Abstract
According to recent neuroimaging studies, temporal fluctuations in functional connectivity patterns can be clustered into dynamic functional connectivity (DFC) states and correspond to fluctuations in vigilance. However, whether there consistently exist DFC states associated with wakefulness and sleep stages and what are the characteristics and electrophysiological origin of these states remain unclear. The aims of the current study were to investigate the properties of DFC in different sleep stages and to explore the relationship between the characteristics of DFC and slow‐wave activity. We collected both eyes‐closed wakefulness and sleep data from 48 healthy young volunteers with simultaneous electroencephalography (EEG) and functional magnetic resonance imaging (fMRI) recordings. EEG data were employed as the gold standard of sleep stage scoring, and DFC states were estimated based on fMRI data. The results demonstrated that DFC states of the fMRI signals consistently corresponded to wakefulness and nonrapid eye movement sleep stages independent of the number of clusters. Furthermore, the mean dwell time of these states significantly correlated with slow‐wave activity. The inclusion or omission of regression of the global signal and the selection of parcellation schemes exerted minimal effects on the current findings. These results provide strong evidence that DFC states underlying fMRI signals match the fluctuations of vigilance and suggest a possible electrophysiological source of DFC states corresponding to vigilance states.
Keywords: DFC, EEG, fMRI, sleep, slow‐wave activity
1. INTRODUCTION
The human brain is organized into large scale networks with complex patterns of functional connectivity (Biswal et al., 2010; Buckner, Krienen, & Yeo, 2013; Fox et al., 2005), and the temporal fluctuations in functional connectivity are considered to dynamically adjust to the constantly changing environment (Cohen, 2018). These abilities are crucial for cognitive operations and the binding of information, which allow humans and animals to adaptively change their cognition and behaviors to successfully live in a complex and changing world (Rabinovich & Varona, 2011). Although previous studies primarily focused on characterizing stationary connectivity patterns throughout the duration of functional magnetic resonance imaging (fMRI) scanning, fluctuations in functional connectivity over shorter timescales might contain more information about adaptive cognition and behavior (Betzel, Fukushima, HeY, & Sporns, 2016; Hutchison et al., 2013; Krienen, Yeo, & Buckner, 2014; Laumann et al., 2017; Wang, Ong, Patanaik, Zhou, & Chee, 2016).
Sleep is one of the most fundamental physiological states of humans and plays a key role in human health and function (Spiegel, Leproult, & Van Cauter, 1999). The human sleep cycle consists of three nonrapid eye movement (NREM) sleep stages of increasing sleep depth defined as N1, N2, and N3 sleep and rapid eye movement (REM) sleep (Iber, Ancoli‐Israel, Chesson, & Quan, 2007). Sleep involves a regular evolution through a series of global brain activities, that are characterized by marked changes in electrophysiological signal and behavior (Duyn, 2012; Iber et al., 2007; Siclari et al., 2017). Each sleep stage has been indicated to be associated with a specific stationary functional connectivity pattern (Tagliazucchi et al., 2013; Tagliazucchi & Laufs, 2014). Moreover, previous studies have reported that loss of consciousness during NREM sleep is associated with increased modularity of brain activity (Boly et al., 2012).
Dynamic functional connectivity (DFC), commonly evaluated using sliding temporal windows correlation (Allen et al., 2014; Tian, Li, Wang, & Yu, 2018), has been adopted to qualify the time‐varying functional connectivity patterns of blood oxygenation level‐dependent (BOLD) signals across spatially separate brain regions. Fluctuations in functional connectivity likely include physiological signals, head motion, and noise in the acquired data (Chang et al., 2013; Handwerker, Roopchansingh, Gonzalez‐Castillo, & Bandettini, 2012; Laumann et al., 2017). However, converging evidence has indicated the neurobehavioral significance of DFC in fluctuations of vigilance, sustained attention, and cognitive vulnerabilities (Chang, Liu, Chen, Liu, & Duyn, 2013; Hutchison et al., 2013; Patanaik et al., 2018; Rosenberg et al., 2016). The association of DFC with neural oscillations, such as alpha and beta power, quantified using simultaneous electroencephalography (EEG) (Chang, Liu, et al., 2013; Laufs et al., 2003; Tagliazucchi, von Wegner, Morzelewski, Brodbeck, & Laufs, 2012) supports a neurobiological origin of DFC during the resting state. Similar to the transient quasi‐stable patterns detected in EEG data (microstates), DFC can be clustered into recurring patterns, that is, DFC states. One recent study showed that DFC states could capture variability across brain states during resting wakefulness to stages of sleep (Haimovici, Tagliazucchi, Balenzuela, & Laufs, 2017). However, whether DFC states consistently exist during NREM sleep and wakefulness is unclear. The relationship between the properties of DFC states and the electrophysiological signatures of deep sleep, that is, slow‐wave activity, is unknown.
In this study, we applied a sliding window approach to characterize the properties of DFC states during eyes‐closed wakefulness and different sleep stages, and explored the relationships between the characteristics of DFC states and slow‐wave activity. Simultaneous EEG‐fMRI scanning was performed during participants’ regular sleep time without sleep deprivation. In the experiment, volunteers fell into NREM sleep, ranging from light (N1 and N2) sleep to deep (N3) sleep. We adopted EEG as the gold standard for sleep stage scoring and estimated DFC states based on fMRI data recorded during wakefulness and different sleep stages to investigate the properties of DFC with different parameter settings during preprocessing and DFC states clustering. We further computed the slow‐wave activity based on EEG data to explore the relationship between these DFC states and slow‐wave activity.
2. MATERIALS AND METHODS
2.1. Participants
Forty‐eight healthy participants (26 males and 22 females; age 22.50 ± 2.64 years old) were recruited from the campus of Peking University. They were all right‐handed and free from the following conditions: (a) history of psychiatric or neurological illness, (b) history of a medically documented brain injury, (c) history of psychoactive drug consumption, or (d) current or previous drug or alcohol abuse. All participants were asked to refrain from alcohol and caffeine during the experiment days. This study was approved by the Institutional Review Board of Peking University Sixth Hospital, and informed consent was obtained from each participant.
2.2. Experimental design
Participants were asked to follow a regular sleep schedule for 2 weeks before MRI data collection, and their habitual sleep patterns were monitored through actigraphy and sleep diaries.
An adaptation session to our MR scanner (3T Prisma Scanner, Siemens Healthineers, Erlangen, Germany) was conducted at the Center for MRI Research, Peking University, within a week after the 2‐week sleep schedule was maintained. Participants were asked to lie in the scanner wearing an EEG cap (64‐channel MR‐compatible EEG system, Brain Products, Munich, Germany) and underwent 6 min of T1‐weighted scanning and 30 min of BOLD fMRI scanning. Within a week after the adaptation session, simultaneous EEG‐fMRI recordings were conducted at the participants’ usual bedtime. Notably, no sleep deprivation was involved, and natural sleep under normal homeostatic sleep pressure was investigated in this study.
2.3. Data acquisition
EEG‐fMRI data were acquired during sleep using a 3T Prisma Scanner and a 64‐channel MR‐compatible EEG system from Brain Products with a sampling rate of 5,000 Hz. The recording montage included 57 EEG channels positioned according to the international 10/20 system, two reference channels (A1, A2), two electrooculography (EOG) channels, two electromyography (EMG) channels, and one electrocardiogram (ECG) channel. The resistance of the reference and ground channels was maintained as less than 10 kΩ, while the resistance of the other channels was maintained as less than 20 kΩ. The resistance of all the channels was confirmed before starting and after ending the MR scanning. During EEG‐fMRI data acquisition, participants laid quietly, and cushions were used to restrain head movements.
Simultaneous EEG‐fMRI sessions were performed using a gradient echo‐planar imaging sequence with following parameters: Repetition time (TR) = 2,000 ms, echo time (TE) = 30 ms, flip angle (FA) = 90°, number of slices = 33, slice thickness = 3.5 mm, gap = 0.7 mm, matrix = 64 × 64, and in‐plane resolution = 3.5 × 3.5 mm2. During the sleep session, participants were instructed to close their eyes and try to sleep. The session ended when participants were completely awake and were unable to fall asleep again or when all 4,096 volumes (the largest number of volumes that can be acquired in a run for the BOLD fMRI sequence) were recorded.
For registration purposes, high‐resolution anatomical images were acquired for each participant using a three‐dimensional magnetization‐prepared rapid gradient echo T1‐weighted sequence (TR = 2,530 ms, TE = 2.98 ms, inversion time = 1,100 ms, FA = 7°, number of slices = 192, matrix = 512 × 448, voxel resolution = 0.5 × 0.5 × 1 mm3). During data acquisition, participants were instructed to lie quietly in the scanner.
2.4. EEG preprocessing and sleep stage scoring
EEG data preprocessing was performed using BrainVision Analyzer 2.1 (Brain Products, Munich, Germany). MR gradient artifacts in the EEG data were removed using the average artifact subtraction method (Allen, Josephs, & Turner, 2000). For the ballistocardiogram artifacts, the R peaks were detected semiautomatically with manual adjustment for peaks misidentified by the software. The R peaks were transferred from the ECG to the EEG over a selectable time delay and the average artifacts were then subtracted from the EEG data (Allen et al., 2000; Allen, Polizzi, Krakow, Fish, & Lemieux, 1998). Then, the data were re‐referenced to the mean values for channels A1 and A2, temporally filtered (0.3–35 Hz) and down‐sampled to 250 Hz. The sleep stage was scored for every 30‐s frame of preprocessed EEG data and was performed visually by an experienced technician and double‐checked by another experienced technician according to American Academy of Sleep Medicine criteria (Iber et al., 2007). Sleep recordings were divided into five discrete stages: (a) eyes‐closed wakefulness (Stage W), (b) non‐REM stage 1 (Stage N1), (c) non‐REM stage 2 (Stage N2), (d) non‐REM stage 3 (Stage N3), and (e) REM.
EEG data acquired from the frontal electrode (F3 and F4) were used to compute slow‐wave activity. The power spectral density was computed using a fast Fourier transform on the 5‐min EEG data after artifact removal based on 4 s Hanning window, overlapped by 2 s, using Welch's method based on custom script (“pwelch” function in MATLAB 2016b). Slow‐wave activity was calculated as the activity within the 0.75–4 Hz band (Leger et al., 2018; Ly et al., 2016).
2.5. fMRI data processing
Sleep fMRI data were divided into sessions with a duration of 5 min based on EEG sleep stage scoring. Each session corresponding to the wake state or a specific sleep stage. For example, we defined a 5‐min session as N1 when the N1 stage occupied the entire 5‐min session. A similar procedure was applied to divide the other sleep stages and wakefulness. For each session, fMRI data were preprocessed using the FMRIB Software Library (FSL, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki, version 5.0.9) tools (Smith et al., 2004). Preprocessing steps included: (a) a correction for the slice acquisition time shift; (b) a correction for head motion; (c) the removal of nonbrain tissue; (d) the projection of each fMRI dataset to the MNI152 standard space and resampling to 2 × 2 × 2 mm3 voxels; (e) the regression of realigned data using 6 head motion parameters and autoregressive models of motion: 6 head motion parameters, 6 head motion parameters at one time point before the volume, and the 12 corresponding squared items (Friston 24‐parameter model; Friston, Williams, Howard, Frackowiak, & Turner, 1996; Yan et al., 2013), the global signal, white matter signal, ventricular signal and their first derivatives; (f) the removal of linear tends and temporal filtering to retain frequencies between 0.01 and 0.1 Hz, and (g) spatial smoothing with a 6‐mm full‐width half‐maximum Gaussian kernel.
2.6. Computation of DFC states
DFC states were computed using MATLAB (2016b, The MathWorks Inc., Natick, MA). Whole‐brain dynamic functional networks were constructed based on the preprocessed fMRI data. We extracted signals from 264 regions defined using a functional atlas (Power et al., 2011), and constructed dynamic functional networks using a sliding window approach (Allen et al., 2014; Hutchison et al., 2013). Specifically, Pearson's correlation coefficients were calculated between each pair of regions based on the time course within each sliding window. The sliding window had a temporal width of 30 TRs (60 s) with a sliding step of 1 TR (2 s). We chose this window to obtain sufficient data to estimate functional connectivity at the low‐frequency band of interest (0.01–0.1 Hz) and to capture temporal variations in functional connectivity over shorter timescales (Betzel et al., 2016; Leonardi & Van De Ville, 2015; Liao et al., 2015). For each 5‐min session, we obtained 121 windows of a 264 × 264 symmetric correlation matrix and transformed the correlation coefficients into z values by Fisher's r‐to‐z transformation.
Correlation matrices from all 5‐min sessions were concatenated together, and k‐means clustering was performed to identify DFC states. Each windowed matrix was regarded as one observation, and each element of the 264 × 264 matrix was regarded as one variable. Cityblock distance was used as the distance measure, and the k‐means++ algorithm was used for cluster center initialization. The number of clusters was set by varying k‐value (k ranges from 2 to 8) to avoid possible bias caused by the use of an inappropriate k‐value. The k‐means clustering was repeated 100 times to increase chances of escaping local minima (Allen et al., 2014).
Three dynamic network metrics were computed based on the clustered states. (a) Mean dwell time (MDT) was measured as the average number of consecutive windows classified as instances of the same states and computed separately for each state. (b) Frequency of expression was measured as the proportion of all windows classified as instances of a particular state, also computed separately for each state. (c) Number of transitions was measured as the number of state transitions (Hutchison & Morton, 2015).
2.7. Statistical analysis
The Kruskal–Wallis test, a nonparametric statistical method used for one‐way ANOVA, was employed to investigate significant differences in dynamic network metrics among wakefulness and N1, N2, and N3 sleep stages. Spearman's correlation was computed to investigate the relationship between the dynamic network metrics and slow‐wave activity. We obtained the corrected p value from the Kruskal–Wallis test by dividing the original p value by the number of clusters (k‐value). The results with p values less than the corrected p values were considered statistically significant after Bonferroni's correction. For example, for k = 4, if the p value from Kruskal–Wallis test was less than 0.0001/4, we concluded that the difference was significant after correction, that is, p < .0001, Bonferroni corrected.
2.8. Validation analysis
Global signal regression in preprocessing is a debatable issue, and some researchers argue that global signal regression introduces artificial anti‐correlations into connectivity networks (Chen et al., 2018; Fox, Zhang, Snyder, & Raichle, 2009; Murphy, Birn, Handwerker, Jones, & Bandettini, 2009). We explored the effects of the global signal on dynamic network metrics and the results of the statistical analyses by eliminating the global signal regression step during preprocessing. We replicated all the analyses with an automatic anatomic labeling (AAL) atlas (Tzourio‐Mazoyer et al., 2002) and compared the results to data based on the Power264 atlas to assess whether the selection of whole‐brain parcellation schemes affected the current findings.
3. RESULTS
3.1. DFC states correspond to wakefulness and N1, N2, and N3 sleep
Thirty‐six sessions of wakefulness (9 males and 3 females; age 22.33 ± 3.52 years old), 25 sessions of N1 sleep (8 males and 10 females; age 21.72 ± 3.01 years old), 101 sessions of N2 sleep (11 males and 12 females; age 21.61 ± 3.15 years old), and 104 sessions of N3 sleep (12 males and 12 females; age 21.46 ± 2.96 years old) were analyzed in this study.
Figure 1a shows DFC states obtained by k‐means clustering with k = 4. State 1 displayed high within‐network connectivity in the somatomotor network, the cingulo‐opercular task control network, the auditory network and the default mode network. Strong anti‐correlations between the cingulo‐opercular task control network and the default mode network and between the auditory network and the default mode network were also observed. State 2 displayed high within‐network connectivity in the frontoparietal task control network and strong anti‐correlations between the visual network and the default mode network and between the visual network and the frontoparietal task control network. State 3 and State 4 displayed intermediate connectivity patterns between State 1 and State 2.
Figure 1.
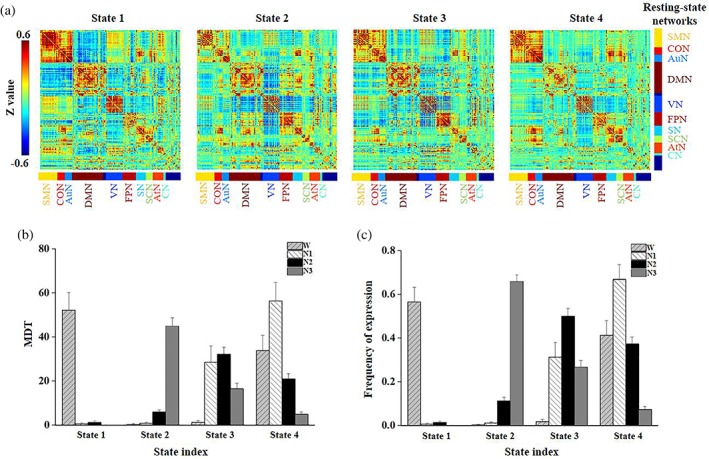
Dynamic functional connectivity states of k‐means clustering when k = 4. (a) The cluster centroids from k‐means clustering. The (b) mean dwell time (MDT) and (c) frequency of expression for the four states during wakefulness, N1, N2, and N3 sleep (error bars refer to standard errors). AtN, attention network; AuN, auditory network; CN, cerebellar network; CON, cingulo‐opercular task control network; DMN, default mode network; FPN, frontoparietal task control network; SCN, subcortical network; SMN, somatomotor network; SN, salience network; VN, visual network [Color figure can be viewed at http://wileyonlinelibrary.com]
Significant differences in the MDT among wakefulness, N1, N2, and N3 were observed for all four DFC states (p < .0001, Bonferroni corrected). State 1 exhibited a shorter MDT and a lower frequency of expression during N1, N2, and N3 than during wakefulness (Figure 1b,c). In contrast, State 2 exhibited a longer MDT and a higher frequency of expression during N3 sleep than during N1, N2 and wakefulness and during N2 than during wakefulness. State 3 exhibited a longer MDT and a higher frequency of expression during N1, N2, and N3 compared with wakefulness and during N2 than during N3. In addition, State 4 exhibited a longer MDT and a higher frequency of expression during N1 than during N2 and N3 and during N2 and wakefulness than during N3.
Figure 2 shows the results for the Kruskal–Wallis test results of both the MDT and frequency of expression during wakefulness, N1, N2, and N3 across varying k values (k = 2–8). The four states obtained when k = 4 were established as the reference states, and the states with other k values were designated based on the strength of the correlations between their connectivity patterns and these four reference states. Significant differences in the MDT and frequency of expression were consistently observed among wakefulness, N1, N2, and N3 (p < .0001, Bonferroni corrected) when k ranged from 2 to 4, in the first five states when k ranged from 5 to 7, and in the first four states and the eighth state when k = 8. There was at least one state with generally weak connectivity across the brain that temporally dominated during N3 sleep and another state with strong connection that dominated during wakefulness for varying k values (k = 2–8).
Figure 2.
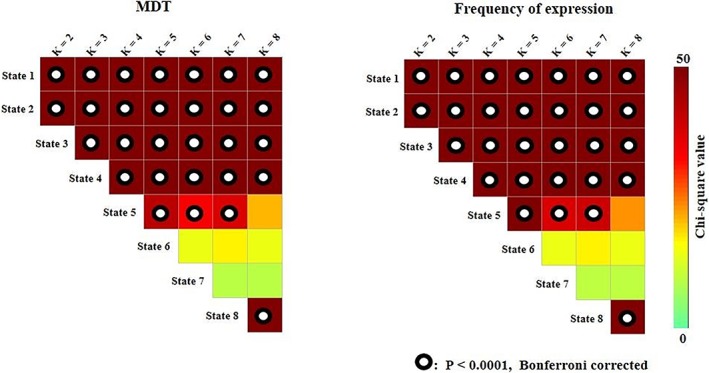
Results of the Kruskal–Wallis test of MDT and frequency of expression across k values [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3 shows the number of state transitions across varying k values (k = 2–8) for wakefulness, N1, N2, and N3. Significant differences in the number of state transitions were observed among wakefulness, N1, N2, and N3 (p < .01, k = 2–8). A consistently greater number of DFC state switches was observed in N2 sleep than in wakefulness (p < .05), and larger numbers were observed in N3 sleep than in wakefulness when k ranged from 3 to 6 (p < .05), in N2 than in N1 sleep when k ranged from 2 to 5 (p < .05), and in N2 than in N3 sleep when k = 2 and k = 3 (p < .05).
Figure 3.
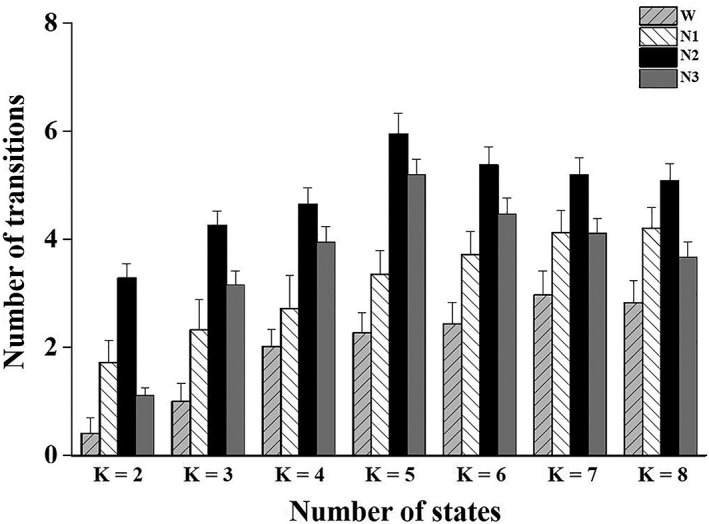
The number of transitions between DFC states across k values (error bars refer to standard errors)
3.2. Slow‐wave activity correlated with the MDT and frequency of expression
We further assessed the relationship between the slow‐wave activity and the MDT of the DFC states (Figure 4). For k = 4, the MDT of State 1 was negatively correlated with slow‐wave activity (R = −.51, p = 1.28 × 10−18), while the MDT of State 2 was positively correlated with slow‐wave activity (R = .66, p = 5.88 × 10−34). Similarly, the frequency of expression of State 1 was negatively correlated with slow‐wave activity (R = −.51, p = 9.76 × 10−19), while that of State 2 was positively correlated with slow‐wave activity (R = .67, p = 4.29 × 10−36). Corresponding to the results in Figure 1b,c, State 2 with k = 4 clusters had a significantly larger MDT and frequency of expression in N3 sleep than in wakefulness, N1 and N2 sleep, which was also marked by a high amplitude of slow‐wave activity. State 1 with k = 4 displayed a significant larger MDT and frequency of expression in wakefulness than in N1, N2, and N3 sleep, which had the lowest slow‐wave activity. No significant correlations were found between the number of transitions and slow‐wave activity.
Figure 4.
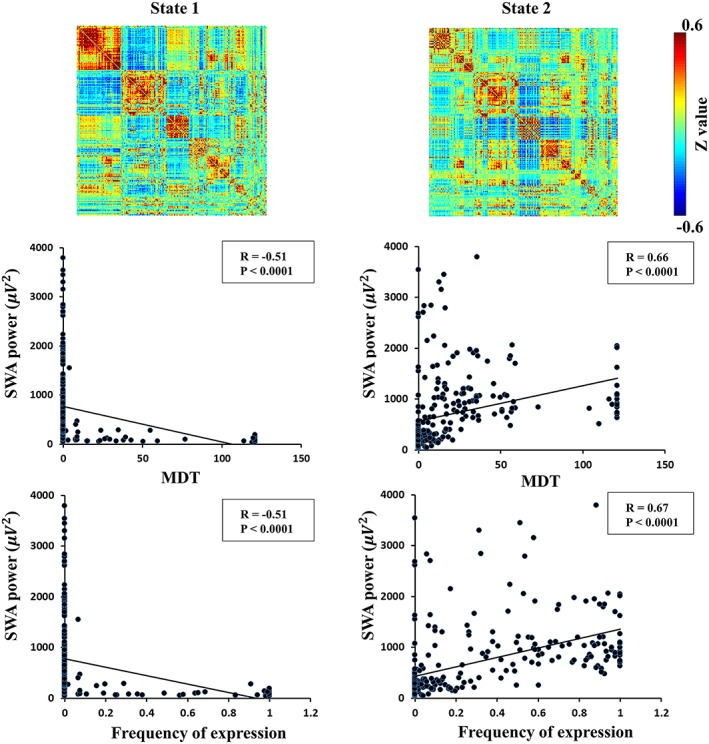
State 1 and State 2 inferred from k‐means clustering (k = 4) temporally predominate during wakefulness and N3 sleep stages and significantly correlate with slow‐wave activity [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 5 shows the strength of the correlations between MDT and frequency of expression with slow‐wave activity across varying k values. When k ranged from 2 to 8, State 1 had a significantly larger MDT and a higher frequency of expression in wakefulness than in N1, N2, and N3 sleep that was consistently negatively correlated with slow‐wave activity, while when k ranged from 2 to 8, State 2 showed a significantly larger MDT and a higher frequency of expression in N3 sleep than in wakefulness, and N1 and N2 sleep were consistently positively correlated with slow‐wave activity. In addition, State 4 with a k‐value ranging from 4 to 8 showed a significantly smaller MDT and frequency of expression in N3 sleep than in wakefulness, and N1 and N2 sleep were consistently negatively correlated with slow‐wave activity. Thus, the number of clusters during k‐means clustering showed minimal influence on the relationship between slow‐wave activity and dynamic network metrics. No significant correlations were found between the number of transitions and slow‐wave activity with other k values.
Figure 5.
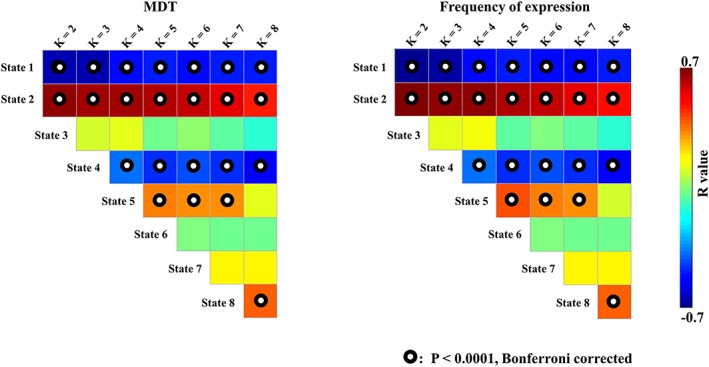
The correlations between MDT/frequency of expression and slow‐wave activity across k values [Color figure can be viewed at http://wileyonlinelibrary.com]
3.3. Validation results
Figure 6 shows the results of the Kruskal–Wallis test for both the MDT and frequency of expression among N1, N2, N3, and wakefulness when the global signal was reserved using the Power264 template and the global signal was removed using the AAL template. The results for the first two states were consistent with the data shown in Figure 2, as these states showed the largest MDT and frequency of expression in wakefulness or N3 sleep compared with other stages. The results were also consistent for the third state, which displayed a smaller MDT and frequency of expression in wakefulness than in N1, N2, and N3 sleep, and for the fourth state, which had a smaller MDT and frequency of expression in N3 sleep than in wakefulness and N1 and N2 sleep (Figure 2 vs. Figure 6).
Figure 6.
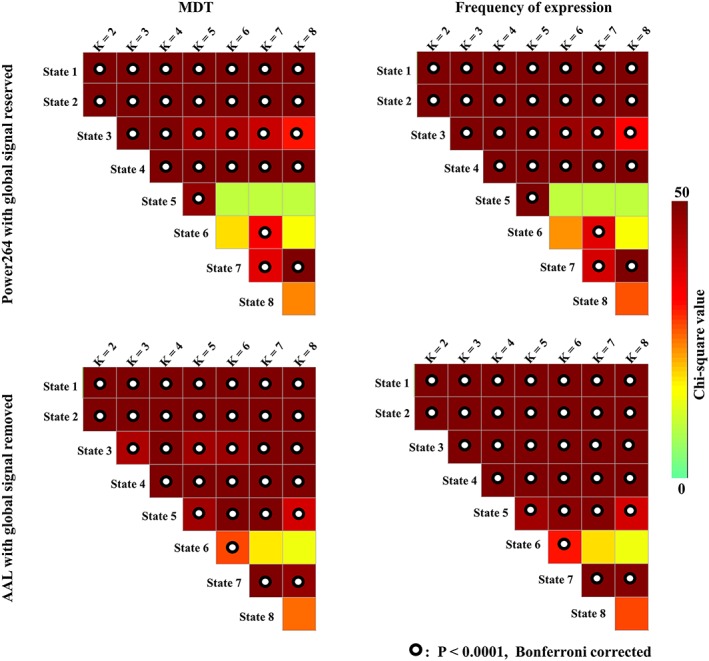
Validation of the results obtained from the Kruskal–Wallis test of MDT/frequency of expression across k‐values using different preprocessing parameters and parcellation schemes [Color figure can be viewed at http://wileyonlinelibrary.com]
The number of transitions was significantly larger in N2 sleep than in wakefulness when k ranged from 2 to 7, larger in N2 sleep than in wakefulness and N1 and N3 sleep when k = 3, larger in N3 sleep than in wakefulness when k ranged from 3 to 6, larger in N1 sleep than in wakefulness when k = 2 and k = 3, and larger in N1 and N2 sleep than in wakefulness and N3 sleep when k = 2 (p < .05) with the global signal reserved using the Power264 template. In addition, the number of transitions was significantly larger in N2 and N3 sleep than in wakefulness when k ranged from 2 to 8, and larger in N1, N2, and N3 sleep than in wakefulness when k ranged from 4 to 8 except k = 6 (p < .05) with the global signal removed using the AAL template. The results were similar to those obtained using the Power264 template with the global signal removed (Figure 7 vs. Figure 3).
Figure 7.
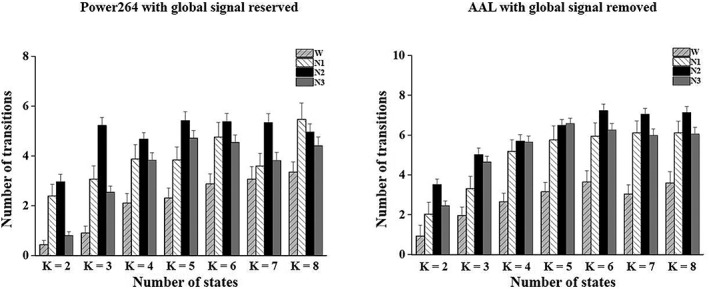
Validation of the number of transitions between DFC states across k values using different preprocessing parameters and parcellation schemes (error bars refer to standard errors)
Figure 8 shows the strength of the correlations between of MDT and frequency of expression with slow‐wave activity when the global signal was reserved using the Power264 template and when the global signal was removed using the AAL template. These results were consistent with the data shown in Figure 5 for the first two states, which had the largest MDT and frequency of expression in wakefulness or N3 sleep compared with the other stages, and for the fourth state which had a smaller MDT and frequency of expression in N3 sleep than in wakefulness and N1 and N2 sleep.
Figure 8.
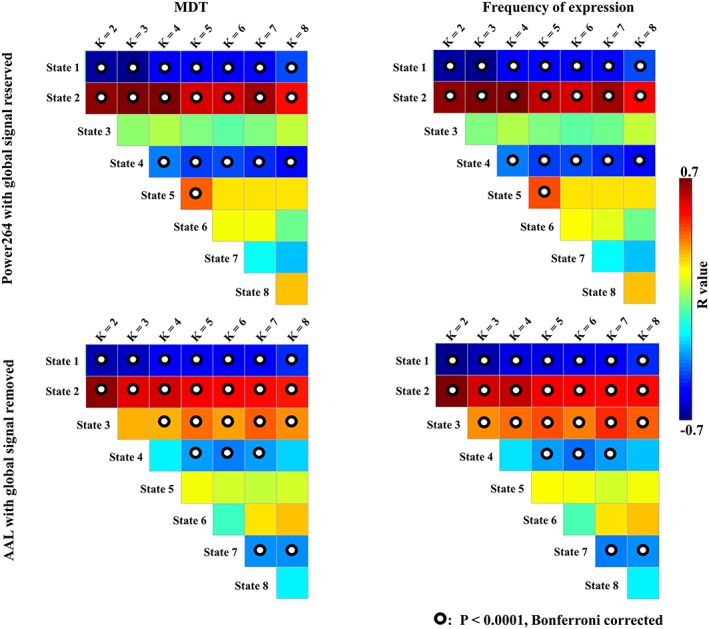
Validation of the correlations between MDT/frequency of expression and slow‐wave activity using different preprocessing parameters and parcellation schemes [Color figure can be viewed at http://wileyonlinelibrary.com]
These results from the validation analyses suggested that global signal removal and selection of the whole‐brain atlas had minimal influence on the characteristics of DFC among wakefulness, N1, N2, and N3, and their relationships with slow‐wave activity.
4. DISCUSSION
In this study, we investigated the properties of DFC in wakefulness and different sleep stages based on the sliding window approach. Our results showed that dynamic connectivity states consistently corresponded to specific sleep stages or wakefulness, independent of the number of clustered states or preprocessing parameters. These results indicated that wakefulness and different sleep stages have specific recurring connectivity patterns. We further explored the relationship between the characteristics of these dynamic connectivity states and slow‐wave activity and found that states with a larger MDT in N3 sleep sessions showed significant positive correlations with slow‐wave activity, while states with a larger MDT in wakefulness sessions showed significant negative correlations with slow‐wave activity. These results revealed the characteristics of dynamic connectivity patterns in wakefulness and different sleep stages and provide a potential neural electrophysiology basis for these dynamic connectivity patterns.
4.1. Dynamic connectivity properties of NREM sleep and wakefulness
Sleep has been associated with reduced functional connectivity within the default mode network and reduced anti‐correlation between task‐positive networks and the default mode network (De Havas, Parimal, Soon, & Chee, 2012) compared with wakefulness. These alterations in functional connectivity have also been observed during mind wandering (Christoff, Gordon, Smallwood, Smith, & Schooler, 2009) and during eyes‐closed rest compared with eyes‐open rest (Van Dijk et al., 2010). It has been proposed that sleep is facilitated by both reduced thalamocortical connectivity at sleep onset (Poudel, Innes, Bones, Watts, & Jones, 2014) and a breakdown of general connectivity associated with deep sleep (Spoormaker et al., 2010). Both of these processes reduce the brain's capacity to integrate information across separated regions (Horovitz et al., 2009; Samann et al., 2011).
DFC states during wakefulness have been reported to exhibit a richer repertoire of functional connectivity compared with states of anesthesia (Barttfeld et al., 2015). It has also been reported that clustering or hierarchical approaches can reveal dynamic connectivity states that could be linked with ongoing cognitive fluctuations and functional connectivity changes when people shift between sleep and wakefulness (Laumann et al., 2017). Haimovici and colleagues demonstrated that coarse dynamic connectivity states matched wakefulness and sleep stages by setting the number of clusters equal to the number of stages in the human NREM sleep cycle (Haimovici et al., 2017). In our study, we set a varying k‐value (k ranges from 2 to 8) and found similar dynamic connectivity patterns that characterized wakefulness and NREM sleep across different k‐values during clustering.
Differences in states that dominated during wakefulness and deep sleep were observed in a widespread network of regions. The state that temporally dominated during wakefulness displayed high within‐network connectivity of the somatomotor network and the default mode network, and strong anti‐correlations between the cingulo‐opercular task control network and the default mode network and between the auditory network and the default mode network. This result was consistent with previous findings of a loss of within‐network connectivity in the somatomotor network and the default mode network during reduced consciousness (Spoormaker, Gleiser, & Czisch, 2012; Wu et al., 2012), and were also consistent with the Global Neuronal Workspace theory stating that different streams of information compete for the brain in widespread network regions (Dehaene & Changeux, 2011).
The number of state transitions was consistently larger in N2 sleep than in wakefulness and generally larger in N3 sleep than in wakefulness, independent of the selection of parcellation schemes and preprocessing procedures. One or two DFC states temporally dominated approximately 90% of the duration of the wakefulness state, thus corresponding to a small number of state transitions. In contrast, more DFC states temporally fluctuated and frequently shifted during N2 and N3 sleep, corresponding to more state transitions. These results differ from findings from previous animal studies, which indicated that wakefulness is characterized by a richer repertoire of functional configurations than the state of anesthesia (Barttfeld et al., 2015). However, the definition of wakefulness state in the previous study was not the same as the definition used in the current study, which considered eyes‐closed while trying to fall sleep as wakefulness, and the state of sedation also is not equivalent to the sleep state. Moreover, another study suggested that spontaneous activity should be reduced to the circulation of a more random pattern of neural activity that is shaped and constrained by anatomical connectivity under the nonconscious condition (Deco, Jirsa, & McIntosh, 2011). However, none of these studies provided direct evidence of the potential changes in the number of state switches under varying consciousness conditions. The current study fills this gap by showing more transitions among DFC states during sleep.
These results provide strong evidence that the dynamic connectivity states underlying the BOLD signal match fluctuations of vigilance.
4.2. Slow‐wave activity correlated with the MDT of dynamic connectivity states
The slow‐wave activity of NREM sleep is a well‐established marker of sleep pressure (Tononi & Cirelli, 2014), and the individual dissipation rate of slow‐wave power reflects individual sleep homoeostasis efficacy in the elimination of sleep pressure (Tarokh, Carskadon, & Achermann, 2012). Furthermore, Boly and colleagues demonstrated that brain network integration decreased in proportion to slow‐wave activity, consistent with a breakdown of brain connectivity resulting from the neural response associated with slow oscillations (Boly et al., 2012). Here, we assessed the relationship between the MDT of the dynamic connectivity states and slow‐wave activity, and found that the MDT of the state that dominated in N3 sleep sessions positively correlated with the power of slow‐wave activity, while the MDT of the state that dominated in wakefulness sessions negatively correlated with slow‐wave activity. These results were consistent across varying k‐values during clustering and were independent of global mean signal removal and brain atlas selection during preprocessing, indicating potential electrophysiological mechanisms underlying dynamic connectivity states that correspond to vigilance states.
4.3. DFC states vary with k‐values and global signals
k‐Means clustering identifies DFC states by grouping the network configurations that are more similar to each other among the network configurations of all clusters, thus, the DFC states with larger k‐values will reveal intermediate network configurations between separate DFC states with smaller k‐values. A previous study reported that the instability of clustering increased with the number of estimated brain states for expansion in the solution space (Reinen et al., 2018). We chose a range of k‐values of 2–8 to cover the number of sleep stages and validate the stability of clustering.
Previous studies revealed the clinical effects of global signal fluctuations in subjects with neuropsychiatric conditions (Yang et al., 2014) and the differences in the global signal among participants only contributed to the prediction of the vigilance state after sleep restriction, but not baseline performance (Patanaik et al., 2018). In line with these studies, the global signal did not change the main results of this study.
4.4. Removal of gradient and ballistocardiogram artifacts
Average artifact subtraction is currently the most commonly used correction method for removing gradient and ballistocardiogram artifacts in previous sleep studies based on simultaneous EEG‐fMRI recordings (Hale et al., 2016; Horovitz et al., 2009; Samann et al., 2011; Tsai et al., 2014). Several studies used average subtraction to remove gradient artifacts and an independent component analysis (ICA) to remove ballistocardiogram artifacts (Boly et al., 2012; Lei, Wang, Yuan, & Chen, 2015). We further adopted an ICA to remove residual artifacts from 21 randomly chosen sessions of EEG data and to determine whether the gradient and ballistocardiogram artifacts were satisfactorily corrected after average artifact subtraction preprocessing in our study. We decomposed each session of EEG data into 59 components after average artifact subtraction preprocessing, and removed one or two components that might be related to residual artifacts. The residual artifact components were always present in the last 30 components after ICA, indicating that the residual artifact components accounted for a very small proportion of the variance in the preprocessed EEG data after average artifact subtraction. The removal of these residual artifacts components did not change the scoring of the sleep stages for these sessions. Moreover, we calculated the power of slow‐wave activity in these sessions with and without further ICA denoising, and the results were almost identical.
4.5. Limitations and future work
The present study has several limitations. First, we collected relatively few 5‐min sessions in the N1 sleep stage for dynamic connectivity analysis, because the proportion of data acquired that corresponded to the N1 sleep stage was relatively small, and these periods lasted for a relatively short time. Second, we chose the sliding window approach to construct the dynamic connectivity matrix, and the window length was fixed, which might influence the flexibility to detect non‐regular changes in functional connectivity (Lindquist, Xu, Nebel, & Caffo, 2014). Finally, our study provided evidence of an association between dynamic connectivity states underlying BOLD signals and slow‐wave activity. However, the plots were relatively scattered, and thus the strength of their correlations and the causal links between neurophysiology and dynamic connectivity patterns in the brain require further investigation.
5. CONCLUSIONS
In the present study, the dynamic connectivity states of BOLD signals consistently corresponded to wakefulness and sleep stages, and the MDT of the state significantly correlated with slow‐wave activity. These results provide strong evidence that dynamic connectivity states match the fluctuation of vigilance, and indicate possible electrophysiology mechanisms underlying dynamic connectivity states that correspond to vigilance states.
CONFLICT OF INTEREST
The authors have no financial or competing interests to declare.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2018YFC2000603 and 2017YFC0108900), China's National Strategic Basic Research Program (“973”) grant (2015CB856400), National Natural Science Foundation of China (81871427, 81671765, 81430037, 81727808, 81790650, 81790651 and 31421003), Beijing Municipal Natural Science Foundation (7172121), Beijing Municipal Science & Technology Commission (Z181100001518005, Z161100002616006 and Z171100000117012), Shenzhen Peacock Plan (KQTD2015033016104926), Guangdong Pearl River Talents Plan Innovative and Entrepreneurial Team (2016ZT06S220), Shenzhen Science and Technology Research Funding Program (JCYJ20170412164413575). We thank the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with MRI data acquisition and data analyses.
Zhou S, Zou G, Xu J, et al. Dynamic functional connectivity states characterize NREM sleep and wakefulness. Hum Brain Mapp. 2019;40:5256–5268. 10.1002/hbm.24770
Funding information Beijing Municipal Natural Science Foundation, Grant/Award Number: 7172121; Beijing Municipal Science & Technology Commission, Grant/Award Numbers: Z181100001518005, Z161100002616006, Z1711000001; China's National Strategic Basic Research Program ("973") grant, Grant/Award Number: 2015CB856400; Guangdong Pearl River Talents Plan Innovative and Entrepreneurial Team, Grant/Award Number: 2016ZT06S220; National Key Research and Development Program of China, Grant/Award Numbers: 2018YFC2000603, 2017YFC0108900; National Natural Science Foundation of China, Grant/Award Numbers: 81871427, 81671765, 81430037, 81727808, 81790650; Shenzhen Peacock Plan, Grant/Award Number: KQTD2015033016104926; Shenzhen Science and Technology Research Funding Program, Grant/Award Number: JCYJ20170412164413575
Contributor Information
Qihong Zou, Email: zouqihong@pku.edu.cn.
Jia‐Hong Gao, Email: jgao@pku.edu.cn.
DATA AVAILABILITY STATEMENT
Data are available on request from the authors. The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Allen, E. A. , Damaraju, E. , Plis, S. M. , Erhardt, E. B. , Eichele, T. , & Calhoun, V. D. (2014). Tracking whole‐brain connectivity dynamics in the resting state. Cerebral Cortex, 24, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, P. J. , Josephs, O. , & Turner, R. (2000). A method for removing imaging artifact from continuous EEG recorded during functional MRI. NeuroImage, 12, 230–239. [DOI] [PubMed] [Google Scholar]
- Allen, P. J. , Polizzi, G. , Krakow, K. , Fish, D. R. , & Lemieux, L. (1998). Identification of EEG events in the MR scanner: The problem of pulse artifact and a method for its subtraction. NeuroImage, 8, 229–239. [DOI] [PubMed] [Google Scholar]
- Barttfeld, P. , Uhrig, L. , Sitt, J. D. , Sigman, M. , Jarraya, B. , & Dehaene, S. (2015). Signature of consciousness in the dynamics of resting‐state brain activity. Proceedings of the National Academy of Sciences of the United States of America, 112, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel, R. F. , Fukushima, M. , HeY, Z. X. N. , & Sporns, O. (2016). Dynamic fluctuations coincide with periods of high and low modularity in resting‐state functional brain networks. NeuroImage, 127, 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal, B. B. , Mennes, M. , Zuo, X. N. , Gohel, S. , Kelly, C. , Smith, S. M. , … Milham, M. P. (2010). Toward discovery science of human brain function. Proceedings of the National Academy of Sciences of the United States of America, 107, 4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boly, M. , Perlbarg, V. , Marrelec, G. , Schabus, M. , Laureys, S. , Doyon, J. , … Benali, H. (2012). Hierarchical clustering of brain activity during human nonrapid eye movement sleep. Proceedings of the National Academy of Sciences of the United States of America, 109, 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner, R. L. , Krienen, F. M. , & Yeo, B. T. (2013). Opportunities and limitations of intrinsic functional connectivity MRI. Nature Neuroscience, 16, 832–837. [DOI] [PubMed] [Google Scholar]
- Chang, C. , Liu, Z. , Chen, M. C. , Liu, X. , & Duyn, J. H. (2013). EEG correlates of time‐varying BOLD functional connectivity. NeuroImage, 72, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. , Metzger, C. D. , Glover, G. H. , Duyn, J. H. , Heinze, H. J. , & Walter, M. (2013). Association between heart rate variability and fluctuations in resting‐state functional connectivity. NeuroImage, 68, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Liao, X. , Dai, Z. , Lin, Q. , Wang, Z. , Li, K. , & He, Y. (2018). Topological analyses of functional connectomics: A crucial role of global signal removal, brain parcellation, and null models. Human Brain Mapping, 39, 4545–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoff, K. , Gordon, A. M. , Smallwood, J. , Smith, R. , & Schooler, J. W. (2009). Experience sampling during fMRI reveals default network and executive system contributions to mind wandering. Proceedings of the National Academy of Sciences of the United States of America, 106, 8719–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. R. (2018). The behavioral and cognitive relevance of time‐varying, dynamic changes in functional connectivity. NeuroImage, 180, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Havas, J. A. , Parimal, S. , Soon, C. S. , & Chee, M. W. (2012). Sleep deprivation reduces default mode network connectivity and anti‐correlation during rest and task performance. NeuroImage, 59, 1745–1751. [DOI] [PubMed] [Google Scholar]
- Deco, G. , Jirsa, V. K. , & McIntosh, A. R. (2011). Emerging concepts for the dynamical organization of resting‐state activity in the brain. Nature Reviews. Neuroscience, 12, 43–56. [DOI] [PubMed] [Google Scholar]
- Dehaene, S. , & Changeux, J. P. (2011). Experimental and theoretical approaches to conscious processing. Neuron, 70, 200–227. [DOI] [PubMed] [Google Scholar]
- Duyn, J. H. (2012). EEG‐fMRI methods for the study of brain networks during sleep. Frontiers in Neurology, 3, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. D. , Snyder, A. Z. , Vincent, J. L. , Corbetta, M. , Van Essen, D. C. , & Raichle, M. E. (2005). The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences of the United States of America, 102, 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. D. , Zhang, D. , Snyder, A. Z. , & Raichle, M. E. (2009). The global signal and observed anticorrelated resting state brain networks. Journal of Neurophysiology, 101, 3270–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston, K. J. , Williams, S. , Howard, R. , Frackowiak, R. S. , & Turner, R. (1996). Movement‐related effects in fMRI time‐series. Magnetic Resonance in Medicine, 35, 346–355. [DOI] [PubMed] [Google Scholar]
- Haimovici, A. , Tagliazucchi, E. , Balenzuela, P. , & Laufs, H. (2017). On wakefulness fluctuations as a source of BOLD functional connectivity dynamics. Scientific Reports, 7, 5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, J. R. , White, T. P. , Mayhew, S. D. , Wilson, R. S. , Rollings, D. T. , Khalsa, S. , … Bagshaw, A. P. (2016). Altered thalamocortical and intra‐thalamic functional connectivity during light sleep compared with wake. NeuroImage, 125, 657–667. [DOI] [PubMed] [Google Scholar]
- Handwerker, D. A. , Roopchansingh, V. , Gonzalez‐Castillo, J. , & Bandettini, P. A. (2012). Periodic changes in fMRI connectivity. NeuroImage, 63, 1712–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz, S. G. , Braun, A. R. , Carr, W. S. , Picchioni, D. , Balkin, T. J. , Fukunaga, M. , & Duyn, J. H. (2009). Decoupling of the brain's default mode network during deep sleep. Proceedings of the National Academy of Sciences of the United States of America, 106, 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, R. M. , & Morton, J. B. (2015). Tracking the brain's functional coupling dynamics over development. The Journal of Neuroscience, 35, 6849–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, R. M. , Womelsdorf, T. , Allen, E. A. , Bandettini, P. A. , Calhoun, V. D. , Corbetta, M. , … Chang, C. (2013). Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage, 80, 360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iber, C. , Ancoli‐Israel, S. S. , Chesson, A. , & Quan, S. F. (2007). The AASM manual for the scoring of sleep and associated events: Rules, terminology, and technical specifications. Westchester, IL: American Academy of Sleep Medicine. [Google Scholar]
- Krienen, F. M. , Yeo, B. T. , & Buckner, R. L. (2014). Reconfigurable task‐dependent functional coupling modes cluster around a core functional architecture. Philosophical Transactions of the Royal Society B: Biological Sciences, 369, 20130526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, H. , Krakow, K. , Sterzer, P. , Eger, E. , Beyerle, A. , Salek‐Haddadi, A. , & Kleinschmidt, A. (2003). Electroencephalographic signatures of attentional and cognitive default modes in spontaneous brain activity fluctuations at rest. Proceedings of the National Academy of Sciences of the United States of America, 100, 11053–11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann, T. O. , Snyder, A. Z. , Mitra, A. , Gordon, E. M. , Gratton, C. , Adeyemo, B. , … Petersen, S. E. (2017). On the stability of BOLD fMRI correlations. Cerebral Cortex, 27, 4719–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger, D. , Debellemaniere, E. , Rabat, A. , Bayon, V. , Benchenane, K. , & Chennaoui, M. (2018). Slow‐wave sleep: From the cell to the clinic. Sleep Medicine Reviews, 41, 113–132. [DOI] [PubMed] [Google Scholar]
- Lei, X. , Wang, Y. , Yuan, H. , & Chen, A. (2015). Brain scale‐free properties in awake rest and NREM sleep: A simultaneous EEG/fMRI study. Brain Topography, 28, 292–304. [DOI] [PubMed] [Google Scholar]
- Leonardi, N. , & Van De Ville, D. (2015). On spurious and real fluctuations of dynamic functional connectivity during rest. NeuroImage, 104, 430–436. [DOI] [PubMed] [Google Scholar]
- Liao, X. , Yuan, L. , Zhao, T. , Dai, Z. , Shu, N. , Xia, M. , … He, Y. (2015). Spontaneous functional network dynamics and associated structural substrates in the human brain. Frontiers in Human Neuroscience, 9, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist, M. A. , Xu, Y. , Nebel, M. B. , & Caffo, B. S. (2014). Evaluating dynamic bivariate correlations in resting‐state fMRI: A comparison study and a new approach. NeuroImage, 101, 531–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly, J. Q. , Gaggioni, G. , Chellappa, S. L. , Papachilleos, S. , Brzozowski, A. , Borsu, C. , … Vandewalle, G. (2016). Circadian regulation of human cortical excitability. Nature Communications, 7, 11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K. , Birn, R. M. , Handwerker, D. A. , Jones, T. B. , & Bandettini, P. A. (2009). The impact of global signal regression on resting state correlations: Are anti‐correlated networks introduced? NeuroImage, 44, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanaik, A. , Tandi, J. , Ong, J. L. , Wang, C. , Zhou, J. , & Chee, M. W. L. (2018). Dynamic functional connectivity and its behavioral correlates beyond vigilance. NeuroImage, 177, 1–10. [DOI] [PubMed] [Google Scholar]
- Poudel, G. R. , Innes, C. R. , Bones, P. J. , Watts, R. , & Jones, R. D. (2014). Losing the struggle to stay awake: Divergent thalamic and cortical activity during microsleeps. Human Brain Mapping, 35, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Cohen, A. L. , Nelson, S. M. , Wig, G. S. , Barnes, K. A. , Church, J. A. , … Petersen, S. E. (2011). Functional network organization of the human brain. Neuron, 72, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich, M. I. , & Varona, P. (2011). Robust transient dynamics and brain functions. Frontiers in Computational Neuroscience, 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinen, J. M. , Chen, O. Y. , Hutchison, R. M. , Yeo, B. T. T. , Anderson, K. M. , Sabuncu, M. R. , … Holmes, A. J. (2018). The human cortex possesses a reconfigurable dynamic network architecture that is disrupted in psychosis. Nature Communications, 9, 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, M. D. , Finn, E. S. , Scheinost, D. , Papademetris, X. , Shen, X. , Constable, R. T. , & Chun, M. M. (2016). A neuromarker of sustained attention from whole‐brain functional connectivity. Nature Neuroscience, 19, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samann, P. G. , Wehrle, R. , Hoehn, D. , Spoormaker, V. I. , Peters, H. , Tully, C. , … Czisch, M. (2011). Development of the brain's default mode network from wakefulness to slow wave sleep. Cerebral Cortex, 21, 2082–2093. [DOI] [PubMed] [Google Scholar]
- Siclari, F. , Baird, B. , Perogamvros, L. , Bernardi, G. , LaRocque, J. J. , Riedner, B. , … Tononi, G. (2017). The neural correlates of dreaming. Nature Neuroscience, 20, 872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Jenkinson, M. , Woolrich, M. W. , Beckmann, C. F. , Behrens, T. E. , Johansen‐Berg, H. , … Matthews, P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23(Suppl 1), S208–S219. [DOI] [PubMed] [Google Scholar]
- Spiegel, K. , Leproult, R. , & Van Cauter, E. (1999). Impact of sleep debt on metabolic and endocrine function. Lancet, 354, 1435–1439. [DOI] [PubMed] [Google Scholar]
- Spoormaker, V. I. , Gleiser, P. M. , & Czisch, M. (2012). Frontoparietal connectivity and hierarchical structure of the brain's functional network during sleep. Frontiers in Neurology, 3, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoormaker, V. I. , Schroter, M. S. , Gleiser, P. M. , Andrade, K. C. , Dresler, M. , Wehrle, R. , … Czisch, M. (2010). Development of a large‐scale functional brain network during human non‐rapid eye movement sleep. The Journal of Neuroscience, 30, 11379–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi, E. , & Laufs, H. (2014). Decoding wakefulness levels from typical fMRI resting‐state data reveals reliable drifts between wakefulness and sleep. Neuron, 82, 695–708. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi, E. , von Wegner, F. , Morzelewski, A. , Brodbeck, V. , Jahnke, K. , & Laufs, H. (2013). Breakdown of long‐range temporal dependence in default mode and attention networks during deep sleep. Proceedings of the National Academy of Sciences of the United States of America, 110, 15419–15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi, E. , von Wegner, F. , Morzelewski, A. , Brodbeck, V. , & Laufs, H. (2012). Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Frontiers in Human Neuroscience, 6, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarokh, L. , Carskadon, M. A. , & Achermann, P. (2012). Dissipation of sleep pressure is stable across adolescence. Neuroscience, 216, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L. , Li, Q. , Wang, C. , & Yu, J. (2018). Changes in dynamic functional connections with aging. NeuroImage, 172, 31–39. [DOI] [PubMed] [Google Scholar]
- Tononi, G. , & Cirelli, C. (2014). Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron, 81, 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, P. J. , Chen, S. C. , Hsu, C. Y. , Wu, C. W. , Wu, Y. C. , Hung, C. S. , … Lin, C. P. (2014). Local awakening: Regional reorganizations of brain oscillations after sleep. NeuroImage, 102(Pt 2), 894–903. [DOI] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer, N. , Landeau, B. , Papathanassiou, D. , Crivello, F. , Etard, O. , Delcroix, N. , … Joliot, M. (2002). Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. NeuroImage, 15, 273–289. [DOI] [PubMed] [Google Scholar]
- Van Dijk, K. R. , Hedden, T. , Venkataraman, A. , Evans, K. C. , Lazar, S. W. , & Buckner, R. L. (2010). Intrinsic functional connectivity as a tool for human connectomics: Theory, properties, and optimization. Journal of Neurophysiology, 103, 297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Ong, J. L. , Patanaik, A. , Zhou, J. , & Chee, M. W. (2016). Spontaneous eyelid closures link vigilance fluctuation with fMRI dynamic connectivity states. Proceedings of the National Academy of Sciences of the United States of America, 113, 9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. W. , Liu, P. Y. , Tsai, P. J. , Wu, Y. C. , Hung, C. S. , Tsai, Y. C. , … Lin, C. P. (2012). Variations in connectivity in the sensorimotor and default‐mode networks during the first nocturnal sleep cycle. Brain Connectivity, 2, 177–190. [DOI] [PubMed] [Google Scholar]
- Yan, C. G. , Cheung, B. , Kelly, C. , Colcombe, S. , Craddock, R. C. , Di Martino, A. , … Milham, M. P. (2013). A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. NeuroImage, 76, 183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G. J. , Murray, J. D. , Repovs, G. , Cole, M. W. , Savic, A. , Glasser, M. F. , … Anticevic, A. (2014). Altered global brain signal in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America, 111, 7438–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available on request from the authors. The data that support the findings of this study are available from the corresponding author upon reasonable request.


