Summary
The genomic control (GC) approach is extensively used to effectively control false positive signals due to population stratification in genome-wide association studies (GWAS). However, GC affects the statistical power of GWAS. The loss of power depends on the magnitude of the inflation factor (λ) that is used for GC. We simulated meta-analyses of different GWAS. Minor allele frequency (MAF) ranged from 0·001 to 0·5 and λ was sampled from two scenarios: (i) random scenario (empirically-derived distribution of real λ values) and (ii) selected scenario from simulation parameter modification. Adjustment for λ was considered under single correction (within study corrected standard errors) and double correction (additional λ corrected summary estimate). MAF was a pivotal determinant of observed power. In random λ scenario, double correction induced a symmetric power reduction in comparison to single correction. For MAF <5%, GC significantly reduced power for genetic risks ranging from 1·2 to 1·4 (n = 10–20). Rising MAF attenuated the correction effect of λ adjustment. Moderate λ approach yielded more conservative results for population stratification adjustment, especially for MAF <5%. Large λ approach yielded an approximate two fold decrease in power when compared to moderate λ approach and almost four fold when the original random λ scenario was considered. Meta-analysis power can be adequate to detect significant variants even for double GC correction when effect size exceeds >1·2 and MAF >5%. Our results provide a quick but detailed index for power considerations of future meta-analyses of GWAS that enables a more flexible design from early steps based on the number of studies accumulated in different groups and the λ values observed in the single studies.
1. Introduction
A variety of common human diseases have a multi-factorial genetic basis and recent advances have enabled large-scale association studies. Under this prism, genome-wide association studies and exome- and whole-genome sequencing studies (GWAS hereafter for simplicity) have been increasingly used to identify polymorphisms underlying complex traits using meta-analytical techniques (Hindorff et al., 2009; Begum et al., 2012). The vast majority of GWAS use population-based samples for the assessment of underlying associations in complex diseases. A potential problem for every population-based association study is the presence of population structure that can mimic the signal of a positive association (Cardon & Bell, 2004). Failure to control for this factor could lead to systematic inflation of the observed magnitude of the effect sizes leading to an increased number of false positive signals (Marchini et al., 2004).
A widely accepted method for taking into account the population stratification is genomic control (GC). GC uses independent marker loci typed in cases and controls to adjust the distribution of a standard association test statistic by an appropriate factor (Devlin & Roeder, 1999; Pritchard & Rosenberg, 1999). Inflation factor (λ) is commonly used for the controlling of ancestry effects by yielding more conservative standard errors and wider confidence intervals. Meta-analysis is a technique where single-study effects that measure the same outcome are synthesized to provide a summary effect estimate. The approach has been widely adopted in genetic epidemiology where the signals are mostly small or at least moderate. Thus by synthesizing all the available information someone can boost the power and reduce potential false positive signals (Evangelou & Ioannidis, 2013). However, it may be argued that using such techniques to increase the proportion of true signals could lead to recruitment of heterogeneous populations, a phenomenon that could be magnified if discrete population structure exists among different studies. Moreover, besides diverse populations, accumulating evidence suggests that population stratification may also be present in apparently homogeneous populations (Clayton et al., 2005; Tian et al., 2008). Detecting realistic effect sizes requires large-scale studies (translated in thousands of individuals) and residual population stratification cannot be excluded. Therefore researchers usually synthesize estimates that are already corrected for the population stratification and in most cases a second correction for the summary results is applied.
On the other hand, the magnitude of λ affects statistical power. A large λ will reduce the statistical power requiring a larger sample size, whereas a relative smaller λ will not require significant adjustment to the sample size. It is obvious that for low frequency and rare variants, λ will significantly affect the number of cases needed in order to identify a novel variant (Panagiotou et al., 2010; Day-Williams & Zeggini, 2011; Koboldt et al., 2013). These power considerations indicate that the expected magnitude of the effect size, the minor allele frequency (MAF) and the value of the λ should be taken into account when designing a meta-analysis of GWAS.
In this study, we simulated GWAS with different effect sizes and MAFs. Subsequently, we meta-analysed the simulated GWAS and we calculated the sample size requirements in order to identify genome-wide significant signals at the level of p < 5 × 10−8. Meta-analysis is a powerful method for synthesizing individual studies that have insufficient power to detect an association. Given the small risk effects usually observed in genetic associations, meta-analysis has been increasingly used in the field of genetic epidemiology. Therefore, we assessed the effect of various λ values in the estimated power of meta-analysed GWAS under different scenarios of MAFs, odds ratios (OR) and number of incorporated studies.
2. Methods
(i). Simulation of single studies
Single case–control studies (case to control ratio equal to 1) were simulated using multinomial approaches as discussed in detail elsewhere (Pereira et al., 2011). We considered a scenario with an autosomal bi-allelic variant, where Hardy–Weinberg equilibrium is assumed to hold for the whole population. The susceptibility alleles are considered the causal variants or surrogate markers in tight linkage disequilibrium (r2 = 1·0). Studies were generated as from a case–control design. Under these settings, sample sizes were uniformly distributed between 1000 and 3000 using a 1:1 proportion for cases and controls.
(ii). Simulation parameters
Parameters for each scenario were chosen to be in a plausible range of values, consistent with insights from recent studies (Lango et al., 2010; Ehret et al., 2011; Estrada et al., 2012). In this respect, we considered genetic effects under a log-additive model (i.e., multiplicative model) in an OR scale, encompassing genetic variants with small (OR = 1·05–1·3), moderate (OR = 1·4–1·6) and high (OR = 1·7–2·0) effects. We also assumed different MAFs. Specifically, we estimated the power assuming MAFs ranging from 0·01% (rare variants) to 50% (common polymorphisms).
We allowed λ to be sampled from two scenarios.
(1) Random λ scenario, where values for λ were sampled from an empirically-derived distribution of real λ values. To estimate the distribution of λ among GWAS data sets, we systematically extracted λ values from all meta-analyses of GWAS published from January 2010 to May 2012 from major genetic journals (Supplementary Index 1).
(2) Selected λ scenario that was subdivided into two approaches on the basis of empirical values of λ values and their distribution in lowest and higher quartiles. In the first approach, values for λ were sampled from a modified gamma distribution that generated values in the range of 1·1 to 1·2 (moderate λ approach), while in the latter, λ values were estimated as previously, from an alternatively modified gamma distribution that sampled values larger than 1·2 until approximately 1·55 (large λ scenario). The selected λ scenario 2 was applied to specific ORs (1·05–1·4) and MAFs (0·1–20%). The compatible simulation parameters for scenario 2 were selected on the basis of the results from the initial screening scenario showing a negligible effect of λ on ORs exceeding 1·4 and MAFs over 20%, either alone or in combination. Gamma distribution modifications were performed through EasyFit5·5 software (MathWave Technologies).
(iii). λ adjustments
We computed the effect of the correction for λ considering two strategies: (i) Estimates from single studies were corrected prior to the meta-analysis and the effect sizes along with the corrected standard errors (SEs) were synthesized (single correction-within study corrected SEs). Corrected SEs were computed by multiplying the original SEs (in log scale) with the squared root of λ. The correction was performed when λ exceeded 1, otherwise original λ were retained. (ii) Estimates from single studies were corrected, the meta-analysis was performed, and a second correction was performed at the meta-analysis level (double correction-within study corrected SEs plus the λ corrected summary estimate). The second, consecutive correction was performed to the SE (in log scale) of the summary effect size identically using the method described in (i).
(iv). Meta-analysis methods
Meta-analyses were carried out under fixed-effects models (inverse-variance method), since this meta-analytical framework is considered the most powerful approach to identify novel loci during discovery screenings (Pereira et al., 2009). We deemed significance at α = 5 × 10−8, which is commensurate with genome-wide cutoffs usually used in a variety of GWAS settings (Panagiotou & Ioannidis, 2012). All simulations were performed in the Stata 11·1 package (Stata Corporation).
3. Results
(i). Random λ scenario
Based on 390 observations, a gamma distribution with shape = 3·45, scale = 0·03 and location parameter = 0·945 provided a good fit for the empirical distribution of real λ values, as discussed in the Methods section. Corresponding mean and standard deviation for random λ was 1·049 and 0·056 respectively. Pooled effect size of meta-analysis and power calculations were performed on a range of five to 30 aggregating simulated studies with a median size of 1500 ± 50 subjects. In all cases, MAF was a pivotal determinant of observed power (Figure S1, Table 1 and Table 2). Results are further analysed on the basis of MAF. We considered common variants SNPs with MAF >5%, low frequency variants those with 1% < MAF < 5% and rare variants SNPs with MAF <1%.
Table 1.
Achieved power for different genetic risks and correction methodology in meta-analysis of GWAS with common SNPs (MAF: 5, 10 and 20%) under random λ approach.
| OR f1, f2, f3 Correction method | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR = 1·1 | OR = 1·2 | |||||||||||||||||
| Studies in meta-analysis | 0·05 | 0·1 | 0·2 | 0·05 | 0·1 | 0·2 | ||||||||||||
| No | Single | Double | No | Single | Double | No | Single | Double | No | Single | Double | No | Single | Double | No | Single | Double | |
| 5 | 0 | 0 | 0 | 0·04 | 0·01 | 0·01 | 0·35 | 0·24 | 0·21 | 0·47 | 0·36 | 0·22 | 7·16 | 5·7 | 4·4 | 42·14 | 37·88 | 33·33 |
| 10 | 0·02 | 0·02 | 0 | 0·51 | 0·34 | 0·25 | 5·76 | 4·41 | 3·44 | 8·65 | 6·72 | 5·12 | 55·69 | 50·73 | 45·35 | 97·08 | 96·07 | 94·59 |
| 15 | 0·22 | 0·12 | 0·1 | 3·05 | 2·34 | 1·77 | 23·42 | 19·37 | 15·91 | 32·57 | 28·38 | 23·88 | 91·64 | 89·5 | 86·51 | 99·96 | 99·96 | 99·94 |
| 20 | 0·73 | 0·51 | 0·38 | 8·87 | 7·05 | 5·41 | 50·02 | 45 | 39·52 | 62·13 | 57·19 | 51·79 | 99·03 | 98·76 | 98·09 | 100 | 100 | 100 |
| 25 | 1·82 | 1·38 | 1·01 | 19·55 | 16·07 | 12·8 | 73·81 | 69·54 | 64·2 | 83·7 | 80·05 | 75·72 | 99·95 | 99·94 | 99·88 | 100 | 100 | 100 |
| 30 | 3·68 | 2·75 | 2·25 | 32·8 | 28·7 | 24·9 | 88·18 | 85·69 | 82·51 | 94·33 | 92·86 | 90·46 | 99·99 | 99·99 | 99·99 | 100 | 100 | 100 |
Table 2.
Achieved power for different genetic risks and correction methodology in meta-analysis of GWAS with uncommon SNPs (MAF <5%) under random λ approach.
| OR f1, f2, f3 Correction method | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A(OR = 1·1–1·2) | ||||||||||||||||||
| OR = 1·1 | ||||||||||||||||||
| 0·001 | 0·005 | 0·01 | 0·02 | 0·03 | 0·04 | |||||||||||||
| n of studies | No | Single | Double | No | Single | Double | No | Single | Double | No | Single | Double | No | Single | Double | No | Single | Double |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0·01 | 0·01 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0·04 | 0·04 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0·02 | 0·02 | 0·01 | 0·09 | 0·07 | 0·06 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0·07 | 0·05 | 0·04 | 0·27 | 0·21 | 0·14 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0·02 | 0 | 0 | 0·25 | 0·13 | 0·1 | 0·68 | 0·45 | 0·31 |
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0·07 | 0 | 0 | 048 | 0·24 | 0·2 | 1·37 | 0·98 | 0·73 |
(a). Common SNPs (MAF ⩾5%)
For MAF = 5%, power exceeded 80% upon the use of 24 studies for a minimum effect size of 1·2 (Table 1). Incremental values for MAF (>5%) led to progressive increase in observed power even when combined with smaller effect sizes (i.e., power 84·15% for OR = 1·15 and identical number of studies, data not shown). The number of studies in the meta-analysis tended to exert a minor effect on yielding adequate power after exceeding the threshold of 80% and under various combinations of MAF and OR. In detail, for a range of number of studies from 5 (OR = 1·4/MAF = 10%) to 30 (OR = 1·1/MAF = 20%), adequate power was observed and fluctuated for the same effect size (OR) under different MAF settings or vice versa (Table 1). The power reached 99% for MAFs exceeding 40% and OR = 1·1.
(b). Low frequency and rare SNPs (MAF <5%)
For all scenarios using MAF <5%, a minimum effect size of OR = 1·3 and the maximum number of studies (n = 30) were required in order to achieve 80% power (MAF = 2%; Table 2(a)). When genetic risk was substantially elevated (OR >1·4), the number of studies required for 80% power for the same MAF was halved to 15. For the same effect size (OR >1·4) power exhibited a flattened region over 99% for MAFs exceeding 4%, independently of the number of studies, whereas for ORs >1·5 this was observed from MAFs as small as 2% (Table 2(b)). For MAFs below 1%, significant power could not be yielded by potential combinations of genetic risk and number of studies (Table 2(a)).
(c). Correction and power modification
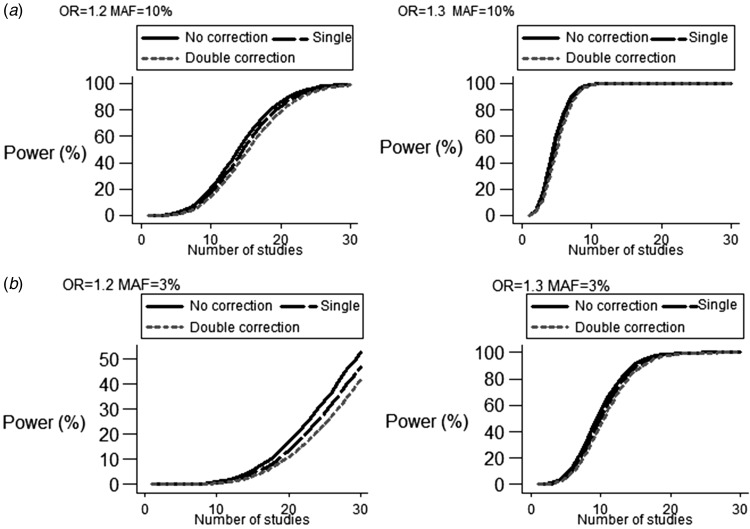
As expected, double correction induced a symmetric reduction in power of meta-analysis in comparison to single correction (Table 1 and Table 2). Double correction strategy exerted a subtle effect on flattened power >90% for combinations of OR (⩾1·2) and number of studies (n ⩾15; Fig. 1(a)). For low frequency and rare SNPs (MAF <5%), λ based correction reduced the power for genetic risks ranging from 1·2 to 1·4 and for aggregating studies between 10 and 20 (Table 2(a) and Fig. 1(b)). For rare variants (MAF <1%), even the combination of maximum studies and large genetic effect (OR >2) could not account for power >80%. For both scenarios of genetic frequencies, elevated MAF attenuated the correction effect of λ adjustment in the observed spectrum of ORs (Figures S2–S4).
Fig. 1.
Power modifications induced by no, single and double correction strategy under different genetic risks, MAF and number of studies for (a) MAF ⩾5% and (b) MAF <5%.
(ii). Selected λ scenario
Selected λ scenario results were extracted from meta-analysed simulated studies (maximum number of studies 30, mean size 1480 ± 28). Results are reported under two major settings, moderate and large λ approach as described in detail in the Methods section.
(a). Moderate λ approach
Common SNPs (MAF ⩾5%)
When λ values were selected from a less skewed distribution (moderate λ approach (MLA), mean λ = 1·14 and standard deviation = 0·017) and MAF surpassed 5%, power considerations were relevant to the random λ scenario (Table S1).
Low frequency and rare SNPs (MAF <5%)
When MAF did not exceed 5%, sufficient power could not be established for moderate OR (<1·3; Table S2). Adequate power (~80%) was first reported for uncommon SNPs (i.e., MAF = 2%) for OR = 1·3 and maximum number of studies. Marginal differences over the number of studies needed before flattening of power (i.e., 30 studies in MLA and OR = 1·15 vs. 25 studies in a random scenario with identical OR) should be evaluated under the prism of simulation-driven data.
Correction and power modification
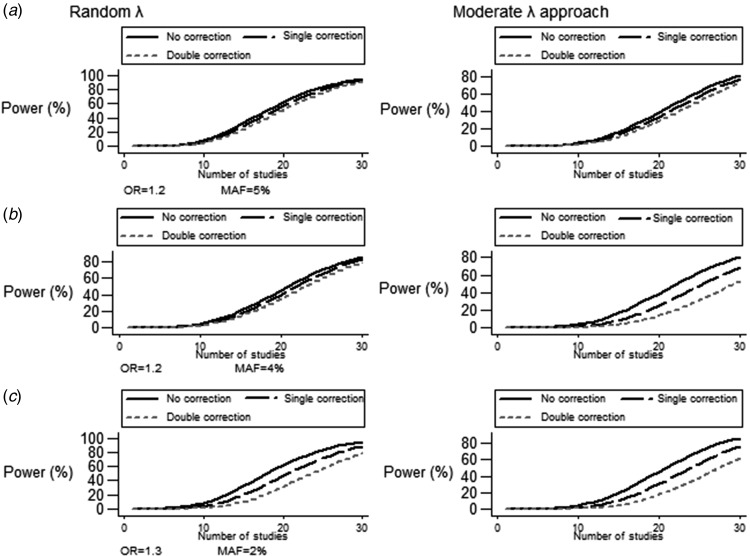
In terms of correction strategy sequelae, MLA tended to yield more conservative results for double adjustment of population stratification across ORs between 1·2 and 1·3 and MAF ⩾5% (Figure 2(a) and Table S1). Discrepancies in power adjustment for λ were magnified for MAF <5% (Table S2). Albeit beginning from almost identical power under unadjusted settings, MLA led to a steeper reduction in power as compared to a random λ approach for a range of ORs (1·2–1·4), MAFs (2–4%) and number of studies (15–25; Figure 2(b) and (c)). Sufficient power for low frequency variants (MAF = 2%) was only established in the upper limit of the studied effect size (OR = 1·4) along with the contribution of 22 studies.
Fig. 2.
Power modifications induced by double correction strategy under different genetic risks, MAFs ⩽5% and number of studies in random λ approach and MLA: (a) OR = 1·2, MAF = 5%; (b) OR = 1·2, MAF = 4%; and (c) OR = 1·3, MAF = 2%. Closer lines to each other corresponding to no correction/single/double correction indicate a less pronounced effect of power tapering.
(b). Large λ approach
Common SNPs (MAF ⩾5%)
When λ factor was inflated (large λ approach (LLA), mean λ = 1·35 and standard deviation = 0·047) and with common SNPs, power >80% was initially observed for a small genetic risk (OR = 1·15) combined though with elevated MAF (20%) and increased number of studies (25 studies aggregated). By contrast, comparable power in the random λ approach would precipitate the same number of studies and genetic risk but with a significantly smaller MAF of 10%. In subsequent scenarios of LLA with MAF >5%, relevant associations were derived about the increased number of meta-analysed studies or MAF needed to achieve equivalent power to the original approach (Table S3).
Low frequency and rare SNPs (MAF <5%)
When MAF was restrained below 5%, significant power (~80%) was established at the cost of maximum OR = 1·4 and increased number of studies (n = 14; Table S4). However, in all comparisons between empirically derived λ and LLA for double correction strategy, power was underestimated in the latter.
Correction and power modification
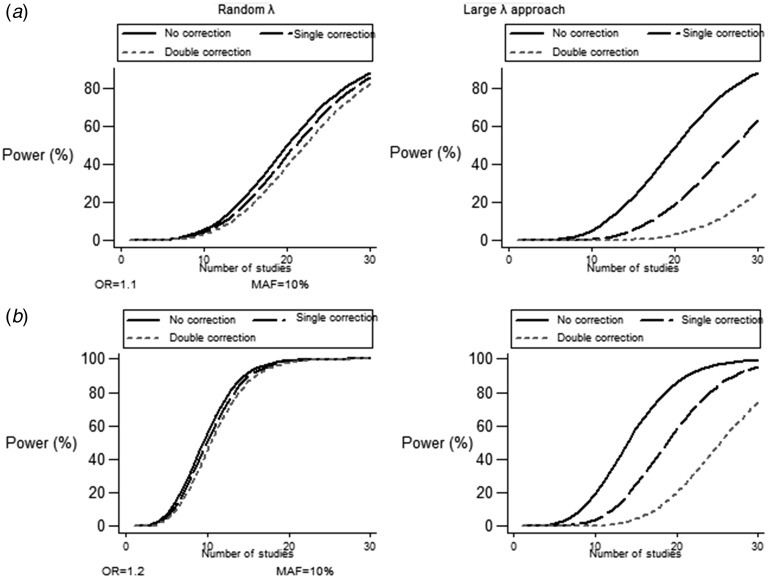
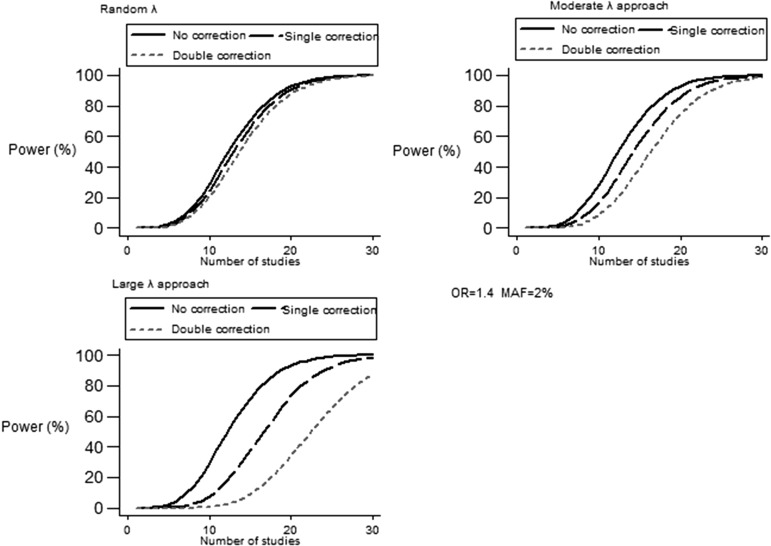
In line with previous analysis, correction strategy attenuated power estimates in LLA (Tables S3 and S4). For MAF ⩾5%, coarse tapering of power was observed, especially when either genetic risk (OR <1·2) or number of studies was low. Figure 3 depicts power adjustment for random λ scenario and LLA under different settings. When MAF was set below the threshold of 5%, doubly corrected power was repeatedly lower in comparison to random λ scenario as well as MLA with identical settings. For rare variants (MAF ⩽2%), maximum OR = 1·4 and maximum number of studies had to be recruited in order to achieve sufficient power (~80%). Intensity of power correction was the main characteristic of LLA: for ORs ranging from 1·2–1·4 and for MAFs of 2–4%, LLA yielded an approximate two fold decrease in power when compared to MLA and almost four fold when the original random λ scenario was considered (Fig. 4). Sigmoid curves are established for power in Fig. 4, indicating prominent modification under a realistic number of studies (10–20) and the subtle effect of this parameter in more extreme values.
Fig. 3.
Power modifications induced by double correction strategy under different genetic risks, MAFs >5% and number of studies in random λ approach and LLA: (a) OR = 1·1, MAF = 10%; (b) OR = 1·2, MAF = 10%. Closer lines to each other corresponding to no correction/single/double correction indicate a less pronounced effect of power tapering.
Fig. 4.
Power modifications induced by double correction strategy under selected scenarios of λ magnitude and number of studies for uncommon variant (MAF = 2%) with predetermined effect size (OR = 1.4).
Table S5 provides a review of major differences between common (MAF ⩾5%) and low frequency or rare SNPs (MAF <5%) in terms of effect size needed for adequate power (⩾80%) across the three scenarios (random λ approach, MLA and LLA) of our study. A ten fold difference in MAF between common and uncommon SNPs was selected for demonstration reasons (i.e., uncommon SNP = 1% vs. common SNP = 10%). Differences between unadjusted estimates and double -corrected counterparts are also reported. Table 3 focuses on uncommon variants and depicts the impact of alternative correction methods (no correction vs. single and double λ correction) on achieved power for varying genetic effects and fixed number of studies in meta-analysis.
Table 3.
Power considerations under selected scenarios of λ magnitude, correction approach and effect size for uncommon variants and predetermined number of studies in meta-analysis.
| RLA | MLA | LLA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No correction | Single correction | Double correction | No correction | Single correction | Double correction | No correction | Single correction | Double correction | ||
| (n = 25 studies) | Power | Power | Power | Power | Power | Power | Power | Power | Power | |
| OR = 1·1 | MAF = 0·01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MAF = 0·03 | 0·25 | 0·13 | 0·1 | 0·1 | 0·03 | 0 | 0·17 | 0 | 0 | |
| OR = 1·2 | MAF = 0·01 | 0·39 | 0·19 | 0·13 | 0·43 | 0·14 | 0·04 | 0·39 | 0·04 | 0 |
| MAF = 0·03 | 33·2 | 28·58 | 23·93 | 33·81 | 20·71 | 10·67 | 32·52 | 8·95 | 0·81 | |
| OR = 1·3 | MAF = 0·01 | 9·26 | 7·43 | 5·77 | 9·53 | 4·38 | 1·74 | 9·58 | 1·29 | 0·04 |
| MAF = 0·03 | 96·85 | 95·74 | 94·01 | 97·12 | 92·89 | 85·55 | 96·94 | 83·54 | 47·98 | |
| OR = 1·4 | MAF = 0·01 | 48·48 | 42·89 | 37·33 | 49·4 | 33·38 | 19·02 | 48·41 | 16·56 | 2·4 |
| MAF = 0·03 | 100 | 100 | 99·99 | 100 | 99·96 | 99·94 | 99·98 | 99·88 | 98·14 | |
(iii). Working example
Here we present an example of the successful AMDGene Consortium (Fritsche et al., 2013) that discovered seven new loci associated with age-related macular degeneration and confirmed older findings. The consortium synthesized results from 17 groups in the discovery stage and, in total, 33 groups were used in the two-stage design, contributing 17 181 cases. The overall λ was 1·06 fitting in our random λ scenario. The smallest OR detected was 1·10. For our working example we assume an overall number of 33 contributing groups. Based on our calculations, the group would have adequate power to detect signals with an OR of >1·3 and/or MAF exceeding 5%; however, the consortium has limited power to discover additional loci for low frequency variants (MAF <5%) and moderate genetic effects (Table S6). Figures S5–S7 depict the power modifications induced by GC for uncommon SNPs under different settings of genetic effects/number of aggregated studies and further emphasize the need for augmentation of the sample size in relevant analyses. From the baseline status of 33 studies with 17 181 cases, an additional sample size of approximately 10 500 subjects should be recruited in order to yield acceptable power (>90%) under double GC correction for uncommon SNPs (MAF = 4%) and a moderate effect size (OR = 1·2). Therefore, the consortium should work towards accumulating larger sample sizes or inviting new groups into the consortium.
4. Discussion
We evaluated the influence of population stratification using the widely applied GC on the power of a meta-analysis under different settings and number of the individual GWAS. Thus, our results provide direct evidence of the power of the discovery efforts to identify new signals. It is clearly shown that the meta-analysis power can be adequate to detect significant variants even if investigators apply the more conservative double GC correction for ORs >1·2 and for MAFs >5%. However, the power of the meta-analysis is greatly influenced when the genetic risk effect is weak or the MAF of the variant under study is rare.
Based on the findings of this simulation study we can describe the different levels of λ regarding their influence on power as: 1–1·10: small λ; 1·1–1·2: moderate λ; and >1·2: large λ. This allows for quick inspection of the data and for power consideration even during the early stages of a consortium being formulated, the participating teams design the study and draft the analysis plan. Moderate and large λ values may substantially increase the sample size requirements to maintain adequate statistical power when compared with weak λ values even if the underlying genetic risk factors are moderate or large and the MAF <5%. Therefore, the calculations presented in the work can be used to estimate the expected power in meta-analyses of GWAS. Power considerations could be taken into account even from the early stages of the study design, based on the number of cases accumulated in different groups and the λ observed in each individual genome-wide association.
In our study we empirically drew a distribution of λ that was subsequently applied in the simulation scenarios. We have shown that in most studies the λ was small (<1·1) indicating that even though stratification cannot be excluded as a possibility in real scenarios, most of the teams continuously monitored for stratification and applied stringent quality control measures to diminish the possibility of confounding by population admixture. This supports previous arguments that unrelated case–control and cohort studies can be effectively compared to other designs such as family-based studies (Risch 1990; Silverman & Palmer, 2000; Cardon & Bell, 2004; Evangelou et al., 2006).
Accumulating evidence from recently published studies suggests that GC may not be effective in controlling population stratification in association studies (Edwards & Gao, 2012; Wang et al., 2012). This problem may be aggravated under meta-analysis settings where a double GC correction method might lead to more prominent inflation of type I error rates at a marker with significant allele frequency differentiation in subpopulations generated by recent strong selection (Bouaziz et al., 2011; Edwards & Gao, 2012; Wang et al., 2012). Conversely, alternative methods, including principal component analysis (PCA) correction and Bayesian semiparametric algorithm for inferring population structure, could control type I error rates and yield much higher power in meta-analyses compared to the double GC correction method (Bouaziz et al., 2011; Edwards & Gao, 2012; Majumdar et al., 2013). Emerging techniques also incorporate linear mixed models to test for association in GWAS, after taking into account relatedness among samples, population stratification and other confounding factors (Zhou & Stephens, 2012). These models present substantial computational challenges but implementation of new algorithms enable exact tests for large GWAS, minimizing the need for approximate methods and subsequent power loss (Zhou & Stephens, 2012; Shin & Lee, 2015). In the presence of subject outliers or markedly admixed populations, methods that are considered more effective than GC for controlling population stratification (i.e., PCA) may need further improvement. Amendments such as robust PCA combined with k-medoids clustering or hybrid approaches with linear mixed models could increase the robustness of techniques designated to adjust for population heterogeneity (Liu et al., 2013; Tucker et al., 2014). Statistical methods in confronting population stratification may include optimal case–control matching through hierarchical clustering or modified spectral clustering (spectral dimensional reduction techniques) in the case of rare variants (Miclaus et al., 2009; Zhang et al., 2013; Lacour et al., 2015).
In our study, we confirmed a prominent decrease in observed power of meta-analysis of GWAS, notably after double GC correction for low frequency and rare variants under small genetic effects (Jiang et al., 2013). Double GC-induced power decrease was aggravated for inflated λ values indicating conservative results in relatively distant populations (Bouaziz et al., 2011; Jiang et al., 2013). However, GC should not be excluded a priori from future analyses on the basis of its computational simplicity and speed. By contrast, adjusted regressions and principal component methods can be very time consuming depending on the algorithm used to infer the population structure.
Certain drawbacks of our study should be acknowledged. First, the need to exhaustively examine a wide range of combinations among potential modifiers of power (MAF/effect size/number of studies) in meta-analyses of GWAS imposed the use of simulated data of independent single studies. Highest number of replication iterations, previously validated methodology for simulation data (Pereira et al., 2009), assessment of λ distribution as derived from published GWAS and biological interpretation of study results partially compensated for the absence of real data. Moreover, genetic associations were analysed under the assessment of a multiplicative genetic model. By contrast, other researchers have suggested the synthetic use of multiplicative and recessive genetic models and the implementation of Bonferroni correction (Salanti & Higgins, 2008). Finally, this study provides descriptive results on the basis of power modifications after adjusting for effect size, MAF and number of studies. Under this prism, quantification of the influence of the above parameters per se on power adjustments could not be ascertained.
Exome- and whole-genome sequencing data for complex traits will exponentially grow in the next few years, therefore the need for power considerations for the next meta-analytical efforts increases. A prior estimate of the number of studies and participants needed will allow international consortia to focus on the recruitment of new partners and will lead to the design of more effective and targeted analyses plans that will lead to novel locus discovery.
We would like to thank Tiago V. Pereira, PhD (Hospital Alemão Oswaldo Cruz Health Technology Assessment Unit, São Paulo, Brazil) for his input in earlier versions of the manuscript.
Declaration of interest
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0016672316000069.
click here to view supplementary material
References
- Begum F., Ghosh D., Tseng G. C. & Feingold E. (2012). Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Research 40(9), 3777–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaziz M., Ambroise C. & Guedj M. (2011). Accounting for population stratification in practice: a comparison of the main strategies dedicated to genome-wide association studies. PLoS One 6(12), e28845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon L. R. & Bell J. I. (2004). Association study designs for complex diseases. Nature Reviews Genetics 2, 91–99. [DOI] [PubMed] [Google Scholar]
- Clayton D. G., Walker N. M., Smyth D. J., Pask R., Cooper J. D., Maier L. M., Smink L. J., Lam A. C., Ovington N. R., Stevens H. E., Nutland S., Howson J. M., Faham M., Moorhead M., Jones H. B., Falkowski M., Hardenbol P., Willis T. D. & Todd J. A. (2005). Population structure, differential bias and genomic control in a large-scale, case–control association study. Nature Genetics 37(11), 1243–1246. [DOI] [PubMed] [Google Scholar]
- Day-Williams A. G. & Zeggini E. (2011). The effect of next-generation sequencing technology on complex trait research. European Journal of Clinical Investigation 41, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B. & Roeder K. (1999). Genomic control for association studies. Biometrics 55, 997–1004. [DOI] [PubMed] [Google Scholar]
- Edwards T. L. & Gao X. (2012). Methods for detecting and correcting for population stratification. Current Protocol in Human Genetics 4(1), Chapter 1: Unit 1·22·1–14. [DOI] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret G. B., Munroe P. B., Rice K. M., Bochud M., Johnson A. D., Chasman D. I., Smith A. V., Tobin M. D., Verwoert G. C., Hwang S. J., Pihur V., Vollenweider P., O'Reilly P. F., Amin N., Bragg-Gresham J. L., Teumer A., Glazer N. L., Launer L., Zhao J. H., Aulchenko Y., Heath S., Sõber S., Parsa A., Luan J., Arora P., Dehghan A., Zhang F., Lucas G., Hicks A. A., Jackson A. U., Peden J. F., Tanaka T., Wild S. H., Rudan I., Igl W., Milaneschi Y., Parker A. N., Fava C., Chambers J. C., Fox E. R., Kumari M., Go M. J., van der Harst P., Kao W. H., Sjögren M., Vinay D. G., Alexander M., Tabara Y., Shaw-Hawkins S., Whincup P. H., Liu Y., Shi G., Kuusisto J., Tayo B., Seielstad M., Sim X., Nguyen K. D., Lehtimäki T., Matullo G., Wu Y., Gaunt T. R., Onland-Moret N. C., Cooper M. N., Platou C. G., Org E., Hardy R., Dahgam S., Palmen J., Vitart V., Braund P. S., Kuznetsova T., Uiterwaal C. S., Adeyemo A., Palmas W., Campbell H., Ludwig B., Tomaszewski M., Tzoulaki I., Palmer ND; CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium, Aspelund T., Garcia M., Chang Y. P., O'Connell J. R., Steinle N. I., Grobbee D. E., Arking D. E., Kardia S. L., Morrison A. C., Hernandez D., Najjar S., McArdle W. L., Hadley D., Brown M. J., Connell J. M., Hingorani A. D., Day I. N., Lawlor D. A., Beilby J. P., Lawrence R. W., Clarke R., Hopewell J. C., Ongen H., Dreisbach A. W., Li Y., Young J. H., Bis J. C., Kähönen M., Viikari J., Adair L. S., Lee N. R., Chen M. H., Olden M., Pattaro C., Bolton J. A., Köttgen A., Bergmann S., Mooser V., Chaturvedi N., Frayling T. M., Islam M., Jafar T. H., Erdmann J., Kulkarni S. R., Bornstein S. R., Grässler J., Groop L., Voight B. F., Kettunen J., Howard P., Taylor A., Guarrera S., Ricceri F., Emilsson V., Plump A., Barroso I., Khaw K. T., Weder A. B., Hunt S. C., Sun Y. V., Bergman R. N., Collins F. S., Bonnycastle L. L., Scott L. J., Stringham H. M., Peltonen L., Perola M., Vartiainen E., Brand S. M., Staessen J. A., Wang T. J., Burton P. R., Soler Artigas M., Dong Y., Snieder H., Wang X., Zhu H., Lohman K. K., Rudock M. E., Heckbert S. R., Smith N. L., Wiggins K. L., Doumatey A., Shriner D., Veldre G., Viigimaa M., Kinra S., Prabhakaran D., Tripathy V., Langefeld C. D., Rosengren A., Thelle D. S., Corsi A. M., Singleton A., Forrester T., Hilton G., McKenzie C. A., Salako T., Iwai N., Kita Y., Ogihara T., Ohkubo T., Okamura T., Ueshima H., Umemura S., Eyheramendy S., Meitinger T., Wichmann H. E., Cho Y. S., Kim H. L., Lee J. Y., Scott J., Sehmi J. S., Zhang W., Hedblad B., Nilsson P., Smith G. D., Wong A., Narisu N., Stančáková A., Raffel L. J., Yao J., Kathiresan S., O'Donnell C. J., Schwartz S. M., Ikram M. A., Longstreth W. T. Jr, Mosley T. H., Seshadri S., Shrine N. R., Wain L. V., Morken M. A., Swift A. J., Laitinen J., Prokopenko I., Zitting P., Cooper J. A., Humphries S. E., Danesh J., Rasheed A., Goel A., Hamsten A., Watkins H., Bakker S. J., van Gilst W. H., Janipalli C. S., Mani K. R., Yajnik C. S., Hofman A., Mattace-Raso F. U., Oostra B. A., Demirkan A., Isaacs A., Rivadeneira F., Lakatta E. G., Orru M., Scuteri A., Ala-Korpela M., Kangas A. J., Lyytikäinen L. P., Soininen P., Tukiainen T., Würtz P., Ong R. T., Dörr M., Kroemer H. K., Völker U., Völzke H., Galan P., Hercberg S., Lathrop M., Zelenika D., Deloukas P., Mangino M., Spector T. D., Zhai G., Meschia J. F., Nalls M. A., Sharma P., Terzic J., Kumar M. V., Denniff M., Zukowska-Szczechowska E., Wagenknecht L. E., Fowkes F. G., Charchar F. J., Schwarz P. E., Hayward C., Guo X., Rotimi C., Bots M. L., Brand E., Samani N. J., Polasek O., Talmud P. J., Nyberg F., Kuh D., Laan M., Hveem K., Palmer L. J., van der Schouw Y. T., Casas J. P., Mohlke K. L., Vineis P., Raitakari O., Ganesh S. K., Wong T. Y., Tai E. S., Cooper R. S., Laakso M., Rao D. C., Harris T. B., Morris R. W., Dominiczak A. F., Kivimaki M., Marmot M. G., Miki T., Saleheen D., Chandak G. R., Coresh J., Navis G., Salomaa V., Han B. G., Zhu X., Kooner J. S., Melander O., Ridker P. M., Bandinelli S., Gyllensten U. B., Wright A. F., Wilson J. F., Ferrucci L., Farrall M., Tuomilehto J., Pramstaller P. P., Elosua R., Soranzo N., Sijbrands E. J., Altshuler D., Loos R. J., Shuldiner A. R., Gieger C., Meneton P., Uitterlinden A. G., Wareham N. J., Gudnason V., Rotter J. I., Rettig R., Uda M., Strachan D. P., Witteman J. C., Hartikainen A. L., Beckmann J. S., Boerwinkle E., Vasan R. S., Boehnke M., Larson M. G., Järvelin M. R., Psaty B. M., Abecasis G. R., Chakravarti A., Elliott P., van Duijn C. M., Newton-Cheh C., Levy D., Caulfield M. J. & Johnson T. (2011). Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478(7367), 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y. H., Duncan E. L., Ntzani E. E., Oei L., Albagha O. M., Amin N., Kemp J. P., Koller D. L., Li G., Liu C. T., Minster R. L., Moayyeri A., Vandenput L., Willner D., Xiao S. M., Yerges-Armstrong L. M., Zheng H. F., Alonso N., Eriksson J., Kammerer C. M., Kaptoge S. K., Leo P. J., Thorleifsson G., Wilson S. G., Wilson J. F., Aalto V., Alen M., Aragaki A. K., Aspelund T., Center J. R., Dailiana Z., Duggan D. J., Garcia M., Garcia-Giralt N., Giroux S., Hallmans G., Hocking L. J., Husted L. B., Jameson K. A., Khusainova R., Kim G. S., Kooperberg C., Koromila T., Kruk M., Laaksonen M., Lacroix A. Z., Lee S. H., Leung P. C., Lewis J. R., Masi L., Mencej-Bedrac S., Nguyen T. V., Nogues X., Patel M. S., Prezelj J., Rose L. M., Scollen S., Siggeirsdottir K., Smith A. V., Svensson O., Trompet S., Trummer O., van Schoor N. M., Woo J., Zhu K., Balcells S., Brandi M. L., Buckley B. M., Cheng S., Christiansen C., Cooper C., Dedoussis G., Ford I., Frost M., Goltzman D., González-Macías J., Kähönen M., Karlsson M., Khusnutdinova E., Koh J. M., Kollia P., Langdahl B. L., Leslie W. D., Lips P., Ljunggren Ö., Lorenc R. S., Marc J., Mellström D., Obermayer-Pietsch B., Olmos J. M., Pettersson-Kymmer U., Reid D. M., Riancho J. A., Ridker P. M., Rousseau F., Slagboom P. E., Tang N. L., Urreizti R., Van Hul W., Viikari J., Zarrabeitia M. T., Aulchenko Y. S., Castano-Betancourt M., Grundberg E., Herrera L., Ingvarsson T., Johannsdottir H., Kwan T., Li R., Luben R., Medina-Gómez C., Palsson S. T., Reppe S., Rotter J. I., Sigurdsson G., van Meurs J. B., Verlaan D., Williams F. M., Wood A. R., Zhou Y., Gautvik K. M., Pastinen T., Raychaudhuri S., Cauley J. A., Chasman D. I., Clark G. R., Cummings S. R., Danoy P., Dennison E. M., Eastell R., Eisman J. A., Gudnason V., Hofman A., Jackson R. D., Jones G., Jukema J. W., Khaw K. T., Lehtimäki T., Liu Y., Lorentzon M., McCloskey E., Mitchell B. D., Nandakumar K., Nicholson G. C., Oostra B. A., Peacock M., Pols H. A., Prince R. L., Raitakari O., Reid I. R., Robbins J., Sambrook P. N., Sham P. C., Shuldiner A. R., Tylavsky F. A., van Duijn C. M., Wareham N. J., Cupples L. A., Econs M. J., Evans D. M., Harris T. B., Kung A. W., Psaty B. M., Reeve J., Spector T. D., Streeten E. A., Zillikens M. C., Thorsteinsdottir U., Ohlsson C., Karasik D., Richards J. B., Brown M. A., Stefansson K., Uitterlinden A. G., Ralston S. H., Ioannidis J. P., Kiel D. P. & Rivadeneira F. (2012). Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nature Genetics 44(5), 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E. & Ioannidis J. P. (2013). Meta-analysis methods for genome-wide association studies and beyond. Nature Reviews Genetics 14(6), 379–389. [DOI] [PubMed] [Google Scholar]
- Evangelou E., Trikalinos T. A., Salanti G. & Ioannidis J. P. (2006). Family-based versus unrelated case–control designs for genetic associations. PLoS Genetics 2(8), e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche L. G., Chen W., Schu M., Yaspan B. L., Yu Y., Thorleifsson G., Zack D. J., Arakawa S., Cipriani V., Ripke S., Igo R. P. Jr, Buitendijk G. H., Sim X., Weeks D. E., Guymer R. H., Merriam J. E., Francis P. J., Hannum G., Agarwal A., Armbrecht A. M., Audo I., Aung T., Barile G. R., Benchaboune M., Bird A. C., Bishop P. N., Branham K. E., Brooks M., Brucker A. J., Cade W. H., Cain M. S., Campochiaro P. A., Chan C. C., Cheng C. Y., Chew E. Y., Chin K. A., Chowers I., Clayton D. G., Cojocaru R., Conley Y. P., Cornes B. K., Daly M. J., Dhillon B., Edwards A. O., Evangelou E., Fagerness J., Ferreyra H. A., Friedman J. S., Geirsdottir A., George R. J., Gieger C., Gupta N., Hagstrom S. A., Harding S. P., Haritoglou C., Heckenlively J. R., Holz F. G., Hughes G., Ioannidis J. P., Ishibashi T., Joseph P., Jun G., Kamatani Y., Katsanis N., N., Keilhauer C., Khan J. C., Kim I. K., Kiyohara Y., Klein B. E., Klein R., Kovach J. L., Kozak I., Lee C. J., Lee K. E., Lichtner P., Lotery A. J., Meitinger T., Mitchell P., Mohand-Saïd S., Moore A. T., Morgan D. J., Morrison M. A., Myers C. E., Naj A. C., Nakamura Y., Okada Y., Orlin A., Ortube M. C., Othman M. I., Pappas C., Park K. H., Pauer G. J., Peachey N. S., Poch O., Priya R. R., Reynolds R., Richardson A. J., Ripp R., Rudolph G., Ryu E., Sahel J. A., Schaumberg D. A., Scholl H. P., Schwartz S. G., Scott W. K., Shahid H., Sigurdsson H., Silvestri G., Sivakumaran T. A., Smith R. T., Sobrin L., Souied E. H., Stambolian D. E., Stefansson H., Sturgill-Short G. M., Takahashi A., Tosakulwong N., Truitt B. J., Tsironi E. E., Uitterlinden A. G., van Duijn C. M., Vijaya L., Vingerling J. R., Vithana E. N., Webster A. R., Wichmann H. E., Winkler T. W., Wong T. Y., Wright A. F., Zelenika D., Zhang M., Zhao L., Zhang K., Klein M. L., Hageman G. S., Lathrop G. M., Stefansson K., Allikmets R., Baird P. N., Gorin M. B., Wang J. J., Klaver C. C., Seddon J. M., Pericak-Vance M. A., Iyengar S. K., Yates J. R., Swaroop A., Weber B. H., Kubo M., Deangelis M. M., Léveillard T., Thorsteinsdottir U., Haines J. L., Farrer L. A., Heid I. M. & Abecasis G. R & AMD Gene Consortium (2013). Seven new loci associated with age-related macular degeneration. Nature Genetics 45(4), 433–439, 439e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., Collins F. S. & Manolio T. A. (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America 106(23), 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Epstein M. P. & Conneely K. N. (2013). Assessing the impact of population stratification on association studies of rare variation. Human Heredity 76(1), 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C., Steinberg K. M., Larson D. E., Wilson R. K. & Mardis E. R. (2013). The next-generation sequencing revolution and its impact on genomics. Cell 155(1), 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacour A., Schuller V., Drichel D., Herold C., Jessen F., Leber M., Maier W., Noethen M. M., Ramirez A., Vaitsiakhovich T. & Becker T. (2015). Novel genetic matching methods for handling population stratification in genome-wide association studies. BMC Bioinformatics 14(16), 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H., Estrada K., Lettre G., Berndt S. I., Weedon M. N., Rivadeneira F., Willer C. J., Jackson A. U., Vedantam S., Raychaudhuri S., Ferreira T., Wood A. R., Weyant R. J., Segrè A. V., Speliotes E. K., Wheeler E., Soranzo N., Park J. H., Yang J., Gudbjartsson D., Heard-Costa N. L., Randall J. C., Qi L., Vernon Smith A., Mägi R., Pastinen T., Liang L., Heid I. M., Luan J., Thorleifsson G., Winkler T. W., Goddard M. E., Sin Lo K., Palmer C., Workalemahu T., Aulchenko Y. S., Johansson A., Zillikens M. C., Feitosa M. F., Esko T., Johnson T., Ketkar S., Kraft P., Mangino M., Prokopenko I., Absher D., Albrecht E., Ernst F., Glazer N. L., Hayward C., Hottenga J. J., Jacobs K. B., Knowles J. W., Kutalik Z., Monda K. L., Polasek O., Preuss M., Rayner N. W., Robertson N. R., Steinthorsdottir V., Tyrer J. P., Voight B. F., Wiklund F., Xu J., Zhao J. H., Nyholt D. R., Pellikka N., Perola M., Perry J. R., Surakka I., Tammesoo M. L., Altmaier E. L., Amin N., Aspelund T., Bhangale T., Boucher G., Chasman D. I., Chen C., Coin L., Cooper M. N., Dixon A. L., Gibson Q., Grundberg E., Hao K., Juhani Junttila M., Kaplan L. M., Kettunen J., König I. R., Kwan T., Lawrence R. W., Levinson D. F., Lorentzon M., McKnight B., Morris A. P., Müller M., Suh Ngwa J., Purcell S., Rafelt S., Salem R. M., Salvi E., Sanna S., Shi J., Sovio U., Thompson J. R., Turchin M. C., Vandenput L., Verlaan D. J., Vitart V., White C. C., Ziegler A., Almgren P., Balmforth A. J., Campbell H., Citterio L., De Grandi A., Dominiczak A., Duan J., Elliott P., Elosua R., Eriksson J. G., Freimer N. B., Geus E. J., Glorioso N., Haiqing S., Hartikainen A. L., Havulinna A. S., Hicks A. A., Hui J., Igl W., Illig T., Jula A., Kajantie E., Kilpeläinen T. O., Koiranen M., Kolcic I., Koskinen S., Kovacs P., Laitinen J., Liu J., Lokki M. L., Marusic A., Maschio A., Meitinger T., Mulas A., Paré G., Parker A. N., Peden J. F., Petersmann A., Pichler I., Pietiläinen K. H., Pouta A., Ridderstråle M., Rotter J. I., Sambrook J. G., Sanders A. R., Schmidt C. O., Sinisalo J., Smit J. H., Stringham H. M., Bragi Walters G., Widen E., Wild S. H., Willemsen G., Zagato L., Zgaga L., Zitting P., Alavere H., Farrall M., McArdle W. L., Nelis M., Peters M. J., Ripatti S., van Meurs J. B., Aben K. K., Ardlie K. G., Beckmann J. S., Beilby J. P., Bergman R. N., Bergmann S., Collins F. S., Cusi D., den Heijer M., Eiriksdottir G., Gejman P. V., Hall A. S., Hamsten A., Huikuri H. V., Iribarren C., Kähönen M., Kaprio J., Kathiresan S., Kiemeney L., Kocher T., Launer L. J., Lehtimäki T., Melander O., Mosley T. H. Jr, Musk A. W., Nieminen M. S., O'Donnell C. J., Ohlsson C., Oostra B., Palmer L. J., Raitakari O., Ridker P. M., Rioux J. D., Rissanen A., Rivolta C., Schunkert H., Shuldiner A. R., Siscovick D. S., Stumvoll M., Tönjes A., Tuomilehto J., van Ommen G. J., Viikari J., Heath A. C., Martin N. G., Montgomery G. W., Province M. A., Kayser M., Arnold A. M., Atwood L. D., Boerwinkle E., Chanock S. J., Deloukas P., Gieger C., Grönberg H., Hall P., Hattersley A. T., Hengstenberg C., Hoffman W., Lathrop G. M., Salomaa V., Schreiber S., Uda M., Waterworth D., Wright A. F., Assimes T. L., Barroso I., Hofman A., Mohlke K. L., Boomsma D. I., Caulfield M. J., Cupples L. A., Erdmann J., Fox C. S., Gudnason V., Gyllensten U., Harris T. B., Hayes R. B., Jarvelin M. R., Mooser V., Munroe P. B., Ouwehand W. H., Penninx B. W., Pramstaller P. P., Quertermous T., Rudan I., Samani N. J., Spector T. D., Völzke H., Watkins H., Wilson J. F., Groop L. C., Haritunians T., Hu F. B., Kaplan R. C., Metspalu A., North K. E., Schlessinger D., Wareham N. J., Hunter D. J., O'Connell J. R., Strachan D. P., Wichmann H. E., Borecki I. B., van Duijn C. M., Schadt E. E., Thorsteinsdottir U., Peltonen L., Uitterlinden A. G., Visscher P. M., Chatterjee N., Loos R. J., Boehnke M., McCarthy M. I., Ingelsson E., Lindgren C. M., Abecasis G. R., Stefansson K., Frayling T. M. & Hirschhorn J. N. (2010). Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467(7317), 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang D., Liu H. & Arendt C. (2013). Robust methods for population stratification in genome wide association studies. BMC Bioinformatics 19(14), 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Bhattacharya S., Basu A. & Ghosh S. (2013). A novel Bayesian semiparametric algorithm for inferring population structure and adjusting for case–control association tests. Biometrics 69(1), 164–173. [DOI] [PubMed] [Google Scholar]
- Marchini J., Cardon L. R., Phillips M. S. & Donnelly P. (2004). The effects of human population structure on large genetic association studies. Nature Genetics 36(5), 512–517. [DOI] [PubMed] [Google Scholar]
- Miclaus K., Wolfinger R. & Czika W. (2009). SNP selection and multidimensional scaling to quantify population structure. Genetic Epidemiology 33(6), 488–496. [DOI] [PubMed] [Google Scholar]
- Panagiotou O. A. & Ioannidis J. P. (2012). What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. International Journal of Epidemiology 41, 273–286. [DOI] [PubMed] [Google Scholar]
- Panagiotou O. A., Evangelou E. & Ioannidis J. P. (2010). Genome-wide significant associations for variants with minor allele frequency of 5% or less--an overview: a HuGE review. American Journal of Epidemiology 172(8), 869–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira T. V., Patsopoulos N. A., Pereira A. C. & Krieger J. E. (2011). Strategies for genetic model specification in the screening of genome-wide meta-analysis signals for further replication. International Journal of Epidemiology 40, 457–469. [DOI] [PubMed] [Google Scholar]
- Pereira T. V., Patsopoulos N. A., Salanti G. & Ioannidis J. P. (2009). Discovery properties of genome wide association signals from cumulatively combined data sets. American Journal of Epidemiology 170(10), 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K. & Rosenberg N. A. (1999). Use of unlinked genetic markers to detect population stratification in association studies. American Journal of Human Genetics 65, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N. (1990). Linkage strategies for genetically complex traits. Multilocus models. American Journal of Human Genetics 46, 222–228. [PMC free article] [PubMed] [Google Scholar]
- Salanti G. & Higgins J. P. (2008). Meta-analysis of genetic association studies under different inheritance models using data reported as merged genotypes. Statistics in Medicine 27, 764–777. [DOI] [PubMed] [Google Scholar]
- Shin J. & Lee C. (2015). A mixed model reduces spurious genetic associations produced by population stratification in genome-wide association studies. Genomics 105(4), 191–196. [DOI] [PubMed] [Google Scholar]
- Silverman E. K. & Palmer L. J. (2000). Case control association studies for the genetics of complex respiratory diseases. American Journal of Respiratory Cell and Molecular Biology 22, 645–648. [DOI] [PubMed] [Google Scholar]
- Tian C., Plenge R. M., Ransom M., Lee A., Villoslada P., Selmi C., Klareskog L., Pulver A. E., Qi L., Gregersen P. K. & Seldin M. F. (2008). Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genetics 4(1), e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker G., Price A. L. & Berger B. (2014). Improving the power of GWAS and avoiding confounding from population stratification with PC-Select. Genetics 197(3), 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chen W., Chen X., Hu F., Archer K. J., Liu H. N., Sun S. & Gao G. (2012). Double genomic control is not effective to correct for population stratification in meta-analysis for genome-wide association studies. Frontiers in Genetics 24(3), 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shen X. & Pan W. (2013). Adjusting for population stratification in a fine scale with principal components and sequencing data. Genetic Epidemiology 37(8), 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. & Stephens M. (2012). Genome-wide efficient mixed-model analysis for association studies. Nature Genetics 44(7), 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0016672316000069.
click here to view supplementary material