Abstract
Primary microcephaly (MCPH) is an autosomal recessive sporadic neurodevelopmental ailment with a trivial head size characteristic that is below 3–4 standard deviations. MCPH is the smaller upshot of an architecturally normal brain; a significant decrease in size is seen in the cerebral cortex. At birth MCPH presents with non-progressive mental retardation, while secondary microcephaly (onset after birth) presents with and without other syndromic features. MCPH is a neurogenic mitotic syndrome nevertheless pretentious patients demonstrate normal neuronal migration, neuronal apoptosis and neural function. Eighteen MCPH loci (MCPH1–MCPH18) have been mapped to date from various populations around the world and contain the following genes: Microcephalin, WDR62, CDK5RAP2, CASC5, ASPM, CENPJ, STIL, CEP135, CEP152, ZNF335, PHC1, CDK6, CENPE, SASS6, MFSD2A, ANKLE2, CIT and WDFY3, clarifying our understanding about the molecular basis of microcephaly genetic disorder. It has previously been reported that phenotype disease is caused by MCB gene mutations and the causes of this phenotype are disarrangement of positions and organization of chromosomes during the cell cycle as a result of mutated DNA, centriole duplication, neurogenesis, neuronal migration, microtubule dynamics, transcriptional control and the cell cycle checkpoint having some invisible centrosomal process that can manage the number of neurons that are produced by neuronal precursor cells. Furthermore, researchers inform us about the clinical management of families that are suffering from MCPH. Establishment of both molecular understanding and genetic advocating may help to decrease the rate of this ailment. This current review study examines newly identified genes along with previously identified genes involved in autosomal recessive MCPH.
Key words: MCPH, MCPH1–MCPH18, microcephaly, molecular genetics
1. Introduction
Microcephaly is a rare neurodevelopmental disorder defined by biparietal diameter (BPD) based on a Brazilian study where small head size is below 3–4 standard deviations and the cerebral cortex region of the brain is reduced in size (Araujo Junior et al., 2014). Microcephaly patients may or may not have mild to severe mental retardation as well as squat physique and seizures or hereditary hearing loss (Darvish et al., 2010). It can be classified into two forms; primary microcephaly (at birth) or secondary microcephaly (onset after birth) with and without other syndromic features (Cowie, 1960). Primary microcephaly is a prenatal developmental neurogenic disorder whereas secondary microcephaly is associated with progressive neurodegenerative disease.
Autosomal recessive primary microcephaly (MCPH) is a neurogenic mitotic disorder with normal neuronal migration, neuronal apoptosis and neural function of affected patients. There are various genetic and environmental causes including chromosomal aberrations, dented DNA as a consequence of incorrect mitotic spindle alignment, impulsive chromosomal abridgment, maternal overconsumption of alcohol, congenital infections, drugs taken during pregnancy, brain injury, metabolic disorders such as alaninuria or reaction to teratogenic remedies and substances taken during pregnancy (Darvish et al., 2010; Faheem et al. 2015). Metabolic disorders often cause secondary rather than primary microcephaly and are often associated with additional symptoms and clinical signs (Von der Hagen et al., 2014). Inquiries of metabolic screening, if necessary, must primarily focus on maternal phenylketonuria, phosphoglycerate dehydrogenase paucity and Amish fatal microcephaly (2-ketoglutaric aciduria) as a secondary basis of microcephaly (Kelley et al. 2002). Rare metabolic causes of primary microcephaly include serine biosynthesis defects, sterol biosynthesis disorders, mitochondriopathies and congenital disorders of glycosylation (Von der Hagen et al., 2014).
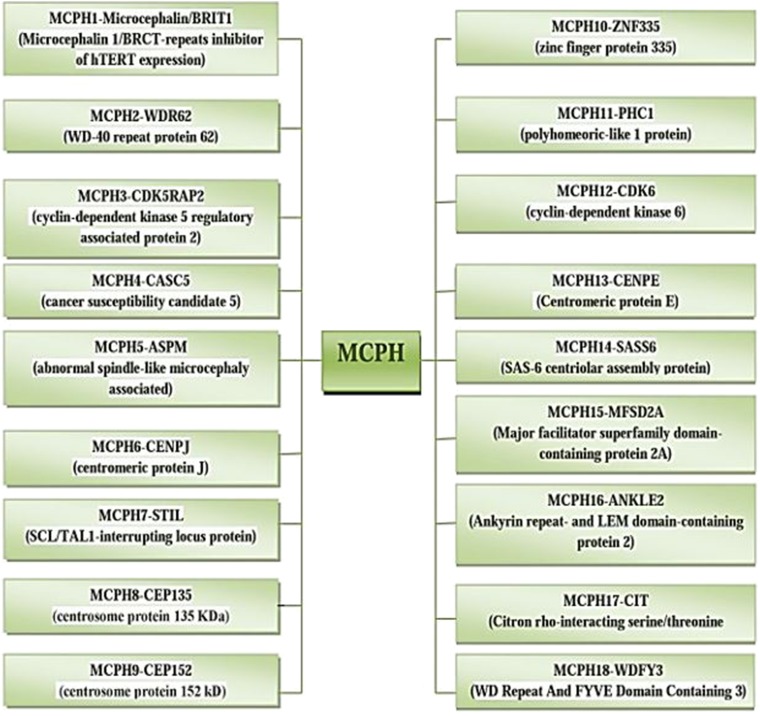
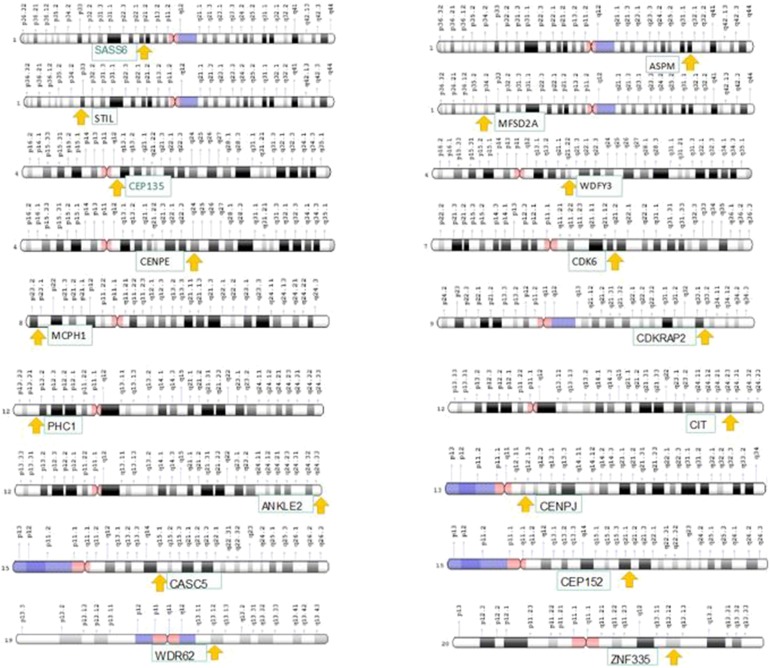
The incidence of MCPH ranges from 1:30,000–1:250,000 per live-birth depending on the population (Zaqout et al. 2017). In Asian and Arab populations where consanguineous marriages are mutual, MCPH is more common than in whites (Muhammad et al., 2009). Worldwide MCPH has been reported in excess of 300 families and distinct patients; often with only sparse phenotype descriptions. Separate from intellectual infirmity (IQ between 30 and 70–80), hyperactivity and devotion deficit, dialog deferral, and a tapered slanting forehead, MCPH patients ordinarily do not have any auxiliary neurological ciphers (Passemard et al., 2009; Kaindl et al., 2010; Bhat et al., 2011; Kraemer et al., 2016). To date, for MCPH 18 loci and residing genes are found to have mutations and are described in Figure 1, and their cytogenic locations are summarized in Figure 2 (Faheem et al., 2015; Zaqout et al., 2017). The most common causes of MCPH are biallelic mutations in ASPM (68.6%), followed by those in the WDR62 gene (14.1%) and MCPH1 gene (8%). More genetic loci are still expected to exist given the lack of mutations in known loci in approximately 50–75% of western Europeans or North Americans with MCPH and approximately 20–30% of Indians or Pakistanis with MCPH (Verloes et al., 1993; Sajid Hussain et al., 2013).
Fig. 1.
MCPH loci and residing genes responsible for microcephaly.
Fig. 2.
Cytogenic location of MCPH1–18 genes.
The up-to-date knowledge on the molecular genetics of MCPH, including the newly notorious locus with its gene (MCPH1–MCPH18), is expansively discussed in this review article. We review the corresponding genes and the proteins encoded by these genes, their probable role in the emerging brain (Table 1) and discuss mutations of these genes found in the Pakistani population (Table 2). In addition, the potential for these genes to perform various cognitive roles during human brain evolutionary processes is discussed.
Table 1.
Functions of MCPH genes causing microcephaly.
| Locus | Chromosome | Genes | Inheritance | Function |
|---|---|---|---|---|
| MCPH1 | 8p23.1 | Microcephalin | AR | Involved in chromosomal condensation, cell cycle checkpoint and DNA damage response Reduced MCPH1 enhances the production of early born neurons, which comprise deep layers (IV–VI), and reduces late-born neurons, which produce the thinner outer cortex layer (II–III). |
| MCPH2 | 19q13.12 | WDR62 | AR | Microtubule, spindle organization and kinetochore association Neurogenesis, spindle orientation and cerebral cortical development, proliferation and migration of neuronal precursors, mutation in WDR62 affects its role in proliferating and migrating neural precursors and causes severe brain malformations |
| MCPH3 | 9q33.2 | CDKRAP2 | AR | Centriole duplication, regulates microtubule function, spindle organization and kinetochore association Mutation in CDK5RAP2 reduces the progenitor pool/decreases the number of neurons and reduces cell survival |
| MCPH4 | 15q15.1 | CASC5 | AR | Vital for the spindle checkpoint of the mitotic cycle, CASC5 underscores the role of kinetochore integrity in the proper volumetric development of the human brain |
| MCPH5 | 1q31.3 | ASPM | AR | Orientation of mitotic spindles during embryonic neurogenesis, centriole duplication, spindle organization and kinetochore association ASPM mutations can decrease the size of the brain by influencing the orientation of the mitotic spindle |
| MCPH6 | 13q12.12-q12.13 | CENPJ | AR | Controls centriole length/microtubule function, spindle organization and kinetochore association Its deletion causes an increased incidence of multiple spindle poles, apoptosis and mitosis arrest |
| MCPH7 | 1p33 | STIL | AR | Apoptosis regulator/cell cycle progression, centriole duplication, and ciliogenesis, its mutation in Zebra fish causes an embryonic lethal defect, and STIL knockout mice (Sil-/-) exhibit numerous developmental abnormalities Decreased size/defective midline neural tube |
| MCPH8 | 4q12 | CEP135 | AR | Maintains organization/structure of the centrosome, centriole duplication, CEP135 Knockdown showed decreased growth rate/disorganized microtubules |
| MCPH9 | 15q21.1 | CEP152 | AR | Centriole duplication/shape and cell/polarity/motility, conversion of glutamine into proline disturbs potential coiled-coiled protein domain/reduced head size |
| MCPH10 | 20q13.12 | ZNF335 | AR | Progenitor cell division/differentiation, neurogenesis, neuronal migration and spindle orientation Mutated ZNF335 gene causes degeneration of neurons, knockdown of ZNF335 caused a small brain size with an absent cortex/disrupted proliferation and differentiation of neuronal cells |
| MCPH11 | 12p13.31 | PHC1 | AR | Regulates cell cycle, and controls DNA damage response PHC1 mutation highlights the role of chromatin remodelling in the pathogenesis of primary microcephaly |
| MCPH12 | 7q21.2 | CDK6 | AR | Controls cell cycle/organizes microtubules, spindle organization and kinetochore association CDK6 mutation affects apical neuronal precursor cells proliferation, reduces the progenitor pool, decreases neuronal production and causes primary microcephaly |
| MCPH13 | 4q24 | CENPE | AR | Microtubule, spindle organization and kinetochore association Mutation in these gene leads to the dysregulation of a large number of genes involved in cell cycle, causes defective DNA repair and cell cycle with reduced proliferative activity |
| MCPH14 | 1p21.2 | SASS6 | AR | Centrioles formation through procentriole complex In the PISA domain of SAS-6 gene, the replacement of threonine with isoleucine at position 62 causes primary microcephaly |
| MCPH15 | 1p34.2 | MFSD2A | AR | Protein encoded by MFSD2A gene is transmembrane, the encoded protein plays important role in blood–brain barrier and is also important for normal brain function and growth When the homozygous mutation occurs in the MSFD2A gene at the location of 1p34 then it causes problems in the activity of the blood–brain barrier, which leads to severe primary microcephaly |
| MCPH16 | 12q24.33 | ANKLE2 | AR | Highly involved in proliferation of neuroprogenitor cells during initial brain development and biogenesis of CNS ANKLE2 mutated gene leads to impaired brain development that reduces the brain size as a result of decreased number of neuroblasts, reduced mitosis, less cell proliferation and increased apoptosis This improper development of brain contributes to MCPH16 due to recessive mutation in gene |
| MCPH17 | 12q24.23 | CIT | AR | CIT is critical for the development of normal brain size, and it is also essential for the successful completion of cytokinesis Mutation in CIT causes MCPH by homozygous missense variants in the kinase domain that can lead to the loss or activation of citron kinase protein |
| MCPH18 | 4q21.23 | WDFY3 | AD | Required for the organization of misfolded proteins that are degraded by autophagy in body Acts as a ligand for some molecules involved in the selective macroautophagy in the degradation of the midbody ring Mutation in gene leads to abnormal cerebral cortex size and other neurodevelopmental disorders |
AD: Autosomal dominant; AR: Autosomal recessive; MCPH: Autosomal recessive primary microcephaly.
Table 2.
Reported mutations of MCPH1–MCPH18 genes.
| MCPH1–Microcephalin | |||
|---|---|---|---|
| 74C-G transversion | Ser25-to-ter (S25X) substitution | Exon2 | Jackson et al. (2002) |
| 1-bp insertion (427insA) | Truncated protein of 146 a.a | Exon5 | Trimborn et al. (2004) |
| 150- to 200-kb deletion | – | Exon1–6 | Garshasbi et al. (2006) |
| 1-bp insertion (566insA) | Frameshift mutation | Exon6 | Darvish et al. (2010) |
| 147C-G transversion | His49-to-gln (H49Q) substitution | Exon3 | Darvish et al. (2010) |
| 215C-T transition | Ser72-to-leu (S72L) substitution | Exon3 | Darvish et al. (2010) |
| 302C-G transversion | Ser101-to-ter (S101X) substitution | Exon4 | Farooq et al. (2010) |
| MCPH2–WDR62 | |||
| 4-bp deletion (c.4205delTGCC) | Val1402GlyfsTer12 | Exon31 | Bilguvar et al. (2010) |
| c.1576G-A substitution | Glu52-to-Lys (E526 K) substitution | Exon12 | Bilguvar et al. (2010) |
| G-to-C transversion | Try224-to-Ser (W224S) substitution | Exon6 | Bilguvar et al. (2010) |
| C-to-T transition | Glu to termination substitution (Q470X) | Exon11 | Bilguvar et al. (2010) |
| 17-bp deletion | Gly1280AlafsTer21 | Exon30 | Bilguvar et al. (2010) |
| 1313G-A transition | Arg438-to-His (R438H) substitution | Exon10 | Nicholas et al. (2010) |
| 1-bp duplication (4241dupT) | Premature termination | Exon31 | Nicholas et al. (2010) |
| 1531G-A transition | Asp511-to-Asn (D511N) substitution | Exon11 | Nicholas et al. (2010) |
| 1-bp insertion (3936insC) | Premature termination | Exon30 | Yu et al. (2010) |
| 1-bp deletion (363delT) | Premature termination | Exon4 | Yu et al. (2010) |
| 193G-A transition | Val65-to-Met (V65M) substitution | Exon2 | Yu et al. (2010) |
| 1-bp deletion (2083delA) | – | Exon17 | Murdock et al. (2011) |
| 2-bp deletion (2472_2473delAG) | – | Exon23 | Murdock et al. (2011) |
| MCPH3–CDK5RAP2 | |||
| c.243 T-A transversion | Ser81-to-Ter (S81X) substitution | Exon4 | Bond et al. (2005) |
| c.246 T-A transversion | Tyr82-to-Ter (Y82X) substitution | – | Hassan et al. (2007) |
| c.15A-G transition | – | Intron26 | Bond et al. (2005) |
| c.700G-T transversion | Glu234-to-Ter (E234X) substitution | Exon8 | Pagnamenta et al. (2012) |
| c.4546G-T transversion | Glu1516-to-Ter (E1516X) substitution | – | Lancaster et al. (2013) |
| c.4672C-T transition | Arg1558-to-Ter (R1558X) substitution | – | Lancaster et al. (2013) |
| 5-bp deletion (c.524_528del) | Gln175ArgfsTer42 | Exon7 | Tan et al. (2014) |
| MCPH4–CASC5 | |||
| 6125G-A transition | Met2041-to-Ile (M2041I) substitution | Exon18 | Genin et al. (2012) |
| MCPH5–ASPM | |||
| 2-bp deletion (719-720delCT) | Premature termination 15 codons downstream | Exon3 | Bond et al. (2002) |
| 7-bp deletion (1258-1264delTCTCAAG) | Premature termination 31 codons downstream | Exon3 | Bond et al. (2002) |
| 7761 T-G transversion | Immediate truncation | Exon18 | Bond et al. (2002) |
| 1-bp deletion (9159delA) | Premature termination four codons downstream | Exon21 | Bond et al. (2002) |
| 9178C-T transition | Gln3060-to-Ter (Q3060X) substitution | Exon21 | Kumar et al. (2004) |
| 3978G-A transition | Trp1326-to-Ter (W1326X) substitution | Exon17 | Kumar et al. (2004) |
| 349C-T transition | Arg117-to-Ter (R117X) substitution | – | Kumar et al. (2004) |
| 6189 T-G transversion | Tyr2063-to-Ter (Y2063X) substitution | Exon18 | Shen et al. (2005) |
| 1-bp insertion (4195insA) | Premature termination | Exon6 | Saadi et al. (2009) |
| 2389C-T transition | Arg797-to-Ter (R797X) substitution | Exon18 | Saadi et al. (2009) |
| 2-bp deletion | 7781delAG | – | Saadi et al. (2009) |
| MCPH6–CENPJ | |||
| 1-bp deletion (17delC) | Thr6fsTer3 | – | Leal et al. (2003); Bond et al. (2005) |
| 1-bp deletion (c.18delC) | Ser7ProfsTer2 | Exon 2 | Sajid Hussain et al. (2013) |
| 3704A-T transversion | Glu1235-to-Val (E1235 V) substitution | Exon16 | Bond et al. (2005) |
| 4-bp deletion (3243delTCAG) | – | Exon11 | Gul et al. (2006 b) |
| IVS11-1G-C | – | Intron11 | Al-Dosari et al. (2010) |
| 2462C-T transition | Thr821-to-Met (T821M) substitution | Exon7 | Darvish et al. (2010) |
| MCPH7–STIL | |||
| 3715C-T transition | Gln1239-to-Ter (Q1239X) substitution | Exon18 | Kumar et al. (2009) |
| 1-bp deletion (3655delG) | – | Exon18 | Kumar et al. (2009) |
| G-to-A transition | – | Intron16 | Kumar et al. (2009) |
| c.2392 T-G transversion | Leu798-to-Trp (L798W) substitution | Exon14 | Papari et al. (2013) |
| c.453 + 5G-A transition | Asp89GlyfsTer8 | Intron5 | Kakar et al. (2015) |
| MCPH8–CEP135 | |||
| 1-bp deletion (970delC) | Gln324SerfsTer2 | Exon8 | Hussain et al. (2012) |
| c.1473 + 1G-A transition | Glu417GlyfsTer2 | Intron11 | Farooq et al. (2016) |
| MCPH9–CEP152 | |||
| A-to-C transversion | Gln265-to-Pro (Q265P) substitution | – | Guernsey et al. (2010) |
| C-to-T transition | Arg987-to-Ter (R987X) substitution | – | Guernsey et al. (2010) |
| MCPH10–ZNF335 | |||
| 3332G-A transition | Arg1111-to-His (R1111H) substitution | Exon20 | Yang et al. (2012) |
| MCPH11–PHC1 | |||
| c.2974C-T transition | Leu992-to-Phe (L992 F) substitution | Awad et al. (2013) | |
| MCPH12–CDK6 | |||
| c.589G-A transition | Ala197-to-Thr (A197 T) substitution | Exon5 | Hussain et al. (2013) |
| MCPH13–CENPE | |||
| c.2797G-A transition | Asp933-to-Asn (D933N) substitution | – | Mirzaa et al. (2014) |
| c.4063A-G transition | Lys1355-to-Glu (K1355E) substitution | Mirzaa et al. (2014) | |
| MCPH14–SASS6 | |||
| c.185 T-C transition | Ile62-to-Thr (I62 T) substitution | Exon3 | Khan et al. (2014) |
| MCPH15–MFSD2A | |||
| c.476C-T transition | Thr159-to-Met (T159M) substitution | – | Guemez-Gamboa et al. (2015) |
| c.497C-T transition | Ser166-to-Leu (S166L) substitution | Exon 10 | Guemez-Gamboa et al. (2015) |
| c.1016C-T transition | Ser339-to-Leu (S339L) substitution | – | Alakbarzade et al. (2015) |
| MCPH16–ANKLE2 | |||
| c.2344C-T transition | Gln782-to-Ter (Q782X) substitution | Exon11 | Yamamoto et al. (2014) |
| c.1717C-G transversion | Leu573-to-Val (L573 V) substitution | Exon10 | |
| MCPH17–CIT | |||
| c.317G-T transversion | Gly106-to-Val (G106 V) substitution | Exon4 | Li et al. (2016) |
| c.376A-C transversion | Lys126-to-Gln (K126Q) substitution | – | Li et al. (2016) |
| c.689A-T transversion | Asp230-to-Val (D230 V) substitution | – | Li et al. (2016) |
| c.1111 + 1G-A transition | Gly353_371delinsAla | Intron9 | Shaheen et al. (2016) |
| c.29_38delATCCTTTGGA | Asn10MetfsTer15 | Exon2 | Harding et al. (2016) |
| c.753 + 3A-T | Asp221Ter | Intron7 | Basit et al. (2016) |
| MCPH18–WDFY3 | |||
| c.7909C-T transition | Arg2637-to-trp (R2637W) substitution | – | Kadir et al. (2016) |
2. Molecular genetics
(i). MCPH1 (microcephalin)
MCPH1 encodes the important regulator of chromosome condensation (BRCT–BRCA1 C-terminus) the ‘Microcephalin protein’. The Microcephalin protein consists of three BCRT domains and conserved tandem repeats of phospho-peptide interacting amino acids (Faheem et al., 2015; Pulvers et al., 2015). This gene is located on chromosome 8p23 comprising 14 exons, 835 amino acids, and with a genome size and molecular weight of 241,905 bp and 92,877 Da, respectively. It exists in three isoforms obtained after splicing and an open reading frame (ORF) of approximately 8032 bp (Venkatesh et al., 2013; Faheem et al., 2015). The Microcephalin protein, being a pleiotropic factor, imparts its significant effect in neurogenesis; it regulates the division of neuroprogenitor cells and prevents them from exhaustion, that is, from microcephaly. It is the significant regulator of telomere integrity and involved in the DNA damage repair mechanism. It also acts as a tumour suppressor in several human cancers, in germline functions and performs its function in brain development and in the regulation of cerebral cortex size (Venkatesh et al., 2013; Pulvers et al., 2015). Some studies have reported that the MCPH1 gene is involved in brain size determination and as a positive selector for primate lineage (Montgomery et al., 2014). It was considered that MCPH1 may be a common denominator in the pathway of causing microcephaly enclosing the spectrum of both environmental and genetic causes. It leads to the drastic reduction of brain size, especially in the region of cerebral cortex, and short stature. Magnetic resonance imaging (MRI) of patients revealed the existence of cerebral deformations, such as the gyral pattern observed in the brain and corpus callosum hypoplasia. It is caused by premature switching of symmetric neuroprogenitors to asymmetric division. Mutation in MCPH1 causes mis-regulated chromosomal condensation, genomic instability and delayed de-condensation post mitosis (Liu et al., 2016). Successful experiments for MCPH1 mouse models reported mis-regulated mitotic chromosome condensation, deficiency in DNA repair, defective spindle orientation and a reduced skull size with approximately 20% reduction in body weight (Zhou et al., 2013; Faheem et al., 2015).
(ii). MCPH2 (WDR62)
The WDR62 gene encodes the ‘WD repeat-containing protein 62’, located on chromosome 19q13.12, comprising 35 exons, 1518 amino acids, with a genomic size of 50,230 bp and a molecular mass of 165,954 Da, it has four known spliced isoforms (Faheem et al., 2015; Pervaiz & Abbasi, 2016). WDR62 consists of a WD40 domain, a JNK docking domain and a MKK7 binding domain. It is a scaffold protein involved in the pathway of the c-Jun N-terminal kinase (Bastaki et al., 2016) being highly expressed in neuronal precursors and within post mitotic neurons of the developing brain and in the ventricular and sub ventricular zone in the forebrain region (Pervaiz & Abbasi, 2016). This protein plays a significant part in the formation of several cellular layers in the cerebral cortex region during embryogenesis. It plays a compelling role in the proliferation and migration of neurons and in the duplication of centrioles that are dependent on mother centrioles (Sgourdou et al., 2017). Several findings revealed that WDR62 functions are somehow similar to ASPM which is another MCPH gene. Moreover, studies revealed that the WDR62 gene, as it plays an important part in brain cortical development, is involved in human brain evolutions indicted by dramatic inflation in the size of the cerebral cortex (Pervaiz et al., 2016). Mutation in WDR62 can lead to a wide range of disorders including: microcephaly, cognitive disability, cortical malformations and multiple transcript variants by alternative splicing. The majority of mutations found in WDR62 revealed that these are responsible for approximately 10% of cases of microcephaly. Homozygous or heterozygous mutations in WDR62 both cause MCPH with or without cortical malformations (Naseer et al., 2017). Patients with these mutations have a head circumference ranging from normal to severe. MRI of patients with these mutations showed numerous cortical malformations such as pachygyria, impulsivity, polymicrogyria, aggression, hypoplasia of corpus callosum, delayed psychomotor development, simplified gyral patterns, mental retardation with reduced head size and lissencephaly (Farag et al., 2013; Faheem et al., 2015; Bastaki et al., 2016). Disruption of WDR62 in a mouse model altered the late neurogenesis neocortical progenitors proliferation process which further depicts asymmetric centrosome inheritance abnormalities leading to microcephaly in mice, and impaired mitotic cycle progression, causing temporary arrest at pro metaphase, as well as defects in spindle pole localization of WDR62 (Farag et al., 2013; Sgourdou et al., 2017).
(iii). MCPH3 (CDKRAP2)
The CDK5RAP2 gene is located on chromosome 9q33.2, comprising 39 exons, 1893 amino acids with a molecular mass of 215,038 Da (Graser et al., 2007). This gene is also known as C48, Cep215 and MCPH3. Cnn-1N is a small motif present at the N-terminal of a group of centrosome or spindle pole body associated proteins, such as Mto1 and Pcp1 from Schizosaccharomyces pombe, centrosomin from flies and CDK5RAP2 from mammals (Megraw et al., 1999; Verde et al., 2001; Flory et al., 2002; Venkatram et al., 2004; Conduit et al., 2014). The CDK5RAP2 gene encodes a regulator of CDK5 activity which is localized in the Golgi complex and centrosome in cells and in the cerebral cortex of the brain (Faheem et al., 2015). It interacts with pericentrin and CDK5R1 and plays an important role in microtubule nucleation and centriole engagement. CDK5RAP2 being a part of the pericentriolar material is crucial for the microtubule organization capacity of the centrosome. Patients exhibiting the primary fibroblasts had a drastic reduction in the amount of CDKRAP2 and showed nuclear and centrosomal abnormalities and an increased frequency of alteration in cell size and migration. In addition, researchers also identified interplay of CDK5RAP2 with the constituents of the Hippo pathway, the transcriptional regulator TAZ and MASTI kinase. This finding enabled us to understand the mechanism of the Hippo pathway, revealing its role in the regulation of centrosome number. Higher levels of TAZ and Yup in fibroblast patients are observed but none of the other genes involved in the Hippo pathway have been seen to be downregulated. Modifications observed in the Hippo pathway constituents could consequently alter cellular properties and centrosomal deficiencies in patients with affected fibroblasts. This could further be relevant to MCPH controlling brain size and development (Sukumaran et al., 2017). Robustly connected with microtubules, centrosome and Golgi apparatus, CDK5RAP2 is mainly found inside the neural progenitors of the ventricular and sub ventricular areas of the developing brain; it has also been discovered in glial cells and early born neurons and is gradually downregulated when brain maturation occurs. In spindle checkpoint regulation CDK5RAP2 also imparts its effect. The damage to CDK5RAP2 shows chromosomal segregation and spindle checkpoint protein expression issues via binding in HeLa cells (Faheem et al., 2015).
Scientist have determined that the ‘an’ homozygous mutation in Hertwig's anemia within the CDK5RAP2 gene results in deletion of exon four. Mutant mice exhibited microcephaly symptoms along with hypoplasia of brain regions – cortex and hippocampus – and hematopoietic phenotype. Mutant mice neuronal progenitors confirmed proliferative and survival defects and underwent apoptosis. The impaired centrosomal characteristic and altered mitotic spindle orientation in neuronal progenitors are consequences of CDK5RAP2 mutation (Lizarraga et al., 2010).
(iv). MCPH4 (CASC5)
CASC5 is a protein coding gene, located on chromosome 15q15.1, comprising 27 exons, 2342 amino acids, with a molecular mass of 265 kDa. It is also known as hKNL-1, D40, Spc7, MCPH4, CT29, AF15Q14, PPP1R55 and hSpc105. In cells, the CASC5 gene is present in the nucleoplasm and in the brain the CASC5 gene is present in the cerebral cortex (Petrovic et al., 2014). The protein encoded by this gene is an important part of the multiprotein assembly that is used for the proper development of kinetochore–microtubule coupling and chromosome separation. It's a conserved scaffold protein that is used for proper kinetochore assembly, checkpoint functioning and spindle assembly. Unlike other MCPH proteins that are present around the centrosome, the CASC5 protein is used for coupling of chromatin with the mitotic apparatus and also interacts with BUB1, which properly manages the spindle assembly checkpoint. Any reduction in CASC5 protein causes chromosome misalignment and drives the cell into the mitosis (Saadi et al., 2016). Mutated CASC5 disrupts hMIS12, which is vital for correct chromosome alignment and segregation. Frequently this gene is part of the metaphase chromosome kinetochore and elevated mitotic index in patient cells designated mitotic arrest within the cells carrying the mutation. Lobulated and fragmented nuclei have also been identified in addition to micronuclei inside the affected cells. Moreover, altered DNA harms reactions with high peaks of γH2AX and 53BP1 seen in mutated cells as compared to control fibroblasts. These studies confirm the role of CASC5 in primary microcephaly (Szczepanski et al., 2016).
(v). MCPH5 (ASPM)
ASPM is located on chromosome 1q31.3, comprising 62,567 bp, 3477 amino acids, 28 exons divided into 10,906 ORFs (Saunders et al., 1997; Ponting, 2006). It is present on spindle poles and centrosomes during mitosis. The domains of ASPM includes an N-terminus, an 81 IQ (isoleucine–glutamine) domain and a Calponin homology domain (Craig & Norbury, 1998; Bond et al., 2002; Kouprina et al., 2005) and a C-terminus without any domain weighing about 220 kDa (Bond et al., 2003). The orthologs of ASPM include almost 20 different species. The cerebral cortex, the ganglia and the mouse neocortex show the expression of the gene during the process of neurogenesis (Bond et al., 2003; Paramasivam et al., 2007). ASPM downregulation is a result of neurosphere differentiation (Thornton & Woods, 2009). ASPM plays a role in pole organization through activation of kinesin-14 and CDK5RAP2 in normal cells (Tungadi et al., 2017). The most important and crucial role of the ASPM gene is to perform cytokinesis during meiosis. ASPM plays a fundamental role in organizing microtubules and focusing spindle poles during mitosis (do Carmo Avides & Glover, 1999). ASPM knockdown mediated by Morpholino results in the reduction of head size in different species such as Zebra fish (Kim et al., 2011). The reduced surface area and increased thickness of white matter are consequences of loss of ASPM, while preserving the memory of patients in contrast to its intellectual disabilities (Passemard et al., 2016). ASPM knockdown effects could be reduced by CITK overexpression and CITK microcephaly phenotype is a result of spindle orientation (Gai et al., 2016). The similarities of WDR62 and ASPM includes the same location and physical interaction during interphase mediated by CEP63. Loss of WDR62 and ASPM in the developing brain induces shortage of cilia and centrosomes. Both genes determine the fate of cells and localize CPAP to the centrosome (Jayaraman et al., 2017). Different types of mutations have been observed in the ASPM gene in 33 families from Pakistan (Gul et al., 2006 a). These mutation types include deletion, substitution, duplication and variation in the intrinsic region (Muhammad et al., 2009; Saadi et al., 2009). The ASP gene in mutant form was first discovered in Drosophila (do Carmo Avides & Glover, 1999). Different experiments were performed to study mutations in ASPM. One study examined the developing cerebral cortex in two mutant mouse lines and concluded that the reduced size of the brain was a consequence of ASPM mutation (Riparbelli et al., 2002). Using whole-genome sequencing, a mutation in exon 16 in the ASPM gene was seen, and non-syndromic microcephaly with altered IQ number was reported.
(vi). MCPH6 (CENPJ)
CENPJ is present on chromosome 13q12.2, is comprised of 40,672 bp, 1338 amino acids, 17 exons distributed in 5187 bp ORFs, weighing approximately 153 kDa (Saunders et al., 1997). The domains of CENPJ include protein phosphorylation domains, five coiled-coil domains (CCDs), and the C-terminal domain has 21 G-box repeats and a leucine zipper motif (Hung et al., 2000; Bond et al., 2005). The gene holds its position within the centriole. The CENPJ gene has almost 204 orthologs. CENPJ carries out microtubule assembly in the centrosome by causing nucleation and depolymerization of microtubules (Hung et al., 2004). CENPJ also maintains centriole integrity with rearrangement of microtubules (Kirkham et al., 2003). During neurogenesis, the frontal cortex neuro-epithelium expresses the CENPJ gene (Bond et al., 2005). Damage to the structure of the centrosome causes cell arrest in mitosis and is caused by low levels of the CENPJ protein (Cho et al., 2006). CENPJ knockdown increases the rate of multiple spindle poles, apoptosis and mitosis arrest and loss of centrioles in Drosophila (Koyanagi et al., 2005; Basto et al., 2006). Drosophila flies without centrioles die at an early age due to loss of cilia or flagella (Stevens et al., 2007). The total number of mutations in the CENPJ gene never exceeded five. The first mutation was discovered in a Brazilian family, the second and third mutations belonged to a Pakistani family (Bond et al., 2005) and the forth was discovered in people suffering from Seckel syndrome (Al-Dosari et al., 2010). Studies showed that in the third mutation, four consecutive nucleotide units (TCAG) are deleted from around 19 bp downstream of exon 11 resulting in frameshifting and premature termination of protein (Gul et al., 2006b).
(vii). MCPH7 (STIL)
STIL is located on chromosome 1p33, comprising 63,018 bp with an ORF of 5225 bp, 20 exons, 1287 amino acids and a molecular weight of 150 kDa (Kumar et al., 2009; Kaindl et al., 2010). It has recently been reported that the human oncogene SCL/TAL1 locus is thoroughly conserved in vertebrate species, and is involved in regulation of toxic susceptibility in PC12 cells through the sonic hedgehog (Shh) pathway. Knockdown of STIL expression by RNAi showed no effect on survival of proliferating PC12 cells following increased cell death in differentiated neurons after drug analysis. In case of overexpression of STIL in proliferating cells toxic susceptibility is increased but it causes no effect in mature neurons (Szczepanski et al., 2016). Biochemical, cell biology and biophysical analysis reported that STIL retains a central short CCD, which suggests a critical role in oligomerization, centrosomal localization and protein interaction. Protein interaction is mediated by the central intrinsically disordered region of STIL, comprising 400–700 residues, just like CPAP interaction during centriole duplication. Generally, STIL is a disordered protein retaining three structured regions: N-terminal domain possessing 1–370 residues, 1062–1148 residues identified as a STIL ANA-2 motif and a short segment of 718–750 that give rise to a CCD. Size exclusion chromatography suggested that the CCD peptide comprises α helices that form a quaternary structure. Repeating patterns of hydrophobic, hydrophilic and charged residues present at specified locations to form a CCD. Recent studies revealed that two regions retaining eight hydrophobic residues exist in this domain and that these regions mediate oligomerization of CCD. Within this domain hydrophobic residues form axes occupied by hydrophilic residues promoting hydrophobic interactions that are an important part of protein structure and stability. Two leucine residues, L718 and L736, identified in this hydrophobic region lead to oligomerization of the CCD, and L736 has a greater role in this process. Further inspection revealed that mutations in these residues did not affect the secondary structure but alter the quaternary configuration which leads to malfunction of STIL. The CCD has a key role in oligomerization and promoting STIL function in self-interaction, centriolar replication, embryogenesis and in development of cilia. Current studies report that interaction of one helix of STIL with a PLK4 PB3 domain occurs via hydrophobic residues in the CCD and promotes centriole duplication. Phosphorylation of STIL by PLK4 facilitates STIL and SAS-6 protein interaction. Note that the L736 residue is critically involved in the interaction with PLK4. The hydrophobic core in the CCD of STIL imparts oligomerization building dimers and tetramers, which are analogues of its Drosophila ortholog Ana-2 and Caenorhabditis elegans centrosomal protein SAS-6 (David et al., 2016). Clinical and molecular genetic studies revealed that MCPH is rarely caused by STIL mutation (2.2%). The expression study demonstrated that STIL is critically involved in early forebrain development that may be associated with the Shh signalling pathway (Mouden et al., 2015).
(viii). MCPH8 (CEP135)
The CEP135 gene is located on chromosome 4q12, comprising 26 exons and 1140 amino acids, it encodes a centrosomal protein that is a reserve helical protein detected throughout the cell cycle at the centrosomes giving greater strength to the centrosomes. Recent studies revealed that CEP135 in Drosophila plays a crucial role in central microtubule pair assembly in sperm axoneme and asymmetric cell division of neuroblasts. Intriguingly, in Homo sapiens and Drosophila microtubule binding sites have been mapped to the N-terminal. Fluorescence microscopy, cryo-electron analysis, including biochemical analysis, elucidated that in vitro development of microtubule bundles is induced by the interaction of CEP135 with tubulin, protofilaments and microtubules. A microtubule binding site has been detected between 96–108 residues and this segment integrated with positively charged surface patch 2; this was discovered in an atomic CEP135 model study, which suggested that the basic amino acids of the microtubule binding domain mediate interactions with the outer surface of the microtubule that is negatively charged (Hilbert et al., 2016). Further studies of major microtubule binding sites discovered a segment of 13 amino acids comprising 96–108 residues leading to microtubule binding activity of CEP135-N. Within this segment three lysine residues, K101, K104 and K108, were identified that contribute highly positive electrostatic surface potential of patch 2. These three lysine residues play a critical role in efficient microtubule bundling or cross linking by CEP135-N. Binding ability of two or more microtubules of CEP135-N leads to the development of microtubule triplets or joining of adjacent triplets within the microtubule wall. In such a way, the centrosomal protein CEP135 plays a key role in the biosynthesis of centrosomes which control the cell (Hilbert et al., 2016).
(ix). MCPH9 (CEP152)
CEP152 is a protein coding gene located on chromosome 15q21.1, comprising 149,368 bp, 38 exons, 1710 amino acids, with a molecular mass of 195,626 Da (Jamieson et al., 2000). The CEP152 gene is also known as Asterless, KIAA0912, SCKL5 and MCPH9 (Dzhindzhev et al., 2010). In cells this gene is localized in centrosomes and in the brain it is localized in the cerebral cortex (Faheem et al., 2015). Centrosomal protein 152 contains (KW-0175) a CCD (KW-9994), also known as a Heptad Repeat pattern including five different conserved protein sequences that lead to the coiled-coil protein domain formation, these are formed of 234–490, 615–664, 700–772, 902–993 and 1170–1241 amino acids (Kalay et al., 2011). The protein of this gene organizes the microtubules of cells and has a very important role in shaping the cell, polarity, receptivity and cellular division. The protein localized in the microtubule and centrosome interacts with PLK4, CENPJ (via N-terminus), CINP, CDKRAP2, WDR62, CEP63, CEP131 and DEUP1 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Kalay et al., 2011; Fırat-Karalar & Stearns, 2014). The interaction of CEP152 with CEP63, CDK5RAP2 and WDR62 form a bit by bit assembled complex at the centrosome creating a ring close to the parental centriole. CEP152 plays an important role in centrosome duplication/shape and cell/polarity/motility, and also functions as a molecular scaffold that facilitates the interaction of CENPJ and PLK4, two molecules that play an important role in centriole configuration (Cizmecioglu et al., 2010). It is suggested to take PLK4 away from PLK4:CEP92 complexes in early G1 daughter centrioles and to reposition PLK4 at the outer boundary of a new forming CEP152 ring structure. It also plays an essential role in deutrosome-mediated centriole amplification that can form many centrioles. Overexpression of CEP152 can drive amplification of centrioles (Dzhindzhev et al., 2010). The CEP152 (human) gene is an ortholog of the Drosophila asterless (asl) gene, comprising 72,835 bp and 1710 amino acids and with a protein with a molecular weight of 152 kDa. A mutation in the CEP152 protein that converts glutamine into proline is assumed to be pathogenic and can disrupt the potential coiled-coiled protein domain, which results in reduction of head size. Similarly, a greater head size reduction was also seen in compound heterozygous females compared with missense homozygous females. The truncated protein suggests a nonsense-mediated decay of the mutated transcript; these findings have also shown that throughout a functional assay to determine subcellular localization that the wild-type CEP152–GFP fusion protein can be found in γ-tubulin co-structures. Mutant CEP152 flagged with GFP failed to co-localize with the γ-tubulin, which furthermore substantiate pathogenicity caused by this mutation (Guernsey et al., 2010).
(x). MCPH10 (ZNF335)
ZNF335 is a protein coding gene located on chromosome 20q13.12, comprising 24,258 bp, 28 exons, 1342 amino acids, with molecular mass of 144,893 Da (Deloukas et al., 2001). The ZNF335 gene is also known as NIF-1, NIF-2 and MCPH10 (Mahajan et al., 2002; Garapaty et al., 2008). In the cell this gene is localized in the nucleus and in the brain it is localized in the cerebral cortex (Faheem et al., 2015). ZNF335 codes for the zinc finger protein 335 and has a repeat domain and a zinc finger C2H2-type (IPR013087) domain (Klug, 1999). The ZNF335 protein is a part or an associated component of some histone methyltransferase complexes and is also involved in regulating transcription through recruitment of these complexes to gene promoters and via the nuclear hormone receptor it also increases ligand dependent transcriptional activation. In addition, it is also very important for proliferation and self-renewal of neural progenitor cells by controlling the regulation of specific genes that are involved in brain development, including REST, and it is also involved in regulating the expression of genes that are involved in somatic development, for example, lymphoblast proliferation. This gene also encodes a component of the vertebrate-specific trithorax H3K4-methylation chromatin remodelling complex that controls central nervous system (CNS) gene expression and cell fate (Yang et al., 2012). ZNF335 is very important for progenitor cell division/differentiation; mutation in this gene leads to the degeneration of neurons, and knockdown of this gene causes a reduction in the size of the brain with a lack of cortex formation as well as disrupted differentiation and proliferation of neuronal cells (McLean et al., 2017). Due to a mutation in ZNF335, MCPH10 was identified in an affected member of an Arab Israeli family. The mutation was identified in exon 20 as a homozygous 3332G-A transition, which resulted in an Arg1111 to His (R1111H) substitution at the conserved residue in the 13th zinc-finger domain. This mutation was found at the final point of a splice donor site that disrupts normal splicing, which leads to the formation of unusually large transcripts that contain intron 19 and 20 with a proposed premature termination sequence (Yang et al., 2012). Levels of ZNF335 protein is severely reduced in patient cells. Various studies regarding this gene have revealed that ZNF335 is attached to the chromatin remodelling complex (analogue to the TrxG (trithorax) complex in Drosophila) involving H3K4 methyltransferase, which can control the expression of important genes in various pathways. nBAF is a neural-specific chromatin regulatory complex, any mutation in this complex disrupts the proliferation of cells, indicating that it can also lead to microcephaly. Deficiency of the ZNF335 genes leads to the degeneration of neurons, which makes it much more critical in contrast to the other microcephaly syndromes that are related to postnatal survival. Studies of in vitro and in vivo mouse models revealed that the knockdown of ZNF335 causes a severe reduction in the size of the brain, which has no cortex, and disturbs the differentiation and proliferation of nerve cells (Yang et al., 2012).
(xi). MCPH11 (PHC1)
The PHC1 gene is located on chromosome12p13.31 and encodes a protein containing 1004 amino acids with a molecular mass of 105,534 Da. The PHC1 gene is also known as Early Developmental Regulator 1, HPH1, EDR1, RAE28, PH1 and MCPH11 (Chavali et al., 2017). MCPH11 is a disease that is caused by a homozygous mutation in the PHC1 gene on chromosome 12p13. PHC1 takes part in the regulation of the cell cycle, and its mutation highlights the role of chromatin remodelling in the pathogenesis of primary microcephaly (Awad et al., 2013). Enhanced geminin expression was observed in cells with a PHC1 mutation. A core component of canonical PRC1 is PHC1 as a genetic basis for MCPH11 in a family with reported consanguinous homozygous missense PHC1 variants. The functional analysis of this pathogenic variant in patient cells revealed lower PHC1 expression with lower genome wide H2AUb1 levels and impaired recruitment of PHC1 to loci of DNA damage and repair (Srivastava et al., 2017). Interestingly, the MCPH11 PHC1 variant essentially had an impact on neural development in affected individuals, potentially highlighting a sensitivity of the developing brain to dysregulation of H2AUb1 modification exchange. As PHC1 regulates the cell cycle, PHC1 mutation highlights the role of chromatin remodelling in the pathogenesis of primary microcephaly (Srivastava et al., 2017; Triglia et al., 2017).
(xii). MCPH12 (CDK6)
CDK6 encodes a protein of the cyclin dependent protein kinase (CDK) family, located on chromosome 7q21.2 with a genome size of 231,707 bp, 326 amino acids, 10 exons and a molecular weight of 36,983 Da. The CDK6 protein controls the mechanism of the cell cycle and plays a very significant role in differentiation of different cell types. A mutated CDK6 gene or knockdown cells cause distorted nuclei disorganization of the spindles and microtubule and supernumerary centrosomes, whereas a decrease in proliferation was also observed in patients. Mutation in cells also disturbs apical neuronal precursor cell proliferation, which leads to the imbalance between symmetric and asymmetric cell division that causes progenitor cell depletion that could be the basis for reduction in neuronal cell production that finally leads to primary microcephaly (Hussain et al., 2013). In a recent study, most related genes that code for CDK6 were found to have a homozygous single nucleotide substitution, 589G > A. This mutation leads to altered mitosis because during the mitosis process patient primary fibroblasts failed to recruit CDK6 to the centrosome and it also affects cellular localization (Grossel et al., 1999). Moreover, this study was also substantiated by another study that revealed reduced proliferation capacity of CDK6 patient primary fibroblasts and knockdown cells (Grossel & Hinds, 2006).
(xiii). MCPH13 (CENPE)
CENPE is a protein coding gene located on chromosome 4q24, comprising 492,604 bp and 2701 amino acids. This gene encodes a protein that has signal transducer activity and sequence specific DNA binding activity. It also has transcription factor activity. Domains of this gene include Ploop_NTPase, Kinesin_motor_dom, CENPE, Kinesin_motor_CS and Kinesin-like FAM. MCPH is a heterogeneous autosomal recessive disorder. In last four years studies there has been increased knowledge about the new mutated genes involved in MCPH and intense work is done at both the clinical and cellular level to determine disease mechanisms. The functions of proteins encoded by WDR62, CASC5, PHC1, CDK6, CENP-E, CENP-F, CEP63, ZNF335, PLK4 and TUBGPC genes, have been added to the complex network of critical cellular processes that play an important role in brain growth and size. In a male child with the mutated CENPE gene and who had microcephaly, his head was seen to start squeezing at the age of 5 and after some years he died. The boy also had dysmorphic facial features, including, prominent nose and sloping forehead. His sister of 3 years old also had microcephaly and also showed the disease phenotype. Up until now various mutations have been found in CENPE (Ahmad et al., 2017).
(xiv). MCPH14 (SASS6)
The SAS-6 gene is located on chromosome-1p21.2, comprising 49,552 bp, 657 amino acids, 17 exons and a mass of 74,397 Da. The SAS-6 gene encodes a protein known as Spindle assembly abnormal protein-6 homolog, which is a coiled-coil protein. The gene is found within the cytoplasm, cytoskeleton, microtubule organizing centre, centrosome, cytoskeleton and centrioles. The most critical and fundamental role of the SAS-6 gene is to help in centriole formation by producing a procentriole. Centrioles play a crucial role in cilia and flagella and so are of much importance and their formation is strongly linked to cell duplication and centrosome replication (Gupta et al., 2015; Arquint & Nigg, 2016). To ensure genome integrity, centriole replication is essential. SAS-6 self-assembles into a cartwheel structure, and is able to undergo dimerization and oligomerization thus supporting centriole formation. The most primitive pathway for assembling centrioles involves seven major constituents: STIL, γ-tubulin, Plk4, CPAP, Cep135, Cep135 and hSAS6.
After attachment of P1k4, Cep135 assembles at the parental centriole position, then phosphorylated STIL and hsSAS-6 gather and through a positive feedback mechanism form a complex with CPAP and initiate the formation of the procentriole (Ohta et al., 2002; Gopalakrishnan et al., 2010). HsSAS-6 located in the cytoplasm gathers around the centriole and forms a complex with Cep135 to initiate cartwheel structure organization by oligomerization by passing through G1/S phase. G2/S phase leads to the stability of hsSAS-6 (Strnad et al., 2007; Keller et al., 2014). A family was selected from Dera Ismail Khan, an urban area in the province of Pakistan, Khyber Pakhtunkhwa. A pedigree analysis of five generations provided a result of four persons diagnosed with several physiological and mental issues resulting from consanguineous marriages. Out of four, two were girls, aged 3.5 and 5 years, and two were men, aged 42 and 50. All four experienced mental retardation, low level IQ (between 20–40), pronunciation issues and even walking problems. By performing several tests including tomography, STR, SNP, genome-wide linkage analysis and whole-genome analysis etc., it was concluded that the SAS-6 gene was responsible for this condition, which was determined to be primary microcephaly. In the PISA domain of the SAS-6 gene, the replacement of threonine with isoleucine at position 62 causes primary microcephaly. Microcephaly linked with hsSAS-6 is an autosomal recessive disease. The drastic impact of hsSAS-6 deficiency on centriole formation also effects cell division thus disturbing normal neurogenesis and ultimately affecting brain nourishment (Khan et al., 2014).
(xv). MCPH15 (MFSD2A)
MFSD2A, also known as MCPH15 and NLS1, encodes a transmembrane protein required for brain uptake of omega-3 (Docosahexaenoic acid) and different long-chain fatty acids (lysophosphatidylcholine) (Alakbarzade et al., 2015; Guemez-Gamboa et al., 2015). This gene is localized on chromosome 1p34.2, comprising 14 exons, 543 amino acids, with a genome size of 14,857 bp and a molecular weight of 60,170 Da. MFSD2A is mainly localized to the cytosol, plasma membrane and cytoplasmic bodies in the human brain as well as other cells. The MFSD2A gene is also present in the cerebral cortex (endothelial cells, neuronal cells, etc) and is an essential component of the blood–brain barrier (Reiling et al., 2011). MFSD2A is evolutionarily conserved from teleost fish to humans. Mutation in the MFSD2A gene causes microcephaly leading to a brain volume that is smaller than usual. High expression of MFSD2A is observed in the blood–brain barrier of both humans and mice. Reduced brain DHA levels along with reduced LPC uptake and microcephaly as the most prominent phenotype is reported in MFSD2A-deficient mice (Nguyen et al., 2014). As supported by these studies, LPC is essential for normal development of human and mice brains (Quek et al., 2016). Most affected people have delayed speech and language skills, and delayed motor skills (standing, sitting and walking). Slim, sloping forehead, behavioural issues and short stature compared to others in their family was also observed. Two distinctive homozygous missense mutations were seen to occur in the MFSD2A gene in affected individuals from two consanguineous families from northern Africa with autosomal recessive microcephaly causing early death (Guemez-Gamboa et al., 2015). Homozygous missense mutations occurring in the MFSD2A gene were also seen in affected individuals belonging to a consanguineous Pakistani family. It was also seen that morpholino knockdown of orthologous MFSD2A in Zebra fish brought about early postnatal lethality, microcephaly and blood–brain barrier interruption. Likewise, it was also found that MFSD2A-null mice had a roughly 40% expansion in plasma levels of LPC compared to controls with debilitated take-up of LPC into the brain (cerebrum) (Guemez-Gamboa et al., 2015).
(xvi). MCPH16 (ANKLE2)
Several mutated genes have been identified to play an important role in the regulation of the cell cycle and cell proliferation. One of them is the ANKLE gene encoding the LEM4 protein, which plays a critical role in nuclear envelope formation through the mitotic phosphorylation of BAF protein during mitosis. Upon mitotic entry, the LEM protein interacts with a chromatin-binding protein known as Barrier to Autointegration. BAF regulates chromatin structure and chromosome segregation in gene expression and development. Recent studies have revealed that the ANKLE2 gene mapped to chromosome 12q24.33, has a size of 938 amino acids and a molecular mass of approximately 10,000 Da. According to current studies its N-terminal tail is followed by a transmembrane domain of approximately 40 amino acids, which is known as the LEM domain and retains two central ankyrin repeats and a cytoplasmic C-terminus. Mutation in this gene leads to abnormal development of the nuclear envelope during mitosis. Mutated ANKLE2 gene leads to impaired brain development and reduces brain size as a result of a decreased number of neuroblasts, reduced mitosis, less cell proliferation and increased apoptosis (Faheem et al., 2015). This improper development of the brain contributes to MCPH16, which is predominantly caused by consanguineous marriages. Clinical review or neuroimaging of patients with MCPH16 reveals symptoms such as reduced brain size, intellectual disability, knee contractures, enlarged posterior horns of the lateral ventricles, adducted thumbs and epilepsy (von der Hagen et al., 2014). Recently, whole-exome sequencing and analysis of genes that are potentially involved in neurological disorders identified compound heterozygous mutations in the ANKLE2 gene in exon 11 (c.23344C-T transition as a result of Gln782 to Ter (Q782X) substitution), and in exon 10 (c.1717C-T as a result of Leu 573 to Val (L573 V) substitution). Analyses of brains from Drosophila identified a mutation in l (l) G0222, the homolog of the ANKLE2 gene in humans. This mutation results in loss of thoracic bristles and impairs development of sensory organs in clones. Such evidence suggests that genes identified in Drosophila clones and variant alleles in Homo sapien homologs in families with Mendelian diseases play a critical role in the discovery of disease causing genes and the mechanisms involved in these disorders (Yamamoto et al., 2014).
(xvii). MCPH17 (CIT)
The CIT gene encoding citron rho-interacting serine/threonine kinase is critical for the development of a normal sized brain. CIT has an N-terminal kinase domain and many C-terminal domains that aid interaction between the components of its contractile ring (i.e., Rho A, aniline, actin and myosin) (Harding et al., 2016). This gene is located on chromosome 12q24.23, comprising 50 exons, 2027 amino acids, with a genome size and molecular weight of 191,501 bp and 231,431 Da, respectively. It has four already known isoforms and a 6207 bp ORF. The CIT protein is located in the mitotic cells midbody and cleavage furrow. This gene is conserved in many species including H. sapiens, M. musculus, D. melanogaster, M .mulatta, P. troglodytes, X. tropicalis, X. tropicalis and A. gambiae. CIT is important in phosphorylation of many compounds, for the maintenance of the structure of the midbody during cell division and in the completion of cytokinesis due to kinase activity (Di Cunto et al., 2000; Li et al., 2016). Homozygous mutation in the kinase domain of CIT causes MCPH by the loss or activation of protein citron kinase (Basit et al., 2016; Harding et al., 2016). Individuals with these mutations exhibit abnormal cytokinesis characterized by deferred mitosis, spindles having multiple poles, chromosomal instability, aneuploidy, PF3 cycle arrest initiation, massive apoptosis and have multinucleated neurons throughout the cerebral cortex (Li et al., 2016). CITK deficient cells have heightened levels of sensitivity to ionizing radiation and inoperative rehabilitation from radiation-induced DNA damage (Bianchi et al., 2017). Mutations in the CIT gene causes a severe neurological disorder characterized by very small head and individuals with this microcephaly have intellectually disability, axia hypotonia, brisk reflexes, social impairment, thin corpus callosum and reduced cerebral volume and sometimes have a sloping forehead, dysmorphic features and their brain shows gyral patterns (Harding et al., 2016; Li et al., 2016; Shaheen et al., 2016). In rodents CITK is important for the proliferation of neural progenitor cells and male germ cell precursors. CIT knockout mice show ataxia, testicular hypoplasia, lethal seizures, growth deficiencies, severe reduction in brain size and often show simplified gyral patterns and temperaments linked to cytokinesis defects, that is, they have multinucleated neurons throughout the cerebrum and cortex and show massive apoptosis (Harding et al., 2016; Li et al., 2016; Bianchi et al., 2017).
(xviii). MCPH18 (WDFY3)
WDFY3, encoding Alfy protein, performs binding and beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity. Alfy protein domains are: WD40_repeat domain, WD40/YVTN domain and FYVE type domain. This gene is located on chromosome 8p23, comprising 3526 amino acids, with a genome size and molecular weight of 296,855 bp and 395,258 Da, respectively, and two already known isoforms. WDFY3 is used as a scaffold protein required for the selective self-breakdown of macromolecules such as aggregation-prone proteins. Alfy protein is highly expressed in the developing CNS and crucial for the development of axonal tracts. The protein breakdown is mandatory to provide the proper atmosphere in which to coordinate complicated cell signalling events and to enhance cellular remodelling. The Wdfy3 gene is suspected to be involved in neurodevelopmental ataxia such as microcephaly and autism. Its homolog is Blue Cheese (bchs), which is abundantly expressed in the developing and adult fly CNS. Wdfy3 dysfunction in mice has subtle effects on migration and proliferation of neural progenitor cells. The affected mice display larger brains as a repercussion of an analogous alteration in the mechanism of radial glial mitoses to symmetric from asymmetric. The other 17 genes of microcephaly follow the autosomal recessive mode of inheritance; however, the WDFY3 gene follows the autosomal dominant mode of inheritance. Heterozygous mutation has been reported in the WDFY3 gene (R2637W), a c.7909C-T transition means tryptophan replaces arginine at the 2637 position. In vitro expression studies on Drosophila have revealed increased levels of DVL3 upon transfection of Drosophila cells with mutant human WDFY3. Increased levels of DVL3 cause abnormal activation of Wnt signalling as well as continued generation of apical progenitor cells without transition to differentiation and generation of the basal progenitor cell layers in the cerebral cortex, thus resulting in impaired cortical development and microcephaly (Jayaraman et al., 2018).
3. Conclusion
Microcephaly is considered to be a rare neurodevelopmental disorder with many underlying causes. Pathogenicity analysis led to the discovery of 18 genes being a possible reason for this primary neurogenic mitotic syndrome. Recent experiments for successful generation of model animals for MCPH genes opened the door for researchers to further comprehend the pathophysiology and aetiology of MCPH. Improvement in the genotypic and imaging techniques along with organizing patients according to the basis of their genotypic homogeneity would enable us to better predict a correlation between genotype–phenotype. Mutational analysis of patients from different regions of the Middle East, and especially Pakistan, would aid in counselling and diagnostic approaches for MCPH ensuring better health quality for patients. This study has allowed us a better understanding of neuronal production processes by stem cells. The ongoing research projects on MCPH genes will lead us towards better understanding of this rare nonprogressive neuropediatric disorder. Moreover, MCPH genes are strong candidates for brain development and evolutionary studies.
Declaration of interest
None.
References
- Ahmad I, Baig SM, Abdulkareem AR, Hussain MS, Sur I, Toliat MR, Nürnberg G, Dalibor N, Moawia A, Waseem SS & Asif M (2017). Genetic heterogeneity in Pakistani microcephaly families revisited. Clinical Genetics 92(1), 62–68. [DOI] [PubMed] [Google Scholar]
- Alakbarzade V, Hameed A, Quek DQ, Chioza BA, Baple EL, Cazenave-Gassiot A, Nguyen LN, Wenk MR, Ahmad AQ, Sreekantan-Nair A & Weedon MN (2015). A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nature Genetics 47(7), 814–817. [DOI] [PubMed] [Google Scholar]
- Al-Dosari MS, Shaheen R, Colak D & Alkuraya FS (2010). Novel CENPJ mutation causes Seckel syndrome. Journal of Medical Genetics 47(6), 411–414. [DOI] [PubMed] [Google Scholar]
- Araujo Junior E, Martins Santana EF, Martins WP, Junior JE, Ruano R, Pires CR & Filho SM (2014). Reference charts of fetal biometric parameters in 31,476 Brazilian singleton pregnancies. Journal of Ultrasound in Medicine 33(7), 1185–1191. [DOI] [PubMed] [Google Scholar]
- Arquint C & Nigg EA (2016). The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochemical Society Transactions 44(5), 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad S, Al-Dosari MS, Al-Yacoub N, Colak D, Salih MA, Alkuraya FS & Poizat C (2013). Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Human Molecular Genetics 22(11), 2200–2213. [DOI] [PubMed] [Google Scholar]
- Basit S, Al-Harbi KM, Alhijji SA, Albalawi AM, Alharby E, Eldardear A & Samman MI (2016). CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Human Genetics 135(10), 1199–1207. [DOI] [PubMed] [Google Scholar]
- Bastaki F, Mohamed M, Nair P, Saif F, Tawfiq N, Aithala G, El-Halik M, Al-Ali M & Hamzeh AR (2016). Novel splice-site mutation in WDR62 revealed by whole-exome sequencing in a Sudanese family with primary microcephaly. Congenital Anomalies 56(3), 135–137. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A & Raff JW (2006) Flies without centrioles. Cell 125(7), 1375–1386. [DOI] [PubMed] [Google Scholar]
- Bhat V, Girimaji S, Mohan G, Arvinda H, Singhmar P, Duvvari M & Kumar A (2011). Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations. Clinical Genetics 80(6), 532–540. [DOI] [PubMed] [Google Scholar]
- Bianchi FT, Tocco C, Pallavicini G, Liu Y, Vernì F, Merigliano C, Bonaccorsi S, El-Assawy N, Priano L, Gai M & Berto GE (2017). Citron kinase deficiency leads to chromosomal instability and TP53-sensitive microcephaly. Cell Reports 18(7), 1674–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgüvar K, Öztürk AK, Louvi A, Kwan KY, Choi M, Tatlı B, Yalnızoğlu D, Tüysüz B, Çağlayan AO, Gökben S & Kaymakçalan H (2010). Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467(7312), 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF & Walsh CA (2002). ASPM is a major determinant of cerebral cortical size. Nature Genetics 32(2), 316–320. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga S, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO & Costa SM (2005). A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nature Genetics 37(4), 353–355. [DOI] [PubMed] [Google Scholar]
- Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida GH, Hennekam RC, Maher ER, Fryns JP & Alswaid A (2003). Protein-truncating mutations in ASPM cause variable reduction in brain size. American Journal of Human Genetics 73(5), 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali PL, Stojic L, Meredith LW, Joseph N, Nahorski MS, Sanford TJ, Sweeney TR, Krishna BA, Hosmillo M, Firth AE & Bayliss R (2017). Neurodevelopmental protein Musashi 1 interacts with the Zika genome and promotes viral replication. Science 357(6346), 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J-H, Chang C-J, Chen C-Y & Tang TK (2006). Depletion of CPAP by RNAi disrupts centrosome integrity and induces multipolar spindles. Biochemical and Biophysical Research Communications 339(3), 742–747. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiß U, Antony C & Hoffmann I (2010). Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. The Journal of Cell Biology 191(4), 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Feng Z, Richens JH, Baumbach J, Wainman A, Bakshi SD, Dobbelaere J, Johnson S, Lea SM & Raff JW (2014). The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Developmental Cell 28(6), 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie V (1960). The genetics and sub-classification of microcephaly. Journal of Intellectual Disability Research 4(1), 42–47. [DOI] [PubMed] [Google Scholar]
- Craig R & Norbury C (1998). The novel murine calmodulin-binding protein Sha1 disrupts mitotic spindle and replication checkpoint functions in fission yeast. Journal of Cell Science 111(24), 3609–3619. [DOI] [PubMed] [Google Scholar]
- Darvish H, Esmaeeli-Nieh S, Monajemi GB, Mohseni M, Ghasemi-Firouzabadi S, Abedini SS, Bahman I, Jamali P, Azimi S, Mojahedi F, Dehghan A, Shafeghati Y, Jankhah A, Falah M, Soltani, Banavandi MJ, Ghani M, Garshasbi M, Rakhshani F, Naghavi A, Tzschach A, Neitzel H, Ropers HH, Kuss AW, Behjati F, Kahrizi K & Najmabadi H (2010). A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. Journal of Medical Genetics 47(12), 823–828. [DOI] [PubMed] [Google Scholar]
- David A, Amartely H, Rabinowicz N, Shamir M, Friedler A & Izraeli S (2016). Molecular basis of the STIL coiled coil oligomerization explains its requirement for de-novo formation of centrosomes in mammalian cells. Scientific Reports 6, 24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Matthews LH, Ashurst J, Burton J, Gilbert JGR, Jones M, Stavrides G, Almeida JP, Babbage AK, Bagguley CL & Bailey J (2001). The DNA sequence and comparative analysis of human chromosome 20. Nature 414(6866), 865–871. [DOI] [PubMed] [Google Scholar]
- Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A, Atzori C, Turco E, Triolo R, Dotto GP & Silengo L (2000). Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28(1), 115–127. [DOI] [PubMed] [Google Scholar]
- do Carmo Avides M & Glover DM (1999). Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science 283(5408), 1733–1735. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Quan DY, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A & Bettencourt-Dias M (2010). Asterless is a scaffold for the onset of centriole assembly. Nature 467(7316), 714. [DOI] [PubMed] [Google Scholar]
- Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, Pushparaj PN, Ahmed F, Algahtani HA, Al-Qahtani MH & Jamal HS (2015). Molecular genetics of human primary microcephaly: an overview. BMC Medical Genomics 8(1), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Baig S, Tommerup N & Kjaer KW (2010). Craniosynostosis-microcephaly with chromosomal breakage and other abnormalities is caused by a truncating MCPH1 mutation and is allelic to premature chromosomal condensation syndrome and primary autosomal recessive microcephaly type 1. American Journal of Medical Genetics Part A 152(2), 495–497. [DOI] [PubMed] [Google Scholar]
- Farooq M, Fatima A, Mang Y, Hansen L, Kjaer KW, Baig SM, Larsen LA & Tommerup N (2016). A novel splice site mutation in CEP135 is associated with primary microcephaly in a Pakistani family. Journal of Human Genetics 61(3), 271. [DOI] [PubMed] [Google Scholar]
- Farag HG, Froehler S, Oexle K, Ravindran E, Schindler D, Staab T, Huebner A, Kraemer N, Chen W & Kaindl AM (2013). Abnormal centrosome and spindle morphology in a patient with autosomal recessive primary microcephaly type 2 due to compound heterozygous WDR62 gene mutations. Orphanet Journal of Rare Diseases 8, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fırat-Karalar EN & Stearns T (2014). The centriole duplication cycle. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 369(1650), 20130460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Morphew M, Joseph JD, Means AR & Davis TN (2002). Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth and Differentiation-Publication American Association for Cancer Research 13(2), 47–58. [PubMed] [Google Scholar]
- Gai M, Bianchi FT, Vagnoni C, Vernì F, Bonaccorsi S, Pasquero S, Berto GE, Sgrò F, Chiotto AM, Annaratone L & Sapino A (2016). ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Reports 17(10), 1396–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty S, Mahajan MA & Samuels HH (2008). Components of the CCR4–NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting factor NIF-1. Journal of Biological Chemistry 283(11), 6806–6816. [DOI] [PubMed] [Google Scholar]
- Garshasbi M, Motazacker MM, Kahrizi K, Behjati F, Abedini SS, Nieh SE, Firouzabadi SG, Becker C, Rüschendorf F, Nürnberg P & Tzschach A (2006). SNP array-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Human Genetics 118(6), 708–715. [DOI] [PubMed] [Google Scholar]
- Genin A, Desir J, Lambert N, Biervliet M, Van Der Aa N, Pierquin G, Killian A, Tosi M, Urbina M, Lefort A & Libert F (2012). Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Human Molecular Genetics 21(24), 5306–5317. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S & Avidor-Reiss T (2010). Self-assembling SAS-6 multimer is a core centriole building block. Journal of Biological Chemistry 285(12), 8759–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof Y-D & Nigg EA (2007). Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. Journal of Cell Science 120(24), 4321–4331. [DOI] [PubMed] [Google Scholar]
- Grossel MJ & Hinds PW (2006). From cell cycle to differentiation: an expanding role for cdk6. Cell Cycle 5(3), 266–270. [DOI] [PubMed] [Google Scholar]
- Grossel MJ, Baker GL & Hinds PW (1999). cdk6 can shorten G(1) phase dependent upon the N-terminal INK4 interaction domain. Journal of Biological Chemistry 274(42), 29960–29967 [DOI] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Nguyen LN, Yang H, Zaki MS, Kara M, Ben-Omran T, Akizu N, Rosti RO, Rosti B, Scott E & Schroth J (2015). Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nature Genetics 47(7), 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC & Ferguson M (2010). Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. American Journal of Human Genetics 87(1), 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul A, Hassan MJ, Mahmood S, Chen W, Rahmani S, Naseer MI, Dellefave L, Muhammad N, Rafiq MA, Ansar M, Chishti MS, Ali G, Siddique T & Ahmad W (2006. a). Genetic studies of autosomal recessive primary microcephaly in 33 Pakistani families: novel sequence variants in ASPM gene. Neurogenetics 7(2), 105–110. [DOI] [PubMed] [Google Scholar]
- Gul A, Hassan MJ, Hussain S, Raza SI, Chishti MS & Ahmad W (2006. b). A novel deletion mutation in CENPJ gene in a Pakistani family with autosomal recessive primary microcephaly. Journal of Human Genetics 51(9), 760–764. [DOI] [PubMed] [Google Scholar]
- Gupta H, Badarudeen B, George A, Thomas GE, Gireesh KK & Manna TK (2015). Human SAS-6 C-terminus nucleates and promotes microtubule assembly in vitro by binding to microtubules. Biochemistry 54(41), 6413–6422. [DOI] [PubMed] [Google Scholar]
- Harding BN, Moccia A, Drunat S, Soukarieh O, Tubeuf H, Chitty LS, Verloes A, Gressens P, El Ghouzzi V, Joriot S & Di Cunto F (2016). Mutations in citron kinase cause recessive microlissencephaly with multinucleated neurons. American Journal of Human Genetics 99(2), 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MJ, Khurshid M, Azeem Z, John P, Ali G, Chishti MS & Ahmad W (2007). Previously described sequence variant in CDK5RAP2 gene in a Pakistani family with autosomal recessive primary microcephaly. BMC Medical Genetics 8(1), 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert M, Noga A, Frey D, Hamel V, Guichard P, Kraatz SH, Pfreundschuh M, Hosner S, Flückiger I, Jaussi R & Wieser MM (2016). SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nature Cell Biology 18(4), 393. [DOI] [PubMed] [Google Scholar]
- Hung L-Y, Chen H-L, Chang C-W, Li B-R & Tang TK (2004). Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Molecular Biology of the Cell 15(6), 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L-Y, Tang C-J. C & Tang TK (2000). Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the γ-tubulin complex. Molecular and Cellular Biology 20(20), 7813–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MS, Baig SM, Neumann S, Nürnberg G, Farooq M, Ahmad I, Alef T, Hennies HC, Technau M, Altmüller J & Frommolt P (2012). A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. The American Journal of Human Genetics 90(5), 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MS, Baig SM, Neumann S, Peche VS, Szczepanski S, Nurnberg G, Tariq M, Jameel M, Khan TN, Fatima A, Malik NA, Ahmad I, Altmüller J, Frommolt P, Thiele H, Höhne W, Yigit G, Wollnik B, Neubauer BA, Nürnberg P & Noegel AA (2013). CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly Human Molecular Genetics 22(25), 5199–5214. [DOI] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ & Karbani G (2002). Identification of microcephalin, a protein implicated in determining the size of the human brain. The American Journal of Human Genetics 71(1), 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Fryns J-P, Jacobs J, Matthijs G & Abramowicz MJ (2000). Primary autosomal recessive microcephaly: MCPH5 maps to 1q25-q32. American Journal of Human Genetics 67(6), 1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Bae B-I & Walsh CA (2018). The genetics of primary microcephaly. Annual Review of Genomics and Human Genetics doi: 10.1146/annurev-genom-083117-021441 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- Jayaraman D, Kodani A, Gonzalez D, Mancias J, Mochida G, Vagnoni C, Harper JW, Reiter J, Yu T, Bae BI & Walsh C (2017). Microcephaly proteins Wdr62 and Aspm define a mother centriole complex regulating centriole biogenesis, apical complex and cell fate (S46.001). Neurology 88(16 Supplement). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadir R, Harel T, Markus B, Perez Y, Bakhrat A, Cohen I, Volodarsky M, Feintsein-Linial M, Chervinski E, Zlotogora J & Sivan S (2016). ALFY-controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genetics 12(3), e1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, Gerard B, Verloes A, Mani S & Gressens P (2010). Many roads lead to primary autosomal recessive microcephaly. Progress in Neurobiology 90(3), 363–383. [DOI] [PubMed] [Google Scholar]
- Kakar N, Ahmad J, Morris-Rosendahl DJ, Altmüller J, Friedrich K, Barbi G, Nürnberg P, Kubisch C, Dobyns WB & Borck G (2015). STIL mutation causes autosomal recessive microcephalic lobar holoprosencephaly. Human Genetics 134(1), 45–51. [DOI] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tüysüz B, Nürnberg G, Kiess W, Koegl M, Baessmann I, Buruk K, Toraman B, Kayipmaz S, Kul S, Ikbal M, Turner DJ, Taylor MS, Aerts J, Scott C, Milstein K, Dollfus H, Wieczorek D, Brunner HG, Hurles M, Jackson AP, Rauch A, Nürnberg P, Karagüzel A & Wollnik B (2011). CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nature Genetics 43(1), 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D, Orpinell M, Olivier N, Wachsmuth M, Mahen R, Wyss R, Hachet V, Ellenberg J, Manley S & Gonczy P (2014). Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. Journal of Cell Biology 204(5), 697–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RI, Robinson D, Puffenberger EG, Strauss KA & Morton DH (2002). Amish lethal microcephaly: a new metabolic disorder with severe congenital microcephaly and 2-ketoglutaric aciduria. American Journal of Medical Genetics Part A 112(4), 318–326. [DOI] [PubMed] [Google Scholar]
- Khan MA, Rupp VM, Orpinell M, Hussain MS, Altmüller J, Steinmetz MO, Enzinger C, Thiele H, Höhne W, Nürnberg G & Baig SM (2014). A missense mutation in the PISA domain of HsSAS-6 causes autosomal recessive primary microcephaly in a large consanguineous Pakistani family. Human Molecular Genetics 23(22), 5940–5949. [DOI] [PubMed] [Google Scholar]
- Kim H-T, Lee M-S, Choi J-H, Jung J-Y, Ahn D-G, Yeo S-Y, Choi D-K & Kim C-H (2011). The microcephaly gene aspm is involved in brain development in zebrafish. Biochemical and Biophysical Research Communications 409(4), 640–644. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Müller-Reichert T, Oegema K, Grill S & Hyman AA (2003). SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112(4), 575–587. [DOI] [PubMed] [Google Scholar]
- Klug A (1999). Zinc finger peptides for the regulation of gene expression. Journal of Molecular Biology 293(2), 215–218. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki J-I, Mochida GH, Risinger JI, Goldsmith P, Gunsior M, Solomon G, Gersch W, Kim JH, Barrett JC, Walsh CA, Jurka J, Masumoto H & Larionov V (2005). The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Human Molecular Genetics 14(15), 2155–2165. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Hijikata M, Watashi K, Masui O & Shimotohno K (2005). Centrosomal P4.1-associated protein is a new member of transcriptional coactivators for nuclear factor-κB. Journal of Biological Chemistry 280(13), 12430–12437. [DOI] [PubMed] [Google Scholar]
- Kraemer N, Picker-Minh S, Abbasi AA, Fröhler S, Ninnemann O, Khan MN, Ali G, Chen W & Kaindl AM (2016). Genetic causes of MCPH in consanguineous Pakistani families. Clinical Genetics 89(6), 744–745. [DOI] [PubMed] [Google Scholar]
- Kumar A, Blanton SH, Babu M, Markandaya M & Girimaji SC (2004). Genetic analysis of primary microcephaly in Indian families: novel ASPM mutations. Clinical Genetics 66(4), 341–348. [DOI] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR & Blanton SH (2009). Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. American Journal of Human Genetics 84(2), 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP & Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501(7467), 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal GF, Roberts E, Silva EO, Costa SMR, Hampshire DJ & Woods CG (2003). A novel locus for autosomal recessive primary microcephaly (MCPH6) maps to 13q12. 2. Journal of Medical Genetics 40(7), 540–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bielas SL, Zaki MS, Ismail S, Farfara D, Um K, Rosti RO, Scott EC, Tu S, Chi NC & Gabriel S (2016). Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. American Journal of Human Genetics 99(2), 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhou ZW & Wang ZQ (2016). The DNA damage response molecule MCPH1 in brain development and beyond. Acta Biochimica et Biophysica Sinica (Shanghai) 48(7), 678–685. [DOI] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA & Fleming MD (2010). Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137(11), 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan MA, Murray A & Samuels HH (2002). NRC-interacting factor 1 is a novel cotransducer that interacts with and regulates the activity of the nuclear hormone receptor coactivator NRC. Molecular and Cellular Biology 22(19), 6883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean E, Bhattarai R, Hughes BW, Mahalingam K & Bagasra O (2017). Computational identification of mutually homologous Zika virus miRNAs that target microcephaly genes. Libyan Journal of Medicine 12(1), 1304505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao L-R & Kaufman TC (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126(13), 2829–2839. [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Vitre B, Carpenter G, Abramowicz I, Gleeson JG, Paciorkowski AR, Cleveland DW, Dobyns WB & O'Driscoll M (2014). Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Human Genetics 133(8), 1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SH & Mundy NI (2014). Microcephaly genes evolved adaptively throughout the evolution of eutherian mammals. BMC Evolutionary Biology 14(1), 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouden C, de Tayrac M, Dubourg C, Rose S, Carre W, Hamdi-Roze H, Babron MC, Akloul L, Héron-Longe B, Odent S, Dupé V, Giet R & David V (2015). Homozygous STIL mutation causes holoprosencephaly and microcephaly in two siblings. PLoS One 10(2), e0117418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad F, Mahmood Baig S, Hansen L, Sajid Hussain M, Anjum Inayat I, Aslam M, Anver Qureshi J, Toilat M, Kirst E, Wajid M & Nürnberg P (2009). Compound heterozygous ASPM mutations in Pakistani MCPH families. American Journal of Medical Genetics Part A 149A(5), 926–930. [DOI] [PubMed] [Google Scholar]
- Murdock DR, Clark GD, Bainbridge MN, Newsham I, Wu YQ, Muzny DM, Cheung SW, Gibbs RA & Ramocki MB (2011). Whole-exome sequencing identifies compound heterozygous mutations in WDR62 in siblings with recurrent polymicrogyria. American Journal of Medical Genetics Part A 155(9), 2071–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer MI, Rasool M, Sogaty S, Chaudhary RA, Mansour HM, Chaudhary AG, Abuzenadah AM & Al-Qahtani MH (2017). A novel WDR62 mutation causes primary microcephaly in a large consanguineous Saudi family. Annals of Saudi Medicine 37(2), 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LN, Ma D, Shui G, Wong P, Cazenave-Gassiot A, Zhang X, Wenk MR, Goh EL & Silver DL (2014). Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509(7501), 503–506. [DOI] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Désir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A & Passemard S (2010). WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nature Genetics 42(11), 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y & Kuriyama R (2002). Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. Journal of Cell Biology 156(1), 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta AT, Murray JE, Yoon G, Akha ES, Harrison V, Bicknell LS, Ajilogba K, Stewart H, Kini U, Taylor JC & Keays DA (2012). A novel nonsense CDK5RAP2 mutation in a Somali child with primary microcephaly and sensorineural hearing loss. American Journal of Medical Genetics Part A 158(10), 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papari E, Bastami M, Farhadi A, Abedini SS, Hosseini M, Bahman I, Mohseni M, Garshasbi M, Moheb LA, Behjati F & Kahrizi K (2013). Investigation of primary microcephaly in Bushehr province of Iran: novel STIL and ASPM mutations. Clinical genetics, 83(5), 488–490. [DOI] [PubMed] [Google Scholar]
- Paramasivam M, Chang Y & LoTurco JJ (2007). ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle 6(13), 1605–1612. [DOI] [PubMed] [Google Scholar]
- Passemard S, Titomanlio L, Elmaleh M, Afenjar A, Alessandri JL, Andria G, de Villemeur TB, Boespflug-Tanguy O, Burglen L, Del Giudice E & Guimiot F (2009). Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology 73(12), 962–969. [DOI] [PubMed] [Google Scholar]
- Passemard S, Verloes A, de Villemeur TB, Boespflug-Tanguy O, Hernandez K, Laurent M, Isidor B, Alberti C, Pouvreau N, Drunat S & Gérard B (2016). Abnormal spindle-like microcephaly-associated (ASPM) mutations strongly disrupt neocortical structure but spare the hippocampus and long-term memory. Cortex 74, 158–176. [DOI] [PubMed] [Google Scholar]
- Pervaiz N & Abbasi AA (2016). Molecular evolution of WDR62, a gene that regulates neocorticogenesis. Meta Gene 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Mosalaganti S, Keller J, Mattiuzzo M, Overlack K, Krenn V, De Antoni A, Wohlgemuth S, Cecatiello V, Pasqualato S & Raunser S (2014). Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Molecular Cell 53(4), 591–605. [DOI] [PubMed] [Google Scholar]
- Ponting CP (2006). A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 22(9), 1031–1035. [DOI] [PubMed] [Google Scholar]
- Pulvers JN, Journiac N, Arai Y & Nardelli J (2015). MCPH1: a window into brain development and evolution. Frontiers in Cellular Neuroscience 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek DQ, Nguyen LN, Fan H & Silver DL (2016). Structural insights into the transport mechanism of the human sodium-dependent lysophosphatidylcholine transporter Mfsd2a. Journal of Biological Chemistry 291(18), 9383–9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Clish CB, Carette JE, Varadarajan M, Brummelkamp TR & Sabatini DM (2011). A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proceedings of the National Academy of Sciences 108(29), 11756–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G, Glover DM & Avides M. d. C (2002). A requirement for the Abnormal spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. Journal of Cell Science 115(5), 913–922. [DOI] [PubMed] [Google Scholar]
- Saadi A, Borck G, Boddaert N, Chekkour MC, Imessaoudene B, Munnich A, Colleaux L & Chaouch M (2009). Compound heterozygous ASPM mutations associated with microcephaly and simplified cortical gyration in a consanguineous Algerian family. European Journal of Medical Genetics 52(4), 180–184. [DOI] [PubMed] [Google Scholar]
- Saadi A, Verny F, Siquier-Pernet K, Bole-Feysot C, Nitschke P, Munnich A, Abada-Dendib M, Chaouch M, Abramowicz M & Colleaux L (2016). Refining the phenotype associated with CASC5 mutation. Neurogenetics 17(1), 71–78. [DOI] [PubMed] [Google Scholar]
- Sajid Hussain M, Marriam Bakhtiar S, Farooq M, Anjum I, Janzen E, Reza Toliat M, Eiberg H, Kjaer KW, Tommerup N, Noegel AA, Nürnberg P, Baig SM & Hansen L (2013). Genetic heterogeneity in Pakistani microcephaly families. Clinical Genetics 83(5), 446–451. [DOI] [PubMed] [Google Scholar]
- Saunders RDC, Avides M. d. C, Howard T, Gonzalez C & Glover DM (1997). The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. Journal of Cell Biology 137(4), 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgourdou P, Mishra-Gorur K, Saotome I, Henagariu O, Tuysuz B, Campos C, Ishigame K, Giannikou K, Quon JL, Sestan N, Caglayan AO, Gunel M & Louvi A (2017). Disruptions in asymmetric centrosome inheritance and WDR62-aurora kinase B interactions in primary microcephaly. Scientific Reports 7, 43708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Hashem A, Abdel-Salam GM, Al-Fadhli F, Ewida N & Alkuraya FS (2016). Mutations in CIT, encoding citron rho-interacting serine/threonine kinase, cause severe primary microcephaly in humans. Human Genetics 135(10), 1191–1197. [DOI] [PubMed] [Google Scholar]
- Shen J, Eyaid W, Mochida GH, Al-Moayyad F, Bodell A, Woods CG & Walsh CA (2005). ASPM mutations identified in patients with primary microcephaly and seizures. Journal of Medical Genetics 42(9), 725–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, McGrath B & Bielas SL (2017). Histone H2A monoubiquitination in neurodevelopmental disorders. Trends in Genetics 33(8), 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo A. A. S. F, Basto R, St Johnston D & Raff JW (2007). From stem cell to embryo without centrioles. Current Biology 17(17), 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A & Gonczy P (2007). Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Developmental Cell 13(2), 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran SK, Stumpf M, Salamon S, Ahmad I, Bhattacharya K, Fischer S, Müller R, Altmüller J, Budde B, Thiele H & Tariq M (2017). CDK5RAP2 interaction with components of the Hippo signaling pathway may play a role in primary microcephaly. Molecular Genetics and Genomics 292(2), 365–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanski S, Hussain MS, Sur I, Altmüller J, Thiele H, Abdullah U, Waseem SS, Moawia A, Nürnberg G, Noegel AA & Baig SM (2016). A novel homozygous splicing mutation of CASC5 causes primary microcephaly in a large Pakistani family. Human Genetics 135(2), 157–170. [DOI] [PubMed] [Google Scholar]
- Tan CA, Topper S, Melver CW, Stein J, Reeder A, Arndt K & Das S (2014). The first case of CDK5RAP2-related primary microcephaly in a non-consanguineous patient identified by next generation sequencing. Brain and Development 36(4), 351–355. [DOI] [PubMed] [Google Scholar]
- Thornton GK & Woods CG (2009). Primary microcephaly: do all roads lead to Rome? Trends in Genetics 25(11), 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia ET, Rito T & Pombo A (2017). A finer print than TADs: PRC1-mediated domains. Molecular Cell 65(3), 373–375. [DOI] [PubMed] [Google Scholar]
- Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K & Neitzel H (2004). Mutations in microcephalin cause aberrant regulation of chromosome condensation. The American Journal of Human Genetics 75(2), 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungadi EA, Ito A, Kiyomitsu T & Goshima G (2017). Human microcephaly ASPM protein is a spindle pole-focusing factor that functions redundantly with CDK5RAP2. Journal of Cell Science jcs-203703. [DOI] [PubMed] [Google Scholar]
- Venkatesh T, Nagashri MN, Swamy SS, Mohiyuddin SMA, Gopinath KS & Kumar A (2013). Primary microcephaly gene MCPH1 shows signatures of tumor suppressors and is regulated by miR-27a in oral squamous cell carcinoma. PLOS One 8(3), e54643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram S, Tasto JJ, Feoktistova A, Jennings JL, Link AJ & Gould KL (2004). Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Molecular Biology of the Cell 15(5), 2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I, Pahlke G, Salanova M, Zhang G, Wang S, Coletti D, Onuffer J, Jin CSL & Conti M (2001). Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. Journal of Biological Chemistry 276(14), 11189–11198. [DOI] [PubMed] [Google Scholar]
- Verloes A, Drunat S, Gressens P & Passemard S (1993). Primary autosomal recessive microcephalies and seckel syndrome spectrum disorders In GeneReviews (ed. Pagon R. A., Adam M. P., Ardinger H. H., Wallace S. E., Amemiya A., Bean L. J. H., Bird T. D., Ledbetter N., Mefford H. C., Smith R. J. H. & Stephens K.). Seattle WA: University of Washington, Seattle. [PubMed] [Google Scholar]
- von der Hagen M, Pivarcsi M, Liebe J, von Bernuth H, Didonato N, Hennermann JB, Bührer C, Wieczorek D & Kaindl AM (2014). Diagnostic approach to microcephaly in childhood: a two-center study and review of the literature. Developmental Medicine and Child Neurology 56(8), 732–741. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B & Zhang K (2014). A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159(1), 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Baltus AE, Mathew RS, Murphy EA, Evrony GD, Gonzalez DM, Wang EP, Marshall-Walker CA, Barry BJ, Murn J & Tatarakis A (2012). Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 151(5), 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topçu M, McDonald MT, Barry BJ, Felie JM, Sunu C, Dobyns WB, Folkerth RD, Barkovich AJ & Walsh CA (2010). Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nature Genetics 42(11), 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaqout S, Morris-Rosendahl D & Kaindl AM (2017). Autosomal recessive primary microcephaly (MCPH): an update. Neuropediatrics 48(3), 135–142. [DOI] [PubMed] [Google Scholar]
- Zhou Z-W, Tapias A, Bruhn C, Gruber R, Sukchev M & Wang Z-Q (2013). DNA damage response in microcephaly development of MCPH1 mouse model. DNA Repair 12(8), 645–655. [DOI] [PubMed] [Google Scholar]