Abstract
Owing to the development of new technologies, the epigenome, a second dimensional method for genome analysis has emerged. Epigenetic mechanisms, including DNA methylation, histone modifications and noncoding RNAs, regulate gene expression without changing the genetic sequence. These epigenetic mechanisms normally modulate gene expression, trans-generational effects and inherited expression states in various biological processes. Abnormal epigenetic patterns typically cause pathological conditions, including cancers, age-related diseases, and specific cartilage and bone diseases. Facing the rapidly developing epigenetic field, we reviewed epigenetic mechanisms and their involvement with the skeletal system and their role in skeletal development, homeostasis and degeneration. Finally, we discuss the prospects for the future of epigenetics.
1. Introduction
The epigenome is a multitude of chemical compounds that instruct the genome how to function. These functions include DNA methylation, chromatin modifications, nucleosome positioning and alterations in the noncoding RNA (ncRNA) profile (Verma, 2015), which may affect gene expression and its regulation without altering the DNA sequence. With the rapid development of bio-technologies, many unknown epigenetic phenomena have been revealed, which has led to an increasing interest in epigenetic mechanisms of individual development and of diseases.
Epigenetic modulation exerts either a positive or negative feedback pathway, leading to silencing of one of the two X chromosomes in female cells in early development (Gendrel & Heard, 2014), genomic imprinting (Gupta et al., 2014a) and paramutation (Brink, 1973). The epigenetic pattern and changed phenotype appear after mitosis or meiosis (Huang et al., 2014). Although various epigenetic programs have been identified, only three main categories are widely accepted, which are as follows: DNA methylation, histone modification and ncRNAs. DNA methylation is the most studied; this includes the methylation of the fifth carbon of cytosine. The DNA methylation pattern is dynamically regulated by DNA methyltransferases (DNMTs) during development. DNA methylation pattern includes endogenous transposable elements repression, chromosome alignment and segregation, second X chromosome control via inactivation in females and modulation of imprinted gene expression. Histone modifications are characterized by histones that can be covalently modified at their flexible N- or C-terminal tails, as well as globular domains. This phenomenon is associated with DNA methylation. It is also regarded as one of the key components of chromatin packaging (Henikoff & Shilatifard, 2011). Histones function both positively and negatively in gene expression regulation; histones are also mainly governed by post-translational histone modifications (PTMs) and specific histone variants (Kimura, 2013). PTMs regulate transcription and other DNA-templated functions, which are dynamically mediated by specific modifying enzymes (Fan et al., 2015). PTMs can be classified into several categories, including lysine acetylation, lysine and arginine methylation, arginine citrullination, lysine ubiquitination, phosphorylation, fatty acylation and ADP-ribosylation, which affect DNA function individually or collectively. NcRNAs are comprised of short and long ncRNAs. These molecules have been highlighted in biological processes with the development of deep sequencing and transcriptome analyses; however, these molecules have been previously regarded as junk RNAs.
Therefore, epigenetic programs are essential to basic biologic events that are associated with physiological and pathological processes, including skeletal genesis, bone remodelling and bone metabolic disorders (Fan et al., 2015). This review will discuss the current knowledge of epigenetics in the skeletal system and will strive to shed new light on the understanding of epigenetic roles in bone and cartilage tissues.
2. DNA methylation
DNA methylation is the methylation of the fifth carbon of cytosine, which is known as one of the most important epigenetic modifications. It plays a considerable role in genome stability, gene expression and individual development in both prokaryotes and eukaryotes.
DNA methylation, which occurs on gene promoters, is linked to transcriptional repression (Chae et al., 2013). Although it happens mostly in CpG sites, non-CpG sites have also been observed to be methylated. However, the role of non-CpG methylation remains unclear. Conversely, gene enhancers that have undergone DNA methylation are correlated with active gene expression (Moore et al., 2013). Except for DNA methylation of promoters and enhancers, DNA methylation emerges on different genomic regions with different functions. DNA methylation, which occurs on intergenic regions, represses the expression of potentially harmful genetic elements when the methylation of the CpG islands impairs the binding of transcription factors, the recruiting of repressive methyl-binding proteins and gene silencing. However, more studies are needed to determine how DNA methylation of the gene body contributes to gene regulation.
DNA methylation is accomplished by DNMTs, including these five enzymes: DNMT1, DNMT3A, DNMT3B, DNMT3L and DNMT2. DNMTs have been reported to be involved in the maintenance and de novo of DNA methylation (Uysal et al., 2015). For example, DNMT1 mainly maintains DNA methylation patterns (Elliott et al., 2015). On the other hand, DNMT3A and DNMT3B are essential for de novo methylation in early development (Okano et al., 1999). DNMT3L cooperates with both DNMT3A and DNMT3B and enhances the initiation of DNA methylation (Cheng & Blumenthal, 2008). The fifth DNA methyltransferase, DNMT2, targets RNA methylation in mammals, rather than participating in genome methylation. This phenomenon modifies the 38th cytosine residue in the anticodon loop of certain tRNAs and enhances the stability of tRNAs (Ashapkin et al., 2016). For example, DNMT2 affects polypeptide synthesis during haematopoiesis by modulating the stability and fragmentation of tRNAs (Tuorto et al., 2015).
DNA methylation changes the nucleic acid structure and the gene phenotype when it occurs at a certain gene region, which makes DNA methylation a potential new biomarker in biological research (Lee et al., 2016). The earliest approach that detected DNA methylation was through the quantification of total methylated cytosines in a chunk of DNA (Umer & Herceg, 2013). With development in technology, genome-wide analyses, such as next-generation sequencing (NGS) technologies, have been widely used in genome-wide and locus-specific DNA methylation analyses. Particularly, novel approaches emerge with pertinence, sensitivity and speed. For example, the global estimation of 5-methylcytosine content can be detected by high-performance capillary electrophoresis with UV-V detection, liquid chromatography with electrospray ionization mass spectrometric detection and LUminometric Methylation Assay (Berdasco et al., 2009). These methods provide information about the disease process and progress and can be useful in various clinical settings and in drug screening (Umer & Herceg, 2013). Locus-specific DNA methylation analysis provides insights into early epigenetic reprogramming events and identifies rare cells with unique epigenetic signatures (Cheow et al., 2015). Many methods are available, including methylation-specific PCR, MethyLight, combined bisulfite conversion restriction analysis, bisulfite (Sanger) sequencing, bisulfite pyrosequencing and methylation-sensitive high-resolution melting (Umer & Herceg, 2013). Although genome-wide and locus-specific analyses provide a comprehensive understanding of DNA methylation, higher coverage and accuracy are required to detect DNA methylation. Firstly, restriction landmark genomic scanning, methylation-specific arbitrarily primed PCR, methylation-sensitive representational differential analysis and amplification of intermethylated sites are widely used, with some inevitable limitations for samples with large quantities of DNA. These approaches are labour-intensive and include complex procedures. Furthermore, microarray-based (DMH, CHARM, HELP assays, MeDIP-Chip and Illumina Infinium) and sequencing-based approaches (bisulfite treatments [BS-Seq, MethylC-Seq, reduced representational bisulfite sequencing (RRBS) and methyl CpG-binding domain (MBD)-isolated genome sequencing (MIGS)]) enhance the sensitivity, simplify the experimental setup and reduce the sequence redundancy in genome-scale DNA methylation analysis (Umer & Herceg, 2013). In addition to these widely used DNA methylation detection methods, some novel approaches have subsequently emerged, which include an electrochemical detection system and a methyl-sensitive fluorescence polarization assay. The former technique targets MBD and a glucose dehydrogenase-fused zinc finger protein (Lee et al., 2016). The latter technique recognizes the palindromic target sequence CCGG through restriction endonucleases, namely, MspI and HpaII (Shiratori et al., 2016). The developments in DNA sequencing technologies, as well as methods to identify and map 5-hydroxymethylcytosine are expected to augment our current understanding of epigenomics.
3. Histone modifications
Eukaryotic DNA is tightly packaged within the 2–10 µm nucleus, which requires several metres of DNA to be compact (Rothbart & Strahl, 2014). These packages are mainly comprised of core nucleosome particles, which are formed of 147 DNA base pairs wrapped around an octamer of histones (two copies each of H2A, H2B, H3 and H4). The central histones modulate the function and dynamics of the chromatin by the presence of specific histone variants and PTMs (Biterge & Schneider, 2014).
On the other hand, histone variants are the key players in the shape of chromatin structure and the regulation of fundamental cellular processes, such as chromosome segregation and gene expression (Vardabasso et al., 2014). Histone variants replace the canonical histones to change DNA expression timings (DNA replication independent) and mRNA characteristics (Biterge & Schneider, 2014). This phenomenon alters the stability, dynamics and accessibility of DNA (Weber & Henikoff, 2014), More importantly, histone variants play important roles in disease progression in certain cases. For example, MacroH2A is a histone variant that is overexpressed in patients with Huntington's disease and steatosis-associated hepatocellular carcinoma (Biterge & Schneider, 2014). H3.3 is the variant of H3 that replaces H3K27me3 in paediatric glioma; it is associated with reduced survival in patients with paediatric glioma (Chan et al., 2013). Furthermore, subsequent studies have reported that other core histone variants, namely H2A.X, H2A.Z, CENP-A and linker histone H1 variants, are linked to biological development of diseases (González-Romero et al., 2012).
For PTMs, covalent modification occurs in histones at their flexible N- or C-terminal tails, as well as globular domains. PTMs also play important roles in many biological processes, including in DNA stability and expression. With the advancement of approaches to biochemical systems, PTMs can be classified into the following categories: lysine acetylation, lysine and arginine methylation, arginine citrullination, lysine ubiquitination, phosphorylation, fatty acylation and ADP ribosylation. These processes are precisely modulated by enzymes that transfer specific chemical groups to implement different modification. Histone acetylation is catalysed by diverse enzymes, in which two converse enzymes, namely, histone acetyltransferases and deacetylases, modulate the acetylation status of histones (Bannister & Kouzarides, 2011). Additionally, a large number of histone methyltransferases catalyse the methylation on the ε-amino group of lysine residues and form the most prevalent histone methylation (Fan et al., 2015). PTMs have diverse functions with enzyme involvement. For example, histone acetylation, which is the so-called opening up of chromatin, makes DNA more accessible to other protein factors (Fan et al., 2015). Histone methylation level is associated with activating transcription, resulting in different chromatin states, influencing aging and aging phenotypes (McCauley & Dang, 2014; Berr et al., 2016). Furthermore, cross-talk between different histone modifications, which may constitute a histone code, have been observed, which may help fine tune overall control (Bannister & Kouzarides, 2011).
Thousands of experiments have provided enormous data repositories in terms of the genome-wide binding pattern of modified histones by ChIP-seq (Rivera & Ren, 2013). Although ChIP-seq is the gold standard for mapping PTMs, limited resolution, dependence on antibodies and the need for large amounts of starting material have limited its application. To break the limitations, researchers have proposed a ChIP-exo technique to provide a single bp accuracy, in which an exonuclease precisely trims the ChIP DNA of the cross-linking site into a small fragment (Rhee & Pugh, 2011). Furthermore, a nano-ChIP-seq protocol, which is combined with a high-sensitivity small-scale ChIP assay and a tailored procedure, generates high-throughput sequencing libraries from scarce amounts of ChIP DNA (Adli & Bernstein, 2011). This nano-ChIP-seq does not only decrease the need for starting materials, but it also makes the entire procedure faster. Additionally, many novel approaches have emerged with comprehensiveness, high-resolution and short setup features, including ChIP-bisulfite-sequencing (ChIP-BS-seq) (Brinkman et al., 2012), bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) (Challen et al., 2014) and other high-resolution mass spectrometry assays (Lin & Garcia, 2012).
4. NcRNAs
Previously, mRNA function had been mainly considered to provide protein-coding information only (Kumari & Sampath, 2015). With the development of deep sequencing and transcriptome analysis, only a tiny portion of the biological genome corresponds to protein-coding sequences; in addition, most genomic loci produce large transcripts rather than proteins, which are defined as ncRNAs (Perez et al., 2013). Although these ncRNAs had been regarded as junk RNAs for a long time, they have now been identified to have a role in many physiological processes, including the maintenance of self-renewal, direction of cell lineage (Guan et al., 2013) and expression of hundreds of genes (Varela et al., 2013).
NcRNAs are classified into the following two categories based on their length: small and long ncRNAs (lncRNAs) (Hirose et al., 2014). Small ncRNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). MiRNAs are one of the most widely studied small ncRNAs. A total of 52% of miRNAs are located in human intergenic regions, 40% lie within the intrinsic regions of genes and the final 8% are exonic (Hsu et al., 2006). Mature miRNAs have similar physiological roles to other RNAs, such as transcriptional activation and inhibition, epigenetic repression and controlled degradation rates (Mott & Mohr, 2015). Furthermore, by targeting the complementary sequence of miRNAs, they modulate over 60% of the translation of protein-coding genes (Mott & Mohr, 2015). Not only in the modulation of gene translation, miRNA also present intricate functions in different tissues (Guo et al., 2010). For example, miR-125b up-regulates and displays oncogenic potential in colon cancer and haematopoietic tumours. Conversely, miR-125b down-regulation contributes to malignant transformation in mammary tumours and hepatocellular carcinoma (Banzhaf-Strathmann & Edbauer, 2014). siRNAs are usually paired with mRNAs with perfect complementarity and are directed in their endonucleolytic cleavage and destruction (Hirose et al., 2014). On the other hand, piRNAs play a pivotal role in germline genome protection from transposons (Hirose et al., 2014). lncRNAs are the second class of ncRNAs, which regulate the RNA and protein content of a cell on the transcriptional and post-transcriptional level (Melissari & Grote, 2016). These molecular mechanisms include dosage compensation, chromatin regulation, genomic imprinting and nuclear organization (Yan et al., 2016). In addition, a small number of lncRNAs modulate the recruitment of RNA polymerase II and induce chromatin remodelling in global or local gene expression in trans or cis (Shi et al., 2013). Therefore, lncRNAs are present in pathological conditions such as cancer and cardiovascular disease, which makes lncRNAs a novel potential biomarker for clinical usage (Schmitz et al., 2016).
Moreover, a small part of ncRNAs codes for proteins including early nodulin 40 and MtHAP2-1 in plants. At the same time, some mRNAs may modulate post-transcriptional level gene expression, which is independent of their encoded proteins. These two kinds of RNAs are called cncRNAs (Kumari & Sampath, 2015), which have shed new light on the understanding of ncRNAs. A total of 300 alternatively spliced bifunctional RNAs may be observed in the human genome (Ulveling et al., 2011). The exploration of protein-coding and noncoding functions for cncRNA loci may be highlighted in future studies (Kumari & Sampath, 2015).
NcRNA realization has been expanded with the advent of NGS (Ilott & Ponting, 2013). In particular, lncRNAs have been identified by RNA-seq technology (Li et al., 2014). Moreover, RNA-seq data integration detects active transcription to further understand the reported lncRNAs (Chen et al., 2016 a). These findings may help to discover novel lncRNAs and may improve the characterization of identified lncRNAs.
5. Bone and cartilage diseases associated with epigenetics
Mesenchymal stromal cells (MSCs) are typical adult progenitor cells, which possess multidifferentiation capacity in vitro and in vivo (Takahashi et al., 2015). MSCs are one of the promising candidate cells for skeletal regeneration due to their convenient isolation and immune-modulatory capability. The epigenetic changes of MSCs are essential in the differentiation of MSCs into bone and cartilage. These processes include DNA methylation, histone modifications and miRNAs.
(i). Osteogenic and chondrocyte differentiation of MSCs by epigenetics
A coordinated cascade of transcription factors and epigenetic modifications drive gene transcription, and cause specific cell fate, and this is indispensable for terminal differentiation of multipotent stem cells (Meyer et al., 2016). For example, the osteogenic and adipogenic differentiation of MSCs are partly regulated by transcription factors, including peroxisome proliferator-activated receptor-γ (PPAR-γ) and Runt-related transcription factor 2 (Runx2) (James, 2013), and various epigenetic alterations (Glemžaitė & Navakauskienė, 2016). These processes are accompanied by a loss of the Brachyury gene. Brachyury inactivation is associated with the methylation of its promoter, which represses stem cell-associated genes (Dansranjavin et al., 2009). In addition to the Brachyury gene, the cytosine methylation accumulation at the endogenous thyroid hormone receptor interactor 10 (Trip10) promoter reduces Trip10 expression, which accelerates osteogenic differentiation (Hsiao et al., 2010). Osteocalcin (OC) is a non-collagenous bone matrix protein, which is partly regulated by DNA methylation and histone modifications. In MSC osteogenic differentiation in vitro, OC expression is usually increased with the decrease in DNA methylation. Additionally, OC is activated by the accumulation of H3 and H4 acetylation. Conversely, OC is inhibited by the decrease in H3 and H4 acetylation in the OC promoter and coding regions. This event occurs in the proliferative period of osteogenic differentiation (Takahashi et al., 2015). For the H3 sub-family, the acetylation levels of H3K9Ac and H3K9me2 are associated with the activation and silencing genes, respectively, which are modulated by vitamin D receptors at specific gene promoters (Tan et al., 2009). For the epigenetic modulation of miRNAs, the function of miRNA has been reported in the inhibition of mRNA translation and degradation (Wei et al., 2012; Takahashi et al., 2015). For example, miR-206 overexpression inhibits the differentiation of osteoblasts by the target of connexin 43 (Inose et al., 2009). MiR-34 inhibits mouse osteoblast proliferation and differentiation by targeting SATB2, which is a nuclear matrix protein involved in osteoblast differentiation (Wei et al., 2012). MiR-27a and miR-489 down-regulate the osteoblast differentiation by targeting PEX7; on the other hand, miR-148b up-regulates the differentiation (Schoolmeesters et al., 2009).
The epigenetic mechanisms of chondrocyte differentiation of MSCs are scarce. In cartilage formation, DNA methylation levels of CpG-rich promoters of chondrocyte-specific genes are mostly at a low level (Ezura et al., 2009). SOX9 is one of the master chondrogenic transcription factors, which is involved in chondrocyte differentiation and cartilage formation (Shi et al., 2015). Age-dependent SOX9 expression is regulated by epigenetic mechanisms (Mak et al., 2015). Epigenetic studies revealed that DNA methylation levels increased at specific CpG islands of the Sox9 gene in mice articular chondrocytes (ACs) at 6 and 12 months old (Zhang et al., 2016). Using the supplementation of 5-azacytidine, which is an inhibitor of DNA methylation, the DNA methylation level is reduced in the Sox9 promoter region, which elevates the level of Sox9 expression in ACs. This finding suggests that expression is associated with DNA methylation (Zhang et al., 2016). Moreover, in DNA methylation, many kinds of histone modifications are involved in chondrocyte differentiation. For example, several transcription factors and coactivators, such as Scleraxis/E47 and p300, cooperatively modulate Sox9-dependent transcription through p300-mediated histone acetylation (Furumatsu & Asahara, 2010). And a novel Runx2 enhancer is localized in the primary osteoblasts and is characterized by the presence of the histone variant H2A.Z, which contains sufficient elements to direct Runx2 expression to osteoblasts (Kawane et al., 2014). For the epigenetic modulation of ncRNAs, miR-101 and HOTTIP are up-regulated by targeting DNMT3B, which alters integrin-α1 methylation, which is a key protein of chondrocyte ossification (Kim et al., 2013a).
(ii). Skeletal diseases associated with epigenetics
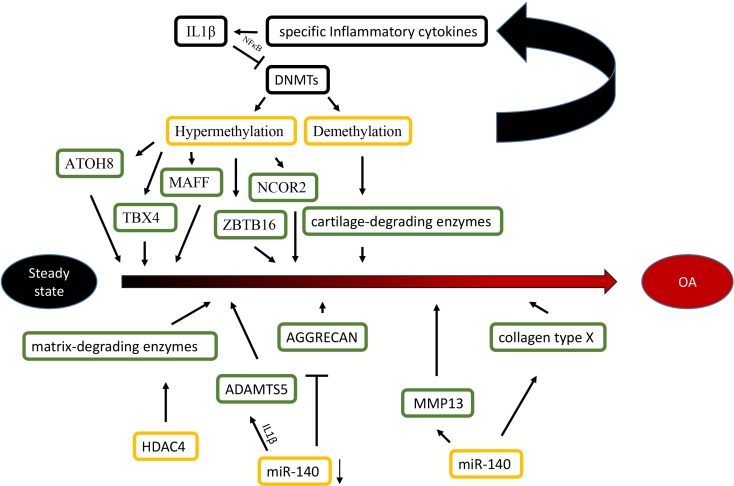
Epigenetic alterations are associated with the aetiology and pathology of bone and cartilage diseases. Osteoarthritis (OA) is a chronic multifactorial disease associated with specific genes. Specific transcription factors (TFs), cartilage-degrading enzymes, pro- and anti-inflammatory cytokines and extracellular matrix proteins contribute to OA development (Zhang & Wang, 2015). More importantly, advanced studies highlight the role of epigenetics in OA, including DNA methylation, histone modifications and miRNAs (Im & Choi, 2013) (Figure 1).
Fig. 1.
Epigenetic mechanisms are associated with OA. Specific inflammatory cytokines were shown to accelerate the development of OA by directly targeting DNA methyltransferases, and the level of DNA methylation modulated the expression of specific inflammatory cytokines in chondrocytes reversely. For histone modifications, the overexpression of HDAC4 significantly led to a release of matrix-degrading enzymes, thus, contributing to bone loss associated with OA. For miRNAs, the reduction of miR-140 played an important role in OA progression by targeting ADAMTS5 and AGGRECAN. Conversely, The overexpression of miR-365 may accelerate the development of OA by targeting MMP13 and collagen type X (Col X).
Differences in the methylome between normal and OA knee articular cartilage have been identified, including 929 differentially methylated sites. Most of the methylated sites in OA (69%) are hypomethylated and enriched among gene enhancers in OA cartilage tissue (Jeffries et al., 2014). For epigenetic changes of TFs, some OA related TFs (ATOH8, MAFF, NCOR2, TBX4, ZBTB16 and ZHX2) are significantly hypermethylated and down-regulated in OA cartilage. These results indicate that the DNA methylation level negatively affects the chondrocyte transcriptome and function in OA pathogenesis (Alvarez-Garcia et al., 2016). For cartilage-degrading enzymes, the demethylation of MMP3, MMP9, MMP13 and ADAMTS4 promoters increase their gene expression in OA cartilage (Reynard, 2016). Additionally, inflammatory mediators modulate DNMTs, DNMT3B and DNMT3A, which target DNA methylation directly (Shen et al., 2017). IL-1β is a proinflammatory cytokine guiding the function of immune and proinflammatory cells and decreases the expression of DNMT3B and DNMT3A through NF-κB signalling (Shen et al., 2017). Another inflammatory chemokine, IL-8, shows increased demethylation in the promoter region in OA chondrocytes, which is correlated with enhanced IL-8 expression (Takahashi et al., 2015). Furthermore, the DNA methylation level modulates the expression of specific inflammatory cytokines in chondrocytes reversely. Significant demethylation of CpG sites in the inflammatory chemokine IL-8 promoter increases the expression of IL-8 in the OA chondrocytes (Takahashi et al., 2015).
With increasing data of genome-wide profiling of DNA methylation in OA, the alterations of DNA methylation affect more gene expression in OA. The demethylation of specific promoter and enhancer sites in GDF5, iNOS and SOST increase their gene expression in OA cartilage and isolated chondrocytes. Conversely, the increased methylation of SOX9, DIO2 and COL9 promoters caused reduced expression in OA cartilage (Iliopoulos et al., 2007; Verma and Dalal, 2011; Kim et al., 2013b; Gupta et al., 2014b; Papathanasiou et al., 2015).
For histone modifications, HDAC4 is one of the key regulators in OA development (Lu et al., 2014). The HDAC4 expression level has a statistically negative correlation with OA severity (Lu et al., 2014). HDAC4 expression reduction significantly leads to a repression of the following matrix-degrading enzymes: MMP1, MMP3, MMP13, ADAMTS4 and ADAMTS5 (Lu et al., 2014). HDAC4 overexpression does not only decrease the expression of IL-1β, Cox2 and iNOS, but it also partially blocks IL-1β mediated effects in catabolic events in human OA chondrocytes (Thompson et al., 2015). However, HDAC4 inhibition has suppressed the expression of inhibited genes (Young et al., 2005). For example, trichostatin A (TSA) and sodium butyrate HDAC inhibitors inhibit cartilage degradation by blocking key MMPs (MMP-1 and MMP-13) and aggrecan-degrading enzymes (ADAMTS4 and ADAMTS5). In addition, HDAC4 inhibitors, including vorinostat, TSA and sodium butyrate, are promising in the potential treatment of OA (Chen et al., 2010; Makki & Haqqi, 2016).
For miRNAs, while normal human articular cartilage expresses miR-140 in chondrocytes, its expression is significantly reduced in OA progression (Zhang et al., 2012). The transfection of miR-140 in chondrocytes down-regulates IL-1β-induced ADAMTS5 expression and alleviates IL-1β-dependent repression of AGGRECAN gene expression (Miyaki et al., 2009). These findings indicate that decreased expression of miR-140 caused the abnormal gene expression profile seen in OA. MiR-365 overexpression in chondrocytes increases the expression of the matrix-degrading enzyme MMP13 and collagen type X, which may accelerate OA development (Yang et al., 2016). Moreover, the interplay of histone modifications and miRNAs have been identified in the aetiology of OA. Subsequently, evidence shows that miR-365 directly targeted HDAC4, which led to HDAC4 expression down-regulation (Yang et al., 2016), which accelerates the development of OA. Furthermore, a study of miR-381 has demonstrated that miR-381 overexpression promotes MMP13 and Runx2 expression by the inhibition of HDAC4, and miR-381 inhibitors increased HDAC4 expression and decreased Runx2 expression (Chen et al., 2016b). This finding provides us with a novel therapeutic method for the treatment of OA. This result may inspire researchers to study inhibitors that block the interaction between histone modifications and miRNAs in order to alleviate the severity of OA.
6. Future direction
This review summarizes the current advances in the study of epigenetics and discusses the epigenetic findings pertaining to the skeletal system. In addition to significant technological developments, emerging epigenetics research may provide new understanding of single genes, specific chromosome regions and the whole genome (Adli & Bernstein, 2011). Moreover, modifications can be induced or inhibited by drugs to alleviate or cure disease; this finding needs further investigation. However, many challenges remain unsolved to fully use this epigenetic information. Thus; these challenges will require further research.
Acknowledgement
The authors thank the National Natural Science Foundation of China (No. 81472078、No. 31670951 and No. 31370992) and Shenzhen Basic Research Grant (No. JCYJ20170307100446585).
References
- Adli M and Bernstein BE (2011) Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nature Protocols 6, 1656–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia O, Fisch KM, Wineinger NE, Akagi R, Saito M, Sasho T, Su AI and Lotz MK (2016) Increased DNA methylation and reduced expression of transcription factors in human osteoarthritis cartilage. Arthritis & Rheumatology 68, 1876–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI and Vanyushin BF (2016) [Dnmt2 is the most evolutionary conserved and enigmatic cytosine DNA methyltransferase in eukaryotes]. Genetika 52, 269–282. [PubMed] [Google Scholar]
- Bannister AJ and Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Research 21, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf-Strathmann J and Edbauer D (2014) Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Communication and Signaling 12, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco MA, Fraga MF and Esteller M (2009) Quantification of global DNA methylation by capillary electrophoresis and mass spectrometry. Methods in Molecular Biology 507, 23. [DOI] [PubMed] [Google Scholar]
- Berr A, Zhang X and Shen WH (2016) [Reciprocity between active transcription and histone methylation]. Biologie Aujourdhui 210, 269–282. [DOI] [PubMed] [Google Scholar]
- Biterge B and Schneider R (2014) Histone variants: key players of chromatin. Cell & Tissue Research 356, 457–466. [DOI] [PubMed] [Google Scholar]
- Brink RA (1973) Paramutation. Annual Review of Genetics 7, 129–152. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A and Stunnenberg HG (2012) Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Research 22, 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H, Park J, Lee SW, Nephew KP and Sun K (2013) Comparative analysis using K-mer and K-flank patterns provides evidence for CpG island sequence evolution in mammalian genomes. Nucleic Acids Research 41, 4783–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, Cullen S, Berg J, Zheng Y, Darlington GJ, Li W and Goodell MA (2014) Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 15, 350–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J and Zhang Z (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes & Development 27, 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJM, Chen LK, Lai YS, Lin YY, Wu DC, Tung YA, Liu KY, Shih HT, Chen YJ, Lin YL, Ma LT, Huang JL, Wu PC, Hong MY, Chu FH, Wu JT, Li WH and Chen CY (2016a) Integrating RNA-seq and ChIP-seq data to characterize long non-coding RNAs in Drosophila melanogaster. BMC Genomics 17, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Sheng P, Huang Z, Meng F, Kang Y, Huang G, Zhang Z, Liao W and Zhang Z (2016b) MicroRNA-381 regulates chondrocyte hypertrophy by inhibiting histone deacetylase 4 expression. International Journal of Molecular Sciences 17, pii: E1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WP, Bao JP, Hu PF, Feng J and Wu LD (2010) Alleviation of osteoarthritis by Trichostatin A, a histone deacetylase inhibitor, in experimental osteoarthritis. Molecular Biology Reports 37, 3967–3972. [DOI] [PubMed] [Google Scholar]
- Cheng X and Blumenthal RM (2008) Mammalian DNA methyltransferases: a structural perspective. Structure 16, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheow LF, Quake SR, Burkholder WF and Messerschmidt DM (2015) Multiplexed locus-specific analysis of DNA methylation in single cells. Nature Protocols 10, 619. [DOI] [PubMed] [Google Scholar]
- Dansranjavin T, Krehl S, Mueller T, Mueller LP, Schmoll HJ and Dammann RH (2009) The role of promoter CpG methylation in the epigenetic control of stem cell related genes during differentiation. Cell Cycle 8, 916–924. [DOI] [PubMed] [Google Scholar]
- Elliott EN, Sheaffer KL, Schug J, Stappenbeck TS and Kaestner KH (2015) Dnmt1 is essential to maintain progenitors in the perinatal intestinal epithelium. Development 142, 2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezura Y, Sekiya I, Koga H, Muneta T and Noda M (2009) Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis & Rheumatism 60, 1416. [DOI] [PubMed] [Google Scholar]
- Fan J, Krautkramer KA, Feldman JL and Denu JM (2015) Metabolic regulation of histone post-translational modifications. ACS Chemical Biology 10, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumatsu T and Asahara H (2010) Histone acetylation influences the activity of Sox9-related transcriptional complex. Acta Medica Okayama 64, 351–357. [DOI] [PubMed] [Google Scholar]
- Gendrel AV and Heard E (2014) Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annual Review of Cell & Developmental Biology 30, 561–580. [DOI] [PubMed] [Google Scholar]
- Glemžaitė M and Navakauskienė R (2016) Osteogenic differentiation of human amniotic fluid mesenchymal stem cells is determined by epigenetic changes. Stem Cells International 2016, 6465307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Romero R, Riveracasas C, Frehlick LJ, Méndez J, Ausió J and Eirínlópez JM (2012) Histone H2A (H2A.X and H2A.Z) variants in molluscs: molecular characterization and potential implications for chromatin dynamics. Plos One 7, e30006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Zhang W, Liu GH and Belmonte JCI (2013) Switching cell fate, ncRNAs coming to play. Cell Death & Disease 4, e464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS and Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Chakraborty D, Taggar R, Kumar D, Sharma R and Singh VP (2014a) Genomic imprinting in mammals – a review. Agricultural Reviews 35, 148–152. [Google Scholar]
- Gupta A, Niger C, Buo AM, Eidelman ER, Chen RJ and Stains JP (2014b) Connexin43 enhances the expression of osteoarthritis-associated genes in synovial fibroblasts in culture. BMC Musculoskeletal Disorders 15, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S and Shilatifard A (2011) Histone modification: cause or cog? Trends in Genetics 27, 389–396. [DOI] [PubMed] [Google Scholar]
- Hirose T, Mishima Y and Tomari Y (2014) Elements and machinery of non-coding RNAs: toward their taxonomy. EMBO Reports 15, 489–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SH, Lee KD, Hsu CC, Tseng MJ, Jin VX, Sun WS, Hung YC, Yeh KT, Yan PS, Lai YY, Sun HS, Lu YJ, Chang YS, Tsai SJ, Huang TH and Leu YW (2010) DNA methylation of the Trip10 promoter accelerates mesenchymal stem cell lineage determination. Biochemical & Biophysical Research Communications 400, 305–312. [DOI] [PubMed] [Google Scholar]
- Hsu PWC, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S and Hofacker IL (2006) miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Research 34(Database issue), D135–D139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jiang C and Zhang R (2014) Epigenetics: the language of the cell? Epigenomics 6, 73–88. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Malizos KN and Tsezou A (2007) Epigenetic regulation of leptin affects MMP-13 expression in osteoarthritic chondrocytes: possible molecular target for osteoarthritis therapeutic intervention. Annals of the Rheumatic Diseases 66, 1616–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott NE and Ponting CP (2013) Predicting long non-coding RNAs using RNA sequencing. Methods 63, 50–59. [DOI] [PubMed] [Google Scholar]
- Im GI and Choi YJ (2013) Epigenetics in osteoarthritis and its implication for future therapeutics. Expert Opinion on Biological Therapy 13, 713–721. [DOI] [PubMed] [Google Scholar]
- Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, Saito K, Nakamura T, Siomi H, Ito H, Arai Y, Shinomiya K and Takeda S (2009) A microRNA regulatory mechanism of osteoblast differentiation. Proceedings of the National Academy of Sciences of the United States of America 106, 20794–20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AW (2013) Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica 2013, 684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, James JA and Sawalha AH (2014) Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis & Rheumatology 66, 2804–2815. [DOI] [PubMed] [Google Scholar]
- Kawane T, Komori H, Liu W, Moriishi T, Miyazaki T, Mori M, Matsuo Y, Takada Y, Izumi S, Jiang Q, Nishimura R, Kawai Y and Komori T (2014) Dlx5 and Mef2 regulate a novel Runx2 enhancer for osteoblast-specific expression. Journal of Bone & Mineral Research 29, 1960–1969. [DOI] [PubMed] [Google Scholar]
- Kim D, Song J, Han J, Kim Y, Chun CH and Jin EJ (2013a) Two non-coding RNAs, microRNA-101 and HOTTIP contribute cartilage integrity by epigenetic and homeotic regulation of integrin-α1. Cellular Signalling 25, 2878–2887. [DOI] [PubMed] [Google Scholar]
- Kim KI, Park YS and Im GI (2013b) Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. Journal of Bone & Mineral Research 28, 1050–1060. [DOI] [PubMed] [Google Scholar]
- Kimura H (2013) Histone modifications for human epigenome analysis. Journal of Human Genetics 58, 439–445. [DOI] [PubMed] [Google Scholar]
- Kumari P and Sampath K (2015) cncRNAs: bi-functional RNAs with protein coding and non-coding functions. Seminars in Cell & Developmental Biology 47–48, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yoshida W, Abe K, Nakabayashi K, Wakeda H, Hata K, Marquette CA, Blum LJ, Sode K and Ikebukuro K (2016) Development of an electrochemical detection system for measuring DNA methylation levels using methyl CpG-binding protein and glucose dehydrogenase-fused zinc finger protein. Biosensors & Bioelectronics 93, 118–123. [DOI] [PubMed] [Google Scholar]
- Li A, Zhang J and Zhou Z (2014) PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 15, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S and Garcia BA (2012) Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods in Enzymology 512, 3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Sun Y, Ge Q, Teng H and Jiang Q (2014) Histone deacetylase 4 alters cartilage homeostasis in human osteoarthritis. BMC Musculoskeletal Disorders 15, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Singh S, Turcotte R and Ghert M (2015) The epigenetic regulation of SOX9 by miR-145 in human chondrosarcoma. Journal of Cellular Biochemistry 116, 37–44. [DOI] [PubMed] [Google Scholar]
- McCauley BS and Dang W (2014) Histone methylation and aging: lessons learned from model systems. Biochimica et Biophysica Acta 1839, 1454–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melissari MT and Grote P (2016) Roles for long non-coding RNAs in physiology and disease. Pflugers Archiv European Journal of Physiology 468, 1–14. [DOI] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Sen B, Rubin J and Pike JW (2016) Epigenetic plasticity drives adipogenic and osteogenic differentiation of marrow-derived mesenchymal stem cells. Journal of Biological Chemistry 291, 17829–17847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK and Asahara H (2009) MicroRNA0 is expressed in differentiated human articular chondrocytes and modulates interleukin responses. Arthritis & Rheumatism 60, 2723–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T and Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology 38, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL and Mohr AM (2015) Overview of microRNA biology. Seminars in Liver Disease 35, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki MS and Haqqi TM (2016) Histone deacetylase inhibitor vorinostat (SAHA, MK0683) perturb miR-9-MCPIP1 axis to block IL-1β-induced IL-6 expression in human OA chondrocytes. Connective Tissue Research 58, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA and Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Papathanasiou I, Kostopoulou F, Malizos KN and Tsezou A (2015) DNA methylation regulates sclerostin (SOST) expression in osteoarthritic chondrocytes by bone morphogenetic protein 2 (BMP-2) induced changes in Smads binding affinity to the CpG region of SOST promoter. Arthritis Research & Therapy 17, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez P, Jang SI and Alevizos I (2013) Emerging landscape of non-coding RNAs in oral health and disease. Oral Diseases 20, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard LN (2016) Analysis of genetics and DNA methylation in osteoarthritis: what have we learnt about the disease? Seminars in Cell & Developmental Biology 62, 57–66. [DOI] [PubMed] [Google Scholar]
- Rhee HS and Pugh BF (2011) Comprehensive genome-wide protein–DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera CM and Ren B (2013) Mapping human epigenomes. Cell 155, 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB and Strahl BD (2014) Interpreting the language of histone and DNA modifications. Biochimica et Biophysica Acta 1839, 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Phillip G and Herrmann BG (2016) Mechanisms of long noncoding RNA function in development and disease. Cellular & Molecular Life Sciences 73, 2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Force AS and Fedorov Y (2009) Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. Plos One 4, e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Abu-Amer Y, O'Keefe RJ and McAlinden A (2017) Inflammation and epigenetic regulation in osteoarthritis. Connective Tissue Research 58, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Wang C, Acton AJ, Eckert GJ and Trippel SB (2015) Role of Sox9 in growth factor regulation of articular chondrocytes. Journal of Cellular Biochemistry 116, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Sun M, Liu H, Yao Y and Song Y (2013) Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Letters 339, 159–166. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Feinweber C, Knothe C, Lötsch J, Thomas D, Geisslinger G, Parnham MJ and Resch E (2016) High-throughput analysis of global DNA methylation using methyl-sensitive digestion. Plos One 11, e0163184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, de Andrés MC, Hashimoto K, Itoi E and Oreffo RO (2015) Epigenetic regulation of interleukin-8, an inflammatory chemokine, in osteoarthritis. Osteoarthritis & Cartilage 23, 1946–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Lu J, Huang W, Dong Z, Kong C, Li L, Gao L, Guo J and Huang B (2009) Genome-wide analysis of histone H3 lysine 9 modifications in human mesenchymal stem cell osteogenic differentiation. Plos One 4, e6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Patel R, Kelly TAN, Wann AKT, Hung CT, Chapple JP and Knight MM (2015) Hedgehog signalling does not stimulate cartilage catabolism and is inhibited by interleukin-1β. Arthritis Research & Therapy 17, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gröne HJ, Dittmar G, Glimm H and Lyko F (2015) The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO Journal 34, 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulveling D, Francastel C and Hubé F (2011) Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie 93, 2024–2027. [DOI] [PubMed] [Google Scholar]
- Umer M and Herceg Z (2013) Deciphering the epigenetic code: an overview of DNA methylation analysis methods. Antioxidants & Redox Signaling 18, 1972–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal F, Akkoyunlu G and Ozturk S (2015) Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos. Biochimie 116, 103–113. [DOI] [PubMed] [Google Scholar]
- Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF and Bernstein E (2014) Histone variants: emerging players in cancer biology. Cellular & Molecular Life Sciences 71, 379–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela MA, Roberts TC and Wood MJ (2013) Epigenetics and ncRNAs in brain function and disease: mechanisms and prospects for therapy. Neurotherapeutics 10, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M (2015) Cancer epigenetics: risk assessment, diagnosis, treatment, and prognosis. Methods in Molecular Biology 1238, v. [PubMed] [Google Scholar]
- Verma P and Dalal K (2011) ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. Journal of Cellular Biochemistry 112, 3507–3514. [DOI] [PubMed] [Google Scholar]
- Weber CM and Henikoff S (2014) Histone variants: dynamic punctuation in transcription. Genes & Development 28, 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R and Karsenty G (2012) miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. Journal of Cell Biology 197, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Arfat Y, Li D, Zhao F, Chen Z, Yin C, Sun Y, Hu L, Yang T and Qian A (2016) Structure prediction: new insights into decrypting long noncoding RNAs. International Journal of Molecular Sciences 17, pii: E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guan Y, Tian S, Wang Y, Sun K and Chen Q (2016) Mechanical and IL-1β responsive miR-365 contributes to osteoarthritis development by targeting histone deacetylase 4. International Journal of Molecular Sciences 17, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DA, Lakey RL, Pennington CJ, Jones D, Kevorkian L, Edwards DR, Cawston TE and Clark IM (2005) Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Research & Therapy 7, R503–R512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu L, Xiao T and Guo W (2012) [Detection of the expression level of miR-140 using realtime fluorescent quantitative PCR in knee synovial fluid of osteoarthritis patients]. Journal of Central South University 37, 1210–1214. [DOI] [PubMed] [Google Scholar]
- Zhang M, Lu Q, Miller AH, Barnthouse NC and Wang J (2016) Dynamic epigenetic mechanisms regulate age-dependent SOX9 expression in mouse articular cartilage. International Journal of Biochemistry & Cell Biology 72, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M and Wang J (2015) Epigenetics and osteoarthritis. Genes & Diseases 2, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]