Abstract
The characteristic lengths of molecular arrangement in primary cilia are below the diffraction limit of light, challenging structural and functional studies of ciliary proteins. Superresolution microscopy can reach up to a 20 nm resolution, significantly improving the ability to map molecules in primary cilia. Here we describe detailed experimental procedure of STED microscopy imaging and dSTORM imaging, two of the most powerful superresolution imaging techniques. Specifically, we emphasize the use of these two methods on imaging proteins in primary cilia.
Keywords: Primary cilium, Ciliary protein, Superresolution microscopy, Diffraction limit, Fluorophore, STED, STORM
1. Introduction
Primary cilium is an organelle with a complex structure confined in a small volume. The diameter of the ciliary membrane is approximately 250–300 nm and its length is about 2–10 gm. It is composed of different elements and compartments, including the axoneme, the ciliary membrane, the ciliary pocket, the transition zone, the transition fibers or distal appendages, the inversin region, and the ciliary tip region, each containing numerous structural and functional proteins. The relative localization of ciliary proteins is essential for understanding interactions and active roles of each ciliary protein, as well as their potential association with different ciliopathic disease phenotypes. However, the packed arrangement of these proteins largely challenges the resolution limit of conventional fluorescent microscopy. For example, many transition zone proteins have been identified at the ciliary base [1–5], but which of them forms the Y-links remains unknown. Many intraflagellar transport (IFT) proteins are involved in ciliary cargo transport, but where IFT-A and IFT-B proteins are assembled is unclear. Key elements of distal appendages have recently been identified [6–8], but how they coordinate ciliogenesis initiation remains elusive. All these knowledge gaps require a geometric framework of ciliary proteins to determine their potential interactions. We have previously shown that superresolution microscopy can be a suitable approach to address some of these problems [9, 10]. Others have also used superresolution imaging for primary cilium studies [4, 6, 11–16]. Much more efforts will be needed to reconstruct the interaction map upon the ciliary molecular architecture.
Superresolution microscopy breaks the diffraction limit of light for far-field imaging, which includes widefield and confocal fluorescent microscopy techniques we routinely use for biological sample imaging. The diffraction limit is defined by Abbe’s law: d=λ/2NA, where d is the resolution, λ is the wavelength, and NA is the numerical aperture of the optical system. For a green fluorescent dye or protein emitting at ~520 nm wavelength and a high-end oil immersion objective with an NA of 1.4, a conventional microscopy can only reach ~190 nm resolution. Various super-resolution techniques have been invented in the past two decades to reach a subdiffraction resolution including STED [17, 18], SIM [19], RESOLFT [20], SSIM [21], PALM [22], STORM [23], FPALM [24], PAINT [25], GSD [26], PALMIRA [27], dSTORM [28], GSDIM [29], SOFI [30], gSTED [31], BaLM [32], Bayesian localization methods [33, 34], and other variations. Among these methods, SIM, STED, and PALM/STORM are three of the most widely used approaches for superresolution imaging of biological samples. For fluorescent imaging of cell cultures, SIM can reach ~110 nm lateral resolution; STED can reach ~50 nm lateral resolution; and PALM/STORM can reach ~20 nm lateral resolution. SIM is the easiest to use, with imaging protocols similar to those of epifluorescent imaging. PALM and STORM both fall into the category of localization microscopy, with PALM used for photoactivatable or photoconvertible fluorescent proteins and STORM used for photoswitchable organic fluorophores. STED is a confocal-based method that can reach a relatively high imaging speed. In this Chapter, we will only focus on the methods of STED and STORM (specifically, dSTORM) for ciliary imaging.
Stimulated emission depletion (STED) microscopy achieves supersolution by directing neighboring molecules to another photon release path, so that the center region emitting fluorescent signals is much smaller than the diffraction limited spot [17, 18]. It is a point-by-point scanning method to achieve subdiffractive imaging one point at a time. For each imaging spot, a diffraction-limited focused laser light (excitation laser) illuminates a volume of the point spread function (PSF) to drive molecules to the excited state. Another laser of a different wavelength (depletion laser) focusing at the same focal spot passes through a special lens (e.g., spiral phase plate) which creates a doughnut-shaped PSF, so that the peripheral molecules are forced to emit photons through stimulated emission at a wavelength distinct from the fluorescent wavelength. By tuning the power of the depletion laser, one can control how large a peripheral region is, and thus adjust how small the volume at the center is where molecules emit photons through fluorescence instead of stimulated emission. Therefore, neighboring molecules that cannot be resolved by conventional microscopy can now be distinguished by different wavelength emissions at the central and peripheral regions of the focal spot. In principle, one can arbitrarily increase the power of the depletion laser to reach an arbitrarily high resolution. In fact, a lateral resolution as high as 5.8 nm has been achieved for nano-diamonds [35]. For imaging organic dyes and fluorescent proteins in biological samples, the signal-to-noise ratio remains as one of the major issues, limiting the lateral resolution to ~50 nm. Deconvolution is often, then, applied to the raw data to improve image quality of STED microscopy.
A variety of STED microscopes are available for different applications. The original STED microscope was implemented with a mode-locked pulsed laser, requiring complex optical manipulation to operate [18]. Later, a continuous wave (CW) laser-based STED microscope was implemented [36]. The CW STED system is much easier to build than the other versions of STED microscopes and is able to achieve a ~60 nm resolution. Dual-color CW STED imaging was also demonstrated [37]. One drawback of this system is its need of high depletion power. Despite its relative early release, the CW STED microscopy system is considered as a good option when the fluorescent signals of the molecule of interest are dim, such as MKSI in the transition zone of primary cilia. Time-gated STED (gSTED) microscopy has later been developed [31], reaching ~50 nm resolution with a much lower laser power, and thus reducing the photodamage of samples. One drawback of gSTED microscopy is that the photon count per image frame may be low due to the time gating if a fluorophore is dim. The other drawback of a gSTED system compared to a CW STED system is its higher cost. Currently gSTED microscopy is considered as the primary STED imaging method. 3D STED imaging has also been achieved [38], as well as two-photon STED imaging [39]. Available commercial STED systems (e.g., Leica and Abberior) include CW STED, gSTED, 3D CW STED, and 3D gSTED systems, all of which are compatible with two-color imaging.
Live-cell STED imaging has been shown [40], where the sampling rate is limited by photon counts and the imaging duration is limited by photobleaching and phototoxicity. For intraflagellar transport imaging of IFT88-EYFP in IMCD cells, we have achieved 4 frames/s to collect enough photons for each frame (unpublished). Only a limit number of frames can be imaged due to photobleaching (<100 frames for CW STED). It is possible that gSTED can achieve a longer imaging duration due to its low depletion laser power, although we have not verified it. Two-color live-cell STED imaging has also been reported [41].
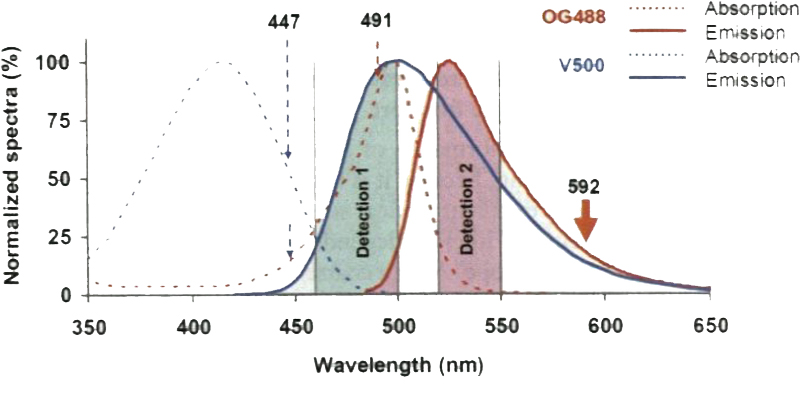
The choice of fluorophores depends on the available laser sources and their STED performance. With a 592-nm depletion laser for a CW STED system, Oregon Green 488 and EYEP as an organic dye and a fluorescent protein, respectively, perform well for single-color STED microscopy. For two-color STED imaging, the combination of Oregon Green 488 and BD Horizon V500 is one of the best pairs of organic dyes. BD Horizon V500 has a large Stokes shift, so that one can use a single depletion laser (e.g., 592 nm laser) for Oregon Green 488 and V500, both excited with different lasers (e.g., 491 nm and 447 nm lasers). One can also use SNAP-tag and HaloTag [42, 43] together with the Oregon Green 488 and BD V500 for live cell two-color STED imaging. For two-color STED imaging of fluorescent protein pairs, ECFP and EYFP can be a reasonable choice for a 592-nm depletion laser, although organic dyes are preferred due to their higher photon output. The new commercial STED systems provide multiple depletion laser sources and a wide spectrum of excitation laser sources (e.g., super-continuum laser), broadening the possible choices of fluorophores. For example, it is suggested that one can use Oregon Green 488 with a 592-nm depletion laser, TRITC with a 660-nm depletion laser, and STAR635P with a 775-nm depletion laser for three-color STED superresolution imaging using a commercial system (Leica). In this chapter we describe the protocol of using the combination of Oregon Green 488 and BD Horizon V500 for two-color STED microscopy, but the procedure of using other combinations of fluorophores can be similar.
Stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM) achieve superresolution by preventing most of the molecules from emitting light and then determining the intensity center of those isolated molecules that do emit fluorescent signals by fitting [22, 23]. Knowing the intensity distribution of photons from a single molecule forms a Gaussian-like distribution (Airy disk) in lateral dimensions, one can fit the distribution function and find the point of the intensity peak with a resolution limited by the signal-to-noise ratio. The subset of fluorophores that light up in a frame will turn dark and a random set of other molecules will be excited in the next frame. Thus, every frame of images contains a portion of molecules shown in a very high resolution. By overlapping a large number of frames, one can then observe most of the fluorescent molecules in superresolution. The key to result in isolated fluorescent molecules is to make them alternate between bright and dark states randomly. This is achieved by creating an environment to promote photo-switchable behaviors of fluorophores.
Various photoactivatable and photoconvertible fluorescent proteins have been used for PALM superresolution imaging [22]. For single-color PALM imaging, mEos3.2 can be one of the best choices with its high photon output [44]. Recent development of mMaple3 can be a good alternative of mEos3.2 [45]. For two-color PALM imaging, combinations of PAGFP/PAmCherry1 and Dronpa/PAmCherry1 are relatively good in performance [46]. The combination of mEos3.2/PSCFP2 has also been used [47].
A variety of photoswitchable organic fluorophores are available for STORM imaging [23, 48]. Mostly these organic dyes are for antibody staining of fixed cells, although they can also be used for live cell imaging when using with target specific labeling techniques including SNAP-tag and HaloTag [42, 43]. STORM was originally implemented using photoswitching activities ofCy3-Cy5 dye pairs [23]. Many related methods based on photoswitching features of fluorescent dyes were later developed. Among them direct STORM (dSTORM) is currently the most popular method due to its compatibility with conventional organic dyes such as Alexa Fluor and ATTO-dyes [28]. By adding a finite amount of a reducing thiol compound (e.g., mercaptoethylamine) and an oxygen scavenging system of catalase/glucose/glucose oxidase to the sample, molecules are populated to a stable non-fluorescent reduced state, i.e., a dark state. A small population of molecules can return to the ground state by spontaneous or light-induced (405 nm light) oxidation to reenter the fluorescent path. Among all organic dyes, Alexa 647 performs the best for dSTORM superresolution imaging due to its high photon yield per switch and low duty cycle. For two-color dSTORM imaging, many choices have been suggested for the second dye, including Alexa Fluor 488, ATTO 488, Alexa Fluor 568, cy3B, CF568, and Alexa 750, although none of them has a performance close to that of Alexa 647. In our hands, Alexa 647/Cy3B is the best combination for two-color dSTORM imaging thus far.
Several methods have been developed for 3D STORM imaging the astigmatism method [49], the biplane method [50], the double-helix point spread function method [51], and the opposing objective method [52]. The astigmatism method may be the most widely used among them so far due to its ease of implementation, although other methods may be more precise in the axial localization. For the astigmatism method, the ellipticity and orientation of the image from a single molecule allow one to quantitatively determine how far the molecule is away from the focal plane, and thus each image spot provides coordinate information in all three dimensions.
Here we describe detailed protocols of CW STED and dSTORM methods for ciliary protein imaging. Many tricks and solutions can also be used for other STED and localization based methods such as gSTED and PALM.
2. Materials
2.1. Cell Culture
Sterilized solutions and reagents should be used for cell culture.
Cells: Here we use retinal pigment epithelial cells (hTERT RPE-I, ATCC CRL-4000) as an example. Cells are incubated in an incubator at 37 °c in 5% CO2. Thin tissue samples (<10 μm, preferably thinner) may also be used.
Growth medium: Dulbecco’s Modified Eagle Medium: Nutrient Mixture F12 (DMEM/F12, Life Technologies, 11330–032) with 10% fetal bovine serum (FBS, Life Technologies, 10437–028) and 1% penicillin-streptomycin (Life Technologies, 15140–122).
0.25% Trypsin-EDTA (Life Technologies, 25200–056).
10-cm tissue culture plates and other basic cell culture consumables.
2.2. Stock Solutions or Common Reagents for Immuno-fluorescence Staining
Poly-L-lysine solution: 0.1 mg/ml (0.01 %) poly-L-lysine hydrobromide (Sigma-Aldrich, P 1274) dissolved in water, sterilized with a 0.22 μm filter, and stored at 4 °c.
1 M hydrogen chloride (HCI).
#1.5 glass coverslips.
Phosphate buffer saline (PBS, pH 7.0, Amresco, K813).
Bovine serum albumin (BSA, Sigma-Aldrich, A9647).
Permeabilizing buffer/washing buffer: PBST, i.e., 0.1 % Triton X-100 (Fisher Scientific, BP151–100) in PBS at pH = 7.0.
Blocking buffer: 3% BSA in PBST.
4% paraformaldehyde (PFA): four times diluted solution of the 16% aqueous paraformaldehyde (Electron Microscopy Sciences, 15710).
100% methanol (Sigma-Aldrich, 32213).
0.01 % sodium azide.
Nail polish.
2.3. Reagents for dSTORM Imaging
Primary antibodies for ciliary markers and proteins of interest. For example, for ciliary markers, one may use anti-acetylated tubulin (Abcam, ab24610), anti-IFT88 (Proteintech, 139671-AP), or anti-Arl13b (Abcam, ab136648).
Secondary antibodies: Alexa Fluor 647 conjugated IgG antibody (Life Technologies) and Cy3B maleimide (GE, PA6313) (conjugation protocol for Cy3B is described below).
Whole IgG affnity-purified antibodies (Jackson ImmunoResearch).
Borate buffer (0.67 M, Thermo Scientific, 1859833).
Dimethylsulfoxide (DMSO, Sigma-Aldrich, D8418).
Dimethylformamide (DMF).
Purification resin (Thermo Scientific, 1860513).
Pierce spin column (Thermo Scientific, 69705).
TetraSpeck™ microspheres (Life Technologies, T7279).
Chamlide magnetic chamber, 35-mm dish (Live Cell Instrument).
- Imaging buffer for oxygen scavenging:
-
(a)Glucose (Sigma-Aldrich, G5767).
-
(b)Glucose oxidase (Sigma-Aldrich, G2133).
-
(c)Catalase (Sigma-Aldrich, C40).
-
(d)Mercaptoethylamine (MEA or Cysteamine, Sigma-Aldrich, 30070).
-
(e)TN buffer : Tris–HCI, pH 8.0 (10 mM), NaCl (150 mM).
-
(a)
2.4. Reagents for STED Imaging
Secondary antibodies: Oregon Green 488 conjugated IgG(Life Technologies) and biotin conjugated antibody (Sigma-Aldrich).
Primary antibodies (see Subheading 2.3).
BD V500 streptavidin (BD, 561419): streptavidin conjugated to BD-Horizon V500 fluorescent dye.
Prolong Antifade Kit (Life Technologies, P-7481 ) or glycerol.
2.5. A microscopy System for STORM Imaging
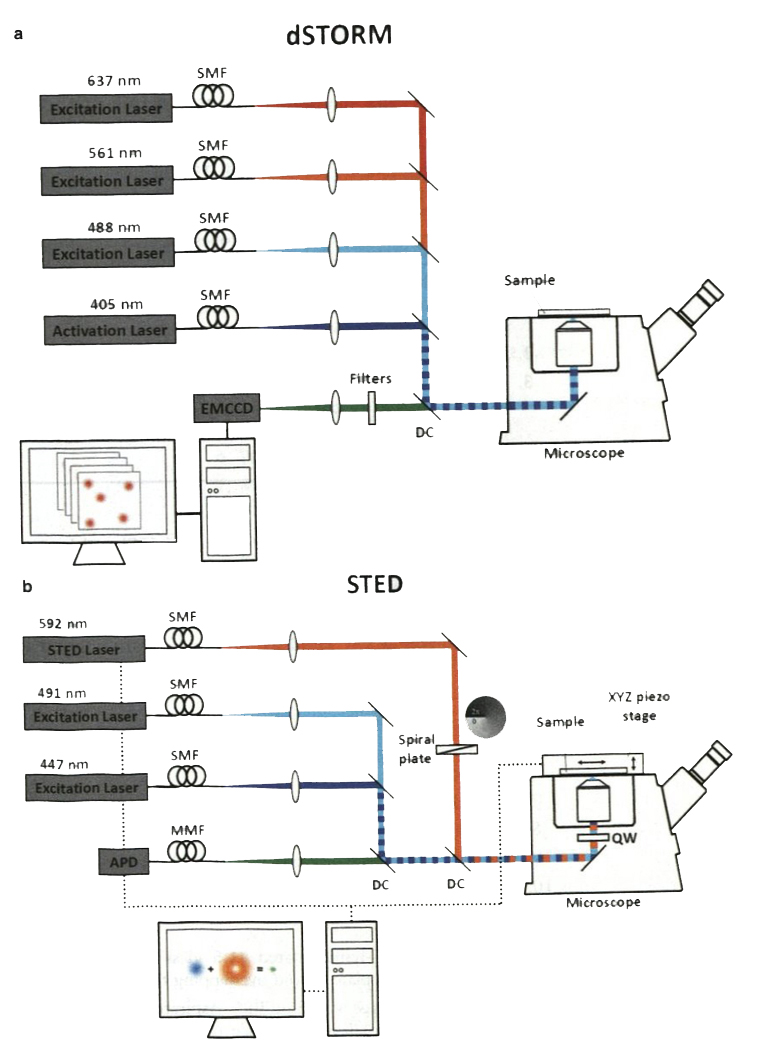
A STORM/PALM system equipped with a high NA objective for (NA≥1.4), an autofocus system, an activation laser (405 nm), multiple excitation lasers depending on the fluorophores in use (e.g., ~640 nm laser for Alexa Fluor 647), and a high-speed, high-sensitivity EMCCD or sCMOS camera (Fig. 1a). As an example, our STORM/PALM system is composed of four lasers ( 100 mW 405 nm solid state optically pumped semiconductor laser (OPSL), 150 mW 488 nm solid State OPSL, 150 mW 561 nm pumped diode laser, and 140 mW 637 nm solid state OPSL, all integrated to a laser unit by Spectral Applied Research), a light path integration platform (Diskovery Platform, Spectral Applied Research), an EMCCD camera (Evolve 512 Delta, Photometrics), a microscope (Eclipse Ti-E, Nikon) equipped with an NA 1.49 100× objective (CFI APO TIRF 100× OIL, Nikon) and an autofocus system (Perfect Focus System, Nikon). Zeiss, Nikon, and Leica all have integrated superresolution microscopy systems with equivalent elements convenient for STORM/PALM imaging.
A computer with a large data storage system.
STORM/PALM image analysis software. Multiple STORM/ PALM image analysis software packages to find single molecule centers and to merge multiple images together are available (e.g., QuickPaIm [53]). We use the MetaMorph Microscopy Automation & Image Analysis Software with a superresolution module, which uses wavelet fitting to allow real-time localization of single molecules [54]. A computer with CPU/GPU integration is used to expedite wavelet fitting.
Vibration isolation system. The vibration isolation system such as a high-performance optical table is important for superresolution imaging. To push the limit of mechanical stability, we use negative stiffness vibration isolators (two 250BM-1, Minus K, and one breadboard, SG-35–2, Newport), which have a better performance than optical tables. The choice Of Minus K systems depends on the total weight of the optical setup.
Fig. 1.
Schematic diagrams of a dSTORM system and a CW STED system in our lab. (a) The dSTORM system consists of three excitation laser lines (637, 561, and 488 nm) and one activation laser (405 nm). A high-speed EMCCD captures single-molecule emission over 5000–20,000 frames to reconstruct a superresolution image. (b) In the STED microscope, two excitation lasers (447 and 491 nm) are chosen for Oregon Green 488 and BD V500 whose emission fluorescence can both be depleted with a 592-nm laser. The light pattern of the depletion laser is converted to a doughnut-shaped distribution through a spiral phase plate. Fluorescent signals from a sample are detected pixel-by-pixel by an avalanche photodiode (APD), a single photon counting module. SMF single-mode polarization-maintaining fiber, MMF multimode fiber, DC dichroic mirror
2.6. A Microscopy System for STED Imaging
A STED microscopy system equipped with a high NA objective (NA ≥ 1.4), a scanning system, excitation lasers, depletion lasers, an optical system integrating both laser sources, and a highly sensitive photon counter (Fig. 1b). Our STED microscopy system is a system using CW lasers, with two excitation lasers (20 mW 447 nm diode laser, PGL-V-H-447, CNI; 25 mW 491-nm DPSS laser, Cobolt AB) and one depletion laser (1 W 592 nm fiber laser, VFL-P-1000–592, MPB Communications). Either a photomultiplier tube (PMT, MP963, PerkinElmer) or an avalanche photodiode single photon counting module (SPCM-AQR-15, PerkinElmer) is used to detect photons from samples held on a three-axis piezo scanner (Nano-PDQ375HS, Mad City Labs). Similar or gSTED versions of STED microscopes are available commercially (Leica and Abberior), some of them including multiple of excitation lasers (592, 660, and 775 nm) and a wide spectrum of excitation laser source (supercontinuum laser) for a broad selection of fluorophores.
A computer with a large data storage system.
Vibration isolation system. To push the limit of mechanical stability, we use stiffness vibration isolators (two 350BM-l, Minus K, and one breadboard, SG-46–2, Newport).
3. Methods
3.1. Sample Preparation for dSTORM
3.1.1. Preparation of Poly-L-Lysine Coating Glass Coverslips (See Note 1)
Put coverslips in a glass beaker with 1 M HCI and heat to 50–60 °c for more than 4 h or overnight.
Rinse off 1 M HCI with ddH2O for three times.
Incubate coverslips in ddH2O and shake gently using an orbital shaker at low speed for 30 min so as to completely remove the HCI.
Wash once with 100% ethanol.
Incubate with 100% ethanol for over 1 h.
Transfer coverslips into a 10-cm plate inside a biosafety cabinet and air dry overnight.
Add 5–10 ml 0.01 % poly-L-lysine (0.01 %) and shake for 4–6 h.
Wash with ddH2O for three times. Air dry completely overnight.
Leave the plate in the biosafety cabinet and sterilize with UV.
Wrap the plate containing coated coverslips with aluminum foil and store inside the biosafety cabinet.
3.1.2. Primary Cilium Growth Upon Serum Starvation
Place 8–10 poly-L-lysine coated #1.5 glass coverslips into a 10-cm cell culture plate. Avoid overlapping the coverslips.
Wash coverslips once with 5 ml PBS. Aspirate PBS completely with a suction to allow adherence of the coverslips to the plate surface.
Grow hTERT RPE-I in the plate with 10 ml growth medium to ~50% confluency.
Aspirate medium and wash the cells twice with 5–10 ml PBS.
Add 10 ml serum free medium to cells.
Incubate cells for 48 h in this serum starved condition to promote cilia formation.
3.1.3. Conjugation of Cy3B Fluorophore to lgG Antibody (Optional, Only Required for 2-Color dSTORM Imaging)
Prepare a 10 mg/ml solution of Cy3B monofunctional NHS ester in DMSO/ DMF [1:1] by adding 100 μl DMSO/DMF solution to 1 mg Cy3B mono NHS ester.
Prepare IgG solution by adding 8 μl of the Borate Buffer to 100 μl of 1 mg/ml IgG antibody.
Add 1.5 μl of the Cy3B solution into the 100 μl IgG solution, pipette up and down to mix.
Briefly centrifuge the vial to collect the mixture in the bottom of the tube.
Incubate the reaction mixture for 60 min at room temperature protected from light.
Place spin column in a microcentrifuge tube.
Mix the purification resin to ensure uniform suspension and add 300 μl of the suspension into the spin column.
Centrifuge for 1 min at 1000×g to remove the storage solution. Discard the used microcentrifuge tube and place the column into a new tube.
Wash column once with PBS and place the column into a new tube.
Add the labeling reaction mixture to the column and allow the sample to mix with the resin by briefly vortexing.
Centrifuge column for 1 min at 1000×g to collect the purified antibody. Discard the used column.
Store the labeled antibody protected from light at 4 °c.
3.1.4. Immuno fluorescence Staining
All steps are performed under room temperature unless otherwise indicated. Solutions or buffers are added to samples at a volume sufficient to cover the glass coverslips.
Wash cells by adding 5–10 ml PBS to the 10-cm culture plate.
Fix cells 4% PFA (or ice-cold methanol at –20 °c, see Note 2) for 10–15 min. Avoid over fixing or otherwise epitope of the protein of interest will be shielded.
Wash cells twice with PBS to remove the fixative.
Permeabilize cells with PBST for 10 min.
Block cells by incubating samples in the blocking buffer for 30–60 min.
Prepare primary antibodies in the blocking buffer at an optimized dilution. The optimized dilution of each primary antibody for superresolution microscopy may be different from that of conventional microscopy and should be found independently. In general, a volume of 50-μl is suffcient to cover one coverslip. For dual color imaging, dilute and mix the two desired primary antibodies at their corresponding optimized dilutions. Incubate cells in the antibody-containing buffer for 1 h.
Wash cells five times with the washing buffer.
For single color imaging, dilute Alexa Fluor 647 secondary antibody (see Note 3) in the blocking buffer to an optimized concentration. For dual color imaging, prepare a mixture of Alexa Fluor 647 antibody and Cy3B conjugated IgG antibody at their corresponding optimized dilutions. Add the solution to cells and incubate for 1 h. Protect samples from light.
Wash cells with the washing buffer for three times.
Dilute TetraSpeck microspheres in a ratio of 1:200 with PBS. Add microspheres to cells and incubate for 30 min (see Note 4).
Wash once with PBS and store samples in PBS at 4 ° c. For long-term storage, keep stained samples in PBS with 0.01 % sodium azide at 4 ° C.
3.2. dSTORM Imaging Procedure
The immunostained sample prepared on a glass coverslip is transferred into a custom-built or commercial imaging chamber (for example, imaging chamber, Live Cell Instrument). A custom-built imaging chamber can be made by mounting a coverslip on a carved glass microscope slide and then sealing the edge of the coverslip with nail polish.
Prepare the imaging buffer for oxygen scavenging. Imaging buffer consists of 10–100 mM MEA (see Note 5), 0.5 mg/ml glucose oxidase, 40 μg/ml catalase, and 10% glucose solution in TN buffer at pH 8.0 (see Note 6). The concentration of MEA depends on the protein density and labeling effciency and has to be optimized to maintain a condition in which single emitters residing within a diffaction-limited region can be separated.
The freshly prepared imaging buffer should be immediately added to the sample and cover the chamber entirely. For the best performance, the imaging chamber is suggested to be sealed with Parafilm to slow down the degradation of the imaging buffer (see Note 7).
All instrument and devices should be turned on at least 30 min before samples are imaged with dSTORM mode to minimize any temperature drift caused by the warming-up of microscope components.
The samples should be placed in the warmed-up microscope at least for 15 min after they have been pre-focused (see Note 8). To stabilize microscope axial position, an electronically feedback control focusing system (for example, Nikon Perfect Focus System) is highly recommended.
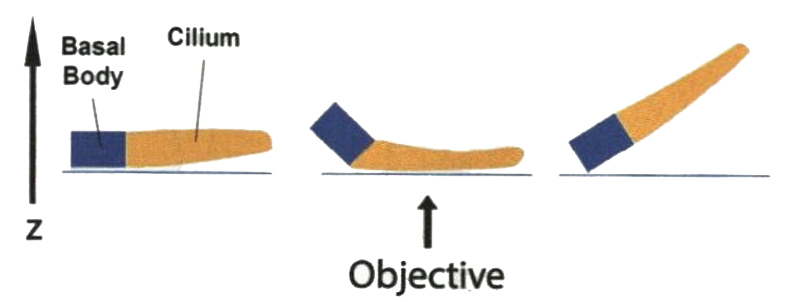
A ciliary marker is required to effectively locate a cilium during imaging. To Start imaging, a ciliary marker (e.g., acetylated tubulin) is first observed in the widefield mode to determine whether the orientation of this cilium is suitable. Perfectly horizontal or vertical cilia usually are not easy to find. Most of the cilia are bent to a certain degree (Fig. 2). In practice, a nearly horizontal cilium can still be found with the use of ciliary markers labeling both the cilium compartment and basal body. If both markers of a cilium have the same focus, this cilium is likely horizontal.
Once a cilium is found using a marker, the target protein is first imaged with the widefield mode to determine if its signal-to noise level is high enough for dSTORM.
Further fine focus adjustment may be needed for the best image contrast of the target protein (see Note 9). After setting the focus, one should switch the imaging channels between the marker and the target protein back and forth to make sure that both signals are correlated with each other.
To successfully achieve superresolution imaging of cilia, fiducial markers (fluorescent beads or gold nanoparticles) should be imaged together with the target protein. Therefore, both signals of proteins of interest and a stable fiducial marker have to appear in the same field of view.
The sample is first illuminated at a gradually increasing power (up to 1–4 kW/cm2) of the excitation laser (e.g., 637 nm laser for Alexa Fluor 647) to drive the dye molecules to a dark state. This step may saturate the EM-CCD camera. To protect CCD, it is suggested to keep the EM gain at a minimal level in the beginning.
The dyes immersed in the imaging buffer start to repetitively photoswitch between on and off state (photoblinking). The EM gain of CCD can be changed to the working level for optimal detection without saturation. It usually takes a few seconds to see an obvious photoswitching mechanism after pumping the intense laser power. However, a good fiducial marker should not show substantial brightness fluctuation and any photoswitching during the imaging procedure.
The exposure time should then be adjusted to match the photoswitching rate, preventing the adjacent emitters from overlapping. The exposure time is usually set to 10–50 ms depending on the density of labeled molecules (see Note 10).
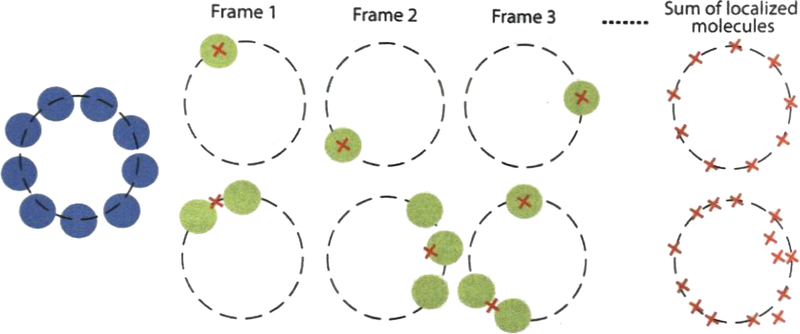
The event from single molecule emission can be confirmed by taking several consecutive snapshot images. A good imaging condition is achieved when only isolated single emitters appear in every single snapshot (Fig. 3) (see Note 11).
To estimate the number of single molecule localization per frame, one can select effective single emitters with a brightness threshold to reject undesired noise (see Note 12).
The ciliary feature may not be easily identified while the dyes are photoblinking. A pre-photoblinking snapshot image would be helpful to recognize the feature. In particular, the size of the transition zone and the basal body are too small to identify because their patterns are similar to nonspecific signals.
After an appropriate threshold level is determined, the stream acquisition of 5000–20,000 frames is performed for the effective dSTORM imaging.
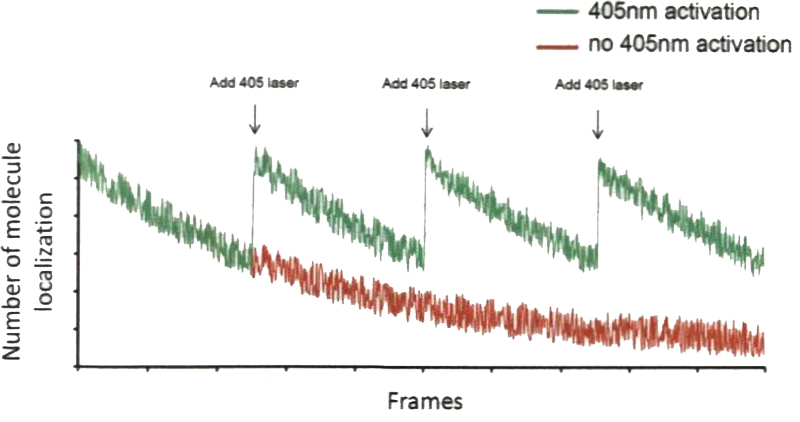
During the acquisition, the total number of single molecule localization should always be monitored to maintain an optimized density of detected localization. The detection number usually decreases with the imaging time (Fig. 4, red). For the best acquisition speed, the detection number has to be maintained approximately constant by adjusting the power of the activation laser (405 nm laser) to activate the transition from the dark state back to the singlet state of a molecule (Fig. 4, green).
In addition to registering photon counts of all pixels, real-time snapshot images every 100 frames, if possible, are highly recommended to monitor that the single molecule detection is still maintained in most of the frames.
The reconstructed dSTORM image extracted from 20,000 frames should well display a nearly complete pattern of target proteins.
For dual-color dSTORM, the first imaging channel is usually assigned to an Alexa Fluor 647-tagged ciliary protein for its good performance. There are multiple options of dyes for the second channel. The working imaging buffer may differ from dye to dye, so switching performance need to be compromised in dual-color imaging. In terms of the brightness and switching properties, the use of red dyes such as Cy3B provides a compromised solution the second channel. For three-color superresolution imaging, it is possible to have the combination of Alexa Fluor 647/Cy3B/AITO 488, although the buffer conditions have to be adjusted and the performance is not optimal.
Basically, a sequential acquisition is conducted in two-color imaging. The Alexa Fluor 647 dye should be imaged first, and then Cy3B dye is imaged (see Note 13). For three-color imaging, ATTO 488 is imaged after Cy3B.
Similar to the first channel, the laser power and exposure time should be optimized for other channels. In particular, the 405 nm laser is required for Cy3B to maintain a reasonable switching rate while imaging. In general, the second dye does not offer the imaging stability as good as that of the first channel using Alexa Fluor 647 (see Note 14).
Continuous images of 5000–20,000 frames for the second dye are also acquired to reconstruct an image for the second protein.
Fig. 2.
Cilia grown at different orientations and with different bending characteristics. Most of are bent to a certain degree, basal body may not be aligned with me ciliary axoneme (middle). Some cilia are not parallel to the surface of coverslip, challenging to focus the entire structure (right). One has to search a horizontal cilium (left) so that its orientation is ideal for Side-view imaging
Fig. 3.
Achieving isolated single emitters for good dSTORM images. Emitter overlapping should be avoided for a better imaging (upper). The overlapping signals from multiple emitters results in mislocalization (lower). In this case, the turn-on frequency should be reduced to meet the requirement of separable single-molecule detections per frame
Fig. 4.
Tuning the power of 405-nm laser line to control the activation efficiency of dyes. When seeing the reduction of number of molecules being localized through the imaging process, one should increase of the power of 405-nm laser to maintain an efficient imaging acquisition, minimizing the required number of frames for reconstructing the whole image. Ideally, number of molecule localization should be a constant during the acquisition
3.2.1. Image Correction for dSTORM
A successful superresolution image usually cannot be reconstructed directly from the raw streaming images because most of the raw data suffer from non-negligible mechanical drift which causes a localization error far larger than the dSTORM resolution ~20 nm. Moreover, the localization deviation between two image channels is significant at this resolution scale. Several steps for alleviating the above issues will be described in the following.
Mechanical drift is induced by an unbalanced sample holding system, immersion oil adhesion force, or temperature change.
To reduce the drift caused by holding system, samples should be clamped only with normal force since any shear force may lead to lateral movement of samples.
Samples should be placed on the microscope stage at least 15 min before imaging to ensure the immersion oil reaches equilibrium.
To actively correct a drifted image, fiducial markers such as fluorescent beads or gold nanoparticles should be immobilized on samples for the tracking of their trajectory during streaming acquisition. All reliable markers should have a nearly identical trajectory (see Note 15).
Based on the pattern correlation analysis of these fiducial markers, drift distance at every single frame can be compensated to generate a drift-free stack image.
Mechanical drift can be easily corrected by subtracting the movement of fiducial markers in each frame. However, nonlinear distortion arising from optical issues such as chromatic aberration has to be corrected by either optimizing the optical system or compensating distortion.
To correct chromatic aberration between two imaging channels, one can first characterize their distance deviation over a full field of view using fiducial markers (Fig. 5, before) and then create a 2D distance deviation function of lateral position x and y.
The error compensation is done by applying this calibration function to a raw image. Therefore, most of fiducial markers of overlap between two channels can be achieved (Fig. 5, after).
A good dual-color dSTORM image can be obtained only after two single-color images are correctly aligned.
Fig. 5.
Chromatic aberration between two imaging channels compensated by calibrating distance deviation of fiducial markers between two channels. The localization error between two channels should be less than optical resolution to ensure an effective superresolution imaging
3.3. Sample Preparation for STED Miroscopy
STED microscopy offers a subdiffraction-limited resolution and is a promising tool for ciliary studies even though optical alignment is very challenging when implementing the STED imaging system. Different from confocal fluorescent microscopy, serious care has to be taken to address multiple sample-specific issues of STED imaging. For imaging proteins in cilia, the primary issues are: (1) fluorescent signals from some ciliary proteins are dim; (2) the depletion phenomenon of STED reduces the light being detected from the readily dim signals; (3) the high power of the depletion laser accelerates photodamage; (4) labeling conditions are sample dependent; and (5) the combination of fluorophores for two-color STED imaging have cross-talks or signal leakages between two channels. Thus, in the following sections several tasks of image acquisition and post-processing toward a STED-based super-resolution imaging of cilia will be described to address issues of dim ciliary signals caused by inherent properties of fluorophores, the depletion effect, photobleaching, and photodamage. This protocol adapts to a system using two excitation lines, i.e., a 447-nm and a 491 -nm line and one 592-nm depletion laser. The setup is suitable for two-color STED imaging for BD Horizon V500 and Oregon Green 488 fluorescent dyes.
3.3.1. Preparation of Poly-L-Lysine coating Glass Coverslips
See Subheading 3.1.1.
3.3.2. Primary Cilium Growth Upon Serum Starvation
See Subheading 3.1.2.
3.3.3. Immuno-fluorescence Staining
All steps are performed under room temperature unless otherwise indicated. Solutions or buffers are added to samples at a volume suffcient to cover the glass coverslips.
Wash cells by adding 5–10 ml PBS to the 10-cm culture plate.
Fix cells with 4% PFA (or ice-cold methanol at –20 ° C) for 10–15 min. Avoid over fixing or otherwise epitope of the target protein may be damaged or shielded.
Wash cells twice with PBS to remove the fixative.
Permeabilize cells with PBST for 10 min.
Block cells by incubating samples in the blocking buffer for 30–60 min.
Prepare primary antibodies in the blocking buffer at an optimized dilution. The optimized dilution of each primary antibody for superresolution microscopy may be different from that of conventional microscopy and should be found independently. In general, 50-μl blocking buffer is sufficient for one coverslip. For dual color imaging, dilute and mix the two desired primary antibodies at their optimized dilutions. Incubate cells in the antibody-containing buffer for 1 h.
Wash cells 5 times with PBST washing buffer.
For single color imaging, dilute Oregon Green 488 secondary antibody with the blocking buffer to an optimized concentration. For dual color imaging, prepare a mixture of Oregon Green 488 antibody and biotin-conjugated antibody at their corresponding optimized dilutions. Add the solution to cells and incubate for 1 h. Protect samples from light.
Wash cells with the washing buffer for three times.
For dual color imaging, incubate cells with BD V500 streptavidin for 30 min, and then wash cells with the washing buffer for three times.
Store samples in PBS at 4 ° c. For long-term storage, keep stained samples in PBS with 0.01 % sodium azide at 4 ° c.
3.4. STED Imaging Procedure
Mount the stained sample on microscope glass slides with 86% glycerol or Prolong Antifade Kit mounting medium for the best match of refractive index.
Seal the sample properly with, for example, nail polish. Let the sample stand for at least 1 h prior to imaging.
The sample should be firmly held on the microscope stage. Although STED microscopy suffers less from mechanical drift than dSTORM during the imaging, placing a sample on the stage 15 min before collecting images is still suggested.
Search for a target area with ciliary proteins of interest as quickly as possible in the confocal mode. During this step, one should always run the microscope at a high scanning speed, a large sampling step size, a minimal working laser power, and a minimal number of repetitive scans. When an area of interest is found, the repetitive scanning should be stopped to protect the sample from pre-photobleached.
The performance of STED microscopy depends strongly on focusing, because it is based on confocal configuration to possess excellent optical sectioning. Therefore, finding the proteins of interest depends highly on the axial position of the microscope. After a coarse focal adjustment to locate signals of a ciliary marker, one should check layer by layer with a small interval in axial direction (e.g., 100 nm) to identify the proteins of interest (see Note 16). Imaging an object at different foci may generate different patterns.
Before STED imaging, a confocal image is obtained by optimizing the scanning rate, the sampling step size, and the excitation laser power. The scanning speed has to be fast in the beginning to minimize photobleaching. The excitation power may be increased to enhance the signal-to-noise ratio due to high-speed scanning.
Once a good confocal image is obtained, STED imaging is then performed first with a low depletion power. The step size of 10–50 nm is used to meet the sampling criterion in STED resolution. When signal-to-noise ratio is high, we use a step size of 10 nm to achieve the resolution as good as possible. Check the improvement in resolution. Next, a moderate STED laser power is used to further improve resolution. As the power increases, the signal-to-noise ratio may be reduced due to signal depletion in the periphery. Use the maximal depletion laser power that remains a reasonable signal-to-noise ratio to maximize the resolution improvement (see Note 17).
For two-color STED imaging, separate channels are acquired sequentially through excitations of 447 nm and 491 nm lasers and a common depletion of 592 nm lasers. As illustrated in Fig. 6, the 447-nm light excites BD V500 as well as Oregon Green 488, thus leading to minor signal cross talk between the two dyes. To address this issue, emission signals from two dyes should be well balanced. A ciliary protein with high brightness should be assigned to the 447-nm channel to minimize signal cross talk. The 491-nm laser only excites Oregon Green 488, thus enabling intrinsically spectrally separable STED imaging without the need of unmixing post-processing.
Fig. 6.
The spectral concept behind the two-color STED system using excitation lasers (447 and 491 nm) and one depletion laser (592 nm). In addition to exciting V500, the 447-nm laser can also excite Oregon Green 488 slightly, causing minor cross talk between channels. Therefore, suitable detection windows to separate the signal of V500 from mat of Oregon Green 488 are necessary. The cross talk can be further reduced by assigning the protein with brighter signals to the V500 channel
3.4.1. Image Processing for STED
Different from dSTORM, the mechanical drift issue of STED imaging of fixed samples is generally negligible because an effective superresolution image can be obtained with a single-frame acquisition. The major problem of STED imaging is the poor signal-to-noise ratio of depleted signals resulting from the STED mechanism. In addition to the optimization of imaging parameters such as the laser power, the sampling step size, and the scanning rate, the resulting raw image can be processed to enhance its signal contrast by methods such as deconvolution. To adequately restore a noisy image with deconvolution, the PSF of the system should be first characterized and utilized in the computation algorithm. An inappropriate PSF could induce incorrect information of images. If the used PSF is too narrow (small full width at half maximum), it may amplify the small and noisy feature, causing image artifacts. In contrast, a wide PSF can remove subtle but real features due to over-filtering. In principle, a qualified deconvolved image should retain most of the original features capt ured in the raw data while improving their contrast.
4. Notes
Poly-L-lysine coating increases the adherence of hTERT RPE-I cells to coverslips. Some cell types may require this coating while others may not, depending on the adherence of each cell type.
The fixation condition may vary for different kinds of proteins. For example, image quality using 4 % PFA fixation is better for membrane-bound proteins as compared to methanol fixation; while methanol fixation is especially good to preserve actin and cytoskeletal proteins. It is advised to optimize the specific fixation condition for different proteins of interest.
Alexa Fluor 647 dye exhibits a photoswitching property better than any other existing dyes for dSTORM superresolution imaging. It is recommended to use Alexa Fluor 647 to mark the protein of higher interest between the two proteins when performing dual-color dSTORM imaging.
Microspheres serve as fiducial markers to calibrate the mechanical drift during imaging. Nano-gold particles can also be used. Because of the stringent requirement of resolution (~20 nm), fiducial markers are essential for a good performance of dSTORM superresolution imaging.
MEA concentration is critical for the control of the photoswitching mechanism and should be adjusted depending on samples. For example, if dyes do not reversibly switch, MEA concentration should be increased.
The stock of MEA, glucose oxidase or catalase can be prepared and kept in a freezer. The stocks such as 1 M MEA, 50 mg/ml glucose oxidase and 40 mg/ml catalase are recommended. These stocks usually can be stored for at least several weeks in a freezer. The fresh MEA within 1 week is recommended after it is thawed.
To maintain the imaging quality, the imaging buffer should be replaced with a fresh one every 1–2 h.
The subtle movement of immersion oil may occur right after the sample is placed, so it is recommended to let samples stand for at least 15 min before imaging.
The power of the light source should be kept at a low level to prevent photobleaching.
Adjustment of the exposure time is always necessary. This parameter can be first set to the upper bound of the range in order to gain the maximal signal to noise ratio. If too many emitters stay in their ON state, exposure should be reduced.
In addition to the camera integration time, the illumination power can significantly affect whether single emitter events are achieved or not. We usually use the full power ~4 kW/cm2 at which we successfully achieve single molecule detection During imaging, the power is kept at the maximum.
If scattered localizations appear randomly all over the image, it indicates that the threshold is too low. A high contrast and continuous image is usually rendered through the optimization of the threshold level.
If one imaged the Cy3B dye first, the Alexa Fluor 647 will be photobleached by the strong power of laser when illuminating Cy3B.
All proteins of interest should be separately studied first with single-color dSTORM imaging (possibly with a ciliary marker using conventional microscopy to locate the protein of interest) to characterize their pattern and spatial distribution prior to two-color imaging.
Some unstable markers may move or dither. They should be excluded from the drift correction procedure.
The fine adjustment at a finer sampling size ~25–50 nm may be needed for some cases. Use a minimal excitation power that is barely enough to differentiate subtle pattern changes in order to avoid photobleaching.
The same field of view is not allowed to be imaged for too many times with an intense STED laser power. Successful STED images are mostly generated from the first three imaging scans.
Acknowledgments
This work was supported by the Ministry of Science and Technology, Taiwan (Grant No. 103-2112-M-001-039-MY3), Academia Sinica Career Development Award, and Academia Sinica Nano Program.
Abbreviations
- BaLM
Bleaching/blinking assisted localization microscopy (BaLM)
- Cw
Continuous wave (CW)
- dSTORM
Direct stochastic optical reconstruction microscopy (dSTORM)
- FPALM
Fluorescence photoactivation localization microscopy (FPALM)
- GSD
Ground state depletion (GSD)
- GSDIM
Ground state depletion microscopy followed by individual molecule return (GSDIM)
- gSTED
Gated stimulated emission depletion (gSTED)
- IFT
Intraflagellar transport (IFT)
- PAINT
Point accumulation for imaging in nanoscale topography (PAINT)
- PALM
Photoactivated localization microscopy (PALM)
- PALMIRA
PALM with independently running acquisition (PALMIRA)
- PSF
Point spread function (PSF)
- RESOLFT
Reversible saturable optical fluorescence transitions (RESOLFT)
- SIM
Structured illumination microscopy (SIM)
- SOFI
Superresolution optical fluctuation imaging (SOFI)
- SSIM
Saturated structured illumination microscopy (SSIM)
- STED
Stimulated emission depletion (STED)
- STORM
Stochastic optical reconstruction microscopy (STORM)
References
- 1.Williams CL, Li C, Kida K, Inglis PN, Mohan S et al. (2011) MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol 192:1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sang L, Miller JJ, Corbit ICC, Giles RH, Brauer MJ et al. (2011) Mapping the NPHPJBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145:513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA et al. (2011) A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet 43:776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG et al. (2011) A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol 14:61–72 [DOI] [PubMed] [Google Scholar]
- 5.Craige B, Tsao C-C, Diener DR, Hou Y, Lechtreck K-F al (2010) CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol 190:927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanos BE, Yang H-J, soni R, wang W-J, Macaluso FP et al. (2013) Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 27:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye X, zeng H, Ning G, Reiter JF, Liu A (2014) C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad sci U S A 111:2164–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke MC, Li F-Q, Cyge B, Arashiro T, Brechbuhl HM et al. (2014) Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J Cell Biol 207:123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang TT, Hampilos PJ, Nathwani B, Miller CH, Sutaria ND et al. (2013) Superresolution STED microscopy reveals differential localization in primary cilia. Cytoskeleton (Hoboken, NJ) 70:54–65 [DOI] [PubMed] [Google Scholar]
- 10.Nathwani B, Yang TT, Liao J-C (2013) Towards a subdiffraction view of motor-mediated transport in primary cilia. Cell Mol Bioeng 6:82–97 [Google Scholar]
- 11.Dorn KV, Hughes CE, Rohatgi R (2012) A smoothened-Evc2 complex transduces the hedgehog signal at primary cilia. Dev Cell 23:823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau L, Lee YL, Sahl SJ, Stearns T, Moerner WE (2012) STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys J 102:2926–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV et al. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329:436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P et al. (2011) Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton 68:619–627 [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Kim S, Lin Y-C, Inoue T, Dynlacht BD (2014) The CPI 10-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J Cell Biol 204:215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA et al. (2015) Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol 17:228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hell S, Wichmann J (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 19:780–782 [DOI] [PubMed] [Google Scholar]
- 18.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW (2000) Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci U S A 97:8206–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustafsson MGL (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198:82–87 [DOI] [PubMed] [Google Scholar]
- 20.Hell SW (2003) Toward fluorescence nanos-copy. Nat Biotechnol 21:1347–1355 [DOI] [PubMed] [Google Scholar]
- 21.Gustafsson MGL (2005) Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A 102:13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645 [DOI] [PubMed] [Google Scholar]
- 23.Rust MJ, Bates M, Zhuang XW (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3:793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess ST, Girirajan TPK, Mason MD (2006) Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J 91:4258–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharonov A, Hochstrasser RM (2006) Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci 103:18911–18916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bretschneider S, Eggeling C, Hell SW (2007) Breaking the barrier in fluorescence microscopy by optical shelving. Phys Rev Lett 98:218103 [DOI] [PubMed] [Google Scholar]
- 27.Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D et al. (2007) Fluorescence nanos-copy in whole cells by asynchronous localization of photoswitching emitters. Biophys J 93:3285–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B et al. (2008) Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed 47:6172–6176 [DOI] [PubMed] [Google Scholar]
- 29.Foiling J, Bossi M, Bock H, Medda R, Wurm CA et al. (2008) Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods 5:943–945 [DOI] [PubMed] [Google Scholar]
- 30.Dertinger T, Colyer R, lyer G, Weiss S, Enderlein J (2009) Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI).Proc Natl Acad Sci 106:22287–22292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicidomini G, Moneron G, Han KY, Westphal V, Ta H et al. (2011) Sharper low-power STED nanoscopy by time gating. Nat Methods 8:571–573 [DOI] [PubMed] [Google Scholar]
- 32.Burnette DT, Sengupta P, Dai Y, Lippincott-Schwartz J, Kachar B (2011) Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc Natl Acad Sci 108:21081–21086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quan T, Zhu H, Liu X, Liu Y, Ding J et al. (2011) High-density localization of active molecules using Structured Sparse Model and Bayesian Information Criterion. Opt Express 19:16963–16974 [DOI] [PubMed] [Google Scholar]
- 34.Cox S, Rosten E, Monypenny J, Jovanovic-Talisman T, Burnette DT et al. (2012) Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat Methods 9:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rittweger E, Han K, Irvine S, Eggeling C, Hell S (2009) STED microscopy reveals crystal colour centres with nanometric Nat Photon 3:144–147 [Google Scholar]
- 36.Willig K, Harke B, Medda R, Hell S (2007) STED microscopy with continuous wave beams. Nat Methods 4:915–918 [DOI] [PubMed] [Google Scholar]
- 37.Meyer L, Wildanger D, Medda R, Punge A, Rizzoli SO et al. (2008) Dual-color STED microscopy at 30-nm focal-plane resolution. Small 4:1095–1100 [DOI] [PubMed] [Google Scholar]
- 38.Wildanger D, Medda R, Kastrup L, Hell SW (2009) A compact STED microscope providing 3D nanoscale resolution. J Microsc 236:35–43 [DOI] [PubMed] [Google Scholar]
- 39.Moneron G, Hell SW (2009) Two-photon excitation STED microscopy. Opt Express 17:14567–14573 [DOI] [PubMed] [Google Scholar]
- 40.Westphal V, Rizzoli S, Lauterbach M, Kamin D, Jahn R et al. (2008) Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science 320:246–249 [DOI] [PubMed] [Google Scholar]
- 41.Pellet PA, Sun X, Gould TJ, Rothman JE, Xu M-Q et al. (2011) Two-color STED microscopy in living cells. Biomed Opt Express 2:2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H et al. (2003) A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21:86–89 [DOI] [PubMed] [Google Scholar]
- 43.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N et al. (2008) HaloTag:.a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3:373–382 [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Chang H, Zhang Y, Yu J, Wu L et al. (2012) Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9:727–729 [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X (2014) Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci 111:8452–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J et al. (2009) Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods 6:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J et al. (2007) Dual-color super-resolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci 104:20308–20313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X (2011) Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods 8:1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang B, Jones SA, Brandenburg B, Zhuang X (2008) Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods 5:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS et al. (2008) Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods 5:527–529 [DOI] [PubMed] [Google Scholar]
- 51.Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N et al. (2009) Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci 106:2995–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM et al. (2009) Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci 106:3125–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henriques R, Lelek M, Fornasiero EF, Valtorta F, Zimmer C et al. (2010) QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat Methods 7:339–340 [DOI] [PubMed] [Google Scholar]
- 54.Izeddin I, Boulanger J, Racine V, Specht CG, Kechkar A et al. (2012) Wavelet analysis for single molecule localization microscopy. Opt Express 20:2081–2095 [DOI] [PubMed] [Google Scholar]