Abstract
Background:
Cannabinoid has long been used for medicinal purposes. Cannabinoid signaling has been considered the therapeutic target for treating pain, addiction, obesity, inflammation, and other diseases. Recent studies have suggested that in addition to CB1 and CB2, there are non-CB1 and non-CB2 cannabinoid-related orphan GPCRs including GPR18, GPR55, and GPR119. In addition, CB1 and CB2 display allosteric binding and biased signaling, revealing correlations between biased signaling and functional outcomes. Interestingly, new investigations have indicated that CB1 is functionally present within the mitochondria of striated and heart muscles directly regulating intramitochondrial signaling and respiration.
Conclusion:
In this review, we summarize the recent progress in cannabinoid-related orphan GPCRs, CB1/CB2 structure, Gi/Gs coupling, allosteric ligands and biased signaling, and mitochondria-localized CB1, and discuss the future promise of this research.
Keywords: Cannabinoid receptor, Structure, Orphan GPCRs, Allosteric ligand, Biased signaling, Mitochondria
1. INTRODUCTION
Cannabis sativa L., commonly known as marijuana, has been used in different civilizations for a variety of medical applications such as appetite stimulation and the treatment of pain, nausea, fever, and gynecological disorders for thousands of years [1, 2]. There are more than 100 different cannabinoids isolated from cannabis plant, exhibiting varied effects by activating cannabinoid receptors. The most notable cannabinoid is △9-tetrahydrocannabinol (THC), the principal psychoactive constituent in cannabis. Cannabidiol (CBD) is another major constituent. Apart from the phytocannabinoids, ligands for cannabinoid receptors also include the endocannabinoids that are produced naturally throughout the body and the synthetic cannabinoids [3]. There are six recognized endocannabinoids currently. Anandamide (AEA) was the first one identified [4]. 2-arachidonoyl glycerol (2-AG), N-arachidonoyl-dopamine (NADA), 2-arachidonyl glyceryl ether (noladin ether), virodhamine (OAE) as well as Lysophosphatidylinositol (LPI) were subsequently discovered [5, 6].
The mechanism of action of cannabinoid drugs became clear with the subsequent identification of two cannabinoid receptors, termed CB1 and CB2 [7, 8]. They are members of the class A G protein-coupled receptor (GPCR) family and possess approximately 44% amino acid similarity [9]. As shown in (Fig. 1), both receptors share a common feature of class A GPCRs, possessing a glycosylated extracellular amino-terminal (N-term) and an intracellular carboxyl-terminal (C-term) domain connected by seven transmembrane domains (7TM), three extracellular loops (ECL1, ECL2, and ECL3) and three intracellular loops (ICL1, ICL2 and ICL3).
Fig. (1).
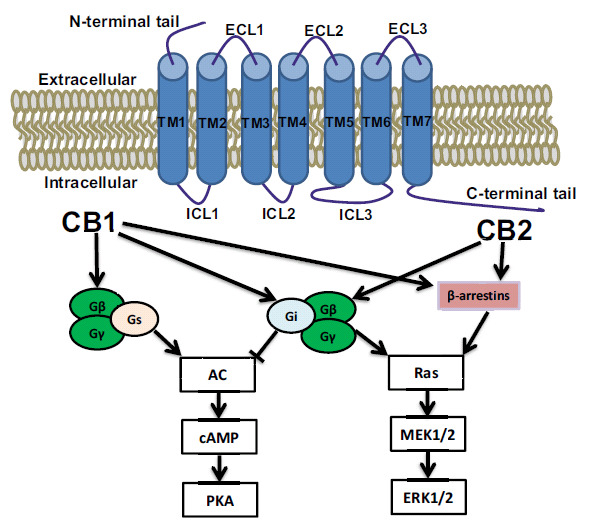
Schematic model of CB1 and CB2, showing the terminal tails, TM helices, intracellular loops, as well as the major intracellular signaling pathways.
CB1 receptors are predominantly expressed in the brain, particularly in cerebral cortex, hippocampus, basal ganglia, and cerebellum [7, 10], where they mediate the majority of the psychotropic and behavioral effects of cannabis. Recent evidence has suggested that CB1 are also expressed in several peripheral tissues [11, 12], including the spleen, lung, thymus, heart, and vasculature. In comparison, CB2 receptors are abundantly expressed in peripheral tissues with immune function, such as leukocytes, spleen, tonsils, thymus, as well as the lung and testes [9, 13, 14]. Subsequent studies have shown the role of the CB2 receptors in a variety of systems including the central nervous systems (CNSs), as well as the cardiovascular and respiratory systems, bone, the gastrointestinal (GI) tract, liver and the reproductive system [15-17]. As shown in (Fig. 1), existing evidence demonstrated that both CB1 and CB2 receptors are coupled to Gi/o protein to inhibit adenylyl cyclase activity leading to a decrease of cAMP levels. Alternatively, CB1 receptors have also been shown to be capable of coupling to Gs protein stimulating cAMP production in some cases [18-20]. It has been reported that a cannabinoid-mediated the increase of cAMP in cultured rat striatal neurons as well as in CB1-expressing CHO cells in the presence of forskolin [14]. The interaction of the CB1 receptor with Gs has also been confirmed in CHO cells transfected with recombinant human CB1 receptors [18, 19]. However, it is not clear whether CB2 receptor could bind with other G proteins [21]. The dual inhibition and activation effects of the CB1 receptor on adenylyl cyclase were verified to be ligand-specific [18, 22]. Our recent studies have revealed that the intracellular loop 2 (ICL2) of both CB1 and CB2 receptors and in particular the residue Leu-222, which resides within a highly conserved DRY(X)5PL motif, played a critical role in Gs and Gi protein coupling and specificity [14, 21].
The cannabinoid receptors have always been the targets of intensive drug development efforts. Accumulating studies suggest that the pharmacologic regulation of the CB1 has been proposed as a promising therapeutic strategy for a wide range of disorders, including pain, inflammation, obesity, neurodegenerative disorders, and cancer [10, 12, 23, 24]. Meanwhile, CB2, predominantly expressed in the peripheral areas, is an attractive therapeutic target for immune-modulators, pain management, osteoporosis, and the treatment of liver diseases [25-27]. This chapter will provide an overview of the recent studies regarding the structure and allosteric modulation of cannabinoid receptors, CB1 and CB2, and characterization of orphan cannabinoid receptors. We also highlight the most recent advances in the biased signaling and intracellular localization of cannabinoid receptors. These findings uncover a previously unexplored framework for cannabinoid system-targeted drug discovery.
2. CANNABINOID RECEPTOR STRUCTURE
The human cannabinoid system plays critical roles in the regulation of human physiology and makes up the targets for intensive drug discovery efforts. On the other hand, the elucidation of the detailed structure of the cannabinoid receptors will facilitate a better understanding of how agonists and antagonists engage to modulate the downstream signaling of the cannabinoid system. Therefore, the human cannabinoid system becomes an attractive area of research. In addition to the improvement of methods including recombinant expression systems and protein engineering, the breakthroughs in key techniques such as the use of a T4 lysozyme insertion, lipidic cubic phase crystallization and microfocus diffraction beamlines lead to a remarkable progress in the elucidation of GPCR structures [28, 29].
The first 2.8 Å crystal structure of the human CB1 receptor in complex with a tight binding AM6538, a stabilizing antagonist, was successfully resolved in 2016 [30]. The structure of the CB1-AM6538 complex reveals key characteristics of the receptor and decisive interactions for antagonist binding. The N terminus of CB1 plays a key role in ligand recognition, which is the same as other receptors. Specifically, the non-truncated part of the N terminus of CB1 forms a V-shaped loop that inserts into the ligand-binding pocket and acts as a plug to restrict access to the pocket from the extracellular side. Meanwhile, the extracellular loop 2 (ECL2) composed of 21 residues folds into a complex structure, projecting four residues (268-271) into the binding pocket. It has been reported that the four residues are essential for mediating interactions with certain kinds of ligands [31] and the two cysteines (Cys257 and Cys264) in ECL2 are pivotal to the function of CB1 [32]. Two months later, the structure of the human CB1 receptor bound to the inhibitor taranabant at 2.6-Å resolution was determined [33]. Consistent with the AM6538-bound structure, the extracellular surface of CB1, including the highly conserved membrane-proximal N-terminal region, forms a critical part of the ligand-binding pocket. However, the taranabant-bound structure showed a stronger electron density for the ligand and the N-terminal region than the former one. It may be important for functional interpretation and prediction.
The structures of CB1 complexed with the antagonist AM6538 and taranabant reveal a molecular understanding of the inactivation state of the receptor, yet they do not show how CB1 elicits its diverse physiological effects. For this reason,two agonist-bound crystal structures of human CB1 in complex with a tetrahydrocannabinol (AM11542) and a hexahydrocannabinol (AM841) at 2.80 Å and 2.95 Å resolution were determined [34]. Comparisons between the agonist- and antagonist-bound CB1 complexes reveal notable structural rearrangements. A notable feature of the CB1 agonist bound structure seems to use an extended molecular toggle switch termed ‘twin toggle switch’, involving a synergistic conformational change between Phe2003.36 and Trp3566.48. In addition, the two CB1-agonist complexes reveal a large (53%) reduction in the volume of the ligand-binding pocket, and subsequent increase in the surface area of the G-protein-binding region. Such plasticity in the orthosteric binding pocket enables CB1 to respond to a variety of ligands with different sizes and shapes.
During the past decade, more than 40 unique receptors have been characterized by X-ray crystallography [28, 35]. However, the atomic level details regarding the CB2 receptor activation remain unanswered. Several research groups tried to construct homology-based comparative CB2 models using some 3D crystal structures of GPCRs, like rhodopsin, β2AR, and A2AAR. The first 3D homology model for the CB2 was reported in 1999, based on the α-helical periodicity in the CB2 sequence [36]. A comparative 3D CB2 model was constructed in 2003 [37]. Three years later, other CB2 comparative models were clarified based on the crystal structure of bovine rhodopsin determined at 2.8 Å as a template, and those models represented the inactivated state [38-40]. During 2011 and 2012, newer 3D CB2 models based on A2AAR were generated [41, 42]. All models above were used to analyze CB2 ligand binding properties and explain the effects of individual biological or pharmacological experiments. However, they are limited to the few crystal structures low sequence identities to CB2.
Although most of CB2 models are built on the reported inactive GPCRs, Xie et al. constructed both active as well as the inactive CB2 models by homology modeling. The active CB2 bound with both the agonist and G protein, while the inactive CB2 bound with inverse agonist [43]. In these models, the inactive CB2 and the inverse agonist remained stable during the molecular dynamics (MD) simulation. While during the assay, dynamical details about the breakdown of the “ionic lock” between R1313.50 and D2406.30 as well as the outward/inward movements of transmembrane (TM) domains that bind with G proteins and agonist (TM5, TM6, and TM7) were observed. Moreover, W2586.48 in TM6 and residues in TM4 (V1644.56−L1694.61) contribute greatly to the binding of the agonist on the basis of the binding energy decomposition, while residues S180-F183 in ECL2 may be essential in recognition of the inverse agonist. Taken together, the increasingly refined homology-based CB2 models provide new insights to a better understanding of the structure and conformation of CB2, useful for virtual screening and drug design.
3. CANNABINOID-RELATED ORPHAN GPCRS
Up to date, there are still around 100 GPCRs with no known endogenous ligand(s), termed orphan GPCRs (oGPCRs) [44]. The deorphanization of oGPCRs opens new possibilities in GPCR-targeted drug discovery. Using CB1-/- and CB2-/- mice, previous studies have suggested the existence of putative orphan cannabinoid receptors, the so-called non-CB1/CB2 receptors, in the vasculature, central nervous system and immune cells [45, 46]. Three oGPCRs, GPR55, GPR18 and GPR119, have emerged as putative non-CB1/CB2 receptors [47, 48].
GPR55 is an orphan GPCR in the purinergic subfamily. GPR55 was first identified from in silico studies and later cloned in 1999 [49]. The first report linking GPR55 to cannabinoids appeared in a patent from GlaxoSmithKline [50]. Subsequently, AstraZeneca published a patent confirmed the association of GPR55 with cannabinoids [51]. However, GPR55 has very low homology to the classical cannabinoid receptors (about 14% and 15% with CB1 and CB2 respectively) [52]. The endogenous ligand of GPR55 L-α- lysophosphatidylinositol (LPI), was first identified by Oka et al [52]. Recently, accumulating data suggest that some synthetic cannabinoid ligands can also activate GPR55, such as CB2 agonists HU210 [53] and CB1 antagonists AM251 and rimonabant [54]. However, in some studies, rimonabant also acted as a GPR55 antagonist [55]. Several groups have pharmacologically determined the downstream signaling of GPR55 in HEK293 cells. In their studies, the GPR55-mediated activation of ERK1/2 signaling [52], Rho activation, calcium ion elevations, and the activation of transcription factors, including nuclear factor of activated T-cells and cAMP response element binding protein (CREB) [56] were characterized. Additionally, GPR55-mediated β-arrestin recruitment has also been demonstrated [57]. Interestingly, GPR55 has been shown to form heteromers with CB2 receptor, which impacts on cell signaling [58], leading to enhanced MAP kinase activation, and reduced transcription factor generation [59].
GPR18 is less homologous to CB1 and CB2 (~13% and 8%), but GPR18 is closely related to the endocannabinoid system (ECS). A number of cannabinoid ligands have been described to be active as agonists or antagonists, such as abn-CBD, O1602, Δ9-THC and N-arachidonoylcyclopropyl amide have all been described as full agonists in GPR18-transfected HEK cells [60]. Several studies have suggested that N-arachidonylglycine (NAGly) is an endogenous ligand for GPR18. However, NAGly has no activity on the classical cannabinoid receptors CB1 and CB2 [61]. Therefore, it is difficult to determine whether GPR18 is a cannabinoid receptor.
GPR119 is an orphan receptor originally identified in genome-sequencing efforts and expressed predominantly in the pancreas and gastrointestinal tract. Many types of research show that GPR119 regulates energy balance in the pancreas [62]. In 2006, Overton et al. identified GPR119 as a cannabinoid receptor [48], and it can be activated by oleoylethanolamine (OEA; the first identified endocannabinoid). Subsequently, 2AG has also been identified as a GPR119 agonist [63]. A third endogenous GPR119 activator, oleoyllysophosphatidylcholine, has also been identified with some structural commonality with the endogenous ligand of GPR55 [64]. In addition, there are many GPR119 small molecule agonists, for example PSN75963 evokes concentration-dependent increases, in cAMP in GPR119-transfected cells by coupling with Gas [48]. AS1269574 is a structurally distinct synthetic GPR119 agonist, which was identified to enhance glucose-stimulated insulin secretion both in vitro and in vivo. To date, no antagonists of GPR119 have been described, this is may be due to the fact that there appears to be little therapeutic potential for them [47]. Studies have shown that GPR119 may be involved in the regulation of type 2-diabetes, metabolic disorders and obesity. In fact, synthetic GPR119 agonists showed positive results in phase II clinical trials of type 2 diabetes [58].
Because of their close phylogenetic relationship with the cannabinoid receptors or their ability to interact with lipids, the orphan receptors GPR3, GPR6, GPR12, GPR23, and GPR92 have been categorized as possible cannabinoid receptor candidates [65, 66]. Orphan receptors GPR3, GPR6, and GPR12 share over 60% of sequence similarity, and are mainly expressed in the brain and the reproductive system [67]. Among them, both GPR3 and GPR6 have been recently found to be activated by the nonpsychoactive phytocannabinoid cannabidiol (CBD) [68]. Using the β-arrestin PathHunter assay system,N-arachidonoyl glycine (NAGly) has been shown to elicit a weak activation of GPR92 [69]. However, to date, there are no published data to show that any known agonists or antagonists for CB1 or CB2 receptors are able to activate orphan receptors GPR12 and GPR23. Therefore, there is a need for these orphan receptors to be examined for their responsiveness to different endogenous, phytogenic, and/or synthetic cannabinoid agonists and antagonists.
4. ALLOSTERIC MODULATORS OF CANNABINOID RECEPTORS
Allosteric ligands, binding to the secondary binding sites, are divided into three groups according to their effects on orthosteric ligand (bind to the same site as the endogenous ligand) responses: Positive Allosteric Modulators (PAMs), Negative Allosteric Modulators (NAMs), and Neutral Allosteric Ligands (NALs) [70, 71]. Pharmacological advantages such as higher specificity and thus reduced side effects have motivated efforts to characterize and develop both positive and negative allosteric modulators targeting GPCR in academia and industry. Here we list both the CB1 and CB2 allosteric modulators.
It has been validated that significant abuse potential and the risk of psychological and mood-altering side effects seriously affect the development of orthosteric agonists and antagonists targeted CB1 [72-74]. This has prompted academia and industry to develop both positive and negative CB1-targeted allosteric small-molecules. Since the first report on a CB1 allosteric ligand-binding site in 2005 [75], both PAM such as PAM1, ZCZ011, Lipoxin A4 and RTI-371 and NAM including Org27569, ABD1027, PSNCBAM-1, GAT358, Pregnenolone, CBD and Fenofibrate, have been developed [76]. The synthetic indole Org27569 is one of the most intensively studied CB1 allosteric modulators. The initial characterization showed that Org27569 displayed contradictory pharmacological behavior as PAMs of orthosteric agonist affinity, but NAMs of agonist efficacy [75-77]. Further profiling demonstrated that Org27569 acts as a biased allosteric agonist showing the potential to inhibit agonist-induced G-protein-mediated inhibition of cAMP production, but to enhance agonist-induced ERK1/2 activation [78]. To date, a number of Org27569 analogs have been developed [79, 80]. Among them, one compound was found to specifically induce β-arrestin1-mediated pathway-biased signaling [81]. Interestingly, the nonpsychoactive cannabidiol (CBD), a phytocannabinoid compound with the potential to interact with many nonendocannabinoid signaling targets including the opioid receptors, the serotonin 5HT1A receptors, the PPARγ receptors, and the transient channel receptor[76, 82], has been demonstrated to act as a negative allosteric modulator in reducing the effect of 2-AG and 19-THC on CB1 internalization and PLCβ3 and ERK1/2 phosphorylation, exerting a wide range of cellular effects through the endocannabinoid system [83, 84].
The CB2 receptor has long been recognized as an attractive therapeutic target for immune-modulators, pain management, osteoporosis, and the treatment of liver diseases [25-27]. However, compared to CB1, few allosteric modulators are developed for CB2. Pepcan-12 (RVD-hemopressin; RVDPVNFKLLSH), the major peptide of a family of endogenous peptide endocannabinoids (pepcans) found to act as Negative Allosteric Modulators (NAM) of cannabinoid CB1 receptors, has recently been shown to be a potent Positive Allosteric Modulator (PAM) for human CB2 receptor [85]. Martinez-Pinilla et al. have provided evidence that CBD at nanomolar concentrations shows the potential not only to slightly but consistently modify the binding of the fluorophore-conjugated CB2-selective compound, CM-157, to HEK-293T cells expressing CB2, but also to significantly reduce the effect of the selective CB2 agonist, JWH133, on forskolin-induced intracellular cAMP levels and on activation of the MAP kinase pathway, suggesting that CBD seems to act as a negative allosteric modulator of the human CB2 receptor [86]. However, more efforts are needed to further characterize small molecules as allosteric modulators of CB2 for future design and synthesis of optimized allosteric modulators.
5. CANNABINOID RECEPTOR BIASED SIGNALING
It has long been believed that most of GPCR couples to multiple Gα proteins (e.g. Gαi/o. Gαs, Gαq, or Gα12/13) and β-arrestins, triggering multiple intracellular signalling pathways in parallel and/or sequentially through different transduction mechanisms [87]. Accumulating evidence has firmly established that distinct GPCR agonists exhibit the potential to selectively activate one specific signaling cascade over another, a phenomenon referred to as “biased agonism” or “functional selectivity”, and trigger distinct physiological responses [88, 89].
Since for the first time introduction of the pioneering concept of biased agonism in the GPCR field by Kenakin [88], amassing number of GPCR agonists have been identified to preferentially signal via either β-arrestin-biased pathway or Gα protein subtype-biased cascade. The first evidence for biased signaling through CB1 came from a study by Glass and Northrup. They demonstrated that agonists HU-210, AEA and WIN55,212-2 evoked maximal Gαi activation, whereas THC induced only partial Gαi activation. In contrast, only HU-210 effected maximal CB1 stimulation of Gαo, with AEA, WIN55,212-2 and THC all partially stimulating [90]. However, agonist-mediated activation of Gαi and Gαo would lead to inhibition of adenyl cyclase. Ligands, HU-210 and CP55,940, were found to differ in potency and efficacy for inhibition of CREB-dependent transcription, leading to opposite effects on AP-1-dependent transcription [91, 92]. Compounds, WIN55,212-2 and HU-210, exhibit the same potential to couple to Gαi and Gαs proteins, whereas both CP55,940 and AEA preferentially activate Gαi-dependent pathway [18, 93]. Based on the structure of indole quinuclidinone (IQD) analogues that act as partial to full CB1 agonists for modulation of Gi-coupled intracellular effector adenylyl cyclase [94], novel agonists PNR-4-20 and PNR-4-02 were developed to act as highly biased Gαi protein agonists with significantly reduced coupling to β-arrestin2 [95]. In contrast, compound ORG27569 has been suggested to selectively activate ERK/12 signaling via β-arrestin1 [96, 97]. Accumulating evidence suggests that development of CB1 agonists with biased signaling toward Gαi-dependent pathway, relative to β-arrestin2 signaling, might provide new avenues for the development of analgesic drugs with fewer and less severe adverse effects both acutely and chronically [95-98]. As shown in (Fig. 2), we summarize the biased signaling at the CB1 receptor.
Fig. (2).
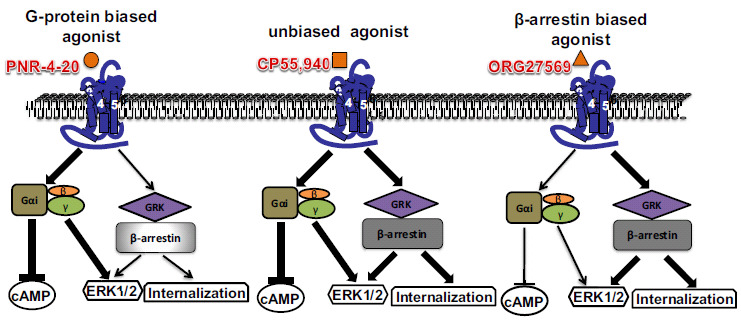
Schematic representation of biased signaling at CB1. Structurally different ligands will induce diverse conformations of the receptor, which may then favor one of the possible signaling pathways over others. In this diagram, the unbiased agonist CP55,940 engages the CB1 and activates both G protein- and β-arrestin-dependent signaling pathways. PNR-4-20 is biased toward the activation of the Gαi heterotrimer over β-arrestin, while ORG27569 favorably activates β-arrestin.
There are few studies to assess signaling bias at CB2 receptor compared to CB1. Recent investigations have demonstrated that compound CP55,940 exhibits the potential to cause robust internalization of rat CB2, whereas WIN55,212-2 shows no activity in promoting receptor internalization [99]. In contrast, assessment using the Black and Leff operational model indicated that CB2 agonists JWH-133 and THC showed efficacious activity in adenylyl cyclase assays, but failed to associate with β-arrestin [100]. A detailed analysis of biased signaling suggested that THC displayed bias toward ERK1/2 signaling compared to arrestin and GTPγS, when acting at CB2 receptor, while another agonist (R,S)-AM1241 was identified to bias toward arrestin coupling and pERK signaling compared to GIRK channel activation [86].
Cannabinoid receptors, CB1 and CB2, respond to a large amount of synthetic, endogenous and plant-derived cannabinoids [101, 102]. In addition to inhibiting the activity of adenylyl cyclase, CB1 has been shown to be capable of coupling to Gαs [14, 18, 103]. Previous studies have demonstrated that both CB1 and CB2 receptors associate with β-arrestins, resulting in receptor internalization [104, 105], and signal to ERK1/2 signaling pathway via either Gβγ or β-arrestin interactions [106]. Additionally, the cannabinoid signaling system has huge potential as a target for therapeutic treatment of many diseases including pain, inflammation, various mental disorders and so on [107]. Therefore, cannabinoid receptors have been regarded as a particularly ideal model to investigate the biased signaling. However, the investigation of biased agonism for the cannabinoid receptors, CB1 and CB2, is still in its infancy, and more efforts are required for identification and characterization of ligands with biased signaling.
6. SIGNALING OF CANNABINOID RECEPTORS FROM INSIDE CELL
It has been traditionally thought that from their position localized on the cell surface, GPCRs transduce external stimuli into a broad range of cellular responses. However, this has been recently challenged by considerable evidence suggesting that various GPCRs have been found to be functionally localized in the intracellular compartment, including endosomes and trans-Golgi network [108-111], as well as intracellular membranes, including the nuclear membrane [112], endoplasmic reticulum [113], and mitochondrial membrane [114].
CB1 receptor is one of the most abundant G protein-coupled receptors in the brain, and control neuronal activity, metabolism, and functions [115]. Early investigation showed that agonist THC could affect mitochondrial functions [116]. Recent evidence also suggests that CB1 receptor-mediated signaling is found to regulate mitochondrial biogenesis in peripheral non-neural tissues [117, 118]. CB1 receptors were identified to be functionally localized to the outer membrane of neuronal mitochondria (termed “mtCB1”), and regulate neuronal energy metabolism [119]. Further characterization demonstrated that upon activation by exogenous cannabinoids and in situ endocannabinoids, mtCB1 induced an intra-mitochondrial signaling pathway involving G proteins, soluble adenylyl cyclase (sAC), and the protein kinase A (PKA), leading to a decrease of complex I enzymatic activity and respiration in neuronal mitochondria [119, 120]. Mutagenesis analysis pointed to the first 22 amino acids of the CB1 protein responsible for mitochondria localization [120]. A very recent study shows that the CB1 receptor distribution in astrocytes in the mice with genetic restoration of CB1 receptor expression completely matches the endogenous CB1 receptor expression and localization, suggesting the localization of CB1 receptors in astrocyte mitochondria for the first time [121, 122]. However, the physiological role of CB1 receptors in astrocyte mitochondria remains to be elucidated.
In addition to mitochondria, the CB1 receptor has been found to localize to endosomal and lysosomal compartments [123, 124]. The second cannabinoid receptor, CB2, well known to play an important role in the peripheral immune system, was also detected in intracellular localization in prefrontal cortex by subcellular fractionation techniques, western blotting, and binding assay, and agonist stimulation led to inositol triphosphate 3 (IP3) receptor-dependent opening of Ca2+-activated chloride channels and decreased neuronal excitability [123, 125, 126]. Both CB1 and CB2 are likely to exert their functions through intracellular localization. However, more studies are needed to further investigate the molecular mechanisms that determine the subcellular distribution and signaling properties.
CONCLUSION
The endocannabinoid system is an attractive area of research owing to the therapeutic potential for the treatment of pain, obesity, inflammation and a variety of psychiatric disorders. This review provides an overview of current findings regarding the cannabinoid receptor structure, orphan cannabinoid receptors, allosteric modulation, biased signaling, and signaling from intracellularly localized receptors. These findings have changed our fundamental understanding of the endocannabinoid system, from the structural and functional basis for receptor activation to the design and optimization of therapeutic modulators with better efficacy and safety profiles. Further research is required for elucidation of mechanisms involved in biased agonism and signaling of intracellular receptors. Since this manuscript was submitted for publication on the 28th of November 2018, very recent reports on the crystal structure of a signaling cannabinoid receptor 1-G protein complex [127] and crystal structure of the human cannabinoid receptor CB2 were not incorporated [128].
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was supported by grants from the National Natural Science Foundation of China (31771543, 81173106, and 31200621 and), the Ministry of Science and Technology of China (2012CB910402).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Adams I.B., Martin B.R. Cannabis: Pharmacology and toxicology in animals and humans. Addiction. 1996;91:1585–1614. [PubMed] [Google Scholar]
- 2.Lambert D.M. Medical use of cannabis through history. J. Pharm. Belg. 2001;56:111–118. [PubMed] [Google Scholar]
- 3.Pacher P., Batkai S., Kunos G. The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacol. Rev. 2006;58:389–462. doi: 10.1124/pr.58.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.di Tomaso E., Beltramo M., Piomelli D. Brain cannabinoids in chocolate. Nature. 1996;382:677–678. doi: 10.1038/382677a0. [DOI] [PubMed] [Google Scholar]
- 5.Lambert D.M., Fowler C.J. The endocannabinoid system: Drug targets, lead compounds, and potential therapeutic applications. J. Med. Chem. 2005;48:5059–5087. doi: 10.1021/jm058183t. [DOI] [PubMed] [Google Scholar]
- 6.Pineiro R., Falasca M. Lysophosphatidylinositol signalling: New wine from an old bottle. Biochim. Biophys. Acta. 2012;1821:694–705. doi: 10.1016/j.bbalip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda L.A., Lolait S.J., Brownstein M.J., Young A.C., Bonner T.I. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 8.Moro O., Lameh J., Hogger P., Sadee W. Hydrophobic amino acid in the i2 loop plays a key role in receptor-G protein coupling. J. Biol. Chem. 1993;268:22273–22276. [PubMed] [Google Scholar]
- 9.Munro S., Thomas K.L., Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 10.Howlett A.C., Barth F., Bonner T.I., Cabral G., Casellas P., Devane W.A., Felder C.C., Herkenham M., Mackie K., Martin B.R., Mechoulam R., Pertwee R.G. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol. Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 11.Bonz A., Laser M., Kullmer S., Kniesch S., Babin-Ebell J., Popp V., Ertl G., Wagner J.A. Cannabinoids acting on CB1 receptors decrease contractile performance in human atrial muscle. J. Cardiovasc. Pharmacol. 2003;41:657–664. doi: 10.1097/00005344-200304000-00020. [DOI] [PubMed] [Google Scholar]
- 12.Feng Z., Alqarni M.H., Yang P., Tong Q., Chowdhury A., Wang L., Xie X.Q. Modeling, molecular dynamics simulation, and mutation validation for structure of cannabinoid receptor 2 based on known crystal structures of GPCRs. J. Chem. Inf. Model. 2014;54:2483–2499. doi: 10.1021/ci5002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown S.M., Wager-Miller J., Mackie K. Cloning and molecular characterization of the rat CB2 cannabinoid receptor. Biochim. Biophys. Acta. 2002;1576:255–264. doi: 10.1016/s0167-4781(02)00341-x. [DOI] [PubMed] [Google Scholar]
- 14.Chen X.P., Yang W., Fan Y., Luo J.S., Hong K., Wang Z., Yan J.F., Chen X., Lu J.X., Benovic J.L., Zhou N.M. Structural determinants in the second intracellular loop of the human cannabinoid CB1 receptor mediate selective coupling to G(s) and G(i). Br. J. Pharmacol. 2010;161:1817–1834. doi: 10.1111/j.1476-5381.2010.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onaivi E.S., Ishiguro H., Gong J.P., Patel S., Perchuk A., Meozzi P.A., Myers L., Mora Z., Tagliaferro P., Gardner E., Brusco A., Akinshola B.E., Liu Q.R., Hope B., Iwasaki S., Arinami T., Teasenfitz L., Uhl G.R. Discovery of the presence and functional expression of cannabinoid CB2 receptors in brain. Ann. N. Y. Acad. Sci. 2006;1074:514–536. doi: 10.1196/annals.1369.052. [DOI] [PubMed] [Google Scholar]
- 16.Bab I., Zimmer A. Cannabinoid receptors and the regulation of bone mass. Br. J. Pharmacol. 2008;153:182–188. doi: 10.1038/sj.bjp.0707593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maccarrone M. CB2 receptors in reproduction. Br. J. Pharmacol. 2008;153:189–198. doi: 10.1038/sj.bjp.0707444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonhaus D.W., Chang L.K., Kwan J., Martin G.R. Dual activation and inhibition of adenylyl cyclase by cannabinoid receptor agonists: evidence for agonist-specific trafficking of intracellular responses. J. Pharmacol. Exp. Ther. 1998;287:884–888. [PubMed] [Google Scholar]
- 19.Calandra B., Portier M., Kerneis A., Delpech M., Carillon C., Le Fur G., Ferrara P., Shire D. Dual intracellular signaling pathways mediated by the human cannabinoid CB1 receptor. Eur. J. Pharmacol. 1999;374:445–455. doi: 10.1016/s0014-2999(99)00349-0. [DOI] [PubMed] [Google Scholar]
- 20.Pertwee R.G. The pharmacology of cannabinoid receptors and their ligands: an overview. Int. J. Obes. 2006;30(Suppl. 1):S13–S18. doi: 10.1038/sj.ijo.0803272. [DOI] [PubMed] [Google Scholar]
- 21.Zheng C., Chen L., Chen X., He X., Yang J., Shi Y., Zhou N. The second intracellular loop of the human cannabinoid CB2 receptor governs G protein coupling in coordination with the carboxyl terminal domain. PLoS One. 2013;8:e63262. doi: 10.1371/journal.pone.0063262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y., Anderson H.D. Cannabinoid signaling in health and disease. Can. J. Physiol. Pharmacol. 2017;95:311–327. doi: 10.1139/cjpp-2016-0346. [DOI] [PubMed] [Google Scholar]
- 23.Pavlopoulos S., Thakur G.A., Nikas S.P., Makriyannis A. Cannabinoid receptors as therapeutic targets. Curr. Pharm. Des. 2006;12:1751–1769. doi: 10.2174/138161206776873743. [DOI] [PubMed] [Google Scholar]
- 24.Lutz B., Marsicano G., Maldonado R., Hillard C.J. The endocannabinoid system in guarding against fear, anxiety and stress. Nat. Rev. Neurosci. 2015;16:705–718. doi: 10.1038/nrn4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mbvundula E.C., Rainsford K.D., Bunning R.A. Cannabinoids in pain and inflammation. Inflammopharmacology. 2004;12:99–114. doi: 10.1163/1568560041352275. [DOI] [PubMed] [Google Scholar]
- 26.Guindon J., Hohmann A.G. Cannabinoid CB2 receptors: a therapeutic target for the treatment of inflammatory and neuropathic pain. Br. J. Pharmacol. 2008;153:319–334. doi: 10.1038/sj.bjp.0707531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng R., Milcarek C.A., Xie X.Q. Antagonism of cannabinoid receptor 2 pathway suppresses IL-6-induced immunoglobulin IgM secretion. BMC Pharmacol. Toxicol. 2014;15:30. doi: 10.1186/2050-6511-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manglik A., Kruse A.C. Structural Basis for G Protein-Coupled Receptor Activation. Biochemistry. 2017;56:5628–5634. doi: 10.1021/acs.biochem.7b00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manglik A., Kobilka B.K., Steyaert J. Nanobodies to Study G Protein-Coupled Receptor Structure and Function. Annu. Rev. Pharmacol. Toxicol. 2017;57:19–37. doi: 10.1146/annurev-pharmtox-010716-104710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua T., Vemuri K., Pu M., Qu L., Han G.W., Wu Y., Zhao S., Shui W., Li S., Korde A., Laprairie R.B., Stahl E.L., Ho J.H., Zvonok N., Zhou H., Kufareva I., Wu B., Zhao Q., Hanson M.A., Bohn L.M., Makriyannis A., Stevens R.C., Liu Z.J. Crystal structure of the human cannabinoid receptor CB1. Cell. 2016;167(3):750–762.e714. doi: 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertalovitz A.C., Ahn K.H., Kendall D.A. Ligand Binding Sensitivity of the Extracellular Loop Two of the Cannabinoid Receptor 1. Drug Dev. Res. 2010;71:404–411. doi: 10.1002/ddr.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fay J.F., Dunham T.D., Farrens D.L. Cysteine residues in the human cannabinoid receptor: only C257 and C264 are required for a functional receptor, and steric bulk at C386 impairs antagonist SR141716A binding. Biochemistry. 2005;44:8757–8769. doi: 10.1021/bi0472651. [DOI] [PubMed] [Google Scholar]
- 33.Shao Z., Yin J., Chapman K., Grzemska M., Clark L., Wang J., Rosenbaum D.M. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature. 2016;540:602–606. doi: 10.1038/nature20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hua T., Vemuri K., Nikas S.P., Laprairie R.B., Wu Y., Qu L., Pu M., Korde A., Jiang S., Ho J.H., Han G.W., Ding K., Li X., Liu H., Hanson M.A., Zhao S., Bohn L.M., Makriyannis A., Stevens R.C., Liu Z.J. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature. 2017;547:468–471. doi: 10.1038/nature23272. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Isberg V., Mordalski S., Munk C., Rataj K., Harpsoe K., Hauser A.S., Vroling B., Bojarski A.J., Vriend G., Gloriam D.E. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2017;45:2936. doi: 10.1093/nar/gkw1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Z.H., Slowey C.A., Hurst D.P., Reggio P.H. The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2). Mol. Pharmacol. 1999;56:834–840. [PubMed] [Google Scholar]
- 37.Xie X.Q., Chen J.Z., Billings E.M. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins. 2003;53:307–319. doi: 10.1002/prot.10511. [DOI] [PubMed] [Google Scholar]
- 38.Tuccinardi T., Ferrarini P.L., Manera C., Ortore G., Saccomanni G., Martinelli A. Cannabinoid CB2/CB1 selectivity. Receptor modeling and automated docking analysis. J. Med. Chem. 2006;49:984–994. doi: 10.1021/jm050875u. [DOI] [PubMed] [Google Scholar]
- 39.Stern E., Muccioli G.G., Millet R., Goossens J.F., Farce A., Chavatte P., Poupaert J.H., Lambert D.M., Depreux P., Henichart J.P. Novel 4-oxo-1, 4-dihydroquinoline-3-carboxamide derivatives as new CB2 cannabinoid receptors agonists: synthesis, pharmacological properties and molecular modeling. J. Med. Chem. 2006;49:70–79. doi: 10.1021/jm050467q. [DOI] [PubMed] [Google Scholar]
- 40.Raduner S., Majewska A., Chen J.Z., Xie X.Q., Hamon J., Faller B., Altmann K.H., Gertsch J. Alkylamides from Echinacea are a new class of cannabinomimetics. Cannabinoid type 2 receptor-dependent and -independent immunomodulatory effects. J. Biol. Chem. 2006;281:14192–14206. doi: 10.1074/jbc.M601074200. [DOI] [PubMed] [Google Scholar]
- 41.Latek D., Kolinski M., Ghoshdastider U., Debinski A., Bombolewski R., Plazinska A., Jozwiak K., Filipek S. Modeling of ligand binding to G protein coupled receptors: cannabinoid CB1, CB2 and adrenergic beta 2 AR. J. Mol. Model. 2011;17:2353–2366. doi: 10.1007/s00894-011-0986-7. [DOI] [PubMed] [Google Scholar]
- 42.Yang P., Myint K.Z., Tong Q., Feng R., Cao H., Almehizia A.A., Alqarni M.H., Wang L., Bartlow P., Gao Y., Gertsch J., Teramachi J., Kurihara N., Roodman G.D., Cheng T., Xie X.Q. Lead discovery, chemistry optimization, and biological evaluation studies of novel biamide derivatives as CB2 receptor inverse agonists and osteoclast inhibitors. J. Med. Chem. 2012;55:9973–9987. doi: 10.1021/jm301212u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J., Feng Z., Ma S., Zhang Y., Tong Q., Alqarni M.H., Gou X., Xie X.Q. Difference and Influence of Inactive and Active States of Cannabinoid Receptor Subtype CB2: From Conformation to Drug Discovery. J. Chem. Inf. Model. 2016;56:1152–1163. doi: 10.1021/acs.jcim.5b00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laschet C., Dupuis N., Hanson J. The G protein-coupled receptors deorphanization landscape. Biochem. Pharmacol. 2018;153:62–74. doi: 10.1016/j.bcp.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Brown A.J. Novel cannabinoid receptors. Br. J. Pharmacol. 2007;152:567–575. doi: 10.1038/sj.bjp.0707481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackie K., Stella N. Cannabinoid receptors and endocannabinoids: Evidence for new players. AAPS J. 2006;8:E298–E306. doi: 10.1007/BF02854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irving A., Abdulrazzaq G., Chan S.L.F., Penman J., Harvey J., Alexander S.P.H. Cannabinoid Receptor-Related Orphan G Protein-Coupled Receptors. Adv. Pharmacol. 2017;80:223–247. doi: 10.1016/bs.apha.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Overton H.A., Babbs A.J., Doel S.M., Fyfe M.C., Gardner L.S., Griffin G., Jackson H.C., Procter M.J., Rasamison C.M., Tang-Christensen M., Widdowson P.S., Williams G.M., Reynet C. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006;3:167–175. doi: 10.1016/j.cmet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Sawzdargo M., Nguyen T., Lee D.K., Lynch K.R., Cheng R., Heng H.H., George S.R., O’Dowd B.F. Identification and cloning of three novel human G protein-coupled receptor genes GPR52, PsiGPR53 and GPR55: GPR55 is extensively expressed in human brain. Brain Res. Mol. Brain Res. 1999;64:193–198. doi: 10.1016/s0169-328x(98)00277-0. [DOI] [PubMed] [Google Scholar]
- 50.Brown A.J. Identification of modulators of GPR55 activity. Assignee: GlaxoSmithKline. 2001;Patent WO00186305 [Google Scholar]
- 51.Drmota E.G.P., Groblewski T. Screening assays for cannabinoid-ligand type modulators. Assignee: Astra Zeneca. 2004;Patent WO2004074844 [Google Scholar]
- 52.Oka S., Nakajima K., Yamashita A., Kishimoto S., Sugiura T. Identification of GPR55 as a lysophosphatidylinositol receptor. Biochem. Biophys. Res. Commun. 2007;362:928–934. doi: 10.1016/j.bbrc.2007.08.078. [DOI] [PubMed] [Google Scholar]
- 53.Anavi-Goffer S., Baillie G., Irving A.J., Gertsch J., Greig I.R., Pertwee R.G., Ross R.A. Modulation of L-alpha-lysophosphatidylinositol/GPR55 mitogen-activated protein kinase (MAPK) signaling by cannabinoids. J. Biol. Chem. 2012;287:91–104. doi: 10.1074/jbc.M111.296020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henstridge C.M., Balenga N.A., Schroder R., Kargl J.K., Platzer W., Martini L., Arthur S., Penman J., Whistler J.L., Kostenis E., Waldhoer M., Irving A.J. GPR55 ligands promote receptor coupling to multiple signalling pathways. Br. J. Pharmacol. 2010;160:604–614. doi: 10.1111/j.1476-5381.2009.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lauckner J.E., Jensen J.B., Chen H.Y., Lu H.C., Hille B., Mackie K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc. Natl. Acad. Sci. USA. 2008;105:2699–2704. doi: 10.1073/pnas.0711278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henstridge C.M., Balenga N.A., Ford L.A., Ross R.A., Waldhoer M., Irving A.J. The GPR55 ligand L-alpha-lysophosphatidylinositol promotes RhoA-dependent Ca2+ signaling and NFAT activation. FASEB J. 2009;23:183–193. doi: 10.1096/fj.08-108670. [DOI] [PubMed] [Google Scholar]
- 57.Kotsikorou E., Sharir H., Shore D.M., Hurst D.P., Lynch D.L., Madrigal K.E., Heynen-Genel S., Milan L.B., Chung T.D., Seltzman H.H., Bai Y., Caron M.G., Barak L.S., Croatt M.P., Abood M.E., Reggio P.H. Identification of the GPR55 antagonist binding site using a novel set of high-potency GPR55 selective ligands. Biochemistry. 2013;52:9456–9469. doi: 10.1021/bi4008885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreno E., Andradas C., Medrano M., Caffarel M.M., Perez-Gomez E., Blasco-Benito S., Gomez-Canas M., Pazos M.R., Irving A.J., Lluis C., Canela E.I., Fernandez-Ruiz J., Guzman M., McCormick P.J., Sanchez C. Targeting CB2-GPR55 receptor heteromers modulates cancer cell signaling. J. Biol. Chem. 2014;289:21960–21972. doi: 10.1074/jbc.M114.561761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balenga N.A., Martinez-Pinilla E., Kargl J., Schroder R., Peinhaupt M., Platzer W., Balint Z., Zamarbide M., Dopeso-Reyes I.G., Ricobaraza A., Perez-Ortiz J.M., Kostenis E., Waldhoer M., Heinemann A., Franco R. Heteromerization of GPR55 and cannabinoid CB2 receptors modulates signalling. Br. J. Pharmacol. 2014;171:5387–5406. doi: 10.1111/bph.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McHugh D., Page J., Dunn E., Bradshaw H.B. Delta(9) -Tetrahydrocannabinol and N-arachidonyl glycine are full agonists at GPR18 receptors and induce migration in human endometrial HEC-1B cells. Br. J. Pharmacol. 2012;165:2414–2424. doi: 10.1111/j.1476-5381.2011.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheskin T., Hanus L., Slager J., Vogel Z., Mechoulam R. Structural requirements for binding of anandamide-type compounds to the brain cannabinoid receptor. J. Med. Chem. 1997;40:659–667. doi: 10.1021/jm960752x. [DOI] [PubMed] [Google Scholar]
- 62.Jones R.M. Discovery of agonists of the glucose dependent insuli-notropic receptor, GPR119, a pancre atic beta-cell oGPCR, for the treatment of NIDDM. Drugs Future 31 (Suppl A). 2006 [Google Scholar]
- 63.Hansen K.B., Rosenkilde M.M., Knop F.K., Wellner N., Diep T.A., Rehfeld J.F., Andersen U.B., Holst J.J., Hansen H.S. 2-Oleoyl glycerol is a GPR119 agonist and signals GLP-1 release in humans. J. Clin. Endocrinol. Metab. 2011;96:E1409–E1417. doi: 10.1210/jc.2011-0647. [DOI] [PubMed] [Google Scholar]
- 64.Soga T., Ohishi T., Matsui T., Saito T., Matsumoto M., Takasaki J., Matsumoto S., Kamohara M., Hiyama H., Yoshida S., Momose K., Ueda Y., Matsushime H., Kobori M., Furuichi K. Lysophosphatidylcholine enhances glucose-dependent insulin secretion via an orphan G-protein-coupled receptor. Biochem. Biophys. Res. Commun. 2005;326:744–751. doi: 10.1016/j.bbrc.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 65.Morales P., Reggio P.H. An Update on Non-CB1, Non-CB2 Cannabinoid Related G-Protein-Coupled Receptors. Cannabis Cannabinoid Res. 2017;2:265–273. doi: 10.1089/can.2017.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertwee R.G., Howlett A.C., Abood M.E., Alexander S.P., Di Marzo V., Elphick M.R., Greasley P.J., Hansen H.S., Kunos G., Mackie K., Mechoulam R., Ross R.A. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: Beyond CB(1) and CB(2). Pharmacol. Rev. 2010;62:588–631. doi: 10.1124/pr.110.003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka S., Ishii K., Kasai K., Yoon S.O., Saeki Y. Neural expression of G protein-coupled receptors GPR3, GPR6, and GPR12 up-regulates cyclic AMP levels and promotes neurite outgrowth. J. Biol. Chem. 2007;282:10506–10515. doi: 10.1074/jbc.M700911200. [DOI] [PubMed] [Google Scholar]
- 68.Laun A.S., Song Z.H. GPR3 and GPR6, novel molecular targets for cannabidiol. Biochem. Biophys. Res. Commun. 2017;490:17–21. doi: 10.1016/j.bbrc.2017.05.165. [DOI] [PubMed] [Google Scholar]
- 69.Yin H., Chu A., Li W., Wang B., Shelton F., Otero F., Nguyen D.G., Caldwell J.S., Chen Y.A. Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. J. Biol. Chem. 2009;284:12328–12338. doi: 10.1074/jbc.M806516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lagerstrom M.C., Schioth H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 71.Christopoulos A., Changeux J.P., Catterall W.A., Fabbro D., Burris T.P., Cidlowski J.A., Olsen R.W., Peters J.A., Neubig R.R., Pin J.P., Sexton P.M., Kenakin T.P., Ehlert F.J., Spedding M., Langmead C.J. International Union of Basic and Clinical Pharmacology. XC. multisite pharmacology: Recommendations for the nomenclature of receptor allosterism and allosteric ligands. Pharmacol. Rev. 2014;66:918–947. doi: 10.1124/pr.114.008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pertwee R.G. Endocannabinoids and Their Pharmacological Actions. Handb. Exp. Pharmacol. 2015;231:1–37. doi: 10.1007/978-3-319-20825-1_1. [DOI] [PubMed] [Google Scholar]
- 73.Pertwee R.G. Targeting the endocannabinoid system with cannabinoid receptor agonists: pharmacological strategies and therapeutic possibilities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:3353–3363. doi: 10.1098/rstb.2011.0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hutcheson D.M., Tzavara E.T., Smadja C., Valjent E., Roques B.P., Hanoune J., Maldonado R. Behavioural and biochemical evidence for signs of abstinence in mice chronically treated with delta-9-tetrahydrocannabinol. Br. J. Pharmacol. 1998;125:1567–1577. doi: 10.1038/sj.bjp.0702228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Price M.R., Baillie G.L., Thomas A., Stevenson L.A., Easson M., Goodwin R., McLean A., McIntosh L., Goodwin G., Walker G., Westwood P., Marrs J., Thomson F., Cowley P., Christopoulos A., Pertwee R.G., Ross R.A. Allosteric modulation of the cannabinoid CB1 receptor. Mol. Pharmacol. 2005;68:1484–1495. doi: 10.1124/mol.105.016162. [DOI] [PubMed] [Google Scholar]
- 76.Morales P., Goya P., Jagerovic N., Hernandez-Folgado L. Allosteric Modulators of the CB1 Cannabinoid Receptor: A Structural Update Review. Cannabis Cannabinoid Res. 2016;1:22–30. doi: 10.1089/can.2015.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horswill J.G., Bali U., Shaaban S., Keily J.F., Jeevaratnam P., Babbs A.J., Reynet C., Wong Kai In P. PSNCBAM-1, a novel allosteric antagonist at cannabinoid CB1 receptors with hypophagic effects in rats. Br. J. Pharmacol. 2007;152:805–814. doi: 10.1038/sj.bjp.0707347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baillie G.L., Horswill J.G., Anavi-Goffer S., Reggio P.H., Bolognini D., Abood M.E., McAllister S., Strange P.G., Stephens G.J., Pertwee R.G., Ross R.A. CB(1) receptor allosteric modulators display both agonist and signaling pathway specificity. Mol. Pharmacol. 2013;83:322–338. doi: 10.1124/mol.112.080879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahn K.H., Mahmoud M.M., Samala S., Lu D., Kendall D.A. Profiling two indole-2-carboxamides for allosteric modulation of the CB1 receptor. J. Neurochem. 2013;124:584–589. doi: 10.1111/jnc.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahmoud M.M., Ali H.I., Ahn K.H., Damaraju A., Samala S., Pulipati V.K., Kolluru S., Kendall D.A., Lu D. Structure-activity relationship study of indole-2-carboxamides identifies a potent allosteric modulator for the cannabinoid receptor 1 (CB1). J. Med. Chem. 2013;56:7965–7975. doi: 10.1021/jm4009828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khurana L., Ali H.I., Olszewska T., Ahn K.H., Damaraju A., Kendall D.A., Lu D. Optimization of chemical functionalities of indole-2-carboxamides to improve allosteric parameters for the cannabinoid receptor 1 (CB1). J. Med. Chem. 2014;57:3040–3052. doi: 10.1021/jm5000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hind W.H., England T.J., O’Sullivan S.E. Cannabidiol protects an in vitro model of the blood-brain barrier from oxygen-glucose deprivation via PPARgamma and 5-HT1A receptors. Br. J. Pharmacol. 2016;173:815–825. doi: 10.1111/bph.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McPartland J.M., Duncan M., Di Marzo V., Pertwee R.G. Are cannabidiol and Delta(9) -tetrahydrocannabivarin negative modulators of the endocannabinoid system? A systematic review. Br. J. Pharmacol. 2015;172:737–753. doi: 10.1111/bph.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laprairie R.B., Bagher A.M., Kelly M.E., Denovan-Wright E.M. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br. J. Pharmacol. 2015;172:4790–4805. doi: 10.1111/bph.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petrucci V., Chicca A., Glasmacher S., Paloczi J., Cao Z., Pacher P., Gertsch J. Pepcan-12 (RVD-hemopressin) is a CB2 receptor positive allosteric modulator constitutively secreted by adrenals and in liver upon tissue damage. Sci. Rep. 2017;7:9560. doi: 10.1038/s41598-017-09808-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soethoudt M., Grether U., Fingerle J., Grim T.W., Fezza F., de Petrocellis L., Ullmer C., Rothenhausler B., Perret C., van Gils N., Finlay D., MacDonald C., Chicca A., Gens M.D., Stuart J., de Vries H., Mastrangelo N., Xia L., Alachouzos G., Baggelaar M.P., Martella A., Mock E.D., Deng H., Heitman L.H., Connor M. Cannabinoid CB2 receptor ligand profiling reveals biased signalling and off-target activity. Nat. Commun. 2017;8:13958. doi: 10.1038/ncomms13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landomiel F., Gallay N., Jegot G., Tranchant T., Durand G., Bourquard T., Crepieux P., Poupon A., Reiter E. Biased signalling in follicle stimulating hormone action. Mol. Cell. Endocrinol. 2014;382:452–459. doi: 10.1016/j.mce.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 88.Kenakin T. Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol. Sci. 1995;16:232–238. doi: 10.1016/s0165-6147(00)89032-x. [DOI] [PubMed] [Google Scholar]
- 89.Violin J.D., Lefkowitz R.J. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol. Sci. 2007;28:416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 90.Glass M., Northup J.K. Agonist selective regulation of G proteins by cannabinoid CB(1) and CB(2) receptors. Mol. Pharmacol. 1999;56:1362–1369. doi: 10.1124/mol.56.6.1362. [DOI] [PubMed] [Google Scholar]
- 91.Bosier B., Hermans E., Lambert D. Differential modulation of AP-1- and CRE-driven transcription by cannabinoid agonists emphasizes functional selectivity at the CB1 receptor. Br. J. Pharmacol. 2008;155:24–33. doi: 10.1038/bjp.2008.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bosier B., Tilleux S., Najimi M., Lambert D.M., Hermans E. Agonist selective modulation of tyrosine hydroxylase expression by cannabinoid ligands in a murine neuroblastoma cell line. J. Neurochem. 2007;102:1996–2007. doi: 10.1111/j.1471-4159.2007.04679.x. [DOI] [PubMed] [Google Scholar]
- 93.Maneuf Y.P., Brotchie J.M. Paradoxical action of the cannabinoid WIN 55, 212-2 in stimulated and basal cyclic AMP accumulation in rat globus pallidus slices. Br. J. Pharmacol. 1997;120:1397–1398. doi: 10.1038/sj.bjp.0701101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franks L.N., Ford B.M., Madadi N.R., Penthala N.R., Crooks P.A., Prather P.L. Characterization of the intrinsic activity for a novel class of cannabinoid receptor ligands: Indole quinuclidine analogs. Eur. J. Pharmacol. 2014;737:140–148. doi: 10.1016/j.ejphar.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ford B.M., Franks L.N., Tai S., Fantegrossi W.E., Stahl E.L., Berquist M.D., Cabanlong C.V., Wilson C.D., Penthala N.R., Crooks P.A., Prather P.L. Characterization of structurally novel G protein biased CB1 agonists: Implications for drug development. Pharmacol. Res. 2017;125:161–177. doi: 10.1016/j.phrs.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahn K.H., Mahmoud M.M., Kendall D.A. Allosteric modulator ORG27569 induces CB1 cannabinoid receptor high affinity agonist binding state, receptor internalization, and Gi protein-independent ERK1/2 kinase activation. J. Biol. Chem. 2012;287:12070–12082. doi: 10.1074/jbc.M111.316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahn K.H., Mahmoud M.M., Shim J.Y., Kendall D.A. Distinct roles of beta-arrestin 1 and beta-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1). J. Biol. Chem. 2013;288:9790–9800. doi: 10.1074/jbc.M112.438804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raehal K.M., Bohn L.M. beta-arrestins: regulatory role and therapeutic potential in opioid and cannabinoid receptor-mediated analgesia. Handb. Exp. Pharmacol. 2014;219:427–443. doi: 10.1007/978-3-642-41199-1_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Atwood B.K., Wager-Miller J., Haskins C., Straiker A., Mackie K. Functional selectivity in CB(2) cannabinoid receptor signaling and regulation: Implications for the therapeutic potential of CB(2) ligands. Mol. Pharmacol. 2012;81:250–263. doi: 10.1124/mol.111.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dhopeshwarkar A., Mackie K. Functional Selectivity of CB2 Cannabinoid Receptor Ligands at a Canonical and Noncanonical Pathway. J. Pharmacol. Exp. Ther. 2016;358:342–351. doi: 10.1124/jpet.116.232561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pertwee R.G. Ligands that target cannabinoid receptors in the brain: From THC to anandamide and beyond. Addict. Biol. 2008;13:147–159. doi: 10.1111/j.1369-1600.2008.00108.x. [DOI] [PubMed] [Google Scholar]
- 102.Laprairie R.B., Bagher A.M., Kelly M.E., Denovan-Wright E.M. Biased Type 1 Cannabinoid Receptor Signaling Influences Neuronal Viability in a Cell Culture Model of Huntington Disease. Mol. Pharmacol. 2016;89:364–375. doi: 10.1124/mol.115.101980. [DOI] [PubMed] [Google Scholar]
- 103.Peters M.F., Scott C.W. Evaluating cellular impedance assays for detection of GPCR pleiotropic signaling and functional selectivity. J. Biomol. Screen. 2009;14:246–255. doi: 10.1177/1087057108330115. [DOI] [PubMed] [Google Scholar]
- 104.Breivogel C.S., Puri V., Lambert J.M., Hill D.K., Huffman J.W., Razdan R.K. The influence of beta-arrestin2 on cannabinoid CB1 receptor coupling to G-proteins and subcellular localization and relative levels of beta-arrestin1 and 2 in mouse brain. J. Recept. Signal Transduct. Res. 2013;33:367–379. doi: 10.3109/10799893.2013.838787. [DOI] [PubMed] [Google Scholar]
- 105.Chen X., Zheng C., Qian J., Sutton S.W., Wang Z., Lv J., Liu C., Zhou N. Involvement of beta-arrestin-2 and clathrin in agonist-mediated internalization of the human cannabinoid CB2 receptor. Curr. Mol. Pharmacol. 2014;7:67–80. doi: 10.2174/1874467207666140714115824. [DOI] [PubMed] [Google Scholar]
- 106.Howlett A.C., Abood M.E. CB1 and CB2 Receptor Pharmacology. Adv. Pharmacol. 2017;80:169–206. doi: 10.1016/bs.apha.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scotter E.L., Abood M.E., Glass M. The endocannabinoid system as a target for the treatment of neurodegenerative disease. Br. J. Pharmacol. 2010;160:480–498. doi: 10.1111/j.1476-5381.2010.00735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Irannejad R., Tomshine J.C., Tomshine J.R., Chevalier M., Mahoney J.P., Steyaert J., Rasmussen S.G., Sunahara R.K., El-Samad H., Huang B., von Zastrow M. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsvetanova N.G., von Zastrow M. Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol. 2014;10:1061–1065. doi: 10.1038/nchembio.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Godbole A., Lyga S., Lohse M.J., Calebiro D. Internalized TSH receptors en route to the TGN induce local Gs-protein signaling and gene transcription. Nat. Commun. 2017;8:443. doi: 10.1038/s41467-017-00357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Irannejad R., Pessino V., Mika D., Huang B., Wedegaertner P.B., Conti M., von Zastrow M. Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol. 2017;13:799–806. doi: 10.1038/nchembio.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boivin B., Vaniotis G., Allen B.G., Hebert T.E. G protein-coupled receptors in and on the cell nucleus: A new signaling paradigm? J. Recept. Signal Transduct. Res. 2008;28:15–28. doi: 10.1080/10799890801941889. [DOI] [PubMed] [Google Scholar]
- 113.Revankar C.M., Cimino D.F., Sklar L.A., Arterburn J.B., Prossnitz E.R. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–1630. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 114.Suofu Y., Li W., Jean-Alphonse F.G., Jia J.Y., Khattar N.K., Li J.T., Baranov S.V., Leronni D., Mihalik A.C., He Y.Q., Cecon E., Wehbi V.L., Kim J., Heath B.E., Baranova O.V., Wang X.M., Gable M.J., Kretz E.S., Di Benedetto G., Lezon T.R., Ferrando L.M., Larkin T.M., Sullivan M., Yablonska S., Wang J.J., Minnigh M.B., Guillaumet G., Suzenet F., Richardson R.M., Poloyac S.M., Stolz D.B., Jockers R., Witt-Enderby P.A., Carlisle D.L., Vilardaga J.P., Friedlander R.M. Dual role of mitochondria in producing melatonin and driving GPCR signaling to block cytochrome c release. Proc. Natl. Acad. Sci. USA. 2017;114:E7997–E8006. doi: 10.1073/pnas.1705768114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marsicano G., Lutz B. Neuromodulatory functions of the endocannabinoid system. J. Endocrinol. Invest. 2006;29:27–46. [PubMed] [Google Scholar]
- 116.Bartova A., Birmingham M.K. Effect of delta9-tetrahydrocannabinol on mitochondrial NADH-oxidase activity. J. Biol. Chem. 1976;251:5002–5006. [PubMed] [Google Scholar]
- 117.Tedesco L., Valerio A., Dossena M., Cardile A., Ragni M., Pagano C., Pagotto U., Carruba M.O., Vettor R., Nisoli E. Cannabinoid receptor stimulation impairs mitochondrial biogenesis in mouse white adipose tissue, muscle, and liver: The role of eNOS, p38 MAPK, and AMPK pathways. Diabetes. 2010;59:2826–2836. doi: 10.2337/db09-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aquila S., Guido C., Santoro A., Perrotta I., Laezza C., Bifulco M., Sebastiano A. Human sperm anatomy: Ultrastructural localization of the cannabinoid1 receptor and a potential role of anandamide in sperm survival and acrosome reaction. Anat. Rec. (Hoboken) 2010;293:298–309. doi: 10.1002/ar.21042. [DOI] [PubMed] [Google Scholar]
- 119.Benard G., Massa F., Puente N., Lourenco J., Bellocchio L., Soria-Gomez E., Matias I., Delamarre A., Metna-Laurent M., Cannich A., Hebert-Chatelain E., Mulle C., Ortega-Gutierrez S., Martin-Fontecha M., Klugmann M., Guggenhuber S., Lutz B., Gertsch J., Chaouloff F., Lopez-Rodriguez M.L., Grandes P., Rossignol R., Marsicano G. Mitochondrial CB(1) receptors regulate neuronal energy metabolism. Nat. Neurosci. 2012;15:558–564. doi: 10.1038/nn.3053. [DOI] [PubMed] [Google Scholar]
- 120.Hebert-Chatelain E., Desprez T., Serrat R., Bellocchio L., Soria-Gomez E., Busquets-Garcia A., Pagano Zottola A.C., Delamarre A., Cannich A., Vincent P., Varilh M., Robin L.M., Terral G., Garcia-Fernandez M.D., Colavita M., Mazier W., Drago F., Puente N., Reguero L., Elezgarai I., Dupuy J.W., Cota D., Lopez-Rodriguez M.L., Barreda-Gomez G., Massa F., Grandes P., Benard G., Marsicano G. A cannabinoid link between mitochondria and memory. Nature. 2016;539:555–559. doi: 10.1038/nature20127. [DOI] [PubMed] [Google Scholar]
- 121.Bosier B., Bellocchio L., Metna-Laurent M., Soria-Gomez E., Matias I., Hebert-Chatelain E., Cannich A., Maitre M., Leste-Lasserre T., Cardinal P., Mendizabal-Zubiaga J., Canduela M.J., Reguero L., Hermans E., Grandes P., Cota D., Marsicano G. Astroglial CB1 cannabinoid receptors regulate leptin signaling in mouse brain astrocytes. Mol. Metab. 2013;2:393–404. doi: 10.1016/j.molmet.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gutierrez-Rodriguez A., Bonilla-Del Rio I., Puente N., Gomez-Urquijo S.M., Fontaine C.J., Egana-Huguet J., Elezgarai I., Ruehle S. Localization of the cannabinoid type-1 receptor in subcellular astrocyte compartments of mutant mouse hippocampus. Glia. 2018;66(7):1417–1431. doi: 10.1002/glia.23314. [DOI] [PubMed] [Google Scholar]
- 123.Brailoiu G.C., Deliu E., Marcu J., Hoffman N.E., Console-Bram L., Zhao P., Madesh M., Abood M.E., Brailoiu E. Differential activation of intracellular versus plasmalemmal CB2 cannabinoid receptors. Biochemistry. 2014;53:4990–4999. doi: 10.1021/bi500632a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hebert-Chatelain E., Reguero L., Puente N., Lutz B., Chaouloff F., Rossignol R., Piazza P.V., Benard G., Grandes P., Marsicano G. Cannabinoid control of brain bioenergetics: Exploring the subcellular localization of the CB1 receptor. Mol. Metab. 2014;3:495–504. doi: 10.1016/j.molmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.den Boon F.S., Chameau P., Schaafsma-Zhao Q., van Aken W., Bari M., Oddi S., Kruse C.G., Maccarrone M., Wadman W.J., Werkman T.R. Excitability of prefrontal cortical pyramidal neurons is modulated by activation of intracellular type-2 cannabinoid receptors. Proc. Natl. Acad. Sci. USA. 2012;109:3534–3539. doi: 10.1073/pnas.1118167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Currie S., Rainbow R.D., Ewart M.A., Kitson S., Pliego E.H., Kane K.A., McCarron J.G. IP(3)R-mediated Ca(2+) release is modulated by anandamide in isolated cardiac nuclei. J. Mol. Cell. Cardiol. 2008;45:804–811. doi: 10.1016/j.yjmcc.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 127.Krishna Kumar K., Shalev-Benami M., Robertson M.J., Hu H., Banister S.D., Hollingsworth S.A., Latorraca N.R., Kato H.E., Hilger D., Maeda S., Weis W.I., Farrens D.L., Dror R.O., Malhotra S.V., Kobilka B.K., Skiniotis G. Structure of a signaling cannabinoid receptor 1-g protein complex. Cell. 2019;176:448–458. doi: 10.1016/j.cell.2018.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li X.T., Hua T., Vemuri K., Ho J.H., Wu Y.R., Wu L.J., Popov P., Benchama O., Zvonok N., Locke K., Qu L., Han G.W., Iyer M.R., Cinar R., Coffey N.J., Wang J.J., Wu M., Katritch V., Zhao S.W., Kunos G., Bohn L.M., Makriyannis A., Stevens R.C., Liu Z.J. Crystal structure of the human cannabinoid receptor cb2. Cell. 2019;176:459–467. doi: 10.1016/j.cell.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


