane fatty acid synthase mutants of probiotic human-derived lactobacillus reuteri are defective in TNF inhibition. Gut Microbes. 2011;2:69–79. doi: 10.4161/gmic.2.2.15282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasonga A., Kruger M.C., Coetzee M. Activation of PPARs modulates signalling pathways and expression of regulatory genes in osteoclasts derived from human CD14+ monocytes. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.A., Morrison A., Hanley D.A., Felsenberg D., McCauley L.K., O’Ryan F., Reid I.R., Ruggiero S.L., Taguchi A., Tetradis S., Watts N.B., Brandi M.L., Peters E., Guise T., Eastell R., Cheung A.M., Morin S.N., Masri B., Cooper C., Morgan S.L., Obermayer-Pietsch B., Langdahl B.L., Al Dabagh R., Davison K.S., Kendler D.L., S??ndor G.K., Josse R.G., Bhandari M., El Rabbany M., Pierroz D.D., Sulimani R., Saunders D.P., Brown J.P., Compston J. Diagnosis and management of osteonecrosis of the jaw: a systematic review and international consensus. J. Bone Miner. Res. 2015;30:3–23. doi: 10.1002/jbmr.2405. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Yoon H.J., Kim B.K., Kang W.Y., Seong S.J., Lim M.S., Kim S.Y., Yoon Y.R. G Protein-Coupled Receptor 120 Signaling Negatively Regulates Osteoclast Differentiation, Survival, and Function. J. Cell. Physiol. 2015:844–851. doi: 10.1002/jcp.25133. [DOI] [PubMed] [Google Scholar]
- Kimble R.B., Bain S., Pacifici R. The functional block of TNF but not of IL-6 prevents bone loss in ovariectomized mice. J. Bone Miner. Res. 1997;12:935–941. doi: 10.1359/jbmr.1997.12.6.935. [DOI] [PubMed] [Google Scholar]
- Krishnan A., Schiöth H.B. The role of G protein-coupled receptors in the early evolution of neurotransmission and the nervous system. J. Exp. Biol. 2015;218:562–571. doi: 10.1242/jeb.110312. [DOI] [PubMed] [Google Scholar]
- Kwon J.-O., Jin W.J., Kim B., Kim H.-H., Lee Z.H. Myristoleic acid inhibits osteoclast formation and bone resorption by suppressing the RANKL activation of Src and Pyk2. Eur. J. Pharmacol. 2015;768:189–198. doi: 10.1016/j.ejphar.2015.10.053. [DOI] [PubMed] [Google Scholar]
- Li J. JAK-STAT and bone metabolism. Jak-Stat. 2013;2 doi: 10.4161/jkst.23930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Chassaing B., Tyagi A.M., Vaccaro C., Luo T., Adams J., Darby T.M., Weitzmann M.N., Mulle J.G., Gewirtz A.T., Jones R.M., Pacifici R. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J. Clin. Invest. 2016;126:2049–2063. doi: 10.1172/JCI86062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.Y., Yang L.C., Guo K., Wang Y.P., Li Y.G. Mitogen-activated protein kinase phosphatase-1: a critical phosphatase manipulating mitogen-activated protein kinase signaling in cardiovascular disease (Review) Int. J. Mol. Med. 2015;35:1095–1102. doi: 10.3892/ijmm.2015.2104. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Hajishengallis G. Topical treatment with probiotic Lactobacillus brevis CD2 inhibits experimental periodontal inflammation and bone loss. J. Periodontal Res. 2014;49:785–791. doi: 10.1111/jre.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marivin A., Leyme A., Parag-Sharma K., DiGiacomo V., Cheung A.Y., Nguyen L.T., Dominguez I., Garcia-Marcos M. Dominant-negative Gα subunits are a mechanism of dysregulated heterotrimeric G protein signaling in human disease. Sci. Signal. 2016;9:ra37. doi: 10.1126/scisignal.aad2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K., Park K.-A., Ito M., Ikeda K., Takeshita S. Osteoclast-derived complement component 3a stimulates osteoblast differentiation. J. Bone Miner. Res. 2014;29:1522–1530. doi: 10.1002/jbmr.2187. [DOI] [PubMed] [Google Scholar]
- McCabe L.R., Irwin R., Schaefer L., Britton R.A. Probiotic use decreases intestinal inflammation and increases bone density in healthy male but not female mice. J. Cell. Physiol. 2013;228:1793–1798. doi: 10.1002/jcp.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H., Katsuyama E., Miyauchi Y., Hoshi H., Miyamoto K., Sato Y., Kobayashi T., Iwasaki R., Yoshida S., Mori T., Kanagawa H., Fujie A., Hao W., Morioka H., Matsumoto M., Toyama Y., Miyamoto T. An essential role for STAT6-STAT1 protein signaling in promoting macrophage cell-cell fusion. J. Biol. Chem. 2012;287:32479–32484. doi: 10.1074/jbc.M112.358226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli L., Capurso L. FAO/WHO Guidelines on Probiotics. J. Clin. Gastroenterol. 2012;46:S1–S2. doi: 10.1097/MCG.0b013e318269fdd5. [DOI] [PubMed] [Google Scholar]
- Mushegian A., Gurevich V.V., Gurevich E.V. The origin and evolution of G protein-coupled receptor kinases. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0033806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutuş R., Kocabagli N., Alp M., Acar N., Eren M., Gezen S.S. The effect of dietary probiotic supplementation on tibial bone characteristics and strength in broilers. Poult. Sci. 2006;85:1621–1625. doi: 10.1093/ps/85.9.1621. [DOI] [PubMed] [Google Scholar]
- Narva M., Collin M., Lamberg-Allardt C., Kärkkäinen M., Poussa T., Vapaatalo H., Korpela R. Effects of long-term intervention with Lactobacillus helveticus-Fermented milk on bone mineral density and bone mineral content in growing rats. Ann. Nutr. Metab. 2004;48:228–234. doi: 10.1159/000080455. [DOI] [PubMed] [Google Scholar]
- Narva M., Rissanen J., Halleen J., Vapaatalo H., Väänänen K., Korpela R. Effects of bioactive peptide, Valyl-Prolyl-Proline (VPP), and Lactobacillus helveticus fermented milk containing VPP on bone loss in ovariectomized rats. Ann. Nutr. Metab. 2007;51:65–74. doi: 10.1159/000100823. [DOI] [PubMed] [Google Scholar]
- Nie S., Xu J., Zhang C., Xu C., Liu M., Yu D. Salicortin inhibits osteoclast differentiation and bone resorption by down-regulating JNK and NF-κB/NFATc1 signaling pathways. Biochem. Biophys. Res. Commun. 2016;470:61–67. doi: 10.1016/j.bbrc.2015.12.115. [DOI] [PubMed] [Google Scholar]
- Oh D.Y., Walenta E., Akiyama T.E., Lagakos W.S., Lackey D., Pessentheiner A.R., Sasik R., Hah N., Chi T.J., Cox J.M., Powels M.A., Di Salvo J., Sinz C., Watkins S.M., Armando A.M., Chung H., Evans R.M., Quehenberger O., McNelis J., Bogner-Strauss J.G., Olefsky J.M. A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nat. Med. 2014;20:942–947. doi: 10.1038/nm.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.Y., Talukdar S., Bae E.J., Imamura T., Morinaga H., Fan W., Li P., Lu W.J., Watkins S.M., Olefsky J.M. GPR120 is an Omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson C., Engdahl C., Fåk F., Andersson A., Windahl S.H., Farman H.H., Movérare-Skrtic S., Islander U., Sjögren K. Probiotics protect mice from ovariectomy-induced cortical bone loss. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C., Wauquier F., Lyan B., Coxam V., Wittrant Y. GPR40, a free fatty acid receptor, differentially impacts osteoblast behavior depending on differentiation stage and environment. Mol. Cell. Biochem. 2016;412:197–208. doi: 10.1007/s11010-015-2626-5. [DOI] [PubMed] [Google Scholar]
- Rahman M.M., Bhattacharya A., Fernandes G. Conjugated linoleic acid inhibits osteoclast differentiation of RAW264.7 cells by modulating RANKL signaling. J. Lipid Res. 2006;47:1739–1748. doi: 10.1194/jlr.M600151-JLR200. [DOI] [PubMed] [Google Scholar]
- Rahman M.M., Halade G.V., Williams P.J., Fernandes G. t10c12-CLA maintains higher bone mineral density during aging by modulating osteoclastogenesis and bone marrow adiposity. J. Cell. Physiol. 2011;226:2406–2414. doi: 10.1002/jcp.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson L., Abtahi J. Bisphosphonate associated osteonecrosis of the jaw: an update on pathophysiology, risk factors, and treatment. Int. J. Dent. 2014;(2014):1–9. doi: 10.1155/2014/471035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., Martino M.E., Balmand S., Hudcovic T., Heddi A., Rieusset J., Kozakova H., Vidal H., Leulier F. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science (80-.) 2016;351:854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- Sharma S.M., Bronisz A., Hu R., Patel K., Mansky K.C., Sif S., Ostrowski M.C. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J. Biol. Chem. 2007;282:15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Weitzmann M.N., Cenci S., Ross F.P., Adler S., Pacifici R. Estrogen decreases TNF gene expression by blocking JNK activity and the resulting production of c-Jun and JunD. J. Clin. Invest. 1999;104:503–513. doi: 10.1172/JCI7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strait K., Li Y., Dillehay D.L., Weitzmann M.N. Suppression of NF-kappaB activation blocks osteoclastic bone resorption during estrogen deficiency. Int. J. Mol. Med. 2008;21:521–525. http://www.ncbi.nlm.nih.gov/pubmed/18360699 [PubMed] [Google Scholar]
- Teitelbaum S.L., Tondravi M.M., Ross F.P. Osteoclasts, macrophages, and the molecular mechanisms of bone resorption. J. Leukoc. Biol. 1997;61:381–388. doi: 10.1002/jlb.61.4.381. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=9103223&retmode=ref&cmd=prlinks%5Cnpapers2://publication/uuid/C7CC78B0-9F3F-4744-801F-18188DC394CC [DOI] [PubMed] [Google Scholar]
- Thomas C.M., Hong T., van Pijkeren J.P., Hemarajata P., Trinh D.V., Hu W., Britton Ra., Kalkum M., Versalovic J. Histamine derived from probiotic lactobacillus reuteri suppresses tnf via modulation of pka and erk signaling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomofuji T., Ekuni D., Azuma T., Irie K., Endo Y., Yamamoto T., Ishikado A., Sato T., Harada K., Suido H., Morita M. Supplementation of broccoli or Bifidobacterium longum-fermented broccoli suppresses serum lipid peroxidation and osteoclast differentiation on alveolar bone surface in rats fed a high-cholesterol diet. Nutr. Res. 2012;32:301–307. doi: 10.1016/j.nutres.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Tu Z., Bu H., Dennis J.E., Lin F. Efficient osteoclast differentiation requires local complement activation. Blood. 2010;116:4456–4463. doi: 10.1182/blood-2010-01-263590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi A.M., Yu M., Darby T.M., Vaccaro C., Li J.-Y., Owens J.A., Hsu E., Adams J., Weitzmann M.N., Jones R.M., Pacifici R. The microbial metabolite butyrate stimulates bone formation via t regulatory cell-mediated regulation of WNT10B expression. Immunity. 2018;49:1116–1131. doi: 10.1016/j.immuni.2018.10.013. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauquier F., Philippe C., Léotoing L., Mercier S., Davicco M.-J., Lebecque P., Guicheux J., Pilet P., Miot-Noirault E., Poitout V., Alquier T., Coxam V., Wittrant Y. The free fatty acid receptor G protein-coupled receptor 40 (GPR40) protects from bone loss through inhibition of osteoclast differentiation. J. Biol. Chem. 2013;288:6542–6551. doi: 10.1074/jbc.M112.429084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J.B., Hobbs M.M., Misukonis M.A. Recombinant human gamma-interferon induces human monocyte polykaryon formation. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4554–4557. doi: 10.1073/pnas.81.14.4554. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=345629&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann M.N., Pacifici R. Estrogen regulation of immune cell bone interactions. Ann. N. Y. Acad. Sci. 2006;1068:256–274. doi: 10.1196/annals.1346.030. [DOI] [PubMed] [Google Scholar]
- Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., Oike Y., Takeya M., Toyama Y., Suda T. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A., Takami M., Kawawa T., Yasuhara R., Zhao B., Mochizuki A., Miyamoto Y., Eto T., Yasuda H., Nakamichi Y., Kim N., Katagiri T., Suda T., Kamijo R. Interleukin-4 inhibition of osteoclast differentiation is stronger than that of interleukin-13 and they are equivalent for induction of osteoprotegerin production from osteoblasts. Immunology. 2007;120:573–579. doi: 10.1111/j.1365-2567.2006.02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Charles J.F. Gut Microbiome and Bone: to Build, Destroy, or Both? Curr. Osteoporos. Rep. 2017;15:376–384. doi: 10.1007/s11914-017-0382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Birnbaum M.J., Mackay Ca., Mason-Savas A., Thompson B., Odgren P.R. Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation. J. Cell. Physiol. 2008;215:497–505. doi: 10.1002/jcp.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Zaidi M., Zhang W., Zhu L.L., Li J., Iqbal J., Varbanov A., Gross G., Phipps R., Troen B.R., Sun L. Functional grouping of osteoclast genes revealed through microarray analysis. Biochem. Biophys. Res. Commun. 2008;366:352–359. doi: 10.1016/j.bbrc.2007.11.106. [DOI] [PubMed] [Google Scholar]
- Yu O.M., Brown J.H. G Protein-coupled Receptor and RhoA-stimulated transcriptional responses: links to inflammation, differentiation, and cell proliferation. Mol. Pharmacol. 2015;88:171–180. doi: 10.1124/mol.115.097857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Geoffroy V., Ridall A.L., Karsenty G., Tracy T., Bonner A.S., Duffy J.B., Gergen J.P., a Karlovich C., Dumstrei K., Banerjee U., Otto F., Zabel B., Mundlos S., Ritchie H.E., Selby P.B., Acampora D., Davidson D.R., Hill R.E., Satokata I., Tribioli C., Frasch M., Lufkin T., Mallo M., Zhang M., Gridley T., Kanzler B., Kuschert S.J., Schinke T., Pierre J.M., Abderrahim L., Debiais F., Lemonnier J., Montero a, Vortkamp a, Karaplis A.C., Hammerschmidt M., Mcmahon A.P., Ducy P., Mundy G.R., Boyce B.F., Yoneda T., Bonewald L.F., Teitelbaum S.L. Bone resorption by osteoclasts. Science (80-.) 2000;289:1504–1508. [Google Scholar]
- Zhang Q., Fong C.-C., Zhang Y., Tzang C.-H., Fong W.-F., Yang M. cDNA microarray analysis of the differentially expressed genes involved in murine pre-osteoclast RAW264.7 cells proliferation stimulated by dexamethasone. Life Sci. 2008;82:135–148. doi: 10.1016/j.lfs.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Zhao H., Gu J., Dai N., Gao Q., Wang D., Song R., Liu W., Yuan Y., Bian J., Liu X., Liu Z. Osteoprotegerin exposure at different stages of osteoclastogenesis differentially affects osteoclast formation and function. Cytotechnology. 2015 doi: 10.1007/s10616-015-9892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Rohatgi N., Chen T.H.-P., Schilling J., Abu-Amer Y., Teitelbaum S.L. PPAR-γ regulates pharmacological but not physiological or pathological osteoclast formation. Nat. Med. 2016;22:1203–1205. doi: 10.1038/nm.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart S.R., Pierson D., Mehta S., Gonda S., Smith S.M. Capacity of omega-3 fatty acids or eicosapentaenoic acid to counteract weightlessness-induced bone loss by inhibiting NF-kappaB activation: from cells to bed rest to astronauts. J. Bone Miner. Res. 2010;25:1049–1057. doi: 10.1359/jbmr.091041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.