Abstract
Correlation between the expression of miR-23a and miR-135 and tumor markers in gastric cancer patients and their significance in diagnosis was investigated. A total of 78 patients with gastric cancer admitted to Dongying People's Hospital from July 2015 to June 2017 were enrolled, and 80 healthy patients were selected as the control group during the same period. The expression levels of miR-23a and miR-135 in the serum of the two groups were detected by RT-qPCR, and the expression levels of tumor markers CEA and carbohydrate antigen 199 (CA199) were detected by ELISA. The receiver operating characteristic curve and area under the curve (AUC) were used to evaluate the diagnostic efficacy of miR-23a and miR-135 as diagnostic indicators. There was no significant difference between the observation and the control group (P>0.05). The expression levels of miR-23a and miR-135 in the observation group were significantly higher than those in the control group (P<0.05). The expression levels of CEA and CA199 in serum of patients in the observation group were significantly higher than those in the control group (P<0.05). Pearson's correlation test showed that the expression levels of miR-23a and miR-135 were positively correlated with CEA and CA199 (P<0.05). The specificity, sensitivity and AUC of miR-23a in the diagnosis of gastric cancer were 67.95, 87.50 and 0.805%, respectively. The specificity, sensitivity and AUC of miR-135 in the diagnosis of gastric cancer were 73.08, 82.50 and 0.824%, respectively. Both could be used in the diagnosis of gastric cancer. In conclusion, miR-23a and miR-135 are highly expressed in gastric cancer patients and positively correlated with tumor markers, which can be used in diagnosis for gastric cancer.
Keywords: miR-23a, miR-135, gastric cancer, tumor marker, correlation, diagnosis
Introduction
Gastric cancer (GC) is the fourth most common malignancy in the world and remains the second leading cause of death for all malignancies worldwide (738,000 deaths per year) (1,2). China is a large country with a high incidence of gastric cancer. As the early symptoms of gastric cancer are relatively hidden, most patients are diagnosed with advanced gastric cancer, and the treatment effect is generally poor (3). The data show that the 5-year relative survival rate of gastric cancer in China is relatively low (4). Therefore, it is of great significance to search for molecular biomarkers related to the diagnosis of gastric cancer, explore the regulatory mechanism of molecular signals during the occurrence and development of gastric cancer, and identify therapeutic targets of biological markers that affect its occurrence and development, so as to improve the early diagnosis and prognosis of gastric cancer patients (5,6).
miRNA is a non-coding RNA with a length of 19–25 nucleotides. It is a novel gene expression regulatory molecule that can participate in the occurrence and development of tumors by regulating the expression of many oncogenes or tumor suppressor genes (7). It has been previously shown that some miRNAs are closely related to the occurrence and development of gastric cancer. For example, Zhu et al (8) showed that plasma microRNAs were potential new biomarkers for early detection of early gastric cancer. Li et al (9) found microRNA-28 promotes the proliferation and invasion of gastric cancer cells through the PTEN/PI3K/AKT signaling pathway. In addition, Peng et al (10) found that microRNA-494 increases the chemosensitivity of doxorubicin in gastric cancer cells by targeting phosphodiesterase 4D. microRNA-23a (miR-23a) has been shown to be upregulated in non-small cell lung cancer (11) and laryngeal cancer (12). Previous findings have shown that microRNA-135 (miR-135) is under-represented in non-small cell lung cancer (13). However, there are few studies on the expression of miR-23a and miR-135 in gastric cancer and the significance of gastric cancer diagnosis.
At present, serum tumor markers such as CEA and carbohydrate antigen 199 (CA199) are mainly used in the clinical diagnosis of gastric cancer, which has the advantages of short detection cycle and small trauma. However, no tumor marker with strong specificity and high sensitivity has been found to be able to detect tumors early (14–16). Therefore, the aim of this study was to compare the serum levels of miR-23a and miR-135 in patients with gastric cancer and normal controls, and explored the correlation between the expression level and cancer markers CEA and CA199 in patients with gastric cancer and their diagnostic significance, so as to provide a reference for clinical search for potential molecular markers of gastric cancer.
Patients and methods
General information
A total of 78 patients with gastric cancer admitted to Dongying People's Hospital from July 2015 to June 2017 were selected as the observation group, and 80 healthy individuals in the same period were selected as the control group. There were 52 males and 26 females in the gastric cancer group, aged 41–76 years, with an average age of 51.57±9.19 years. In the control group, there were 55 males and 25 females, aged 43–68 years, with an average age of 51.39±10.17 years. According to the Borrmann classification (17), there were 23 cases of type I, 22 cases of type II, 16 cases of type III, and 19 cases of type IV in the observation group. The inclusion criteria were: diagnosis of gastric cancer was confirmed by pathology, and the clinical, imaging and histopathological data were complete; the healthy group was examined in the physical examination center of Dongying People's Hospital and the results were normal, without other types of tumors, heart, liver, kidney and other important organ diseases, and no family members had a history of cancer. The exclusion criteria were: patients with immune system diseases; long-term bedridden patients; patients with other malignant tumors; severe hypertension, diabetic, mental and cognitive dysfunction; pregnant or lactation women; using glucocorticoids and antibiotics for up to 2 weeks. Clinical data, including BMI (kg/m2), heart rate(time/min), urea nitrogen, age, tumor stage, smoking history, alcohol abuse history, of the two groups were collected and compared.
The study was approved by the Ethics Committee of Dongying People's Hospital. All the patients agreed to participate in the experiment and signed informed consent.
Main instruments and reagents
CA-199 ELISA kit [Shanghai Jing Kang Bioengineering Co., Ltd., JK-(a)-5894]; CEARIA and ELISA kit [Shanghai Jing Kang Bioengineering Co., Ltd., JK-(a)-6071]; Multi-function microplate reader (BioTek Berten, DLK0001622); desktop high-speed centrifuge (Sichuan Yanke Instrument Co., Ltd., TG-16); PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc. 7500); total RNA extraction kit Easy Pure miRNA kit, PCR + reverse transcription kit TransScript Green miRNA Two-Step qRT-PCR Super Mix (TransGenBiotech Co., Ltd., Beijing, China); UV spectrophotometer (Thermo Fisher Scientific, Inc.; Multiskan Sky); qmiR-23a, miR-135 and U6 internal reference primers were synthesized by Shanghai Harling Biotechnology Co., Ltd. (Table I).
Table I.
miR-23a, miR-135 and U6 primer sequences.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| miR-23a | 5′-GGGGATCACATTGCCAGG-3′ | 5′-AGTGCGTGTCGTGGAGTC-3′ |
| miR-135 | 5′-ACAUAGGAAUAAAAAGCCAUAtt-3′ | 5′-CUAUGGCUUUUUAUUCCUAUGUGA-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
Detection method
Venous blood (5 ml) of two groups collected in the morning was placed in the vacuum collecting vessel and centrifuged for routine separation at a speed of 2,600 × g for 10 min at 4°C. The expression levels of CEA and CA199 in the collected serum samples were determined by ELISA. Blank, sample and standard wells were set up, respectively. The samples were added to the the sample well to be tested, and the standard sample with different concentrations was added to the standard well. The samples were added to the bottom of the well of the enzyme label plate, avoiding touching the wall of the well, then gently shaken and mixed. After sealing the plate with the sealing plate membrane, the samples were incubated at 37°C for 30 min. The 30-fold concentrated detergent was diluted with distilled water 30 times for later use. The sealing plate film was carefully removed, the liquid was discarded, and shaken dry. Each well was filled with the washing solution, and left to stand for 30 sec, then discarded, and repeated 5 times, and patted dry. Enzyme-labeled reagent was added to each well and placed on the sealing plate at 37°C for 30 min, except the blank wells. The sealing film was carefully removed, the liquid was discarded and the film dried. Color reagent A (50 µl) was added in each well, followed by color reagent B 50 µl, gently shaken and mixed at 37°C in the dark for 15 min to develop the color. Termination liquid (50 µl) was added to each well to terminate the reaction. The OD of each well was measured in sequence at 0 and 450 nm wavelength of blank air conditioner, and the determination was carried out within 15 min after the addition of terminating liquid. The calibration curve was drawn and the linear regression equation was obtained. The OD value of the sample was substituted into the equation and the concentration of the sample was calculated.
miR-23a and miR-135 detected by RT-qPCR
Total RNA in serum was extracted according to the instruction of total RNA extraction kit. The purity of the extracted total RNA was detected by ultraviolet spectrophotometer and the concentration was calculated. Total RNA (2 µl) and reverse transcription were detected as per the instruction manual of the kit for cDNA synthesis. The reaction system was: 42°C for 60 min, 95°C for 5 min, and the synthesized cDNA sample was stored at −20°C for later use. The total volume of 20 µl reaction system was: SYBR-Green PCRP remix 10 µl, upstream primer (10X) 2 µl, downstream primer (10X) 2 µl, dd water (Rnase- and Dnase-free) 5 µl. RT-qPCR reaction conditions were: 90°C for 5 min, 90°C for 5 sec, 60°C for 30 sec, 72°C for 5 sec, total of 40 cycles. U6 was the internal reference gene, and 2−∆∆Cq was used to analyze the data (18).
Statistical analysis
SPSS 22.0 (SPSS Inc.) was used to analyze the data. Enumeration data were described by [n (%)], and Chi-square test was used for comparison between groups. Measurement data were expressed as mean ± standard deviation. The t-test was used to compare the measurements between groups. The receiver operating characteristic (ROC) curve was used to evaluate the diagnostic efficacy of miR-23a and miR-135 for gastric cancer. Pearson's correlation analysis was used to test the correlation between miR-23a and miR-135 and CEA and CA-199. P<0.05 was considered to indicate a statistically significant difference.
Results
Comparison of general clinical data
The clinical data of the two groups were collected for comparison, and the results showed that there was no statistical difference between the observation and control groups in BMI (kg/m2), heart rate (time/min), urea nitrogen, age, tumor stage, smoking history, alcohol abuse history (P>0.05), as shown in Table II.
Table II.
Comparison of general clinical data between the two groups (mean ± SD) [n (%)].
| Clinical factors | Control group (n=80) | Observation group (n=78) | t/χ2 value | P-value |
|---|---|---|---|---|
| BMI (kg/m2) | 23.98±1.26 | 24.36±1.57 | 1.68 | 0.095 |
| Heart rate (times/points) | 105.61±11.53 | 105.79±11.66 | 0.098 | 0.922 |
| Urea nitrogen (mmol/l) | 12.97±3.51 | 13.16±3.71 | 0.331 | 0.741 |
| Age | 49.67±5.68 | 50.45±4.71 | 0.938 | 0.35 |
| Tumor staging | 0.239 | 0.625 | ||
| I+II | 39 (48.75) | 35 (44.87) | ||
| IIIa+IIIb | 41 (51.25) | 43 (55.13) | ||
| Borrmann classification | 0.534 | 0.465 | ||
| I+II | 37 (46.25) | 32 (41.03) | ||
| III+IV | 43 (53.75) | 46 (58.97) | ||
| Sex | 0.143 | 0.378 | ||
| Male | 45 (56.25) | 42 (53.85) | ||
| Female | 35 (43.75) | 36 (46.15) | ||
| Drinking history | 0.38 | 0.617 | ||
| Yes | 51 (63.75) | 47 (60.26) | ||
| No | 29 (36.25) | 31 (39.74) | ||
| Degree of differentiation | 0.355 | 0.837 | ||
| Differentiated type | 21 (26.25) | 21 (26.92) | ||
| Poorly differentiated | 32 (40) | 34 (43.59) | ||
| Undifferentiated type | 27 (33 75) | 23 (29.49) | ||
| Lymph node metastasis | 0.894 | 0.345 | ||
| Yes | 46 (57.5) | 39 (50) | ||
| No | 34 (42.5) | 39 (50) | ||
| Degree of differentiation | 0.261 | 0.61 | ||
| High school and below | 11 (13.75) | 13 (16.67) | ||
| Over Senior High School | 69 (86.25) | 65 (83.33) | ||
| History of smoking | 0.23 | 0.632 | ||
| No | 41 (51.25) | 37 (47.44) | ||
| Yes | 39 (48.75) | 41 (52.56) |
Comparison of serum miR-23a and miR-135 expression between the observation and control groups
The expression levels of serum miR-23a and miR-135 in the observation group were significantly higher than those in the control group, with statistically significant differences (P<0.05) (Table III).
Table III.
Comparison of serum miR-23a and miR-135 expression between the two groups (mean ± SD).
| Group | n | miR-23a | miR-135 |
|---|---|---|---|
| Observation | 78 | 13.72±3.27 | 4.50±1.41 |
| Control | 80 | 10.15±2.26 | 3.03±0.74 |
| t value | – | 8.000 | 8.234 |
| P-value | – | <0.001 | <0.001 |
Comparison of the expression of serum CA199 and CEA between the observation and control groups
The levels of serum CA199 and CEA in the observation group were significantly higher than those in the control group (P<0.05) (Table IV).
Table IV.
Comparison of serum CA199 and CEA expression between the two groups (mean ± SD).
| Group | n | CA199 (ng/ml) | CEA (ng/ml) |
|---|---|---|---|
| Observation | 78 | 53.07±38.27 | 16.58±11.18 |
| Control | 80 | 11.34±3.5 | 1.8±1.40 |
| t value | – | 0.877 | 11.730 |
| P-value | – | <0.001 | <0.001 |
CA199, carbohydrate antigen 199.
Diagnostic value of serum miR-23a and miR-135 in gastric cancer
The ROC curve was drawn for the diagnosis of gastric cancer by the expression levels of miR-23a and miR-135. The results showed that the AUC for the diagnosis of gastric cancer by miR-23a was 0.805 (95% CI: 0.735–0.874), the specificity was 67.95%, the sensitivity was 87.50%, and the cut-off value was 12.500. The AUC of gastric cancer diagnosed by miR-135 was 0.824 (95% CI: 0.758–0.890), the specificity was 73.08%, the sensitivity was 82.50%, and the cut-off value was 3.677 (Table V, Fig. 1).
Table V.
Diagnostic value of serum miR-23a and miR-135 in gastric cancer.
| Indicators | AUC | 95% CI | Specificity (%) | Sensitivity (%) | Cut-off |
|---|---|---|---|---|---|
| miR-23a | 0.805 | 0.735–0.874 | 67.95 | 87.50 | 12.500 |
| miR-135 | 0.824 | 0.758–0.890 | 73.08 | 82.50 | 3.677 |
AUC, area under the curve.
Figure 1.
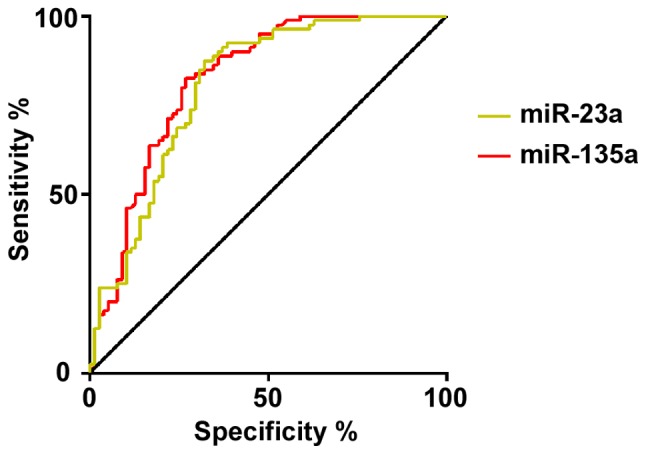
ROC curve of serum miR-23a and miR-135 in the diagnosis of gastric cancer. The AUC for the diagnosis of gastric cancer by miR-23a was 0.805, the specificity was 67.95%, and the sensitivity was 87.50%. The AUC of gastric cancer diagnosed by miR-135 was 0.824, the specificity was 73.08%, and the sensitivity was 82.50%. AUC, area under the curve.
Correlation analysis between serum miR-23a, miR-135 and CA199, CEA in patients with gastric cancer
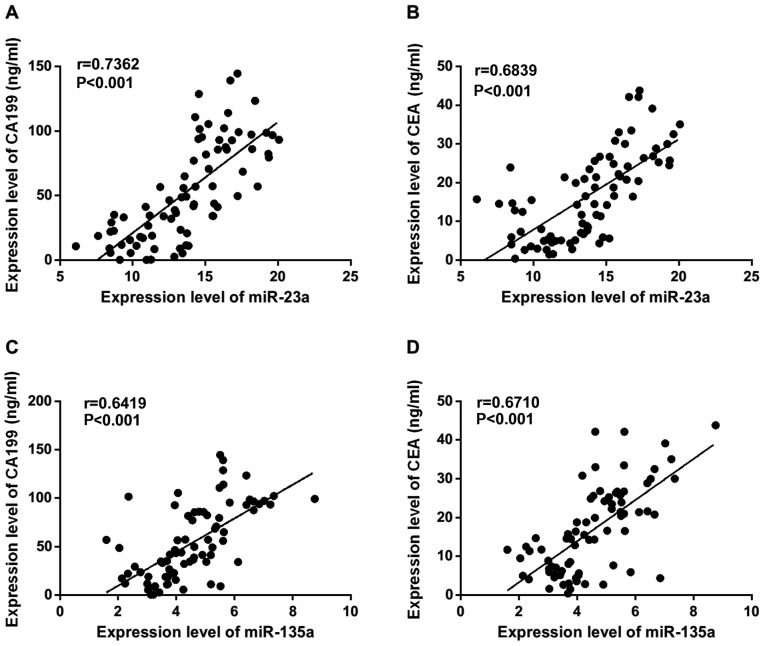
The relationship between miR-23a and miR-135 in serum and CA199 and CEA was analyzed by Pearson's correlation analysis. It was found that miR-23a was positively correlated with the expression of CA199 (r=0.7362, P<0.001) and CEA (r=0.6839, P<0.001); miR-135 was positively correlated with the expression of CA199 (r=0.6419, P<0.001) and CEA (r=0.6710, P<0.001) (Fig. 2).
Figure 2.
Correlation analysis between miR-23a, miR-135, CA199 and CEA. Correlation between the expression of serum (A) miR-23a and CA199, (B) miR-23a and CEA, (C) miR-135 and CA199, and (D) miR-135 and CEA in gastric cancer patients. CA199, carbohydrate antigen 199.
Discussion
Gastric cancer is a relatively common malignant tumor in clinical practice, and its pathogenesis has not been clarified at present (19). Many gastric cancer patients have a lack of specificity in the early stage of the disease, and screening for gastric cancer requires biopsy pathology under gastroscopy. This invasive examination can make patients feel more uncomfortable, which leads to a low diagnostic rate of gastric cancer (20). Many patients are in the middle and late stages of treatment, and it is often difficult to achieve the desired therapeutic effect (21). Therefore, searching for biological indicators that have a strong diagnostic value for gastric cancer is of great significance for improving the prognosis of patients and increasing the survival rate (22).
miRNA is a class of non-coding small RNAs with a wide range of biological effects, which participates in the regulation of the expression of various proto-oncogenes and anti-oncogenes in vivo, thereby affecting the biological behavior of cell proliferation, invasion and angiogenesis (23,24). Although the current understanding of miRNA function is not very clear, relevant studies have shown that its abnormal expression or mutation is closely related to the occurrence and development of many tumors and the efficacy of some antitumor drugs (25), which is a new highlight in the field of tumor biotherapy research (26). The levels of serum miR-23a and miR-135 in the observation group were significantly higher than those in the control group. Previous studies have shown that miR-23a is abnormally expressed in gastric cancer. Zhu et al (27) found that miR-23a is up-regulated in gastric adenocarcinoma and may promote the growth of gastric cancer tissues. Studies on gastric cancer cell line MGC80 showed that the expression of interleukin-6 receptor protein can be reduced by the expression of mRNA-23a, which can promote the growth of gastric adenocarcinoma cell line MGC803. The expression of mRNA-23a in peripheral blood and tumor cells of patients with cancer is increased. Zhou et al (28) found that miR-135 can promote the growth and invasion of colorectal cancer in vitro by transferring inhibitors, while Golubovskaya et al (29) showed that miR-135 can directly target adhesive plaque kinase, inhibit cell invasion and improve the sensitivity of cancer cells to chemotherapy. However, its specific mechanism in gastric cancer is still unclear, but it indicates that miR-23a and miR-135 are very likely to be involved in the occurrence, development, migration and metastasis of gastric cancer, and they are expected to become biological markers for the diagnosis of gastric cancer.
Our further investigation showed that the specificity of serum miR-23a in the diagnosis of gastric cancer was 67.95%, and the sensitivity was 87.50%; the specificity of serum miR-135 in the diagnosis of gastric cancer was 73.08%, and the sensitivity was 82.50%. These results suggest that both can be used as biological indicators with better sensitivity and specificity in the diagnosis of gastric cancer. At the end of the study, Pearson's correlation analysis was used to detect the correlation between the expression levels of miR-21 and miR-124 in serum of patients with gastric cancer and the expression levels of CA199 and CEA. It was found that miR-23a and miR-135 were positively correlated with the expression of CA199 and CEA, but the specific relationship was not explored in depth.
In this study, we compared the expression levels of miR-23a and miR-135 in the serum of patients with gastric cancer and normal controls, and explored the diagnostic value of the detection for gastric cancer. However, the efficacy and prognosis of these markers were not observed, thus, futher studies are still required.
In conclusion, miR-23a and miR-135 are highly expressed in gastric cancer patients and positively correlated with tumor markers. The detection of serum miR-23a and miR-135 has good sensitivity and specificity for the diagnosis of gastric cancer, and show potential as biomarkers for the diagnosis of gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Hospital level Scientific Research Project ‘Application of microRNA-203 combined with TSGF and CEA in diagnosis of early gastric cancer’ (no. 2016DYYZ20).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LY and GX conceived and designed the study, and drafted the manuscript. LY, GX, YZ and YW collected, analyzed and interpreted the experimental data. YZ and YW performed PCR. LY revised the manuscript for important intellectual content and assisted with statistical analysis. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Dongying People's Hospital. Signed informed consents were obtained from the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Shen L, Li J, Xu J, Pan H, Dai G, Qin S, Wang L, Wang J, Yang Z, Shu Y, et al. Bevacizumab plus capecitabine and cisplatin in Chinese patients with inoperable locally advanced or metastatic gastric or gastroesophageal junction cancer: Randomized, double-blind, phase III study (AVATAR study) Gastric Cancer. 2015;18:168–176. doi: 10.1007/s10120-014-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer survival in China, 2003–2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 5.Buckland G, Travier N, Huerta JM, Bueno-de-Mesquita HB, Siersema PD, Skeie G, Weiderpass E, Engeset D, Ericson U, Ohlsson B, et al. Healthy lifestyle index and risk of gastric adenocarcinoma in the EPIC cohort study. Int J Cancer. 2015;137:598–606. doi: 10.1002/ijc.29411. [DOI] [PubMed] [Google Scholar]
- 6.Massarrat S, Stolte M. Development of gastric cancer and its prevention. Arch Iran Med. 2014;17:514–520. [PubMed] [Google Scholar]
- 7.Rao SA, Santosh V, Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol. 2010;23:1404–1417. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 8.Zhu XL, Ren LF, Wang HP, Bai ZT, Zhang L, Meng WB, Zhu KX, Ding FH, Miao L, Yan J, et al. Plasma microRNAs as potential new biomarkers for early detection of early gastric cancer. World J Gastroenterol. 2019;25:1580–1591. doi: 10.3748/wjg.v25.i13.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Zhu X, Shou T, Yang L, Cheng X, Wang J, Deng L, Zheng Y. MicroRNA-28 promotes cell proliferation and invasion in gastric cancer via the PTEN/PI3K/AKT signalling pathway. Mol Med Rep. 2018;17:4003–4010. doi: 10.3892/mmr.2017.8299. [DOI] [PubMed] [Google Scholar]
- 10.Peng QP, Du DB, Ming Q, Hu F, Wu ZB, Qiu S. MicroRNA 494 increases chemosensitivity to doxorubicin in gastric cancer cells by targeting phosphodiesterases 4D. Cell Mol Biol. 2018;64:62–66. doi: 10.14715/cmb/2017.64.15.10. [DOI] [PubMed] [Google Scholar]
- 11.Qu WQ, Liu L, Yu Z. Clinical value of microRNA-23a upregulation in non-small cell lung cancer. Int J Clin Exp Med. 2015;8:13598–13603. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XW, Liu N, Chen S, Wang Y, Zhang ZX, Sun YY, Qiu GB, Fu WN. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn Pathol. 2015;10:22. doi: 10.1186/s13000-015-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, Zhang T. Downregulation of MicroRNA-135 promotes sensitivity of non-small cell lung cancer to Gefitinib by targeting TRIM16. Oncol Res. 2018;26:1005–1014. doi: 10.3727/096504017X15144755633680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Virgilio E, Proietti A, D'Urso R, Cardelli P, Giarnieri E, Montagnini M, Giovagnoli MR, Mercantini P, Balducci G, Cavallini M. Measuring intragastric tumor markers in gastric cancer patients: A systematic literature review on significance and reliability. Anticancer Res. 2017;37:2817–2821. doi: 10.21873/anticanres.11632. [DOI] [PubMed] [Google Scholar]
- 15.Xiao S, Feng F, Sun L, Cai L, Liu Z, Liu S, Fan D, Zhang H. Blood type AB predicts promising prognosis in gastric cancer patients with positive preoperative serum CEA. Medicine (Baltimore) 2017;96:e8496. doi: 10.1097/MD.0000000000008496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu H, Sun L, Dong X, Gong Y, Xu Q, Jing J, Bostick RM, Wu X, Yuan Y. A serological biopsy using five stomach-specific circulating biomarkers for gastric cancer risk assessment: A multi-phase study. Am J Gastroenterol. 2017;112:704–715. doi: 10.1038/ajg.2017.55. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Wang X H, Kou H J, et al. Comparing single oral contrast-enhanced ultrasonography and double contrast-enhanced ultrasonography in the preoperative Borrmann classification of advanced gastric cancer. Oncotarget. 2018;9:8716. doi: 10.18632/oncotarget.23819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Okines A, Verheij M, Allum W, Cunningham D, Cervantes A, ESMO Guidelines Working Group Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v50–v54. doi: 10.1093/annonc/mdq164. [DOI] [PubMed] [Google Scholar]
- 20.Deng G, Qu J, Zhai S, Shi Y, Wang X. Effect of neoadjuvant chemotherapy on nutritional status of locally advanced gastric cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2018;21:331–335. (In Chinese) [PubMed] [Google Scholar]
- 21.Shen L, Shan YS, Hu HM, Price TJ, Sirohi B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al. Management of gastric cancer in Asia: Resource-stratified guidelines. Lancet Oncol. 2013;14:e535–e547. doi: 10.1016/S1470-2045(13)70436-4. [DOI] [PubMed] [Google Scholar]
- 22.Harada K, Mizrak Kaya D, Shimodaira Y, Song S, Baba H, Ajani JA. Proteomics approach to identify biomarkers for upper gastrointestinal cancer. Expert Rev Proteomics. 2016;13:1041–1053. doi: 10.1080/14789450.2016.1246189. [DOI] [PubMed] [Google Scholar]
- 23.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131–140. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K, et al. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433:207–212. doi: 10.1016/j.bbrc.2013.02.084. [DOI] [PubMed] [Google Scholar]
- 25.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, Li X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010;277:3726–3734. doi: 10.1111/j.1742-4658.2010.07773.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W, Li X, Liu F, Xiao Z, He M, Shen S, Liu S. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin (Shanghai) 2012;44:838–846. doi: 10.1093/abbs/gms071. [DOI] [PubMed] [Google Scholar]
- 29.Golubovskaya VM, Sumbler B, Ho B, Yemma M, Cance WG. MiR-138 and MiR-135 directly target focal adhesion kinase, inhibit cell invasion, and increase sensitivity to chemotherapy in cancer cells. Anticancer Agents Med Chem. 2014;14:18–28. doi: 10.2174/187152061401140108113435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.