Abstract.
We employ transcranial wide-field single-photon imaging to compare genetically encoded calcium sensors under transgenic or viral vector expression strategies. Awake, head-fixed animals and brief visual flash stimuli are used to assess function. The use of awake transcranial imaging may reduce confounds attributed to cranial window implantation or anesthesia states. We report differences in wide-field epifluorescence brightness and peak response to visual stimulation between expression strategies. Other metrics for indicator performance include fluctuation analysis (standard deviation) and regional correlation maps made from spontaneous activity. We suggest that multiple measures, such as stimulus-evoked signal-to-noise ratio, brightness, and averaged visual response, may be necessary to characterize indicator sensitivity and methods of expression. Furthermore, we show that strategies using blood brain barrier-permeable viruses, such as PHP.eB, yield comparable expression and function as those derived from transgenic mice. We suggest that testing of new genetically engineered activity sensors could employ a single-photon, wide-field imaging pipeline involving visual stimulation in awake mice that have been intravenously injected with PHP.eB.
Keywords: wide-field imaging, calcium, adeno-associated virus, transgenic, fluorescence, resting state
1. Introduction
Mesoscale functional imaging is a powerful approach to monitor large-scale cortical activity and functional connectivity.1–4 This imaging modality measures cortical neuronal activity directly through the use of fluorescent indicators (e.g., calcium and neurotransmitter indicators or voltage-sensitive dyes), or indirectly through intrinsic optical signals. Intrinsic signal imaging monitors changes in reflected light from the cortex due to hemodynamic activity, and can be used to map functional cortical areas.5,6 However, this imaging method suffers from a low signal-to-noise ratio (SNR) and a lack of specificity and time resolution.2,7 Moreover, this measure is performed through the indirect neurohemodynamic response and can be affected by inconsistency due to neurovascular coupling. The use of ultrasensitive fluorescent indicators, such as calcium sensor, GCaMP6,8 or superfast indicators, such as glutamate sensor and iGluSnFR, can overcome these limitations.9–13 Whatever the signal recorded, optical access to the entire dorsal cortex can be obtained through the intact skull in mice using a transcranial chronic window.14,15
Expression of GCaMP6 can be obtained using transgenic mice where the gene could be inserted under the Thy1 promoter (Thy1-GCaMP6s,16), under the control of cre-recombinase and the tetracycline transactivator (tTA) (Ai94, TITL-GCaMP6s crossed with CamK2a-tTA and Emx1-cre,17–20) or the tTA protein coupled with the tetracycline operator (tetO) (tetO-GCaMP6s crossed with CamK2a-tTA,21). While Thy1-GCaMP6s is a single gene transgenic, tetO must be crossed with a strain expressing tTA (two-mouse crossing) and Ai94 requires a three-mouse crossing. Alternatively, GCaMP6 can be expressed with intracerebral injection of viral vectors. Intracerebral injection of adeno-associated virus (AAV) vectors is an attractive option for gene transfer to neural cells due to its high transduction efficiency and low immunogenicity.22 Expression can also be achieved within weeks in adult animals, allowing for the reduction of costs associated with breeding and maintaining colonies. However, intracerebral injections are invasive, and multiple injections are typically required to achieve adequate coverage over large cortical areas for wide-field imaging.11
As an alternative to intracerebral injection, there have been reports of blood brain barrier (BBB) crossing using intravenous injection of AAV9 serotype.23–25 Based on this observation, new variants, PHP.B and PHP.eB, were generated from this serotype.26,27 These two new capsids provide a strong BBB crossing and neuronal transfection in mice, but also in rats23,28 and marmosets,25 despite results showing a limited interspecies specificity.29 Using this serotype, functional imaging can be performed with two-photon microscopy using GCaMP6,30 or mesoscale wide-field imaging of two specific neuronal populations using green and red calcium indicators under the control of two different promoters.31
Despite the great potential of this innovative BBB crossing viral vector, no comparison was performed in parallel to other classical approaches of expression. In this study, GCaMP6s was expressed in the cortex of negative C57BL/6J mice using AAV-PHP.eB viral vector under a pan-neuronal human synapsin (hSyn) promoter. The fluorescence, responsiveness, and connectivity mapping performance were then compared with three transgenic lines: Thy1-GCaMP6s, TITL-GCaMP6s, and tetO-GCaMP6s. The results showed that, while the TIGRE mouse strategy provided the strongest basal fluorescent level and visual response SNR, evoked signals and the spatial organization of spontaneous activity were comparable with tetO-GCaMP6s and negative mice infected by AAV-PHP.eB-GCaMP6s. Thy1-GCaMP6s, despite its convenience (single gene) was the weakest means of GCAMP6 expression when judged by mesoscale transcranial imaging. Given the strong functional signals presented using intravenous injection of AAV-PHP.eB, and the ability to express genetic material without transgenesis or intracerebral injection, this viral vector will likely become an important experimental tool for the testing of novel fluorescent indicators of neuronal or glial activity in vivo.8,9,12,32–35
2. Methods
2.1. Animals
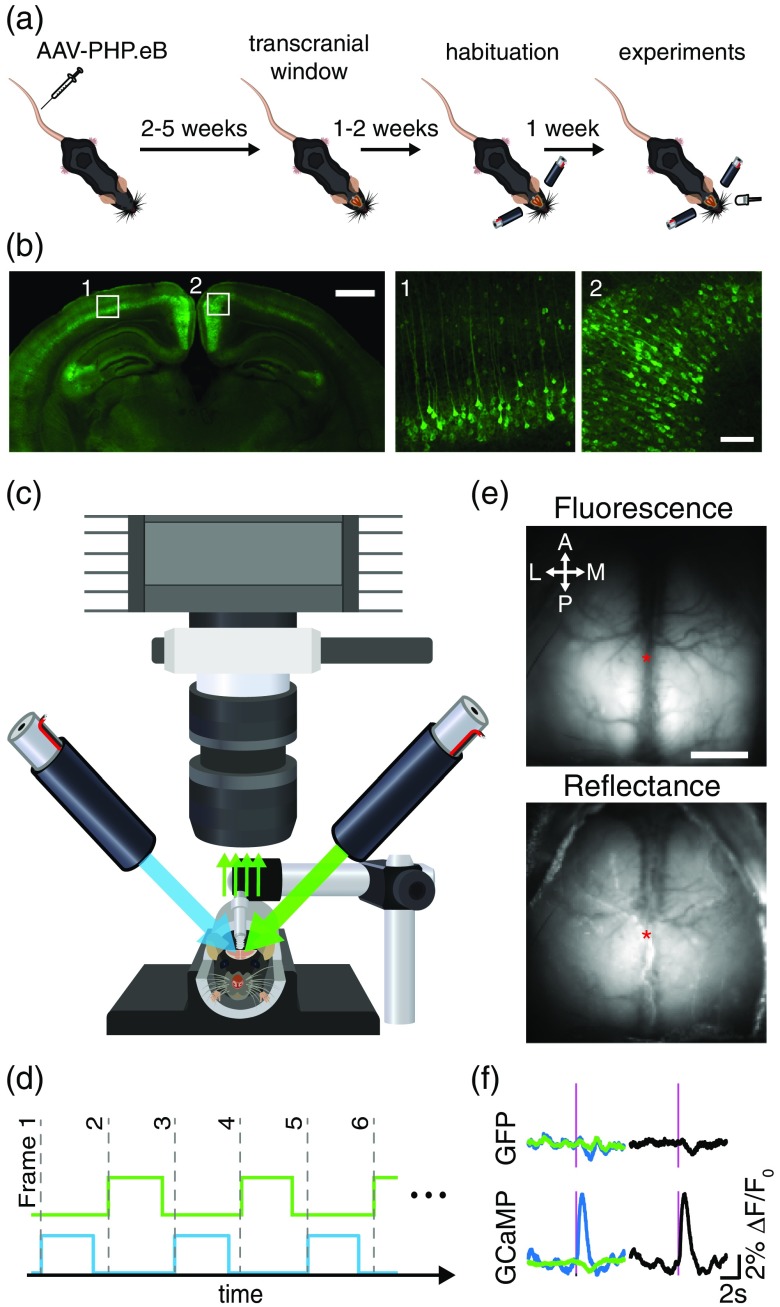
All procedures were approved by the University of British Columbia Animal Care Committee and conformed to the Canadian Council of Animal Care and Use guidelines. Mesoscopic calcium imaging was performed on transgenic mice expressing the calcium indicator GCaMP6s8 or negative/wild-type (WT) mice injected with AAV-PHP.eB virus carrying a plasmid-expressing GCaMP6s.27 All animals examined in this study were males. Three transgenic lines were used for calcium imaging: (1) Thy1-GCaMP6s () (Jackson mouse #024275, GP4.3, C57BL/6-Tg(Thy1-GCaMP6s)GP4.3Dkim/J16), (2) tetO-GCaMP6s () (crossing of two lines: #024742; B6;DBA-Tg(tetO-GCaMP6s)2Niell/J, B6 with #003010 B6;CBA-Tg(Camk2a-tTA)1Mmay/J21), and (3) TIGRE-GCaMP6s () (crossing of three lines: #005628 and #007004 B6.Cg-Tg(Camk2a-tTA)1Mmay/DboJ and #024104; Ai94; strains20 but also18). The expression of GCaMP6s was assessed by polymerase chain reaction (PCR) genotyping on each animal. AAV-PHP.eB capsids were used to deliver plasmid expressing a GCaMP6s reporter driven by the human synapsin promoter (AAV-PHP.eB-hSyn-GCaMP6s-WPRE). 11 genome copies in were injected into the tail vein of WT or fos-cre −/− mice () 4 to 8 weeks before imaging [Fig. 1(a)]. To control the efficiency of blood artifact correction, non-activity-dependent fluorescence was recorded in transgenic mice expressing green fluorescent protein (GFP) in excitatory cortical neurons ( Thy1-GFP mice, B6;CBA-Tg(Thy1-EGFP)SJrs/NdivJ, #01107036).
Fig. 1.
Experimental and imaging setup. (a) Experimental timeline. (b) Low magnification image showing GCaMP expression throughout a coronal histological slice of an AAV-PHP.eB-injected mouse (left). Higher magnification images (right) showing GCaMP-expressing neurons in the primary visual and retrosplenial cortex, outlined by the white boxes (scale bars: 1 mm and 100 μm for low and high magnification images, respectively). (c) Green epifluorescence and reflectance images are acquired at 150 Hz using a CCD camera configured with a tandem lens optical setup and bandpass emission filter. (d) Blue and green LEDs are sequentially illuminated at the beginning of each frame’s 6.67-ms exposure period for a duration of 5.5 ms to produce fluorescence and reflectance images each at 75 Hz. (e) Example transcranial images of green fluorescence (evoked by the blue LED) and green reflectance (evoked by the green LED) from a PHP.eB-GCaMP6s mouse (arrows: anterior/posterior and lateral/medial; asterisk: bregma; ). (f) Example trial-averaged visually evoked responses measured from contralateral V1 (see Sec. 2) for Thy1-GFP mice (top) and tetO-GCaMP6s mice (bottom). Blue LED-evoked epifluorescence is shown in blue, and green reflectance is shown in green. Magenta lines demonstrate 5-ms flash stimulus. Blood artifact-corrected responses for each mouse (right, shown in black) are represented by the subtraction of reflectance signal (green) from the fluorescence signal (blue).
2.2. Surgical Procedures
Large intact skull transcranial windows were installed on mice, as described in Ref. 15. Mice were anesthetized with 2% isoflurane and maintained with 1.5% isoflurane in air, and placed in a stereotactic frame. Body temperature was maintained at 37°C using a feedback-regulated heating pad with a rectal probe. Mice were injected under the scalp with lidocaine (0.1 mL, 0.2%) and subcutaneously with a saline solution containing buprenorphine (), atropine (), and glucose (20 mM). The fur on the top of the head was removed using scissors, and the skin was prepared with a triple scrub of 0.1% betadine in water followed by 70% ethanol. Skin on the top of the head was cut and removed, and fascia and connective tissues on the surface of the skull were removed so that the skull surface was completely clear of debris and dry. Clear dental cement, prepared by mixing 1 scoop of Metabond powder, 6 drops of C&B Metabond Quick Base, and one drop of C&B Universal catalyst (Parkell, Edgewood, New York) was used to glue a titanium head-fixing bar () to the cerebellar plate, directly posterior to the lambda.37 A layer of dental adhesive was applied directly on the skull, and a precut cover glass was placed gently on top of the mixture before it solidified.15,37 For two of the AAV-PHP.eB mice, a stainless steel setscrew15 was used for head-fixing instead of the titanium bar. Animals were monitored daily for 1 week postsurgery.
2.3. Histology
Expression of GCaMP in PHP.eB-injected animals was verified with postmortem histology weeks after experiments were conducted. Animals were sacrificed with an intraperitoneal injection of pentobarbital sodium () and transcardially perfused with 10 mL phosphate-buffered saline (PBS) followed by 10 mL of chilled 4% paraformaldehyde (PFA) in PBS. The brain was removed and immersed in a 4% PFA solution overnight before being transferred to a 0.02% sodium azide in PBS solution for storage at 8°C. Coronal brain sections ( thickness) were cut using a vibratome (Leica VT1000S) and mounted onto coverslips with Fluoromount-G (SouthernBiotech). The 12-bit images were acquired using a confocal microscope (Zeiss LSM 510 Meta) with a magnification/0.25 numerical aperture (NA) objective for the tiled image, or a NA objective for -stack images. GCaMP was excited using a 488-nm argon laser and epifluorescence was filtered using a 505 to 540-nm bandpass filter. Maximum intensity projections of -stack images ( range, interval, per slice, per pixel) were created and automatically adjusted for brightness and contrast, using ImageJ [Fig. 1(b)].
2.4. Imaging
All mice were habituated for 1 week prior to data collection. Awake mice were head-fixed and placed in a dark imaging chamber for data collection. Resting-state and visually evoked activity were recorded during 5-min trials. A behavioral camera (Microsoft Lifecam) and an infrared light were placed inside the imaging chamber to monitor active behaviors, such as grooming or whisking. To evoke visual responses, a white light-emitting diode (LED) was placed from the eye ( azimuth, elevation) and illuminated for a pulse duration of 5 ms with an interpulse interval of 10 s.
A Pantera 1M60 CCD camera (Dalsa) was equipped with two front-to-front lenses (50 mm, , ; Nikon Nikkor) and a bandpass emission filter (, Chroma) [Fig. 1(b)]. The 12-bit images were captured at a frame rate of 150 Hz (exposure time of 6.67 ms) with on-chip spatial binning using EPIX XCAP imaging software. The cortex was sequentially illuminated with alternating blue and green LEDs (Thorlabs) [Fig. 1(c)]. Blue light (473 nm) with a bandpass filter (467 to 499 nm) was used to excite calcium indicators and green light (525 nm) with a bandpass filter () was used to observe changes in cerebral blood volume. The blue and green LEDs were sequentially activated and synchronized to the start of each frame’s exposure period with transistor–transistor logic such that each frame collected only fluorescence or reflectance signals at 75 Hz each [Fig. 1(d)]. This LED strobe frequency of 75 Hz exceeded the critical flicker fusion frequency for mice, which marks the highest temporal frequency that an observer can resolve flicker before it becomes indistinguishable from constant light (estimated with electrophysiological38 and behavioral tests39) and was likely imperceptible to the mice. Reflectance signals were subtracted from fluorescence signals to mitigate the contribution of hemodynamics to fluorescence signals [Fig. 1(e)].
2.5. Brightness Comparison
One mouse from each group was randomly chosen as a representative example. Spontaneous activity was imaged in each mouse under at least three different LED power conditions. LED power was measured immediately before each trial using an optical power meter (Thorlabs PM100D) placed at the same position of the cortex. To benchmark mouse brightness, fluorescence standard solutions of carboxyfluorescein in distilled deionized water were prepared. Fluorescent standard solutions were loaded into borosilicate thin wall glass capillary tubes (1.17-mm inner diameter, 1.5-mm outer diameter) and imaged under the same conditions.
2.6. Image Processing and Analysis
All data were analyzed with MATLAB (Mathworks, Inc. Natick, Massachusetts). Before processing, cortical images were spatially binned by a factor of 2 to improve SNR. Behaviors monitored with the webcam were identified by calculating the temporal gradient within a region of interest (ROI) placed over the mouse’s front paws18 [Fig. S1(a) in the Supplementary Material]. The gradient was convolved with a Gaussian kernel () and a threshold was applied at the to identify periods of active behavior [Fig. S1(c) in the Supplementary Material]. These periods were removed from further analysis to avoid including large magnitude movement-related changes in fluorescence [Figs. S1(b) and S1(c) in the Supplementary Material]. While we have taken the precaution of removing these movement epochs, we acknowledge that this may also be an approach to generate brain activity maps associated with specific behaviors such as hindlimb movement, as shown in Fig. S1(b) in the Supplementary Material.
2.7. Visually Evoked Activity
Stimulus trials that occurred within 8 s of detected movements were discarded before analysis. The baseline fluorescence and reflectance for each flash stimulus was calculated as the average of the respective signal within the 5-s prestimulus period. Fluorescence and reflectance traces within a 10-s window centered on each visual stimulus (5 s before and after stimulus) were then converted to a percentage change from baseline () by subtracting this baseline from each point and then dividing by the baseline. To compensate for changes in absorbance due to increased hemodynamic activity, green reflectance changes () were subtracted from fluorescence images ().21,40 To calculate the visually evoked response, a ROI was placed over right monocular V1 (2.7-mm posterior and 1.6-mm lateral to bregma), and the spatially averaged corrected signal was computed as a function of time. SNR was calculated as the ratio of the peak in the corrected response 1 s after the stimulus to the standard deviation of the 1 s prestimulus interval. For trial-averaged responses, the SNR was calculated in the same way on the trial-averaged corrected fluorescence time series.
2.8. Resting-State Activity
Recordings without visual stimulation were also performed. Images were aligned across trials with a rigid body transformation (translation and rotation), using an intensity-based image registration algorithm. The global signal (average of all pixels within the entire bilateral cortical ROI) was then regressed out of each pixel.7 Images were then spatially smoothed with a two-dimensional Gaussian kernel () and then fluorescence and reflectance images which occurred within 8 s of identified periods of active behavior were removed from the data [Fig. S1(c) in the Supplementary Material]. The resulting image data were separated into 30-s epochs, and converted to a percentage change from baseline by subtracting the mean intensity over the 30-s period (baseline) from each pixel value () and then dividing by the baseline (). Images were then temporally filtered using a second-order Chebyshev bandpass filter (0.3 to 3 Hz). Finally, hemodynamic artifacts were corrected by subtracting reflectance images from fluorescence images.21,40 Seed pixel correlation (SPC) maps were created by calculating Pearson’s correlation coefficient between a seeded pixel’s time series with every other pixel within the masked brain region over the 30-s epoch. To determine the accuracy of resting state mapping, correspondence between clusters of spontaneous brain activity were compared with anatomical regions. We seeded pixels in four regions (M1, HL, RS, and V1), and then compared the SPC maps to binary template maps from the Allen Institute cortical atlas.41 SPC maps were compared to binary template maps by first setting all weakly- or anti-correlated pixels () to 0, and then calculating the correlation between the template map and the thresholded SPC map.
2.9. Statistical Analysis
Box plots show the sample median (horizontal line) and the first and third quartiles (edges of the box). Whiskers extend to the adjacent data point that lies past the first and third quartiles by the interquartile range. Outliers are shown with black circles. Mice are compared across groups using a Kruskal–Wallis test. Significance is assessed with a posthoc Bonferroni correction.
3. Results
3.1. Brightness Comparison
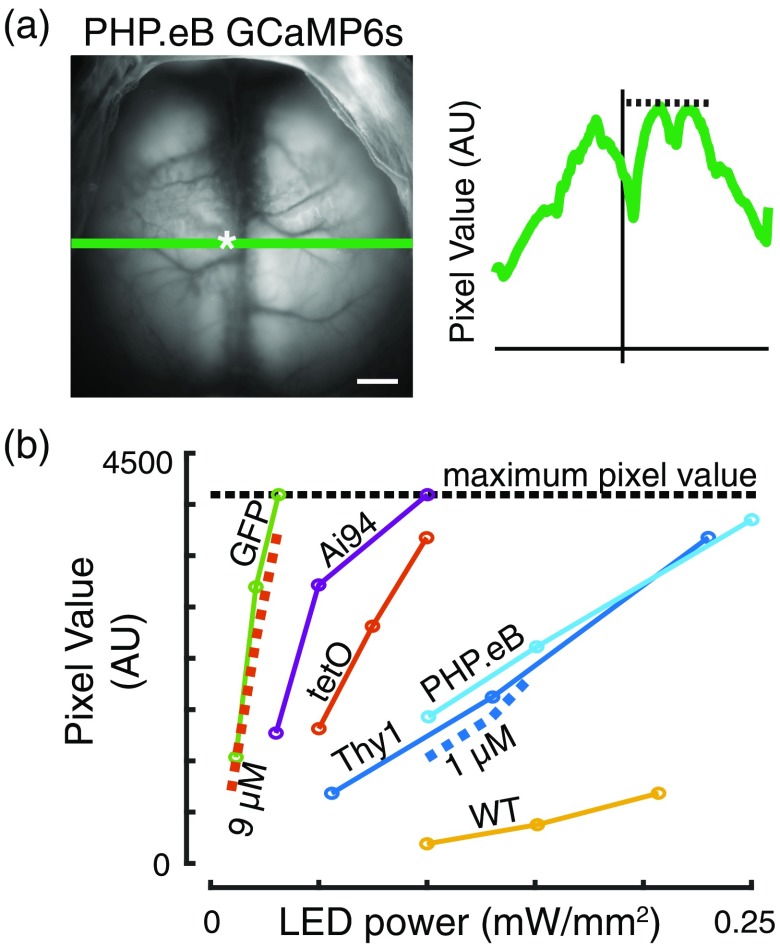
The brightness of fluorescence was compared across mice by examining 15 s of movement-free spontaneous activity, averaged across time. The maximum pixel value obtained along a horizontal line through the bregma was measured [Fig. 2(a)] for multiple LED powers. The Thy1-GFP mice produced the brightest basal level of fluorescence, comparable to the carboxyfluorescein standard solution. In comparison, Ai94 and tetO mice were the brightest GCaMP-expressing mice. Mice infected with AAV-PHP.eB-GCaMP6s were approximately as bright as the Thy1-GCaMP6s mice or carboxyfluorescein standard solution but dimmer than Ai94 and tetO mice [Fig. 2(b)]. This level of fluorescence was also brighter than the WT mice. It should be noted that relatively high LED power (up to ) was required to image the cortex due to the use of a short frame exposure period (6.667 ms, required to accurately strobe between two excitation wavelengths), which was consistent with other experiments in this study.
Fig. 2.
Brightness comparison between different GECI mouse lines and PHP.eB expression. (a) Fluorescence image from a negative mouse infected with AAV-PHP.eB-GCaMP6s showing process for comparing absolute brightness. Bregma is marked by the white asterisk (). The maximum fluorescence intensity is calculated for all pixels along a line through the bregma (green line, bregma denoted by white asterisk). (b) Maximum fluorescence intensity [as described in (a)] versus LED power. The 1 and indicate carboxyfluorescein concentrations used in standard solutions (dashed lines). Black dotted line at 4095 indicates the upper limit for pixel values for the 12-bit imaging system.
3.2. Visual Stimulus
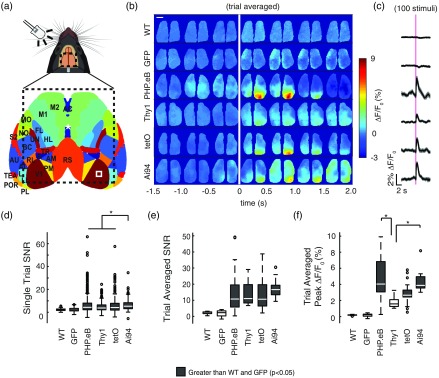
Visually evoked responses were produced by flashing a white LED in the mouse’s left visual field [Fig. 3(a)]. By alternating blue and green LED illumination, we were able to record signals coming from GCaMP fluorescence and blood artifact reflectance, respectively. Visual stimulation produced a strong positive change in fluorescence signal and a slight decrease in reflectance signal in GCaMP mice, while decreases in both fluorescence and reflectance signals were observed in the nonactivity-dependent Thy1-GFP line [Fig. 1(e)]. The negative response corresponded to the reported increase in local cerebral blood volume, which led to increased absorption of both green and blue lights by hemoglobin, limiting the excitation of GCaMP as well as its recorded fluorescence.40 This hemodynamic artifact was mitigated, but not completely eliminated, by subtracting the reflectance signal from the fluorescence signal [Fig. 1(e)]. Robust responses were observed in contralateral visual cortex of all mice expressing GCaMP6s, while WT and Thy1-GFP mice failed to exhibit significant responses [Figs. 3(b)–3(c)]. The strength of the evoked response was compared between mice by calculating the SNR for single-stimulus presentations and trial-averaged responses (see Sec. 2, Figs. 3(d)–3(f)], as well as the peak for trial-averaged responses. While the peak metric informed us about the amplitude of the response, the SNR reported a measure of the magnitude of the event relative to the background fluctuations. While Ai94 was most efficient to report single LED flash events relative to baseline [Fig. 3(d)], in the trial-averaged response, all lines and viral strategies produced comparable signals [Fig. 3(e)]. In terms of signal amplitude (peak ), however, both Ai94 and AAV-PHPeB.GCaMP6s mice displayed stronger signals than those observed from Thy1-GCaMP6s mice (, Kruskal–Wallis with posthoc Bonferroni correction).
Fig. 3.
Visually evoked responses. (a) Top: representation of LED flash stimulus experiments. A LED, facing toward the left eye of the mouse flashed once every 10 s. Bottom: cortical atlas from Allen Institute outlining cortical regions observed in transcranial imaging setup (outlined by dotted line). Bregma is shown by white asterisk. Evoked responses are observed in contralateral monocular V1 (white box). (b) Representative averaged responses over a 5-min trial. Individual cortical images represent the averaged over 0.5-s increments. A 5-ms LED flash occurs at 0 s (vertical white line) (). (c) Responses from contralateral V1 [white box from (a)]. LED flash indicated by magenta line. (d) SNR measurements for single LED flashes (see Sec. 2). (e) SNR measurements and (f) peak for trial-averaged LED flashes (*; Kruskal–Wallis with post hoc Bonferroni correction).
By definition, the SNR calculation was dependent on the variance of the signal during the pre-stimulus interval. However, some trials were removed from the 5-min recordings before analysis due to animal movements. This resulted in an unequal number of trials being averaged during the calculation of the trial-averaged SNR, due to variability in the number of movements between animals. This likely skewed the measured SNR, as the variance in the noise (pre-stimulus signal) was inversely proportional to the number of trials averaged [Fig. S2(a) in the Supplementary Material]. To explore this further, we calculated a bootstrapped estimation of the SNR by compiling all single-trial responses per animal across recording sessions and computing the average of 20 randomly sampled trials with replacement. The SNR was calculated for this 20-trial averaged signal and the process was repeated for 1000 iterations. Ai94 mice had a significantly greater bootstrapped response SNR than the other GCaMP mice, and both tetO and Thy1-GCaMP mice had a greater bootstrapped response SNR than the AAV-PHP.eB-GCaMP mice [Fig. S2(b) in the Supplementary Material]. Nevertheless, the bootstrapped peak for the AAV-PHP.eB and Ai94 mice were significantly greater than those in the tetO and Thy1-GCaMP [Fig. S2(c) in the Supplementary Material]. Ai94 represented, then, the strategy producing the highest level of basal and evoked signals. However, an accurate functional measure can also be observed with mice expressing AAV-PHPeB.GCaMP6s [Figs. 3(b)–3(f), S2(c) in the Supplementary Material], although the basal level of fluorescence was dimmer (see Fig. 2).
3.3. Resting State
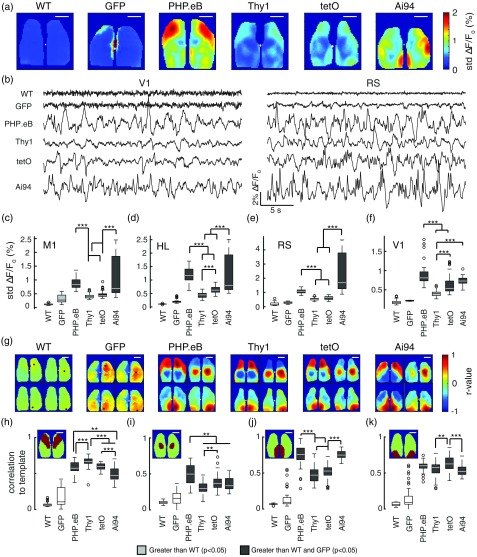
As the SNR of the visually evoked response depends on background fluctuations, the standard deviation of spontaneous activity was calculated. While the standard deviation of spontaneous fluctuations was relatively low and uniform in WT and Thy1-GFP mice, striking differences were observed in the topography of standard deviation maps across GCaMP-expressing mice [Figs. 4(a) and 4(b)]. Mice infected with PHP.eB-GCaMP6s had larger variations in background activity measured in M1, HL, RS, and V1 compared to Thy1-GCaMP6s or tetO-GCaMP6s mice [Figs. 4(c)–4(f)], , Kruskal–Wallis with post hoc Bonferroni correction). Similarly, Ai94 mice had significantly larger background fluctuations compared to both Thy1-GCaMP6s and tetO-GCaMP6s mice in M1, HL, and RS and compared to Thy1-GCaMP6s mice in V1 [Fig. 4(c)–4(f)], ).
Fig. 4.
Resting-state activity. (a) Standard deviation maps of cortical activity (median per pixel over all 30-s epochs in all mice). (b) Example traces (30 s duration) of movement-free spontaneous activity from V1 (left) or RS (right). (c–f) Median standard deviation of resting-state activity in M1, HL, RS, and V1. (g) Median seed pixel correlation maps for seeded pixels (black) located in primary motor cortex (M1), hindlimb cortical area (HL), retrosplenial cortex (RS), and primary visual cortex (V1) (top left, top right, bottom left, and bottom right subpanels, respectively). (h–k) Seed pixel correlation maps are compared to binary template maps for M1, HL, RS, and V1 (insets) (***; **; *; Kruskal–Wallis with posthoc Bonferroni correction). All .
To assess the uniformity of interhemispheric GCaMP expression and the spatial consistency of neural activity, seed pixel correlation maps were created using pixels seeded in M1, HL, RS, and V1 [Fig. 4(b)]. As expected, WT and Thy1-GFP mice did not exhibit the strong, bilateral, and regionally distinct correlations that were observed in the GCaMP-expressing mice [Fig. 4(b)]. Interestingly, SPC maps in Ai94 mice were not as regionally constrained as the other GCaMP mice [Fig. 4(g)]. We further compared these SPC map features between mice by estimating the similarity between each mouse’s SPC map (seed in M1, HL, RS, and V1) and a corresponding binary template for M1, HL, RS, and V1 derived from a cortical atlas from Allen Institute for Brain Science. The cortical atlas is an image where the functional regions of dorsal cortex have been identified [Fig. 3(a)]. The atlas was scaled and cropped to match the size of the brain images acquired in our imaging setup, and then pixels within regions that were chosen for SPC analysis were set to 1, while those outside the chosen region were set to 0 [Figs. 4(h)–4(k), insets]. While all GCaMP-expressing mice had SPC maps with significantly higher correlations to the corresponding binary template map [Figs. 4(h)–4(k)], Ai94 mice had significantly lower correspondence to the M1, HL, and V1 template regions compared to some of the other GCaMP mice.
4. Discussion
We conducted a head-to-head comparison of three transgenic models and one AAV-transduced model expressing GCaMP6s in cortical neurons using in vivo transcranial wide-field imaging. Tail vein injection of AAV-PHP.eB effectively transduced neurons with GCaMP6s, allowing for wide-field functional imaging of the entire dorsal cortex [Fig. 1(b)]. Further information about expression patterns can be found in the original work (e.g., Refs. 16, 20, 21 and 27). Absolute brightness, visually evoked signals, and the variance and regional correlation of spontaneous activity were examined in awake, head-restrained mice. Ai94 mice had the brightest basal fluorescence level among the GCaMP-expressing mice, followed by tetO-GCaMP6s mice, and then the Thy1-GCaMP6s and AAV-PHP.eB-hSyn-GCaMP6s mice, which were both roughly comparable. Although all GCaMP mice were generally comparable in terms of evoked performance compared to background activity, Ai94 mice exhibited a slightly greater SNR than the other GCaMP mice, and both AAV-PHP.eB-transduced mice and Ai94 mice had significantly greater peak responses than the Thy1-GCaMP6s mice. Resting-state activity was generally more dynamic in the AAV-PHP.eB mice and the Ai94 mice compared to the Thy1-GCaMP and tetO-GCaMP mice. Although all GCaMP mice exhibited strong bilateral correlations, SPC analysis revealed that spontaneous activity in Ai94 mice was not as spatially distinct compared to the other GCaMP mice.
We employed methodological strategies to avoid confounding factors that might limit the optical characterization of different mouse lines. For example, although mesoscale sensory responses are commonly examined under anesthesia,7 anesthetics are known to affect cortical sensory processing,42 excitatory-inhibitory tone,43 and brain-wide calcium dynamics.44,45 We, therefore, characterized GECI performance in these mice using a visually evoked paradigm, which allowed for the examination of spontaneous and sensory-evoked activity in an awake condition. Furthermore, GCaMP fluorescence can be affected by gross changes in cerebral blood flow or tissue oxygenation, as both the excitation and emission bands of GCaMP overlap with the absorption bands of oxygenated and deoxygenated hemoglobin.40 We, therefore, used sequential illumination of the cortex with 473- and 525-nm LEDs to evaluate and correct for the hemodynamic contribution to epifluorescence signals.18,19,21,40 Although subtraction of the reflectance signal did not completely eliminate the effects of hemodynamic changes on GFP fluorescence [Fig. 1(e)], the residual artifact was reduced and was unlikely to have major effects on visual performance metrics in the GCaMP mice, since the onset of the hemodynamic negative response typically lags neuronal activation and would therefore have minimal influence on the large positive peak . The use of red fluorescent indicators can also be used to overcome hemodynamic confounds as well as to image deeper into the cortex.46–49
The magnitude of the visually evoked response was significantly larger in the AAV-PHP.eB and Ai94 mice compared to the Thy1-GCaMP6s mice, suggesting differences in GCaMP expression in the responding neuronal population between mouse lines. Moreover, differences between mice were observed in the spatial organization of resting-state activity and absolute brightness. These differences may be expected, as the Thy1-GCaMP6s GP4.3 mice express GCaMP6s in a subset of excitatory cells,16 and mice expressing GCaMP6s under the CaMKIIa promoter (Ai94, tetO) have GCaMP expressed in excitatory neurons,50 while the AAV-PHP.eB-hSyn-GCaMP6s mice express GCaMP in all neurons.51 For this reason, multiple measures of performance besides for evoked SNR were necessary to comprehensively benchmark these mice for wide-field imaging. The use of wide-field imaging would not be appropriate for lines where relatively sparse cellular-level expression is observed. Sparse cellular expression would be ideally resolved with two-photon microscopy52 or methods where individual neurons can be segmented using wide-field imaging.53
Imaging GECIs in vivo is central to the analysis of neural circuit activity.8,54,55–57 We characterized the advantages and disadvantages in performance of three transgenic GCaMP models for use in wide-field imaging applications. While Ai94 mice exhibited desirable performance in brightness and visually evoked SNR, regional correlations were generally less distinct compared to the other mouse lines, and a relatively high level of spontaneous activity was observed. Moreover, a low incidence of epileptiform activity has been reported in this mouse line.58 In comparison, the tetO-GCaMP6s mouse exhibited similar evoked performance, with distinct regional correlations and relatively low levels of spontaneous activity. We also found that mice tail vein injected with AAV-PHP.eB-hSyn-GCaMP6s-WPRE achieved comparable performance to the transgenic models in terms of absolute brightness, visually evoked responses, resting-state activity, and regional correlation. Notably, we did not observe any abnormal physiological activity in the virus-injected mice, and even after 20 weeks postinjection, neurons in V1 and RS have histological features associated with healthy cells [Fig. 1(b)]. The BBB crossing PHP.eB27 serotype, therefore, provides an excellent opportunity to achieve widespread expression in adult animals for mesoscale mapping of cortex, without the need for multiple cortical microinjections. Moreover, this strategy provides the opportunity to express GCaMP6 or other optogenetic tools in adult transgenic disease models.59,60 Finally, this option also avoids any developmental contributions to aberrant electrophysiology, as reported in Ref. 58, because these viruses are injected into adult mice.
While we acknowledge that flash stimulation grossly underrepresents the possibilities for visual systems neuroscience,61 visual stimuli are preferred for dorsal cortical imaging because they can be applied noninvasively and without causing agitation to awake animals. However, based on the ability to produce spontaneous activity correlation maps (Fig. 4), we anticipate that PHP.eB-injected mice will provide robust functional signals outside of V1. Air puff stimuli, for example, can also be effective in mapping barrel cortex in the awake animal. Although whisker stimulation is also noninvasive, piezoelectric transducers necessary to move individual whiskers can be difficult to reproducibly position. Somatosensory stimulation using fore or hind-paw movement or electrical stimulation in awake animals is difficult to perform as they require attachment of transducers or stimulators to an awake animal. Auditory stimulation is also a possibility, but primary auditory cortex is not accessible through the standard transcranial windows. For the reasons described above we suggest that intravenous injection of AAV-PHP.eB and subsequent visual or air puff stimulation may provide a rapid means of assessing the functionally relevant signal from new genetically encoded sensors in vivo.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (CIHR) T.H.M. FDN-143209 and from Brain Canada for the Canadian Neurophotonics Platform to THM. CIHR or Brain Canada had no involvement in the research or decision to publish. We thank Pumin Wang for helping with surgery and Cindy Jiang for helping with animal husbandry. We thank Luis Bolaños for helping with creating figures. We thank Jamie Boyd for the technical help with data collection. We thank Marie-Eve Paquet (Canadian Neurophotonics Platform/University Laval), Viviana Gradinaru (Caltech), and Loren Looger (HHMI Janelia Farm) for providing the GCaMP6s viral vectors. We thank Cris Niell (University of Oregon) for providing the tetO-GCaMP6s mice to Jackson Labs.
Biography
Biographies of the authors are not available.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Silasi G., Murphy T. H., “Stroke and the connectome: how connectivity guides therapeutic intervention,” Neuron 83(6), 1354–1368 (2014). 10.1016/j.neuron.2014.08.052 [DOI] [PubMed] [Google Scholar]
- 2.White B. R., et al. , “Imaging of functional connectivity in the mouse brain,” PLoS One 6(1), e16322 (2011). 10.1371/journal.pone.0016322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song C., et al. , “Cortical signatures of wakeful somatosensory processing,” Sci. Rep. 8, 11977 (2018). 10.1038/s41598-018-30422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy M. C., et al. , “Macroscale variation in resting-state neuronal activity and connectivity assessed by simultaneous calcium imaging, hemodynamic imaging and electrophysiology,” Neuroimage 169, 352–362 (2018). 10.1016/j.neuroimage.2017.12.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grinvald A., et al. , “Functional architecture of cortex revealed by optical imaging of intrinsic signals,” Nature 324, 361–364 (1986). 10.1038/324361a0 [DOI] [PubMed] [Google Scholar]
- 6.Ts’o D. Y., et al. , “Functional organization of primate visual cortex revealed by high resolution optical imaging,” Science 249, 417–420 (1990). 10.1126/science.2165630 [DOI] [PubMed] [Google Scholar]
- 7.Vanni M. P., Murphy T. H., “Mesoscale transcranial spontaneous activity mapping in GCaMP3 transgenic mice reveals extensive reciprocal connections between areas of somatomotor cortex,” J. Neurosci. 34, 15931–15946 (2014). 10.1523/JNEUROSCI.1818-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T.-W., et al. , “Ultrasensitive fluorescent proteins for imaging neuronal activity,” Nature 499, 295–300 (2013). 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marvin J. S., et al. , “An optimized fluorescent probe for visualizing glutamate neurotransmission,” Nat. Methods 10, 162–170 (2013). 10.1038/nmeth.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons M. P., et al. , “Real-time imaging of glutamate clearance reveals normal striatal uptake in Huntington disease mouse models,” Nat. Commun. 7, 11251 (2016). 10.1038/ncomms11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y., et al. , “Resolution of high-frequency mesoscale intracortical maps using the genetically encoded glutamate sensor iGluSnFR,” J. Neurosci. 36, 1261–1272 (2016). 10.1523/JNEUROSCI.2744-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marvin J. S., et al. , “Publisher Correction: stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR,” Nat. Methods 16, 206–206 (2019). 10.1038/s41592-018-0300-z [DOI] [PubMed] [Google Scholar]
- 13.McGirr A., et al. , “Cortical functional hyperconnectivity in a mouse model of depression and selective network effects of ketamine,” Brain 140(8), 2210–2225 (2017). 10.1093/brain/awx142 [DOI] [PubMed] [Google Scholar]
- 14.Silasi G., et al. , “Improved methods for chronic light-based motor mapping in mice: automated movement tracking with accelerometers, and chronic EEG recording in a bilateral thin-skull preparation,” Front. Neural Circuits 7, 123 (2013). 10.3389/fncir.2013.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silasi G., et al. , “Intact skull chronic windows for mesoscopic wide-field imaging in awake mice,” J. Neurosci. Methods 267, 141–149 (2016). 10.1016/j.jneumeth.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dana H., et al. , “Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo,” PLoS One 9, e108697 (2014). 10.1371/journal.pone.0108697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jun J. J., et al. , “Fully integrated silicon probes for high-density recording of neural activity,” Nature 551, 232–236 (2017). 10.1038/nature24636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanni M. P., et al. , “Mesoscale mapping of mouse cortex reveals frequency-dependent cycling between distinct macroscale functional modules,” J. Neurosci. 37, 7513–7533 (2017). 10.1523/JNEUROSCI.3560-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao D., et al. , “Mapping cortical mesoscopic networks of single spiking cortical or sub-cortical neurons,” Elife 6, e19976 (2017). 10.7554/eLife.19976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madisen L., et al. , “Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance,” Neuron 85(5), 942–958 (2015). 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wekselblatt J. B., et al. , “Large-scale imaging of cortical dynamics during sensory perception and behavior,” J. Neurophysiol. 115(6), 2852–2866 (2016). 10.1152/jn.01056.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royo N. C., et al. , “Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity,” Brain Res. 1190, 15–22 (2008). 10.1016/j.brainres.2007.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson K. L., et al. , “Better targeting, better efficiency for wide-scale neuronal transduction with the Synapsin promoter and AAV-PHP.B,” Front. Mol. Neurosci. 9, 116 (2016). 10.3389/fnmol.2016.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saraiva J., Nobre R. J., Pereira de Almeida L., “Gene therapy for the CNS using AAVs: the impact of systemic delivery by AAV9,” J. Control. Release 241, 94–109 (2016). 10.1016/j.jconrel.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki Y., et al. , “Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain,” Neurosci. Lett. 665, 182–188 (2018). 10.1016/j.neulet.2017.11.049 [DOI] [PubMed] [Google Scholar]
- 26.Deverman B. E., et al. , “Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain,” Nat. Biotechnol. 34, 204–209 (2016). 10.1038/nbt.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan K. Y., et al. , “Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems,” Nat. Neurosci. 20, 1172–1179 (2017). 10.1038/nn.4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dayton R. D., Grames M. S., Klein R. L., “More expansive gene transfer to the rat CNS: AAV PHP.EB vector dose-response and comparison to AAV PHP.B,” Gene Ther. 25, 392–400 (2018). 10.1038/s41434-018-0028-5 [DOI] [PubMed] [Google Scholar]
- 29.Hordeaux J., et al. , “The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice,” Mol. Ther. 26, 664–668 (2018). 10.1016/j.ymthe.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillier D., et al. , “Causal evidence for retina-dependent and -independent visual motion computations in mouse cortex,” Nat. Neurosci. 20, 960–968 (2017). 10.1038/nn.4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen W. E., et al. , “Global representations of goal-directed behavior in distinct cell types of mouse neocortex,” Neuron 94, 891–907.e6 (2017). 10.1016/j.neuron.2017.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y., et al. , “Inverse-response Ca indicators for optogenetic visualization of neuronal inhibition,” Sci. Rep. 8, 11758 (2018). 10.1038/s41598-018-30080-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y., Lai T., Campbell R. E., “Red fluorescent proteins (RFPs) and RFP-based biosensors for neuronal imaging applications,” Neurophotonics 2, 031203 (2015). 10.1117/1.NPh.2.3.031203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St-Pierre F., et al. , “High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor,” Nat. Neurosci. 17, 884–889 (2014). 10.1038/nn.3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., et al. , “Astrocytic endfoot Ca2+ correlates with parenchymal vessel responses during 4-AP induced epilepsy: an in vivo two-photon lifetime microscopy study,” J. Cereb. Blood Flow Metab. 39(2), 260–271 (2019). 10.1177/0271678X17725417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng G., et al. , “Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP,” Neuron 28, 41–51 (2000). 10.1016/S0896-6273(00)00084-2 [DOI] [PubMed] [Google Scholar]
- 37.Bolanos F., et al. , “Mesoscale cortical calcium imaging reveals widespread synchronized infraslow activity during social touch in mice,” bioRxiv (2018). 10.1101/430306 [DOI] [Google Scholar]
- 38.Ridder W. H., Nusinowitz S., “The visual evoked potential in the mouse—origins and response characteristics,” Vision Res. 46(6-7), 902–913 (2006). 10.1016/j.visres.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 39.Nomura Y., et al. , “Evaluation of critical flicker-fusion frequency measurement methods using a touchscreen-based visual temporal discrimination task in the behaving mouse,” Neuroscience Research (2018). 10.1016/j.neures.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 40.Ma Y., et al. , “Correction to ‘Wide-field optical mapping of neural activity and brain haemodynamics: considerations and novel approaches’,” Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160539 (2017). 10.1098/rstb.2016.0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh S. W., et al. , “A mesoscale connectome of the mouse brain,” Nature 508, 207–214 (2014). 10.1038/nature13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michelson N. J., Kozai T. D. Y., “Isoflurane and ketamine differentially influence spontaneous and evoked laminar electrophysiology in mouse V1,” J. Neurophysiol. 120(5), 2232–2245 (2018). 10.1152/jn.00299.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haider B., Häusser M., Carandini M., “Inhibition dominates sensory responses in the awake cortex,” Nature 493, 97–100 (2013). 10.1038/nature11665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitra A., et al. , “Spontaneous infra-slow brain activity has unique spatiotemporal dynamics and laminar structure,” Neuron 98, 297–305.e6 (2018). 10.1016/j.neuron.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright P. W., et al. , “Functional connectivity structure of cortical calcium dynamics in anesthetized and awake mice,” PLoS One 12(10), e0185759 (2017). 10.1371/journal.pone.0185759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dana H., et al. , “Sensitive red protein calcium indicators for imaging neural activity,” Elife 5, e12727 (2016). 10.7554/eLife.1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dana H., et al. , “Thy1 transgenic mice expressing the red fluorescent calcium indicator jRGECO1a for neuronal population imaging in vivo,” PLoS One 13, e0205444 (2018). 10.1371/journal.pone.0205444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montagni E., et al. , “Wide-field imaging of cortical neuronal activity with red-shifted functional indicators during motor task execution,” J. Phys. D. Appl. Phys. 52, 074001 (2019). 10.1088/1361-6463/aaf26c [DOI] [Google Scholar]
- 49.Bethge P., et al. , “An R-CaMP1.07 reporter mouse for cell-type-specific expression of a sensitive red fluorescent calcium indicator,” PLoS One 12(6), e0179460 (2017). 10.1371/journal.pone.0179460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X., et al. , “Distribution of CaMKIIα expression in the brain in vivo, studied by CaMKIIα-GFP mice,” Brain Res. 1518, 9–25 (2013). 10.1016/j.brainres.2013.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kügler S., et al. , “Differential transgene expression in brain cells in vivo and in vitro from AAV-2 vectors with small transcriptional control units,” Virology 311, 89–95 (2003). 10.1016/S0042-6822(03)00162-4 [DOI] [PubMed] [Google Scholar]
- 52.Sofroniew N. J., et al. , “A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging,” Elife 5, e14472 (2016). 10.7554/eLife.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim T. H., et al. , “Long-term optical access to an estimated one million neurons in the live mouse cortex,” Cell Rep. 17(12), 3385–3394 (2016). 10.1016/j.celrep.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J. L., et al. , “Behaviour-dependent recruitment of long-range projection neurons in somatosensory cortex,” Nature 499, 336–340 (2013). 10.1038/nature12236 [DOI] [PubMed] [Google Scholar]
- 55.Mohajerani M. H., et al. , “Spontaneous cortical activity alternates between motifs defined by regional axonal projections,” Nat. Neurosci. 16, 1426–1435 (2013). 10.1038/nn.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gunaydin L. A., et al. , “Natural neural projection dynamics underlying social behavior,” Cell 157(7), 1535–1551 (2014). 10.1016/j.cell.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stettler D. D., Axel R., “Representations of Odor in the Piriform Cortex,” Neuron 63(6), 854–864 (2009). 10.1016/j.neuron.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 58.Steinmetz N. A., et al. , “Aberrant cortical activity in multiple GCaMP6-expressing transgenic MOUSE lines,” eNeuro 4, 0207–0217 (2017). 10.1523/ENEURO.0207-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peça J., et al. , “Shank3 mutant mice display autistic-like behaviours and striatal dysfunction,” Nature 472, 437–442 (2011). 10.1038/nature09965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jankowsky J. L., et al. , “Mutant presenilins specifically elevate the levels of the 42 residue -amyloid peptide in vivo: evidence for augmentation of a 42-specific γ secretase,” Hum. Mol. Genet. 13, 159–170 (2003). 10.1093/hmg/ddh019 [DOI] [PubMed] [Google Scholar]
- 61.Zhuang J., et al. , “An extended retinotopic map of mouse cortex,” Elife 6, e18372 (2017). 10.7554/eLife.18372 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.