Abstract
Glioma is one of the most common malignant tumor types of the central nervous system. It is necessary to identify biomarkers and novel therapeutic targets for glioma. The purpose of the present study was to distinguish lipid biomarkers with differential expression patterns in glioma tissues and normal brain tissues by matrix assisted laser desorption/ionization (MALDI)-imaging and MALDI-time of flight (TOF)-mass spectrometry (MS). Additionally, identification of lipid biomarkers was performed to describe novel therapeutic targets for glioma treatment. A total of six tissues from three patients with glioma and three control patients with traumatic brain injury were analyzed using UltrafleXtreme MALDI-TOF/TOF. The expression levels of 15 lipid peaks were higher in the TBT samples compared with in the GBT samples. The expression levels of another 16 lipid peaks were higher in the GBT samples compared with in the TBT samples. 14 peaks were identified as sphingomyelins using MS/MS. Additional results were also obtained from experiments using the glioma cell line U373-MG. These results indicated that treatment with the drug desipramine (Desi) inhibited the accumulation of ceramide on the cell membranes of glioma U373-MG cells. Treatment with Desi inhibited the activation of insulin-like growth factor-1 receptor and inhibited the activation of proteins in the PI3K/Akt signaling pathway.
Keywords: glioma, matrix assisted laser desorption/ionization-imaging, lipid biomarkers, sphingomyelin, PI3K/Akt signaling pathway
Introduction
Glioma is the most common type of malignant brain tumor in adults worldwide (1,2). The principal diagnosis methods for patients with glioma include disease history questionnaires, nervous system physical examination and imaging examination with different modalities (3). Since early glioma prediction and diagnosis is difficult, the majority of patients are diagnosed at a late stage. Typically, treatment for patients with glioma involves maximal safe surgical resection, followed by chemotherapy and radiotherapy. The prognosis of patients with glioma is frequently poor due to its aggressive properties (4,5). Hence, there is an urgent need to identify novel biomarkers for glioma diagnosis and novel therapeutic targets to improve patient survival.
Protein and lipid are two types of diagnostic biomarkers for glioma (6–8). Biomarkers may be identified from patient samples, including serum, cerebrospinal fluid (CSF) and glioma cells (9). Capillary morphogenesis protein 2 (CMG2) has been identified as a prognostic biomarker for glioma, and CMG2 increased expression is associated with increased tumor grade and poor patient survival (10). The effect of protein biomarker T-LAK cell-originating protein kinase (TOPK) expression on the outcome in malignant glioma has been previously reported, revealing that low TOPK expression is an independent predictor of longer overall survival (11). Ribom et al (12) performed a comparative proteomics analysis with CSF samples from glioma cases and revealed that α-2-Heremans-Schmid glycoprotein was highly expressed in the CSF of patients with low-grade disease. Iwadate et al (13) conducted survival prediction research in glioma based on proteomic analysis and identified 37 protein biomarkers.
In addition to protein biomarkers, lipid biomarkers are also required for the diagnosis and treatment of glioma. The matrix assisted laser desorption/ionization (MALDI)-imaging technique has been widely used in lipid biomarker research (14–16). Wildburger et al (15) identified lipids directly from glioblastoma tissues by MALDI-imaging mass spectrometry (MS), and a number of species of signaling lipids were observed. Shrivas et al (17) identified phospholipids in mouse liver and cerebellum tissue sections using an ionic matrix for enhanced MALDI-imaging MS.
In the present study, three glioma brain tissue (GBT) samples and three trauma brain tissue (TBT) samples were collected to identify lipid biomarkers using MALDI-time of flight (TOF)-tandem mass spectrometry (MS/MS). The lipid biomarkers with differential expression were selected and identified. The expression of a number of lipid biomarkers with potential clinical significance was verified using the MALDI-imaging technique. In order to reveal the regulatory mechanisms of lipid biomarkers in glioma signaling pathways, the glioma cell line U373-MG was cultured, and additional research was performed regarding the PI3K/AKT signaling pathway.
Materials and methods
Brain tissue samples
In the present study, three GBT samples from patients with glioma (two women aged 46 and 55 years and one man aged 60 years) and three TBT samples from patients with traumatic brain injury (two men aged 48 and 50 years and one woman aged 56 years) were collected following neurosurgery at The First Affiliated Hospital of Zhejiang University School of Medicine (Hangzhou, China) in December 2013. Informed consent was obtained from participants for the use of their tissues in the present study. All diagnoses of glioma were histopathologically confirmed.
Glioma cell culture
The human glioma cell line U373-MG was purchased from American Type Culture Collection (ATCC). The U373-MG ATCC cell line is a U-251 derivative according to Cellosaurus (https://web.expasy.org/cellosaurus/CVCL_2219). It was directly used in the present study without additional authentication. The cell line was cultured in DMEM (Thermo Fisher Scientific, Inc.), supplemented with 1% penicillin/streptomycin and 10% FBS (Thermo Fisher Scientific, Inc.). The cells were grown at 37°C with 5% CO2 in a humidified incubator.
Biomarker identification with MALDI-TOF-MS/MS
GBTs and TBTs were removed from the −80°C freezer and cut into 10-µm-thick sections with a Leica CM950 freezing microtome (Leica Microsystems GmbH) at −20°C. Each tissue section was pasted on the imaging slide and dried in a vacuum dryer for 45 min at room temperature. The imaging slide was placed into the ImagePrep machine and flushed uniformly with matrix. The imaging slide was placed on the MTP Slide Adapter II and analyzed with MALDI-TOF-MS/MS. The mass scan range was set at m/z: 0–3,000. The mass spectrum data were analyzed using FlexImaging software version 3.0 (Bruker Daltonics). The lipid peaks were identified using MS/MS and results were searched in the Human Metabolome Database (18).
Live staining
U373-MG cells were cultured as aforementioned in small glass dishes overnight so that they reach 60–75% confluence. Subsequent to removal of the medium, the cells were washed once with 500 µl DMEM containing 2% FBS. The fluorescent antibody anti-human insulin-like growth factor-1 receptor (IGF-1R; eBioscience; cat. no. 16-8849; 1:20) was added to the DMEM containing 2% FBS. The cells were cultured in the new medium with antibody at 37°C with 5% CO2 in a humidified incubator for 10 min. The medium was discarded and the cells were washed once with DMEM containing 2% FBS without antibody. Subsequently, DMEM with 2% FBS was added into the dishes. The prepared dishes were placed into the CO2 incubator of a laser scanning confocal fluorescence microscope. IGF-1R signal was recorded using laser scanning confocal microscopy.
Cell immunostaining
U373-MG cells were cultured in small glass dishes as aforementioned. When cells reached 60–75% of the dishes, the medium was discarded and the cells were washed once with 1 ml PBS. A volume of 1 ml fixation and permeabilization solution (BD Biosciences) was added into the dish, which was kept in the dark for 1 h at 4°C. The fixing reagent was discarded and the dish was washed once with 1 ml PBS. Goat serum (5%; Jackson Immuno Research) in PBS was used for blocking at room temperature for 1 h. The cells were incubated with mouse monoclonal primary antibody against ceramide (Enzo Life Sciences; cat. no. ALX-804-196; 1:10) at 4°C overnight. Following three washes with PBS, the cells were incubated with goat anti-mouse IgG secondary antibody (AF647-conjugated; Jackson Immunologicals; cat. no. 115-606-146; 1:300) for 1 h in the dark at room temperature. The dish was washed three times with PBS and visualized using laser scanning confocal microscopy.
For the regulation of IGF-1R expression, U373-MG cells were divided into the DMSO and Desi groups. Cells from the DMSO group represented the control group and were treated with DMSO only and stimulated with insulin for 30 min. Cells from the Desi group were treated with the drug Desi and stimulated with insulin for 30 min.
Western blotting
The glioma cell line U373-MG was lysed with NP40 lysis buffer (Thermo Fisher Scientific, Inc.). The protein density in each sample was determined by ultraviolet spectrometry and adjusted to the same density using loading buffer. Total protein (5 µg) was separated by SDS-PAGE (10% gel) and transferred onto PVDF membranes (EMD Millipore). The membranes were blocked at 4°C overnight with blocking buffer (pH 7.6) containing 5% nonfat dried milk and incubated with rabbit anti-human monoclonal antibody, which was purchased from Cell Signaling Technology, Inc. Immunoreactive proteins were stained using a chemiluminescent detection system. Membranes were subsequently washed with stripping solution for 1 h and treated as aforementioned, with mouse β-actin antibody (ProteinTech Group, Inc.) as an internal control. The relative target protein expression levels were normalized to those of β-actin.
For the regulation of PI3K/Akt pathway, U373-MG cells were divided into the DMSO and Desi groups. Cells from the DMSO group represented the control group and were treated with DMSO only. Cells from the Desi group were treated with the drug Desi. Both groups were then stimulated with insulin for 15, 30 and 60 min. Western blot analysis of these samples was performed to observe the PI3K/Akt pathway.
Statistical analysis
ClinProTools™ software (version 2.2; Bruker Corporation) was used to perform the statistical analysis of the peaks. Wilcoxon rank-sum test was used to compare data from TBT and GBT groups. Data are presented as the means ± standard deviation. Each experiment was repeated three times. P<0.01 was considered to indicate a statistically significant difference.
Results
Screening of lipid biomarkers between GBT and TBT samples
In the present study, three GBT samples and three TBT samples were collected, sliced and analyzed by MALDI-TOF-MS. The mass spectrum peaks were analyzed using FlexImaging software Ver3.0. Increased attention was paid to the differential expression peaks of lipid biomarkers in GBT and TBT samples. A total of 15 lipid peaks exhibited a higher expression intensity in normal control TBT samples compared with GBT samples (Table I). Another 16 lipid peaks exhibited a higher expression intensity in GBT samples compared with normal control TBT samples (Table II).
Table I.
Lipid peaks with higher expression in TBT samples compared with GBT samples.
| Bio-marker (m/z) | P-value | Expression in TBT (mean ± SD) | Expression in GBT (mean ± SD) |
|---|---|---|---|
| 798.57 | <1×10−6 | 198.68±67.29 | 14.34±11.75 |
| 722.55 | <1×10−6 | 122.60±42.17 | 9.37±5.57 |
| 848.64 | <1×10−6 | 26.64±9.18 | 3.44±2 |
| 799.56 | <1×10−6 | 90.11±32.72 | 7.11±5.92 |
| 800.56 | <1×10−6 | 64.7±23.62 | 5.92±4.26 |
| 769.58 (SM) | <1×10−6 | 94.32±34.4 | 9.06±5.37 |
| 828.57 | <1×10−6 | 14.17±4.68 | 3.32±2.06 |
| 849.65 | <1×10−6 | 13.13±4.42 | 2.93±2.09 |
| 770.56 | <1×10−6 | 56.06±20.65 | 7.46±4.99 |
| 773.54 | <1×10−6 | 50.43±18.96 | 5.18±3.47 |
| 755.58 (SM) | <1×10−6 | 14.11±5.83 | 6.11±3.92 |
| 753.60 (SM) | <1×10−6 | 70.94±31.10 | 10.69±7.62 |
| 771.57 (SM) | <1×10−6 | 27.07±10.97 | 6.23±2.62 |
| 783.79 (SM) | <1×10−6 | 99.88±38.85 | 42.26±21.45 |
| 797.60 (SM) | <1×10−6 | 17.00±6.60 | 10.27±3.59 |
GBT, glioma brain tissue; SD, standard deviation; SM, sphingomyelin; TBT, trauma brain tissue.
Table II.
Lipid peaks with higher expression in GBT samples compared TBT samples.
| Bio-marker (m/z) | P-value | Expression in TBT (mean ± SD) | Expression in GBT (mean ± SD) |
|---|---|---|---|
| 758.60 | <1×10−6 | 11.31±4.45 | 142.89±64.64 |
| 759.60 (SM) | <1×10−6 | 6.84±5.15 | 62.44±30.06 |
| 786.64 | <1×10−6 | 11.49±5 | 88.97±43.94 |
| 808.60 | <1×10−6 | 11.79±4.41 | 26.3±13.68 |
| 616.17 | <1×10−6 | 23.52±26.34 | 264.5±274.96 |
| 617.07 | <1×10−6 | 15.61±17.58 | 189.39±211.06 |
| 618.17 | <1×10−6 | 6.52±5.76 | 70.8±82.58 |
| 614.16 | <1×10−6 | 3.6±2.9 | 23.38±25.52 |
| 678.47 | <1×10−6 | 2.91±1.34 | 22.32±13.36 |
| 703.43 (SM) | <1×10−6 | 2.50±1.00 | 40.01±28.42 |
| 729.45 (SM) | <1×10−6 | 3.28±1.60 | 16.56±8.60 |
| 731.48 (SM) | <1×10−6 | 18.51±15.14 | 85.24±45.36 |
| 741.54 (SM) | <1×10−6 | 7.82±4.55 | 10.49±5.70 |
| 757.57 (SM) | <1×10−6 | 12.39±4.88 | 19.34±7.93 |
| 781.59 (SM) | <1×10−6 | 10.40±4.58 | 20.64±8.47 |
| 811.62 (SM) | <1×10−6 | 13.46±6.05 | 21.62±11.03 |
GBT, glioma brain tissue; SD, standard deviation; SM, sphingomyelin; TBT, trauma brain tissue.
The expression intensity of the lipid peaks of 798.57, 722.55, 799.56, 769.58 and 783.79 m/z was markedly increased in the TBT samples compared with the GBT samples (Table I). Taking the peak of 798.57 m/z as an example, its expression intensity in normal control TBT samples was as high as 198.68, while its expression in GBT was decreased to 14.34.
Among these lipid peaks, the 758.60, 786.64, 617.07, 703.43 and 731.48 m/z peaks exhibited markedly higher expression intensity in GBT samples compared with TBT samples. Taking the peak of 758.60 m/z as an example, its expression value in GBT was as high as 142.89, while its expression in normal control TBT was decreased to 11.31. Some of the differentially expressed lipid peaks in Tables I and II have the potential to serve as clinical diagnosis biomarkers for the detection and treatment of glioma.
Identification of lipid peaks
A total of 31 lipid peaks were detected using MALDI-TOF-MS, and 14 of these lipid peaks were identified as sphingomyelin (SM) using the second mass spectrum. These 14 peaks are listed and marked in Tables I and II with the label ‘SM’. The SM peak with the m/z value of 769.58 had an intensity of 94.32 in TBT and the SM peak with the m/z value of 753.60 had an intensity of 70.94 in TBT (Table I). Table III shows the structural data for nine SM peaks.
Table III.
Structural data of various sphingomyelin peaks.
| Biomarker (m/z) | Common name | Chemical composition |
|---|---|---|
| 741.54 | SM(d18:1/16:0) | [C39H79N2O6P+K] 1+ |
| 755.58 | SM(d18:1/17:0) | [C40H81N2O6P+K] 1+ |
| 757.57 | SM(d18:0/17:0) | [C40H83N2O6P+K] 1+ |
| 769.58 | SM(d18:1/18:0) | [C41H83N2O6P+K] 1+ |
| 771.57 | SM(d18:0/18:0) | [C41H85N2O6P+K] 1+ |
| 797.60 | SM(d18:1/20:0) | [C43H87N2O6P+K] 1+ |
| 811.62 | SM(d16:1/23:0) | [C44H89N2O6P+K] 1+ |
| 783.79 | SM(d18:1/19:0) | [C42H85N2O6P+K] 1+ |
| 729.45 | SM(d18:0/15:0) | [C38H79N2O6P+K] 1+ |
Verification of the expression of lipid peaks with MALDI-imaging
A total of 14 peaks were identified as SM. Due to the important role of SM in cell signal transduction, verification of the expression of SM peaks with MALDI-imaging was performed. Fig. 1 shows the MALDI-imaging results of four SM peaks. The sample patterns on the left are the MALDI-imaging results from TBT samples and the sample patterns on the right are the MALDI-imaging results from GBT samples. The expression intensities of the SM peaks with m/z values of 769.58, 771.57, 797.60, 783.79 and 753.60 were higher in the normal control TBT samples compared with in the GBT samples. The expression intensities of the SM peaks with m/z values of 741.54, 781.59, 759.60, 757.57, 731.48 and 703.43 were higher in GBT samples compared with in normal control TBT samples. The results of the MALDI-imaging experiments were consistent with the expression intensity results from the MALDI-TOF-MS experiments.
Figure 1.

MALDI-imaging results of sphingomyelin peaks in GBT and normal control TBT samples. MALDI-imaging results of 741.54, 759.60, 769.58 and 771.57 m/z are shown. GBT, glioma brain tissue; TBT, trauma brain tissue.
Regulation and control of the expression properties of insulin-like growth factor-1 receptor (IGF-1R) and the PI3K/AKT pathway
Firstly, SM biomarkers, which were differentially expressed between GBT and TBT samples, were identified. Subsequently, the regulation of IGF-1R and the PI3K/AKT pathway by ceramide were investigated. The regulatory effect of ceramide on IGF-1R and the PI3K/Akt pathway was investigated using the glioma cell line U373-MG.
Distribution of ceramide and IGF-1R on the cell membrane
The distribution characteristics of ceramide and IGF-1R on the cell membrane were observed by immunostaining. Fig. 2A shows the distribution of ceramide, as revealed by immunostaining. It was identified that ceramide was made up of lipid rafts, and large quantities of ceramide aggregated to form an areatus-like distribution. Fig. 2B shows the live staining pattern of IGF-1R relative to ceramide. It was revealed that IGF-1R formed an areatus-like distribution that was similar to the distribution of ceramide.
Figure 2.
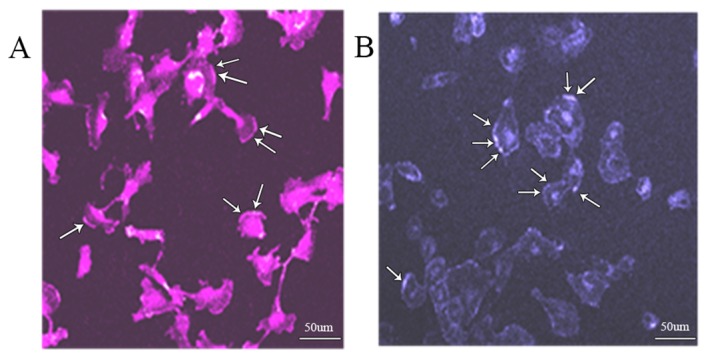
Distribution of ceramide and IGF-1R on the cell membrane. (A) Distribution of ceramide was obtained by immunostaining. (B) Distribution of IGF-1R was obtained by live staining. IGF-1R, insulin-like growth factor-1 receptor. Scale bar, 50 µm.
Observation of the movement of IGF-1R with live staining technology
The movement of IGF-1R, a ceramide receptor (19), on the cell membrane was recorded with live staining technology. Fig. 3 illustrates the movement of IGF-1R, as visualized with live cell staining technology. The cell dish was placed into a CO2 incubator and observed via confocal microscopy at 37°C with 5% CO2. The recording times were 0, 5, 10 and 20 min. At 0 min, IGF-1R fluorescence was observed on the cell membrane, whereas IGF-1R fluorescence in the inner area of some cells was not clear. As the recording time increased, the IGF-1R fluorescence in the inner area of cells increased. When the recording time reached 20 min, the inner area of numerous cells presented high fluorescence intensity. IGF-1R fluorescence was indicated with arrows in Fig. 3.
Figure 3.
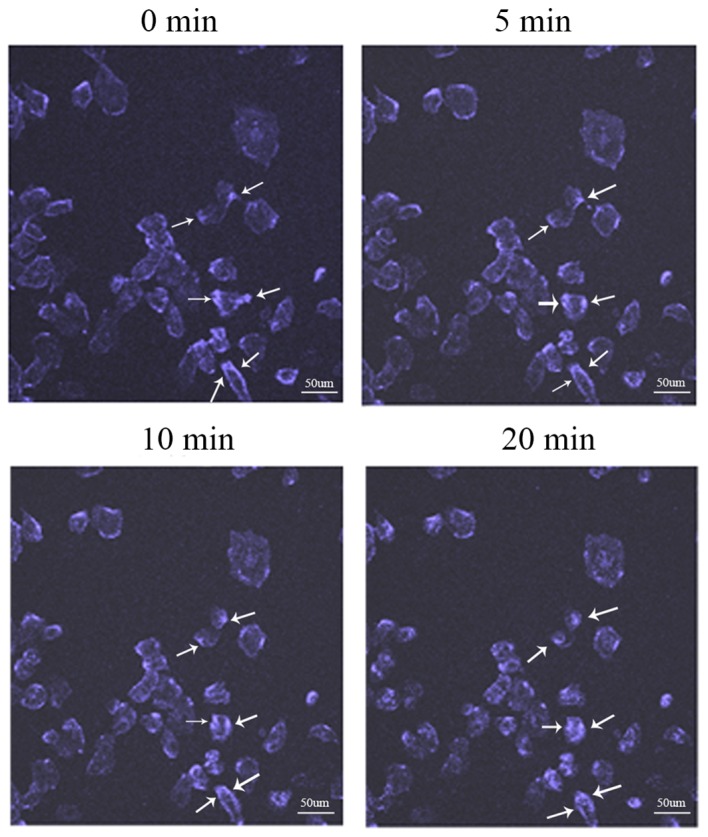
Movement of IGF-1R. The distribution status of IGF-1R on the cell membrane was recorded by live staining technology. The movement track of IGF-1R can be observed with the laser scanning confocal microscopy at 0, 5, 10 and 20 min. IGF-1R, insulin-like growth factor-1 receptor. Scale bar, 50 µm.
Regulation of the expression of IGF-1R
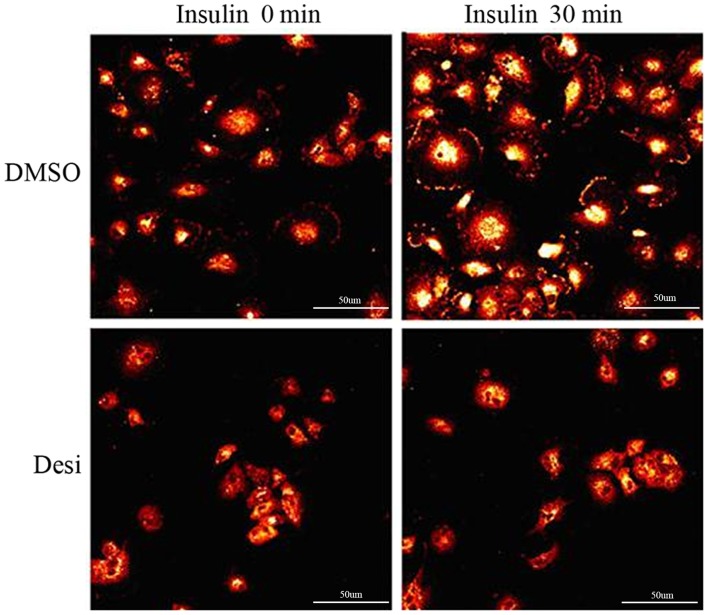
Data from Fig. 4 indicated that the expression level of phosphorylated IGF-1R was markedly stimulated by insulin when the glioma cell line U373-MG was treated with DMSO as a control. Compared with the fluorescence intensity at 0 min in the DMSO group and the fluorescence intensity at 0 min in the Desi group, the expression level of phosphorylated IGF-1R was significantly decreased when U373-MG cells were treated with Desi. In the DMSO group, the fluorescence intensity of phosphorylated IGF-1R was higher when the cells were stimulated with insulin for 30 min. In the Desi group, the fluorescence intensity of phosphorylated IGF-1R was not markedly increased when cells were further stimulated with insulin for 30 min.
Figure 4.
Expression of IGF-1R on the cell surface. The immunostaining results of two groups of glioma U373-MG cells are shown. Cells in the two groups were treated with insulin. The influence of drug Desi on IGF-1R expression was observed by comparing the two groups. Desi, desipramine; IGF-1R, insulin-like growth factor-1 receptor. Scale bar, 50 µm.
Regulation of the PI3K/Akt pathway
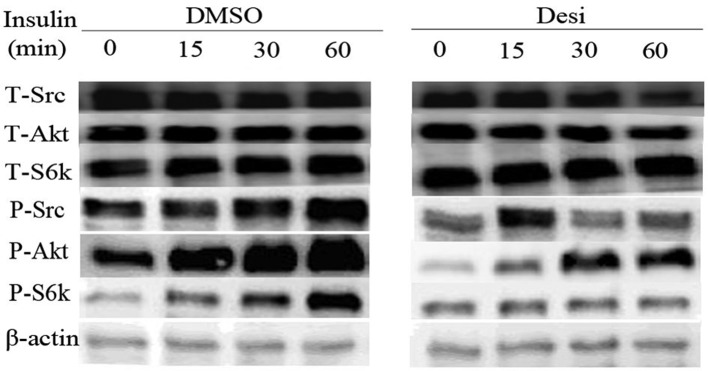
Results on the PI3K/Akt pathway regulation are presented in Fig. 5. The expression intensity of phosphorylated-(P-)Akt, P-SRC proto-oncogene non-receptor tyrosine kinase (Src) and P-ribosomal protein S6 kinase B1 (S6k) in DMSO group increased gradually with increasing time of treatment with insulin. Furthermore, the influence of insulin in Desi group was weakened by the application of Desi. Taking the P-Akt as an example, its quantitative result is shown in Fig. 6. The relative intensity of P-Akt in group DMSO at 0 min is 0.70, while that in group Desi decreases to 0.16. Furthermore, the relative intensity of P-Akt in group Desi are less than that in group DMSO at 15, 30 and 60 min.
Figure 5.
Expression levels of proteins associated with the PI3K/Akt pathway. Western blotting results of phosphorylated and total Src, S6k and Akt levels. U373-MG cells were treated with insulin for 0, 15, 30 and 60 min. One group of cells was treated with DMSO while the other group was treated with Desi. P-, phosphorylated-; S6k, ribosomal protein S6 kinase B1; Src, SRC proto-oncogene non-receptor tyrosine kinase; T-, total-; Desi, desipramine.
Figure 6.
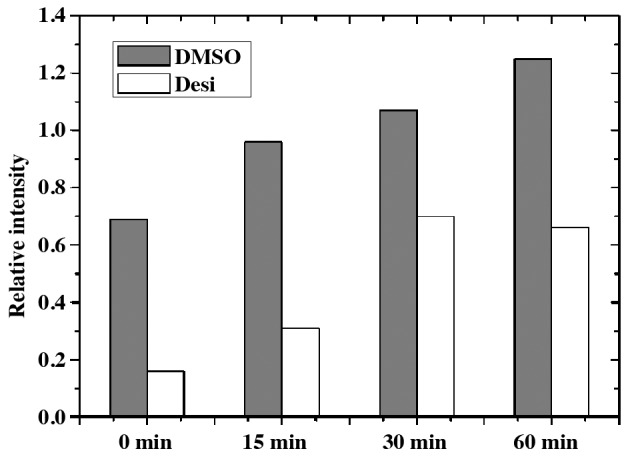
Western blotting quantitative result for P-Akt. U373-MG cells were treated with insulin for 0, 15, 30 and 60 min. One group of cells was treated with DMSO whereas the other group was treated with Desi. Desi, desipramine.
Discussion
In the present study, MALDI-imaging and MALDI-TOF-MS were used to investigate the differences in the expression levels of lipid biomarkers in GBT and TBT samples. A total of 31 lipid peaks with differential expression properties were detected in the GBT and TBT samples. A total of 15 lipid peaks exhibited higher expression intensity in TBT compared with GBT samples. Furthermore, 14 lipid peaks were identified as sphingomyelin peaks. As previously reported, sphingomyelin can be hydrolyzed into ceramide via acid sphingomyelinase (ASM) (20,21). The results demonstrated that ceramide and IGF-1R had similar membrane distribution. The live cell staining results indicated that IGF-1R slowly entered the cell from the cell membrane, resulting in increased IGF-1R fluorescence in the inner area of cell. These results were in accordance with a previous study that reported that ceramide can bind to IGF-1R and is involved in cell signal transduction (22). The drug Desi was able to inhibit the accumulation of ceramide on the cell membrane. Insulin was able to stimulate IGF-1R expression, although IGF-1R expression was inhibited by treatment with Desi. Similar results were also observed for the expression levels of proteins P-Src, P-Akt and P-S6k from the PI3K/Akt signaling pathway. Previous studies reported that the dysregulation of the IGF-1R signaling pathway results in the development of malignant gliomas (19,23,24). In addition, glioma cell senescence and apoptosis can be influenced by the activity of the PI3K/Akt signaling pathway (25,26).
Due to the difficulty of obtaining brain tissues, only three GBT samples and three normal control TBT samples were collected and used in the present study. For MALDI-imaging, it is advantageous to obtain cancer tissues and normal tissues from the same individual; however, it is difficult to perform experiments on glioma tissue and normal brain tissue from the same person. Therefore, the brain tissues for glioma research were usually collected from different individuals (27–29). The results of the present study were based on a limited sample size, and the findings would be more valuable and reliable if they were based on a larger sample size. These first-stage results may serve as the research foundation for further studies. There are several topics requiring further investigation, including how the opposite alterations in the same type of lipid biomarkers may be used to form a diagnostic pattern for patients with glioma. Additionally, it should be investigated how to predict the functional effect of these differentially expressed lipid biomarkers between GBT and normal TBT by pathway enrichment analysis. In the future, these issues will be addressed in a study with a larger sample size.
In conclusion, the present study demonstrated that sphingomyelin peaks may be identified as biomarkers in GBT samples compared with normal control TBT samples. Furthermore, insulin could stimulate the high expression of ceramide and of P-Akt from the PI3K/Akt signaling pathway. However, Desi inhibited the increase in ceramide and P-Akt expression. Future study will investigate the association between these sphingomyelins and the survival rate of patients with glioma, and the expression intensity variation of these sphingomyelin along with the clinical treatment process of patients with glioma.
Acknowledgements
The authors would like to thank Dr Zuogang Guo (China Southern Power Grid Company Limited) for critically evaluating this manuscript and technical assistance.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 30901731) and the Natural Science Foundation of Guangdong Province (grant no. 2016A030310187).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XZ collected the samples and performed the study. JX participated in the data analysis and manuscript preparation. JY provided support in experimental design and manuscript preparation. HS and SZ contributed to the conception and design of this research plan. HS and SZ also participated in the research data discussion, and critically revised this manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang University and The Second Affiliated Hospital of Zhejiang University. Written informed consent was obtained from participants for the use of their tissues in the present study.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 2.Ohashi R, Matsuda Y, Ishiwata T, Naito Z. Downregulation of fibroblast growth factor receptor 2 and its isoforms correlates with a high proliferation rate and poor prognosis in high-grade glioma. Oncol Rep. 2014;32:1163–1169. doi: 10.3892/or.2014.3283. [DOI] [PubMed] [Google Scholar]
- 3.Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: Diagnostic and therapeutic implications. Acta Neuropathol. 2014;128:573–581. doi: 10.1007/s00401-014-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JJ, Wen PY. Emerging targeted therapies for glioma. Expert Opin Emerg Drugs. 2016;21:441–452. doi: 10.1080/14728214.2016.1257609. [DOI] [PubMed] [Google Scholar]
- 5.Yun DP, Wang YQ, Meng DL, Ji YY, Chen JX, Chen HY, Lu DR. Actin-capping protein CapG is associated with prognosis, proliferation and metastasis in human glioma. Oncol Rep. 2018;39:1011–1022. doi: 10.3892/or.2018.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ. Perfusion MRI as a diagnostic biomarker for differentiating glioma from brain metastasis: A systematic review and meta-analysis. Eur Radiol. 2018;28:3819–3831. doi: 10.1007/s00330-018-5335-0. [DOI] [PubMed] [Google Scholar]
- 7.Su L, Yang K, Li S, Liu C, Han J, Zhang Y, Xu G. Enolase-phosphatase 1 as a novel potential malignant glioma indicator promotes cell proliferation and migration. Oncol Rep. 2018;40:2233–2241. doi: 10.3892/or.2018.6592. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Xu L, Bao Z, Xu P, Chang H, Wu J, Bei Y, Xia L, Wu P, Cui G. High expression of PIWIL2 promotes tumor cell proliferation, migration and predicts a poor prognosis in glioma. Oncol Rep. 2017;38:183–192. doi: 10.3892/or.2017.5647. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Wang Y, Zhang YT, Geng YP, Si LS, Wang YL. Proteomics-based analysis of a pair of glioma cell lines with different tumor forming characteristics. Neurosci Lett. 2006;401:59–64. doi: 10.1016/j.neulet.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, Liu M, Zhang JY, Yao YL, Wang YX, Lin Y, Song K, Tan J, Wu JR, Cui YH, et al. Capillary morphogenesis protein 2 is a novel prognostic biomarker and plays oncogenic roles in glioma. J Pathol. 2018;245:160–171. doi: 10.1002/path.5062. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi T, Hayakawa Y, Koh M, Tomita T, Nagai S, Kashiwazaki D, Sugimori M, Origasa H, Kuroda S. Impact of a novel biomarker, T-LAK cell-originating protein kinase (TOPK) expression on outcome in malignant glioma. Neuropathology. 2018;38:144–153. doi: 10.1111/neup.12446. [DOI] [PubMed] [Google Scholar]
- 12.Ribom D, Westman-Brinkmalm A, Smits A, Davidsson P. Elevated levels of alpha-2-Heremans-Schmid glycoprotein in CSF of patients with low-grade gliomas. Tumour Biol. 2003;24:94–99. doi: 10.1159/000071082. [DOI] [PubMed] [Google Scholar]
- 13.Iwadate Y, Sakaida T, Hiwasa T, Nagai Y, Ishikur H, Takiguchi M, Yamaura A. Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 2004;64:2496–2501. doi: 10.1158/0008-5472.CAN-03-1254. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz M, Meyer B, Wirnitzer B, Hopf C. Standardized processing of MALDI imaging raw data for enhancement of weak analyte signals in mouse models of gastric cancer and Alzheimer's disease. Anal Bioanal Chem. 2015;407:2255–2264. doi: 10.1007/s00216-014-8356-9. [DOI] [PubMed] [Google Scholar]
- 15.Wildburger NC, Wood PL, Gumin J, Lichti CF, Emmett MR, Lang FF, Nilsson CL. ESI-MS/MS and MALDI–IMS localization reveal alterations in phosphatidic acid, diacylglycerol, and DHA in glioma stem cell xenografts. J Proteome Res. 2015;14:2511–2519. doi: 10.1021/acs.jproteome.5b00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberlin LS, Liu X, Ferreira CR, Santagata S, Agar NY, Cooks RG. Desorption electrospray ionization then MALDI mass spectrometry imaging of lipid and protein distributions in single tissue sections. Anal Chem. 2011;83:8366–8371. doi: 10.1021/ac202016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrivas K, Hayasaka T, Goto-Inoue N, Sugiura Y, Zaima N, Setou M. Ionic matrix for enhanced MALDI imaging mass spectrometry for identification of phospholipids in mouse liver and cerebellum tissue sections. Anal Chem. 2010;82:8800–8806. doi: 10.1021/ac102422b. [DOI] [PubMed] [Google Scholar]
- 18.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian HW, Zhou Y, Jian ZH, Liu RZ. MiR-323-5p acts as a tumor suppressor by targeting the insulin-like growth factor 1 receptor in human glioma cells. Asian Pac J Cancer Prev. 2014;15:10181–10185. doi: 10.7314/APJCP.2014.15.23.10181. [DOI] [PubMed] [Google Scholar]
- 20.Lin M, Liao W, Dong M, Zhu R, Xiao J, Sun T, Chen Z, Wu B, Jin J. Exosomal neutral sphingomyelinase 1 suppresses hepatocellular carcinoma via decreasing the ratio of sphingomyelin/ceramide. FEBS J. 2018;285:3835–3848. doi: 10.1111/febs.14635. [DOI] [PubMed] [Google Scholar]
- 21.Catapano ER, Natale P, Monroy F, López-Montero I. The enzymatic sphingomyelin to ceramide conversion increases the shear membrane viscosity at the air-water interface. Adv Colloid Interface Sci. 2017;247:555–560. doi: 10.1016/j.cis.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, Lowry PR, Zhou X, Depry C, Wei Z, Wong GW, Zhang J. PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proc Natl Acad Sci USA. 2011;108:14509–14514. doi: 10.1073/pnas.1019386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q, Zhang J, Cui Q, Li X, Gao G, Wang Y, Xu Y, Gao X. GSK1904529A, an insulin-like growth factor-1 receptor inhibitor, inhibits glioma tumor growth, induces apoptosis and inhibits migration. Mol Med Rep. 2015;12:3381–3385. doi: 10.3892/mmr.2015.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arun S, Ravisankar S, Vanisree AJ. Implication of connexin30 on the stemness of glioma: Connexin30 reverses the malignant phenotype of glioma by modulating IGF-1R, CD133 and cMyc. J Neurooncol. 2017;135:473–485. doi: 10.1007/s11060-017-2608-4. [DOI] [PubMed] [Google Scholar]
- 25.Liao YX, Zhang ZP, Zhao J, Liu JP. Effects of fibronectin 1 on cell proliferation, senescence and apoptosis of human glioma cells through the PI3K/Akt signaling pathway. Cell Physiol Biochem. 2018;48:1382–1396. doi: 10.1159/000492096. [DOI] [PubMed] [Google Scholar]
- 26.Nicoletti NF, Erig TC, Zanin RF, Pereira TC, Bogo MR, Campos MM, Morrone FB. Mechanisms involved in kinin-induced glioma cells proliferation: The role of ERK1/2 and PI3K/Akt pathways. J Neurooncol. 2014;120:235–244. doi: 10.1007/s11060-014-1549-4. [DOI] [PubMed] [Google Scholar]
- 27.Scheidegger R, Wong ET, Alsop DC. Contributors to contrast between glioma and brain tissue in chemical exchange saturation transfer sensitive imaging at 3 Tesla. Neuroimage. 2014;99:256–268. doi: 10.1016/j.neuroimage.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Fan Y, He M, Ma X, Song Y, Liu M, Xu J. Accuracy of Raman spectroscopy in differentiating brain tumor from normal brain tissue. Oncotarget. 2017;8:36824–36831. doi: 10.18632/oncotarget.15975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krafft C, Sobottka SB, Schackert G, Salzer R. Analysis of human brain tissue, brain tumors and tumor cells by infrared spectroscopic mapping. Analyst. 2004;129:921–925. doi: 10.1039/b408934k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.