Key Points
Question
Are there differences in genetic risk factors for primary open-angle glaucoma based on ancestry?
Findings
In this multistage, case-control, genome-wide association study that included 26 295 participants, the amyloid-β A4 precursor protein-binding family B member 2 (APBB2) locus was significantly associated with primary open-angle glaucoma among individuals of African ancestry (odds ratio, 1.19 per copy of the risk allele for single-nucleotide polymorphism rs59892895T>C), but not of European or Asian ancestry.
Meaning
This study identified a single-nucleotide polymorphism that demonstrated differential association with primary open-angle glaucoma by ancestry.
Abstract
Importance
Primary open-angle glaucoma presents with increased prevalence and a higher degree of clinical severity in populations of African ancestry compared with European or Asian ancestry. Despite this, individuals of African ancestry remain understudied in genomic research for blinding disorders.
Objectives
To perform a genome-wide association study (GWAS) of African ancestry populations and evaluate potential mechanisms of pathogenesis for loci associated with primary open-angle glaucoma.
Design, Settings, and Participants
A 2-stage GWAS with a discovery data set of 2320 individuals with primary open-angle glaucoma and 2121 control individuals without primary open-angle glaucoma. The validation stage included an additional 6937 affected individuals and 14 917 unaffected individuals using multicenter clinic- and population-based participant recruitment approaches. Study participants were recruited from Ghana, Nigeria, South Africa, the United States, Tanzania, Britain, Cameroon, Saudi Arabia, Brazil, the Democratic Republic of the Congo, Morocco, Peru, and Mali from 2003 to 2018. Individuals with primary open-angle glaucoma had open iridocorneal angles and displayed glaucomatous optic neuropathy with visual field defects. Elevated intraocular pressure was not included in the case definition. Control individuals had no elevated intraocular pressure and no signs of glaucoma.
Exposures
Genetic variants associated with primary open-angle glaucoma.
Main Outcomes and Measures
Presence of primary open-angle glaucoma. Genome-wide significance was defined as P < 5 × 10−8 in the discovery stage and in the meta-analysis of combined discovery and validation data.
Results
A total of 2320 individuals with primary open-angle glaucoma (mean [interquartile range] age, 64.6 [56-74] years; 1055 [45.5%] women) and 2121 individuals without primary open-angle glaucoma (mean [interquartile range] age, 63.4 [55-71] years; 1025 [48.3%] women) were included in the discovery GWAS. The GWAS discovery meta-analysis demonstrated association of variants at amyloid-β A4 precursor protein-binding family B member 2 (APBB2; chromosome 4, rs59892895T>C) with primary open-angle glaucoma (odds ratio [OR], 1.32 [95% CI, 1.20-1.46]; P = 2 × 10−8). The association was validated in an analysis of an additional 6937 affected individuals and 14 917 unaffected individuals (OR, 1.15 [95% CI, 1.09-1.21]; P < .001). Each copy of the rs59892895*C risk allele was associated with increased risk of primary open-angle glaucoma when all data were included in a meta-analysis (OR, 1.19 [95% CI, 1.14-1.25]; P = 4 × 10−13). The rs59892895*C risk allele was present at appreciable frequency only in African ancestry populations. In contrast, the rs59892895*C risk allele had a frequency of less than 0.1% in individuals of European or Asian ancestry.
Conclusions and Relevance
In this genome-wide association study, variants at the APBB2 locus demonstrated differential association with primary open-angle glaucoma by ancestry. If validated in additional populations this finding may have implications for risk assessment and therapeutic strategies.
This genome-wide association study (GWAS) investigates genetic loci associated with primary open-angle glaucoma in individuals in Africa and in the United States with African ancestry.
Introduction
Primary open-angle glaucoma affects millions of people worldwide and is a leading cause of irreversible blindness.1,2 Genome-wide association studies (GWASs) have identified more than 15 genetic loci associated with increased risk of primary open-angle glaucoma in populations with European or Asian ancestry,3,4 and these results have been supported by analyses of glaucoma-associated quantitative traits, such as intraocular pressure (IOP) and vertical cup-to-disc ratio.5 In contrast, although GWASs of individuals of African ancestry have been performed,6,7 no genome-wide significant loci have been identified to date in this disproportionately affected population. Studies have shown that while individuals of European ancestry older than 40 years exhibit a disease prevalence of 1%, the prevalence is markedly higher in individuals with African ancestry older than 40 years (up to 6.8%).2,8,9 Primary open-angle glaucoma also has earlier onset and is more severe in individuals of African ancestry.10,11,12 A 2018 GWAS examining primary open-angle glaucoma in a multiethnic sample, the Genetic Epidemiology Research on Adult Health and Aging cohort, confirmed that there is a higher prevalence of primary open-angle glaucoma in individuals of African ancestry (16.1%) compared with individuals of East Asian (9.9%) and European (7.4%) ancestry.13
The objective of this study was to address this disparity in genomic science research by performing a GWAS of primary open-angle glaucoma via a multicenter research partnership to obtain biological insights into disease pathogenesis in individuals with African ancestry.
Methods
Study Populations
The study populations comprised 2 groups: the GWAS discovery stage followed by a validation stage. The validation stage included 2 separate meta-analyses. All participants were recruited using the same criteria in both stages. Individuals with primary open-angle glaucoma, control individuals without primary open-angle glaucoma, and eye and brain tissue donors (or family members of deceased donors) were enrolled after written informed consent was obtained from each participant, in full adherence to the Declaration of Helsinki. All relevant local and hospital institutional review boards approved the study. Participant ancestry was self-reported.
For the GWAS discovery stage, study participants of African ancestry were recruited from Ghana, Nigeria, South Africa, and Duke University (Durham, NC), where the same phenotype definition was applied to diagnose primary open-angle glaucoma.3,14,15 Individuals with primary open-angle glaucoma were defined by the presence of glaucomatous optic neuropathy (defined as loss of neuroretinal rim with a vertical cup-to-disc ratio of >0.7 or an intereye asymmetry >0.2 and/or notching attributable to glaucoma) with compatible visual field loss, open angles on gonioscopy, and absence of secondary causes of glaucomatous optic neuropathy. Elevation of IOP was not a criterion in inclusion or exclusion of patients. Patients who were unable to give informed consent or those with secondary glaucoma due to trauma, uveitis, neovascularization, exfoliation syndrome, or pigment dispersion were excluded. Control individuals were recruited in a hospital-based or population-based manner. Hospital-based control individuals were older than 40 years and were confirmed to have no sign of glaucoma or other major eye diseases. These participants had an IOP of less than 21 mm Hg with open angles at the time of recruitment, healthy optic nerves, normal visual fields, and no family history of glaucoma. Population-based control individuals were matched by hospital and ancestry and were healthy individuals older than 40 years. The participating institutions are listed in the eAppendix in the Supplement. The sample collections from the discovery GWAS analysis were ascertained or reexamined between January 1, 2012, and December 31, 2017.
The study design and genotyping method for the 7 sample collections analyzed in the first validation meta-analysis have been previously described.6,7,13,16,17,18 The study names of these collections are listed in the eAppendix in the Supplement. The sample collections were ascertained or reexamined between January 1, 2003, and December 31, 2018.
The second validation meta-analysis included individuals with primary open-angle glaucoma and matched control individuals from Mali, Cameroon, Nigeria (Lagos, Kaduna, and Enugu), Brazil, Saudi Arabia, the Democratic Republic of the Congo, Morocco, and Peru. The participating hospital institutions are listed in the eAppendix in the Supplement. These sample collections were ascertained or reexamined between January 1, 2015, and December 31, 2018.
Genotyping and Quality Control Procedures
For the GWAS discovery stage, genome-wide genotyping was performed using the Illumina OmniExpress beadchip, which directly assessed more than 700 000 single-nucleotide polymorphism (SNP) markers across the human genome. This genotyping array has been successfully used in multiple genome-wide scans,19,20 including scans for primary open-angle glaucoma and other forms of glaucoma.14,21 Further details on GWAS genotyping and quality control procedures are included in the eAppendix in the Supplement.
The first validation meta-analysis for amyloid-β A4 precursor protein-binding family B member 2 (APBB2; Refseq NM_004307) rs59892895 was performed with genome-wide genotyping data from previously described matched primary open-angle glaucoma case-control data sets. The second validation meta-analysis for APBB2 rs59892895 was performed with the Sequenom MassArray primer extension system, with the genotypes further verified with Applied Biosystems Taqman assays.
Fine-scale imputation analysis using the 1000 Genomes Project cosmopolitan reference panel was performed to increase the density of the discovery GWAS. The IMPUTE version 2 software package was used to perform the imputation.22 Stringent quality control was applied on the imputed data by only including imputed genotypes with an information score of at least 0.95 and by only including SNPs with minor allele frequency of at least 1%. The imputation accuracy of APBB2 rs59892895T>C was validated with direct genotyping using the MassArray (Sequenom) and Taqman (Applied Biosystems) systems (with >99% concordance).
Immunohistochemistry and Image Analysis of APBB2 and β-Amyloid in Donor Retina Tissues
APBB2 was shown to increase β-amyloid flux through both the amyloidogenic and nonamyloidogenic pathways of amyloid precursor protein (APP) processing.23 Donor eyes were selected from a tissue collection obtained from the Iowa Lions Eye Bank within 8 hours after death (eTable 1 in the Supplement). Retinal tissue sections from the donor eyes were assessed for APBB2 and β-amyloid expression using immunohistochemistry and image analysis. Further details on the experimental methods are included in the eAppendix in the Supplement.
Immunohistochemistry and Image Analysis of APBB2 and β-Amyloid in Primary Visual Cortex Tissues
All donor samples (eTable 2 in the Supplement) were obtained from the Duke Kathleen Price Bryan Brain Bank and Biorepository and were matched for age and Alzheimer disease severity stage using the Braak classification.24,25 Sections from the primary visual cortex of the donor samples were assessed for APBB2 and β-amyloid expression using immunohistochemistry and image analysis. Further details on the experimental methods are included in the eAppendix in the Supplement.
Statistical Analysis
An analysis of the discovery GWAS and follow-up validation stages were prespecified. The evaluation of previously described loci and their consequences on primary open-angle glaucoma risk in the cohort of participants with African ancestry as well as studies exploring potential pathogenic mechanisms related to APBB2 were undertaken in an exploratory, post hoc manner.
For the discovery GWAS analysis, the association between individual SNP genotypes and primary open-angle glaucoma risk was modeled additively for each copy of the minor allele using logistic regression adjusted for the top 3 principal components of population stratification using PLINK, version 1.9 (details on principal component analysis are included in the eAppendix in the Supplement).26 For imputed genotypes, the information content for allele dosage (range, 0-1; 1 indicates perfect information) were included into the association test model to account for and average across imputation uncertainty. The assumptions of this logistic regression model were that the effective sample size was sufficiently large to allow for χ2-distributed test statistics (eg, the Wald statistic for logistic regression) to be valid, for the observations to be independent of one another, and for only a low degree of collinearity to exist between the independent variables. The model assumptions were all met.
Genomic inflation (λgc) in the GWAS discovery stage was estimated using the median regression test statistic. The genomic inflation factor is presented for each of the 4 GWAS discovery sites as well as for the GWAS discovery meta-analysis in eFigure 1 in the Supplement. For the GWAS discovery analysis, P < 5 × 10−8 (2-sided test) was considered statistically significant. In the validation stages, the association between APBB2 rs59892895 and primary open-angle glaucoma risk was measured using logistic regression for an additive model. Because only 1 SNP (APBB2 rs59892895) was tested in the first and second validation stages, P < .05 (2-sided test) was considered statistically significant for each validation stage. Meta-analyses were conducted using the inverse-variance fixed-effects method (eAppendix in the Supplement).27 Intercohort heterogeneity was assessed for the GWAS discovery and validation stages.
Previous case-control studies of primary open-angle glaucoma in individuals with European and Asian ancestry have robustly implicated at least 26 SNPs mapping to 15 distinct gene loci (TMCO1, FMNL2, CADM2, AFAP1, THSD7A, CAV1-CAV2, ANGPT1, CDKN2B-AS, ABCA1, LMX1B, PLCE1, TMTC2, SIX6, TCF12, and GAS7), accompanied by validation in at least 2 independent studies.3,13,14,28,29,30,31 The association between these 26 SNPs and primary open-angle glaucoma risk were tested using logistic regression in the collections from individuals with African ancestry, which included 5153 affected individuals and 10 014 unaffected individuals with available genotyping data. An inverse-variance, fixed-effects meta-analysis was conducted to summarize the estimates.
A case-only quantitative trait locus analysis was performed between APBB2 rs59892895T>C and 2 clinical parameters: maximum IOP and vertical cup-to-disc ratio at the time of examination (using the mean ratio if available for both eyes). The association between APBB2 rs59892895T>C and the clinical parameters was assessed using linear regression with sex and age as covariates. The fluorescence intensity data from the immunohistochemical analysis (eAppendix in the Supplement) of retina and primary visual cortex tissues were analyzed with regards to APBB2 rs59892895T>C carrier status using a linear mixed model incorporating additional random effect terms for individual (for both retina and visual cortex data) and Braak stage (for visual cortex data only).
Results
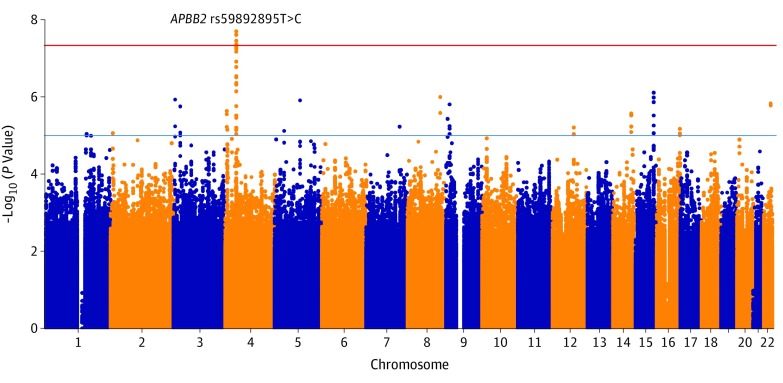
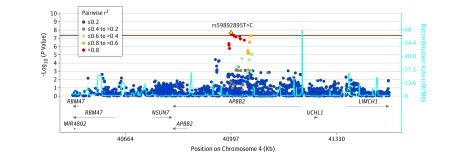
A total of 26 295 individuals were included in the study. The number of individuals with primary open-angle glaucoma and control individuals analyzed in the GWAS and validation stages are presented in the Table (quality control results and handling of population stratification can be found in the eAppendix and eFigures 1, 2, and 3 in the Supplement). The discovery meta-analysis identified a genome-wide significant association at 1 locus on chromosome 4, characterized by multiple SNP markers showing high pairwise linkage disequilibrium (Figure 1; summary statistics for the GWASs are publicly available per the data sharing statement). The association mapped to the APBB2 rs59892895T>C locus (Figure 2), whereby the minor C allele was observed to be associated with increased risk of primary open-angle glaucoma (per-allele odds ratio [OR], 1.32 [95% CI, 1.20-1.46]; P = 2 × 10−8).
Table. Summary of Case-Control Collections in a Study of the Association of Genetic Variants With Primary Open-Angle Glaucoma Among Individuals With African Ancestry.
| Sample Collectiona | rs59892895*C Present, % | Sample Size, No. | ||
|---|---|---|---|---|
| Individuals With Primary Open-Angle Glaucoma | Control Individuals Without Primary Open-Angle Glaucoma | Individuals With Primary Open-Angle Glaucoma | Control Individuals Without Primary Open-Angle Glaucoma | |
| GWAS Discovery | ||||
| Ghana | 26.9 | 23.3 | 833 | 896 |
| Nigeria | 30.4 | 22.6 | 554 | 348 |
| South Africa | 31.1 | 22.9 | 228 | 269 |
| United States | 26.1 | 21.7 | 705 | 608 |
| Total | 2320 | 2121 | ||
| First Validation | ||||
| Women’s Health Initiative | 23.1 | 20.8 | 1720 | 6067 |
| Kaiser Permanente GERA | 23.3 | 18.4 | 300 | 2700 |
| ADAGES | 24.3 | 21.4 | 1890 | 2205 |
| South Africa | 13.0 | 14.4 | 297 | 147 |
| Tanzania | 30.1 | 28.3 | 366 | 329 |
| United States | 23.6 | 22.6 | 450 | 1350 |
| South London | 26.6 | 21.6 | 378 | 217 |
| Total | 5401 | 13 015 | ||
| Second Validation | ||||
| Cameroon | 31.3 | 29.0 | 56 | 57 |
| Nigeria | 28.6 | 26.2 | 231 | 61 |
| Nigeria (Kaduna) | 29.2 | 30.2 | 99 | 88 |
| Nigeria (African Glaucoma Genetics Project) | 32.6 | 31.0 | 131 | 71 |
| Saudi Arabia | 5.5 | 3.1 | 276 | 655 |
| Brazil | 8.8 | 4.5 | 399 | 460 |
| The Democratic Republic of the Congo | 37.7 | 33.9 | 124 | 120 |
| Morocco | 6.8 | 3.5 | 37 | 130 |
| Peru | 2.0 | 1.2 | 51 | 128 |
| Mali | 27.3 | 23.4 | 132 | 132 |
| Total | 1536 | 1902 | ||
| All validation | 6937 | 14 917 | ||
| Total samples | 9257 | 17 038 | ||
Abbreviations: ADAGES, African Descent and Glaucoma Evaluation Study; GERA, Genetic Epidemiology Research on Adult Health and Aging; GWAS, genome-wide association study.
The study site (country) or name of the study that the sample collection was taken from. More information on the sample collections can be found in the eAppendix in the Supplement.
Figure 1. Discovery Analysis of 2320 Individuals With Primary Open-Angle Glaucoma and 2121 Age-Matched Control Individuals.
Single-nucleotide polymorphisms (SNPs) were only considered if they were assessed across all 4 genome-wide association discovery study sites. A total of 6 734 161 SNPs are included in this figure. The blue horizontal line indicates a threshold for suggestive statistical significance commonly used in genome-wide association studies (P = 10−5) and the red horizontal line indicates the threshold for genome-wide significance (P = 5 × 10−8). Multiple SNPs at the gene encoding for amyloid-β A4 precursor protein-binding family B member 2 (APBB2) have a significant association with primary open-angle glaucoma disease risk.
Figure 2. Association at the Amyloid-β A4 Precursor Protein-Binding Family B Member 2 (APBB2) Locus on Chromosome 4.
The association between single-nucleotide polymorphisms (SNPs) and primary open-angle glaucoma were plotted by genomic position on chromosome 4 and degree of statistical significance. The horizontal line shows the threshold for genome-wide significance, P = 5 × 10−8. The dots indicate the extent of linkage disequilibrium (LD) between each tested SNP with rs59892895 (based on pairwise r2 values calculated from the discovery analysis). LD refers to the association between alleles of SNPs located close to one another on the same chromosome; SNPs in strong LD can serve as proxies for one another. Estimated recombination rates were plotted in light blue to reflect the LD structure in individuals with African ancestry. The estimated recombination rate shows the average frequency in which recombination occurs at a particular location. The extent of LD drops with increasing recombination rate. The horizontal lines accompanied by arrows in the lower panel of the plot reflect the genes mapping to the given genomic locations. The arrows indicate the direction of transcription of the genes. The horizontal lines labeled APBB2 and RBM47 reflect 2 different gene transcripts of different lengths for both genes.
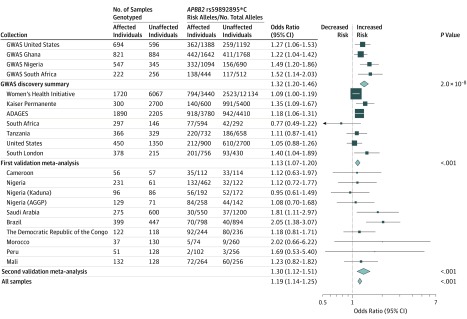
The APBB2 rs59892895 association was tested in the first validation meta-analysis comprising 7 independently ascertained sample collections from participants with African ancestry (Table). This first validation data set with prior genome-wide genotyping data available for 5401 individuals with primary open-angle glaucoma and 13 015 individuals without primary open-angle glaucoma had greater than 90% statistical power to validate an SNP with a minor allele frequency as low as 20% and an OR as low as 1.10 at a 2-sided P value less than .05.32 The association at APBB2 rs59892895 was validated, with the risk C allele associated with increased risk of primary open-angle glaucoma (OR, 1.13 [95% CI, 1.07-1.20]; P < .001; Figure 3). There was no significant heterogeneity across all sites analyzed (P value for heterogeneity = .15).
Figure 3. Association Between Amyloid-β A4 Precursor Protein-Binding Family B Member 2 (APBB2) rs59892895 and Primary Open-Angle Glaucoma Risk.
The oblong data markers represent odds ratios, with the height of the data markers being inversely proportional to the standard error of the odds ratio. The diamonds represent the odds ratios after the meta-analysis, with the width representing the 95% CIs. Collections from the United States were taken from an African American population and the collection from South London was taken from a West African population. ADAGES indicates African Descent and Glaucoma Evaluation Study; AGGP, African Glaucoma Genetics Project; GWAS, genome-wide association study.
A second validation meta-analysis for this association was completed with an additional 1536 affected individuals and 1902 unaffected individuals with African ancestry (Table). A significant association between the rs59892895*C allele and increased risk of primary open-angle glaucoma was observed (OR, 1.30 [95% CI, 1.12-1.51]; P < .001; Figure 3), with no significant heterogeneity across all 10 sites analyzed (P value for heterogeneity = .27). An overall meta-analysis of rs59892895*C involving the 9257 individuals with primary open-angle glaucoma and 17 038 individuals without primary open-angle glaucoma from all cohorts showed consistent association with primary open-angle glaucoma risk (OR, 1.19 [95% CI, 1.14-1.25]; P < .001) (Figure 3). The rs59892895*C allele was observed to not be significantly associated with IOP and vertical cup-to-disc ratio (P = .62) in an analysis of 6179 individuals of African ancestry with available data.
Only individuals of African ancestry or individuals with African ancestry admixture (eg, Brazil, Morocco, Peru, and Saudi Arabia; Table) were polymorphic at APBB2 rs59892895—the rs59892895*C risk allele was not present in European, South Asian, or East Asian individuals (eFigure 4 in the Supplement). The sources of data accessed to ascertain the frequency of rs59892895 in participants with other ancestries were from the 1000 Genomes Project browser33 (eFigure 5 in the Supplement), the Genome Aggregation Database (gNOMAD; eTable 3 in the Supplement),34 as well as previously published GWAS data sets of European3 (eAppendix in the Supplement) and Asian individuals.15,35 No SNP within the broad APBB2 locus was significantly associated with primary open-angle glaucoma in previously studied European or Asian ancestry case-control collections (eFigure 6 in the Supplement). APBB2 rs59892895T>C was not observed to be in linkage disequilibrium (defined as pairwise r2 > 0.8) with other coding genetic variants or with potentially functional regulatory elements (eFigure 7 in the Supplement).
Post Hoc Evaluation of Previously Described Loci and Their Consequences on Risk in the Cohorts
Analysis of 26 well-validated primary open-angle glaucoma SNPs from studies of individuals of European and Asian ancestry showed that for 23 of the 26 SNPs, the ORs in participants with African ancestry were smaller than in individuals with European ancestry. Heterogeneity tests showed 19 of the SNPs had P values for heterogeneity of less than .05 in support of a smaller OR in participants with African ancestry compared with European individuals (eTable 4 and eFigure 8 in the Supplement). Assessment of the relationship between IOP loci13,30 and primary open-angle glaucoma risk showed a weak correlation between IOP and primary open-angle glaucoma risk in individuals with African ancestry (eTable 5 and eFigure 9 in the Supplement). Assessment of the relationship between vertical cup-to-disc ratio loci36 and primary open-angle glaucoma risk showed that all previously reported loci for vertical cup-to-disc ratio were not associated with primary open-angle glaucoma risk in participants with African ancestry (eTable 6 in the Supplement).
Post Hoc Studies Exploring Potential Pathogenic Mechanisms Related to APBB2
All donor retina tissues were from individuals of African ancestry. For primary visual cortex tissues, carriers of the APBB2 rs59892895*C risk allele were of African ancestry, whereas 3 of the 4 individuals with the APBB2 rs59892895*TT homozygous baseline genotype were of European ancestry to match for the Braak stage of Alzheimer disease diagnosis (eTable 2 in the Supplement). Exploratory analyses using immunohistochemistry on donor retina and primary visual cortex tissues (eTable 1 and eTable 2 in the Supplement) suggested that participants carrying the rs59892895*C risk allele had associated higher APBB2 expression as well as associated increased staining for β-amyloid compared with participants homozygous for the baseline rs59892895*T nonrisk allele (eFigures 10, 11, and 12 in the Supplement).
Discussion
This study of 26 295 individuals found a genetic variant in the APBB2 gene that was associated with a higher risk of primary open-angle glaucoma. The genetic association between primary open-angle glaucoma and APBB2 was observed only in individuals of African ancestry. The level of genetic diversity in participants of African ancestry was higher than in individuals of European or Asian ancestry, and the functional risk alleles in APBB2 may not be present in these other populations. The increased risk associated with the APBB2 allele appeared not to be mediated via increased IOP or optic nerve neuropathy associated with an increasing vertical cup-to-disc ratio, thus suggesting a new insight to primary open-angle glaucoma disease pathogenesis.
Because the odds ratio of the APBB2 rs59892895*C risk allele on primary open-angle glaucoma appeared to be larger in populations with African ancestry admixture, such as in Saudi Arabia, Brazil, Peru, and Morocco, this raised the possibility that estimates from these 4 populations could have been confounded by population stratification. However, principal component analysis of ancestry-informative markers from Brazil, Peru, and Morocco, which had sufficient DNA to allow assessment of these additional markers, revealed little evidence of population stratification between affected and unaffected individuals (eFigure 13 in the Supplement).
Because APBB2 was shown to be involved in the amyloidogenic pathway of APP processing, an exploratory analysis of human retinal and primary visual cortex tissues suggested a potential relationship between the APBB2 rs59892895*C risk allele, increased APBB2 expression, and associated increased β-amyloid plaque deposition. Primary open-angle glaucoma neurotoxicity may result from incomplete clearance of amyloid β and other neurotoxins from the interstitial space of the optic nerve.37 However, there has been no conclusive evidence that these pathways contribute to primary open-angle glaucoma in humans.
The present analysis suggests that the majority of open-angle glaucoma genetic loci described in individuals of European or Asian ancestry have a much more modest effect in individuals of African ancestry. Also, in contrast to studies of European individuals, the present data on individuals of African ancestry showed a much weaker correlation between genetic factors contributing to increased IOP and primary open-angle glaucoma risk. It is possible that these differences in genetic architecture are, at least in part, responsible for the increased prevalence and severity of primary open-angle glaucoma in African ancestry populations.
Limitations
This study has several limitations. First, despite the moderately large sample size of participants of African ancestry, there was only sufficient statistical power to detect associations with common genetic variants. Second, although all study sites used well-established clinical protocols to diagnose primary open-angle glaucoma, the heterogeneity of the primary open-angle glaucoma phenotype may have limited the ability to detect some genetic associations by biasing the effect estimates toward the null. Third, while the association observed between APBB2 rs59892895 and primary open-angle glaucoma risk was statistically significant, the causal mechanisms of association have yet to be elucidated. Fourth, because the observations from human retinal and visual cortex tissues in this report are based on limited sample sizes, they are interpreted here as exploratory and hypothesis generating, and would require further validation through future research.
Conclusions
In this GWAS, variants at the APBB2 locus demonstrated differential association with primary open-angle glaucoma by ancestry. If validated in additional populations this may have implications for risk assessment and therapeutic strategies.
eAppendix. Supplemental methods and Addendum
eTable 1. Demographic and diagnostic data from five African American retinal tissue donors.
eTable 2. Demographic and diagnostic data from tissue donors with Alzheimer’s disease
eAppendix.
eTable 3. Frequency distribution of the APBB2 rs59892895-C allele from the genome aggregation database
(gNOMAD).
eTable 4. Association analysis of genetic loci previously reported to show genome-wide significant association with POAG in Europeans. The analysis here compares the effect sizes in participants of African ancestry (5,153 cases and 10,014 controls) and European ancestry (more than 11,000 cases and more than 100,000 controls).
eTable 5. Relationship between 109 genome-wide significant IOP SNP loci with POAG risk in Africans and Europeans.
eTable 6. Association analysis within the Eyes of Africa GWAS discovery study for 15 loci previously reported to be significantly associated with vertical cup-to-disk ratio in a large European and Asian meta-analysis. The POAG association in Europeans is also appended for comparison.
eFigure 1. Quantile-quantile plot of the association test statistics from the Eyes of Africa GWAS discovery stage.
eFigure 2. Principal component analysis of genetic stratification for the four POAG case-control collections
(African Americans, Ghanaians, Nigerians, and South Africans) analyzed in the GWAS discovery stage.
eFigure 3. Principal component analysis of genetic stratification for the individual POAG case-control collections analyzed in the GWAS discovery stage.
eFigure 4. Principal component analysis of individuals of African, East Asian, and European descent, together with Moroccans, Saudi Arabians, Brazilians, and Peruvians contributing to this study.
eFigure 5. Frequency distribution of the APBB2 rs59892895-C allele from the 1000 Genomes Project Browser.
eFigure 6. Regional association analysis for the APBB2 locus within the NEIGHBORHOOD and the Singaporean Chinese POAG studies.
eFigure 7. Genetic variants in linkage-disequilibrium with APBB2 rs59892895.
eFigure 8. Forest plot showing the effect sizes of genetic loci previously reported to show genome-wide significant association with POAG in Europeans.
eFigure 9. Scatter plot showing the correlation between IOP effect sizes for SNPs showing genome-wide significant association with intra-ocular pressure and POAG disease risk in Europeans and African ancestry participants. efigure 10. Immunohistochemical staining of human retina samples.
efigure 10. Immunohistochemical staining of human retina samples.
eFigure 11. Carriers of APBB2 rs59892895-C risk allele show increased β-amyloid in the primary visual cortex.
eFigure 12. Carriers of the APBB2 risk allele show increased APBB2 expression in the primary visual cortex.
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262-267. doi: 10.1136/bjo.2005.081224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081-2090. doi: 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 3.Bailey JN, Loomis SJ, Kang JH, et al. ; ANZRAG Consortium . Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016;48(2):189-194. doi: 10.1038/ng.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiga Y, Akiyama M, Nishiguchi KM, et al. ; Japan Glaucoma Society Omics Group (JGS-OG); NEIGHBORHOOD Consortium . Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum Mol Genet. 2018;27(8):1486-1496. doi: 10.1093/hmg/ddy053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gharahkhani P, Burdon KP, Cooke Bailey JN, et al. ; NEIGHBORHOOD consortium . Analysis combining correlated glaucoma traits identifies five new risk loci for open-angle glaucoma. Sci Rep. 2018;8(1):3124. doi: 10.1038/s41598-018-20435-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor KD, Guo X, Zangwill LM, et al. ; African Descent and Glaucoma Evaluation Study III Genomics Study Group . Genetic architecture of primary open-angle glaucoma in individuals of African descent: the African descent and glaucoma evaluation study III. Ophthalmology. 2019;126(1):38-48. doi: 10.1016/j.ophtha.2018.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnemaijer PWM, Iglesias AI, Nadkarni GN, et al. ; GIGA Study Group; Eyes of Africa Genetics Consortium; NEIGHBORHOOD Consortium . Genome-wide association study of primary open-angle glaucoma in continental and admixed African populations. Hum Genet. 2018;137(10):847-862. doi: 10.1007/s00439-018-1943-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma: the Baltimore Eye Survey. JAMA. 1991;266(3):369-374. doi: 10.1001/jama.1991.03470030069026 [DOI] [PubMed] [Google Scholar]
- 9.Budenz DL, Barton K, Whiteside-de Vos J, et al. ; Tema Eye Survey Study Group . Prevalence of glaucoma in an urban West African population: the Tema Eye Survey. JAMA Ophthalmol. 2013;131(5):651-658. doi: 10.1001/jamaophthalmol.2013.1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukesh BN, McCarty CA, Rait JL, Taylor HR. Five-year incidence of open-angle glaucoma: the visual impairment project. Ophthalmology. 2002;109(6):1047-1051. doi: 10.1016/S0161-6420(02)01040-0 [DOI] [PubMed] [Google Scholar]
- 11.Leske MC, Connell AM, Wu SY, et al. ; The Barbados Eye Studies Group . Incidence of open-angle glaucoma: the Barbados Eye Studies. Arch Ophthalmol. 2001;119(1):89-95. [PubMed] [Google Scholar]
- 12.Bengtsson BO. Incidence of manifest glaucoma. Br J Ophthalmol. 1989;73(7):483-487. doi: 10.1136/bjo.73.7.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choquet H, Paylakhi S, Kneeland SC, et al. . A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat Commun. 2018;9(1):2278. doi: 10.1038/s41467-018-04555-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gharahkhani P, Burdon KP, Fogarty R, et al. ; Wellcome Trust Case Control Consortium 2, NEIGHBORHOOD consortium . Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46(10):1120-1125. doi: 10.1038/ng.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Allingham RR, Nakano M, et al. ; ICAARE-Glaucoma Consortium; NEIGHBORHOOD Consortium . A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet. 2015;24(13):3880-3892. doi: 10.1093/hmg/ddv128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choquet H, Thai KK, Yin J, et al. . A large multi-ethnic genome-wide association study identifies novel genetic loci for intraocular pressure. Nat Commun. 2017;8(1):2108. doi: 10.1038/s41467-017-01913-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann TJ, Tang H, Thornton TA, et al. . Genome-wide association and admixture analysis of glaucoma in the Women’s Health Initiative. Hum Mol Genet. 2014;23(24):6634-6643. doi: 10.1093/hmg/ddu364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zangwill LM, Ayyagari R, Liebmann JM, et al. ; African Descent and Glaucoma Evaluation Study III Genomics Study Group . The African Descent and Glaucoma Evaluation Study (ADAGES) III: contribution of genotype to glaucoma phenotype in African Americans: study design and baseline data. Ophthalmology. 2019;126(1):156-170.doi: 10.1016/j.ophtha.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christophersen IE, Rienstra M, Roselli C, et al. ; METASTROKE Consortium of the ISGC; Neurology Working Group of the CHARGE Consortium; AFGen Consortium . Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017;49(6):946-952. doi: 10.1038/ng.3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGinnis R, Steinthorsdottir V, Williams NO, et al. ; FINNPEC Consortium; GOPEC Consortium . Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat Genet. 2017;49(8):1255-1260. doi: 10.1038/ng.3895 [DOI] [PubMed] [Google Scholar]
- 21.Aung T, Ozaki M, Lee MC, et al. . Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci. Nat Genet. 2017;49(7):993-1004. doi: 10.1038/ng.3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955-959. doi: 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang Y, Tesco G, Jeong WJ, et al. . Generation of the beta-amyloid peptide and the amyloid precursor protein C-terminal fragment gamma are potentiated by FE65L1. J Biol Chem. 2003;278(51):51100-51107. doi: 10.1074/jbc.M309561200 [DOI] [PubMed] [Google Scholar]
- 24.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239-259. doi: 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- 25.Hyman BT, Phelps CH, Beach TG, et al. . National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8(1):1-13. doi: 10.1016/j.jalz.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritsche LG, Igl W, Bailey JN, et al. . A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134-143. doi: 10.1038/ng.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Lin Y, Vithana EN, et al. . Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014;46(10):1115-1119. doi: 10.1038/ng.3078 [DOI] [PubMed] [Google Scholar]
- 29.Hysi PG, Cheng CY, Springelkamp H, et al. ; BMES GWAS Group; NEIGHBORHOOD Consortium; Wellcome Trust Case Control Consortium 2 . Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46(10):1126-1130. doi: 10.1038/ng.3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khawaja AP, Cooke Bailey JN, Wareham NJ, et al. ; UK Biobank Eye and Vision Consortium; NEIGHBORHOOD Consortium . Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018;50(6):778-782. doi: 10.1038/s41588-018-0126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacGregor S, Ong JS, An J, et al. . Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018;50(8):1067-1071. doi: 10.1038/s41588-018-0176-y [DOI] [PubMed] [Google Scholar]
- 32.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149-150. doi: 10.1093/bioinformatics/19.1.149 [DOI] [PubMed] [Google Scholar]
- 33.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285-291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khor CC, Do T, Jia H, et al. . Genome-wide association study identifies five new susceptibility loci for primary angle closure glaucoma. Nat Genet. 2016;48(5):556-562. doi: 10.1038/ng.3540 [DOI] [PubMed] [Google Scholar]
- 36.Springelkamp H, Höhn R, Mishra A, et al. ; Blue Mountains Eye Study—GWAS group; NEIGHBORHOOD Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2) . Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat Commun. 2014;5:4883. doi: 10.1038/ncomms5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wostyn P, De Groot V, Van Dam D, Audenaert K, Killer HE, De Deyn PP. Glaucoma considered as an imbalance between production and clearance of neurotoxins. Invest Ophthalmol Vis Sci. 2014;55(8):5351-5352. doi: 10.1167/iovs.14-15041 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Supplemental methods and Addendum
eTable 1. Demographic and diagnostic data from five African American retinal tissue donors.
eTable 2. Demographic and diagnostic data from tissue donors with Alzheimer’s disease
eAppendix.
eTable 3. Frequency distribution of the APBB2 rs59892895-C allele from the genome aggregation database
(gNOMAD).
eTable 4. Association analysis of genetic loci previously reported to show genome-wide significant association with POAG in Europeans. The analysis here compares the effect sizes in participants of African ancestry (5,153 cases and 10,014 controls) and European ancestry (more than 11,000 cases and more than 100,000 controls).
eTable 5. Relationship between 109 genome-wide significant IOP SNP loci with POAG risk in Africans and Europeans.
eTable 6. Association analysis within the Eyes of Africa GWAS discovery study for 15 loci previously reported to be significantly associated with vertical cup-to-disk ratio in a large European and Asian meta-analysis. The POAG association in Europeans is also appended for comparison.
eFigure 1. Quantile-quantile plot of the association test statistics from the Eyes of Africa GWAS discovery stage.
eFigure 2. Principal component analysis of genetic stratification for the four POAG case-control collections
(African Americans, Ghanaians, Nigerians, and South Africans) analyzed in the GWAS discovery stage.
eFigure 3. Principal component analysis of genetic stratification for the individual POAG case-control collections analyzed in the GWAS discovery stage.
eFigure 4. Principal component analysis of individuals of African, East Asian, and European descent, together with Moroccans, Saudi Arabians, Brazilians, and Peruvians contributing to this study.
eFigure 5. Frequency distribution of the APBB2 rs59892895-C allele from the 1000 Genomes Project Browser.
eFigure 6. Regional association analysis for the APBB2 locus within the NEIGHBORHOOD and the Singaporean Chinese POAG studies.
eFigure 7. Genetic variants in linkage-disequilibrium with APBB2 rs59892895.
eFigure 8. Forest plot showing the effect sizes of genetic loci previously reported to show genome-wide significant association with POAG in Europeans.
eFigure 9. Scatter plot showing the correlation between IOP effect sizes for SNPs showing genome-wide significant association with intra-ocular pressure and POAG disease risk in Europeans and African ancestry participants. efigure 10. Immunohistochemical staining of human retina samples.
efigure 10. Immunohistochemical staining of human retina samples.
eFigure 11. Carriers of APBB2 rs59892895-C risk allele show increased β-amyloid in the primary visual cortex.
eFigure 12. Carriers of the APBB2 risk allele show increased APBB2 expression in the primary visual cortex.