Key Points
Question
To what extent can genomic features revealed by clinical targeted tumor sequencing enable diagnostic accuracy of tumor type?
Findings
This cohort study used machine learning techniques to construct and train an algorithmic classifier on a cohort of 7791 prospectively sequenced tumors representing 22 cancer types to predict cancer type and origin from DNA sequence data obtained at the point of care. In some cases, genome-directed reassessment of diagnosis prompted tumor type reclassification resulting in altered therapy for patients with cancer.
Meaning
The clinical implementation of artificial intelligence to guide tumor type diagnosis at the point of care may complement standard histopathologic testing and imaging to enable improved diagnostic accuracy.
Abstract
Importance
Diagnosing the site of origin for cancer is a pillar of disease classification that has directed clinical care for more than a century. Even in an era of precision oncologic practice, in which treatment is increasingly informed by the presence or absence of mutant genes responsible for cancer growth and progression, tumor origin remains a critical factor in tumor biologic characteristics and therapeutic sensitivity.
Objective
To evaluate whether data derived from routine clinical DNA sequencing of tumors could complement conventional approaches to enable improved diagnostic accuracy.
Design, Setting, and Participants
A machine learning approach was developed to predict tumor type from targeted panel DNA sequence data obtained at the point of care, incorporating both discrete molecular alterations and inferred features such as mutational signatures. This algorithm was trained on 7791 tumors representing 22 cancer types selected from a prospectively sequenced cohort of patients with advanced cancer.
Results
The correct tumor type was predicted for 5748 of the 7791 patients (73.8%) in the training set as well as 8623 of 11 644 patients (74.1%) in an independent cohort. Predictions were assigned probabilities that reflected empirical accuracy, with 3388 cases (43.5%) representing high-confidence predictions (>95% probability). Informative molecular features and feature categories varied widely by tumor type. Genomic analysis of plasma cell-free DNA yielded accurate predictions in 45 of 60 cases (75.0%), suggesting that this approach may be applied in diverse clinical settings including as an adjunct to cancer screening. Likely tissues of origin were predicted from targeted tumor sequencing in 95 of 141 patients (67.4%) with cancers of unknown primary site. Applying this method prospectively to patients under active care enabled genome-directed reassessment of diagnosis in 2 patients initially presumed to have metastatic breast cancer, leading to the selection of more appropriate treatments, which elicited clinical responses.
Conclusions and Relevance
These results suggest that the application of artificial intelligence to predict tissue of origin in oncologic practice can act as a useful complement to conventional histologic review to provide integrated pathologic diagnoses, often with important therapeutic implications.
This cohort study develops a machine learning algorithm, using tumors from patients with advanced cancer, to aid in the prediction and classification of tumor types.
Introduction
The clinical management of cancer is associated with its site of origin, histopathologic subtype, and stage. Even for patients with tumors harboring a therapeutically sensitizing mutation that can guide molecularly targeted therapy, clinical responses are often associated with tumor origin. For example, BRAF V600E mutations are observed in cancers arising from numerous tissue sites, and the likelihood of response to RAF inhibitors varies widely as a function of tumor type.1 Although critical for guiding patient management, histologic-based cancer diagnosis remains challenging in many patients, especially in those initially presenting with metastatic, poorly differentiated neoplasms in which ambiguous or incorrect classification may adversely affect choice of therapy and outcome.2
While conventional cancer diagnosis has benefited from thorough immunohistochemical evaluation coupled with high-quality cross-sectional imaging, molecular alterations highly indicative of the tumor site of origin may further assist in diagnosis when conventional tools fail. Some genomic alterations and mutational signatures are associated with specific individual tumor types, such as APC loss-of-function mutations in colorectal cancers, TMPRSS2-ERG fusions in prostate cancers, and a UV-associated mutational signature of C>T substitutions in cutaneous melanomas. For other cancer types, combinations of genomic alterations may commonly co-occur, such as TP53 and CTNNB1 mutations in endometrial cancer. The absence of highly prevalent alterations in a given tumor type, such as KRAS mutations in pancreatic adenocarcinoma and recurrent gene fusions in certain sarcomas, can also provide evidence against that particular diagnostic classification. Both common and rare genomic alterations across numerous different cancers may, therefore, guide the inference of tumor origin as an adjunct to existing diagnostic approaches.
The feasibility of tumor type classification from genomic data, including mutations, copy number alterations, gene expression, methylation, and nucleosome occupancy, has been demonstrated.3,4,5,6,7,8,9,10,11 Moreover, such molecular reassessment of diagnosis can lead to a change of therapy.12 Yet the systematic application of such approaches to prospectively generated clinical sequencing data from often suboptimal formalin-fixed paraffin-embedded biopsies, as well as their accuracy when applied to the targeted cancer gene panels most commonly used in the clinic to facilitate treatment selection, remain to our knowledge largely unexplored.
Herein, we report a machine learning–based approach to infer the probabilities of each common solid tumor type diagnosis based on a broad array of genomic alterations identified by targeted tumor sequencing. To ensure applicability for clinical care, we trained our model on prospective genomic data from 7791 patients with advanced cancer. Using a population-scale approach allowed us to account for the varying prevalence and co-occurrence of genomic features across all tumor types. The probabilistic genome-based tumor type prediction we establish herein, when considered alongside traditional immunohistochemical and clinical evaluation, may enable improved diagnostic accuracy, with important therapeutic implications.
Methods
Patients
The training data set was derived from the published Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) clinical cohort.13 Patients with rare cancer types or low tumor content were excluded from analysis, resulting in a total training data set of 7791 patients diagnosed with 1 of 22 cancer types (eTable 1 in the Supplement). An additional 11 644 patients subsequently tested by MSK-IMPACT made up an independent test set. Data are deidentifed. All patients undergoing MSK-IMPACT testing provided informed consent with a signed clinical consent form or a consent form for enrollment in a research protocol approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board (NCT01775072). Demographic characteristics of both cohorts are displayed in eTable 2 in the Supplement.
Genomic Analysis
Tumor and matched normal DNAs were sequenced in a Clinical Laboratory Improvement Amendments–compliant laboratory using MSK-IMPACT, a US Food and Drug Administration–authorized clinical sequencing assay targeting up to 468 key cancer-associated genes.13,14 Genomic alterations, including mutations, indels, copy number alterations, structural rearrangements, and selected mutation signatures, were reported to patients and physicians to guide clinical care and aggregated in a Health Insurance Portability and Accountability Act–compliant manner in the cBioPortal for Cancer Genomics for further analysis and visualization.
Random Forest Classifier
To predict tumor site of origin, we constructed a random forest classifier using the training cohort of 7791 patients.15 Prediction accuracy was determined from 5-fold cross-validation of the training data as well as the independent test set. Because many diverse alterations and mutation patterns are associated with different sites of origin, the feature set for classification was drawn from the following categories: mutations and indels (hotspots and gene level), focal amplifications and deletions, broad copy number gains and losses, structural rearrangements, mutation signatures, mutation rate, and sex. Classifier scores were subsequently calibrated using multinomial logistic regression to match empirically observed classification probabilities.16 The source code is available at https://github.com/bergerm1/GenomeDerivedDiagnosis.
Results
Performance of Tumor Type Predictor
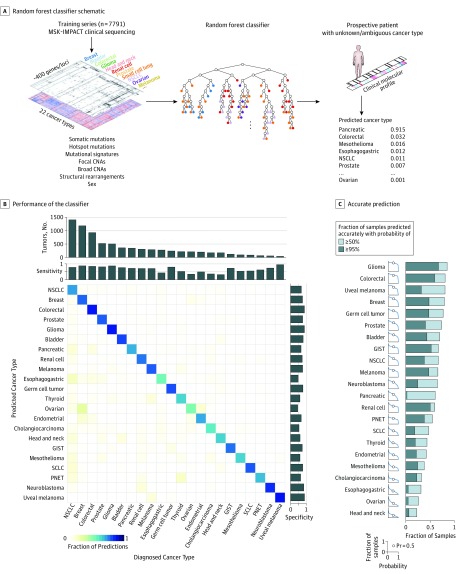
We hypothesized that the information content from clinical targeted tumor genomic profiling would be sufficiently rich to predict the tumor site of origin with high accuracy. We therefore developed a machine learning–based classifier to determine the ability of DNA genomic alterations (specifically, mutations and indels, focal and broad copy number alterations, structural rearrangements, and mutation signatures) to inform the diagnosis in patients with advanced cancer (Figure 1A; eMethods in the Supplement). In our training set of 7791 patients tested by MSK-IMPACT,13,14 the diagnostic cancer type was accurately predicted in 5748 cases (73.8%) of cases based on 5-fold cross-validation (Figure 1B; eTable 3, and eTable 4 in the Supplement). The positive predictive value was highest in tumor types with distinctive molecular profiles, such as uveal melanoma (95%), glioma (87%), and colorectal cancer (85%), with predictions driven by diverse sets of genomic features (eFigure 1 in the Supplement). For other, more heterogeneous, tumor type categories, prediction accuracy varied among detailed histologic subtypes (eTable 5 in the Supplement). Applying the full classifier to predict the site of origin from MSK-IMPACT clinical sequencing in an independent test set of 11 644 additional patients, we observed an equivalent accuracy of 74.1% (n = 8623).
Figure 1. Classifier Performance Across Cancers.
A, Schematic of random forest classifier. Molecular alterations from Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) sequencing of 7791 patients diagnosed with 1 of 22 tumor types were used to train the classifier. For a given combination of genomic features, the classifier returns a calibrated probability of each tumor type. B, Performance of the classifier across 22 cancer types. True (established) cancer types are displayed horizontally and predicted cancer types are displayed vertically. The number of tumors for each cancer type in the cohort is shown at the top, and sensitivity and specificity of predictions are indicated at the top and right. C, The fraction of samples (vertical axis) with the correct prediction made at or above a given probability (horizontal axis) within each cancer type. CNAs indicates copy number alterations; GIST, gastrointestinal stromal tumor; NSCLC, non–small cell lung cancer; PNET, pancreatic neuroendocrine tumor; Pr, probability; and SCLC, small cell lung cancer.
Owing to the importance of high-confidence predictions for clinical decision making in individual patients, we sought to estimate the probability associated with each tumor type prediction. Raw classifier scores were calibrated to match empirically observed classification probabilities from cross-validation (log loss, 0.98; eFigure 2 in the Supplement). In many cancer types, approximately half or more cases were classified with greater than 95% probability (Figure 1C). In other challenging cancer types, such as esophagogastric, ovarian, and head and neck cancer, a minority of cases were predicted with confidence greater than 50% owing to increased molecular heterogeneity among tumors and the lack of distinguishing genomic alterations. Nevertheless, 3388 of all cases (43.5%) were predicted with probability greater than 95% and an empirical accuracy of 96.6%, indicating an abundance of high-confidence, reliable predictions enabled by our classifier (eFigure 3 in the Supplement). Moreover, most of the incorrect predictions were made with low confidence (probability <50%) and are therefore unlikely to be a factor in diagnostic or clinical decisions.
Relative Importance of Molecular Features
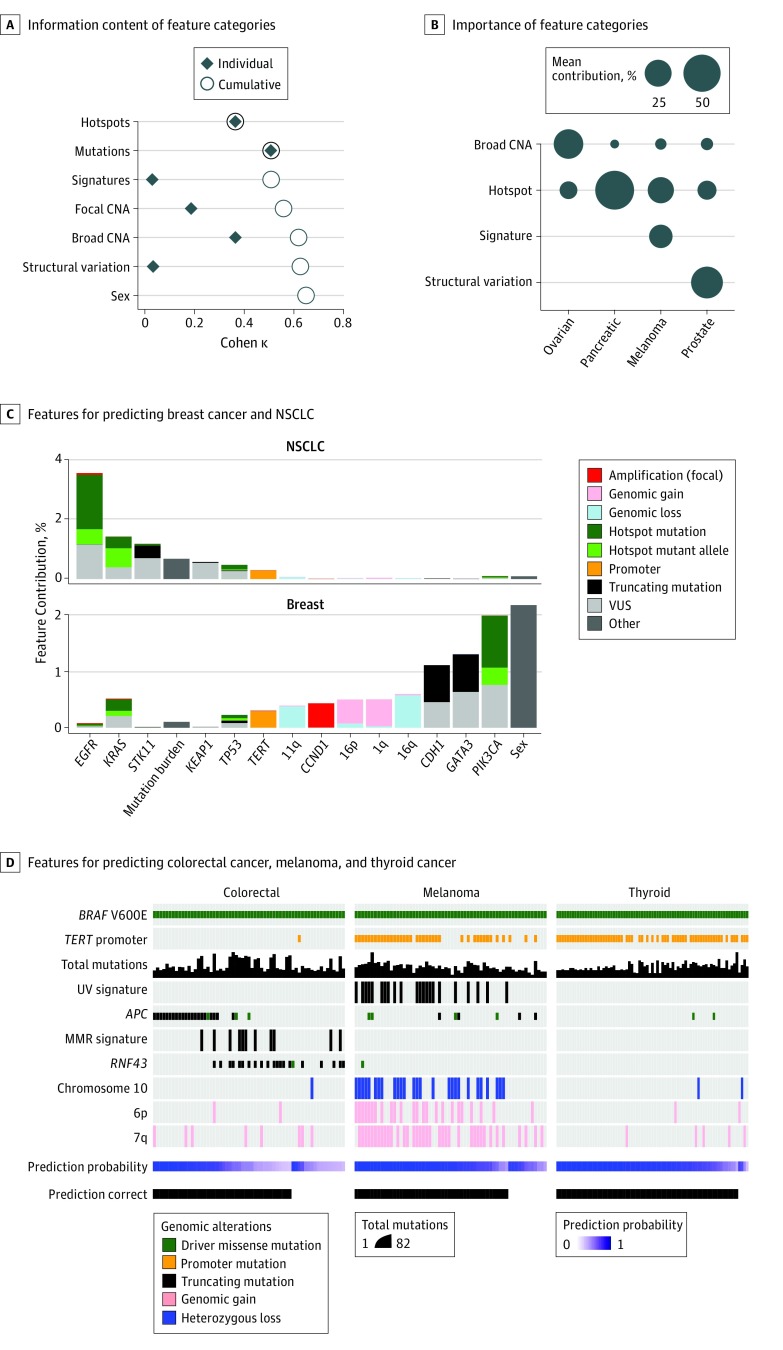
Given the diverse categories of genomic features that we incorporated into our classifier (eTable 6 in the Supplement), we sought to determine the relative importance of each molecular alteration type to the overall classification performance. Using the Cohen κ metric to represent overall accuracy, we found that somatic substitutions and indels had the highest predictive value, followed by chromosome arm-level (broad) copy number alterations (Figure 2A). Broad copy number alterations were especially informative for predicting tumor types with a low mutational burden and few other distinguishing features, such as prostate cancers lacking TMPRSS2-ERG fusions, neuroblastomas, germ cell tumors, and certain gastrointestinal cancers. Moreover, different feature categories contributed to prediction accuracy to differing degrees for individual cancer types, reinforcing the value of diverse feature categories for broad applicability and prediction accuracy (Figure 2B).
Figure 2. Predictive Power of Molecular Features and Feature Classes.
A, Relative information content of different feature categories as shown by the Cohen κ metric as a measure of overall accuracy. Black diamonds represent the accuracy of a classifier built for each feature category as indicated; open circles represent the accuracy on incrementally adding feature categories (top to bottom). Mutations encompass hotspots and non-hotspots. B, Relative importance of different feature categories in different cancer types. Circle size represents the mean contribution of the features in each category to accurate predictions in each cancer type. C, Selected individual features for predicting breast cancer and non–small cell lung cancer (NSCLC) in the study cohort and their relative contribution. Informative features driving correct predictions in all tumor types are shown in eFigure 1 in the Supplement. D, Different features contributing to tumor type predictions in BRAF V600E-mutant colorectal cancer, melanoma, and thyroid cancer, establishing the value of feature interactions to inform tumor type prediction in a cohort of patients that nevertheless share a common molecular alteration. CNA indicates copy number alterations; MMR, mismatch repair; VUS, variants of unknown significance.
Likewise, there was great breadth and variability among the specific features used to predict different cancer types (Figure 2C; eFigure 1 in the Supplement). Among all individual features, truncating APC mutation was the most informative overall owing to its high prevalence in and specificity for colorectal cancer. The TERT promoter mutations occurred at high frequency in multiple tumor types, but in others they were entirely absent, leading to strongly positive and negative associations for different lineages. In other instances, more subtle patterns were evident, such as the position of mutant alleles within genes, as for EGFR-mutant lung cancers and gliomas.17 The absence of common features also contributed to predictions of certain tumor types, such as KRAS mutations and breast cancer (Figure 2C). In summary, these results reveal the diversity of individual genomic features and feature categories that mediate tumor type predictions.
We next sought to determine whether such feature diversity and feature interaction could discriminate among different tumor types that nevertheless share a common molecular feature that is therefore not discriminatory. In BRAF V600E-mutant melanomas, colorectal, and thyroid cancers, where response rates to RAF inhibitor therapies vary, the classifier correctly predicted the tissue of origin in 162 of 195 cases (83.1%). Despite the presence of BRAF V600E in all cases, high confidence predictions were aided by distinct, co-occurring mutations and genomic features, such as TERT promoter mutations in melanoma and thyroid cancer, APC mutations and microsatellite instability in colorectal cancer, and UV-associated signatures in melanoma (Figure 2D). Misclassifications were associated with either low tumor purity or rare atypical genomic profiles (eg, melanomas with APC-truncating mutations). These results highlight the power of incorporating multiple diverse categories of molecular aberrations to guide challenging cancer type classifications when they share individual alterations.
Application to Cell-Free DNA
While this algorithmic approach was established on training data from tissue biopsies of solid tumors, the advent of noninvasive molecular profiling of plasma circulating tumor DNA (ctDNA) raises the possibility of inferring a suggested diagnosis in patients receiving cancer screening or with inaccessible disease. We therefore tested the predictive power of our classifier in 2 independent cohorts: 19 patients with genitourinary cancers and MSK-IMPACT sequencing of ctDNA and a previously published set of 41 patients with metastatic breast or prostate cancer and whole-exome sequencing of ctDNA.18 We correctly predicted the tumor type from MSK-IMPACT in 12 of 19 patients (63.2%) with prostate, bladder, and testicular cancer from among the 22 cancer types included in our classifier, including 8 of 8 predictions with a probability greater than 85%. Only 1 of 10 predictions with probability greater than 75% was inaccurate; a prostate cancer with a single missense mutation in VHL was incorrectly predicted as renal cell carcinoma. We also correctly predicted the tumor type from whole-exome sequencing in 23 of 27 patients (85.2%) with breast cancer and in 10 of 14 patients (71.4%) with prostate cancer, suggesting the general applicability of our classifier to multiple sequencing platforms as well as its suitability for diverse specimen types, such as ctDNA.
Application to Challenging Clinical Scenarios
Given the predictive power of our classifier, we sought to evaluate real-time, molecularly guided classifications in multiple challenging clinical scenarios. One unmet clinical need for such diagnostic resolution is the inference of the tissue of origin for cancers of unknown primary site.2 Refining diagnostic classification in this population can facilitate selection of potentially effective routine and investigational therapies. Using our classifier, we predicted a likely tissue of origin with probability greater than 50% in 95 of 141 patients (67.4%) (eFigure 4 in the Supplement). Although histopathologic assessment was unable to produce a definitive diagnosis for these patients, molecularly guided classifications frequently supported clinical suspicions; for instance, of 29 patients with predicted non–small cell lung cancer (>50%), 28 individuals (96.6%) had a self-reported history of smoking. In a separate example, emphasizing the need for tissue of origin classification even in an era of molecularly targeted therapy, we predicted a colorectal origin for one cancer of unknown primary site with 96% probability based on the presence of BRAF V600E and biallelic inactivating APC mutations (eFigure 5 in the Supplement). Because single-agent RAF inhibition has little activity in colon cancer, the inferred diagnosis suggested that combined BRAF, MEK, and EGFR therapy may be required to elicit a response.1,19,20
We also hypothesized that our classifier could help to resolve the diagnostic uncertainty that often arises between primary brain tumors and metastatic tumors to the central nervous system. Including both cohorts, we sequenced 299 brain metastases of solid tumors originating outside the central nervous system, including 133 non–small cell lung cancers, 56 breast cancers, 43 melanomas, and 67 other tumors. We accurately predicted the correct tumor type in 248 of 299 cases (82.9%). Of 51 incorrect predictions, only 2 were predicted as glioma. These results suggest the diagnostic value of our classifier for central nervous system tumors and its possible promise for noninvasive ctDNA profiling from cerebrospinal fluid.21
Another common and complex diagnostic challenge occurs when patients with a history of cancer present with a new tumor that may represent either a distant metastasis of their prior diagnosis or a second primary tumor. We therefore sought to assess the utility of molecularly guided classifications to clarify such complex diagnostic distinctions. In one representative case, a 67-year-old woman with a history of breast cancer presented with a lymph node lesion 3 years after her initial diagnosis. Histopathologic assessment suggested metastatic, poorly differentiated adenocarcinoma with micropapillary and apocrine cytologic characteristics, and immunohistochemistry showed weak to moderate estrogen receptor staining, collectively leading to a classification of estrogen receptor–positive breast cancer and a planned regimen of hormonal therapy (eFigure 6 in the Supplement). However, concurrent clinical sequencing revealed a high mutational burden, including KRAS G12C and other mutations, producing a high-confidence classification of non–small cell lung cancer (99%). These computational findings, acquired in real time, prompted additional lung cancer–specific immunohistochemistry, leading to a revised diagnosis of metastatic lung adenocarcinoma. To reaffirm the patient’s initial diagnosis, we subsequently obtained and sequenced the original primary breast tumor and identified no shared mutations, a somatic GATA3 truncating mutation, and a predicted classification of breast cancer (99%). The resulting change of diagnosis to metastatic lung cancer prompted a change in the treatment plan from hormonal therapy to chemotherapy for this patient.
Two cancers in a single patient may occasionally share mechanisms of pathogenesis that further complicate the distinction between metastatic progression and independent primary tumors. In a representative case, a 77-year-old woman was referred to our center with lesions in the breast and bladder and a diagnosis of metastatic breast lobular carcinoma (eFigure 6 in the Supplement). Clinical sequencing of the bladder lesion revealed 22 somatic mutations, including in the TERT promoter, CDH1, and RB1, and an APOBEC-associated mutational signature, producing a prediction of bladder cancer (74%). This prediction prompted subsequent histopathologic analysis that confirmed a diagnosis of plasmacytoid bladder cancer with corresponding loss of E-cadherin. Loss-of-function mutations in CDH1, while not generally predictive of bladder cancer (occurring more often in lobular breast and diffuse gastric cancers), are the defining feature of plasmacytoid bladder tumors.22 We subsequently performed MSK-IMPACT sequencing on the breast biopsy, which revealed 10 independent somatic mutations, including a different CDH1 mutation (X765_splice), that together were predictive of breast cancer (92%). The realization that the bladder lesion was a synchronous primary tumor rather than a clonally related metastasis led to consideration of surgical intervention as well as genetic testing for a cancer-predisposing germline mutation in CDH1. The diagnosis of bladder cancer also ultimately facilitated on-label treatment with the immune checkpoint inhibitor nivolumab, to which the patient responded. Taken together, these representative clinical cases suggest how genome-directed diagnosis may provide orthogonal diagnostic resolution that, when integrated with conventional pathologic testing, can lead to different therapeutic modalities, including surgery, hormonal therapy, chemotherapy, immunotherapy, and targeted therapy.
Discussion
We have developed and deployed a systematic computational approach for molecularly guided prediction of the site of origin of tumors based on targeted DNA sequencing. Although tumor sequencing is rapidly being adopted as a routine test in clinical cancer care, its use thus far has been limited to driving new enrollments onto clinical trials and for the identification of biomarkers of treatment response and resistance.13,14,23,24,25,26,27,28 Herein, our findings suggest the potential utility of such sequencing to inform cancer diagnosis as an adjunct to conventional histopathologic assessment. In our approach, we incorporated multifaceted molecular alteration types into a probabilistic prediction and tested its accuracy for identifying therapeutically significant cancer type differences under challenging diagnostic circumstances.
The results of this study have possible wide-ranging clinical implications. Genome-directed diagnosis, as typified by the representative cases presented herein, may be able to alter patient eligibility for various clinical modalities. As liquid biopsy is increasingly used as a screening tool for cancer recurrence and new cancers, our approach possibly can inform the site of origin when ctDNA is detected. There are also many ways in which predictions may be used clinically, especially in light of our development of probability estimates on individual predictions. In cases in which traditional diagnosis is ambiguous or challenging, computational predictions from genomic data apparently can exclude possibilities even if the predictions are not definitive. In other cases, a high-confidence prediction that disagrees with the defined or suspected diagnosis can prompt pathologic and clinical reevaluation, allowing additional testing that may help to support an alternative diagnosis. While messenger RNA–based tissue classification has been used extensively to predict the site of origin for cancers of unknown primary site, an advantage of our approach is its ability to enumerate the discrete genomic features aiding individual predictions, thereby providing pathologists and oncologists an opportunity to rationally interpret discordant results.
Limitations
This study has limitations. Key limitations of our approach will require continued improvement and investigation to expand the clinical utility of such genome-directed diagnosis. First, we initially limited our classifier to training data from only 22 common cancer types. As our prospective cohort grows, we will have the opportunity to include rare cancer types as well as molecularly or histologically distinct cancer subtypes. Second, our work is based on targeted clinical sequencing of established cancer-associated genes but can be extended to unbiased whole-exome or genome sequencing as these methods are introduced into clinical practice. The accuracy of our classifier, trained on MSK-IMPACT data, for predicting tumor type from ctDNA whole-exome sequencing data suggests broad applicability to other panels with shared genomic targets. Third, although the application to cancers of unknown primary site represents one of the greatest clinical opportunities, the absence of a precise histopathologic diagnosis in such cases makes it difficult to benchmark the accuracy of predictions and alter therapeutic decisions. Ultimately, the true clinical importance and frequency with which this approach resolves challenging diagnostic scenarios, alters established diagnoses (via prompting of additional pathologic assessment), and affects therapeutic modalities will require further prospective clinical investigation. Such studies could focus on a broader assessment of the performance and utility of noninvasive, molecularly guided diagnosis from ctDNA.
Conclusions
Overall, as our understanding improves of how lineage is a factor associated with response to the newest generation of therapies in cancer, this systematic approach to molecularly guided diagnosis coupled with conventional clinical histories, histopathologic assessment, and imaging may improve diagnostic and treatment decisions. Our results appear to illustrate the emerging and powerful role of artificial intelligence in medicine for clinical decision support.29,30
eMethods. Detailed Methods
eFigure 1. Most Informative Features for Each Tumor Type
eFigure 2. Calibration of Probability Scores
eFigure 3. Number of Correct and Total Predictions Made Within Each Probability Range
eFigure 4. Classification Performance for Cancers of Unknown Primary
eFigure 5. Prediction of Colorectal Cancer for a Cancer of Unknown Primary
eFigure 6. Molecular Re-Diagnosis Changes Therapeutic Intervention
eReferences
eTable 1. Distinct Tumor Types Considered for Classification
eTable 2. Clinical and Technical Characteristics of the Training and Validation Cohorts
eTable 3. Tumor Type Predictions From Cross Validation for All Tumors Included in the Training Cohort
eTable 4. Sensitivity and Specificity of Predictions for Each Tumor Type
eTable 5. Prediction Accuracy for Detailed Histological Subtypes
eTable 6. Individual Molecular Features Selected by the Classifier
References
- 1.Hyman DM, Puzanov I, Subbiah V, et al. . Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726-736. doi: 10.1056/NEJMoa1502309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varghese AM, Arora A, Capanu M, et al. . Clinical and molecular characterization of patients with cancer of unknown primary in the modern era. Ann Oncol. 2017;28(12):3015-3021. doi: 10.1093/annonc/mdx545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golub TR, Slonim DK, Tamayo P, et al. . Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531-537. doi: 10.1126/science.286.5439.531 [DOI] [PubMed] [Google Scholar]
- 4.Greco FA, Spigel DR, Yardley DA, Erlander MG, Ma XJ, Hainsworth JD. Molecular profiling in unknown primary cancer: accuracy of tissue of origin prediction. Oncologist. 2010;15(5):500-506. doi: 10.1634/theoncologist.2009-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marquard AM, Birkbak NJ, Thomas CE, et al. . TumorTracer: a method to identify the tissue of origin from the somatic mutations of a tumor specimen. BMC Med Genomics. 2015;8:58. doi: 10.1186/s12920-015-0130-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran S, Martínez-Cardús A, Sayols S, et al. . Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17(10):1386-1395. doi: 10.1016/S1470-2045(16)30297-2 [DOI] [PubMed] [Google Scholar]
- 7.Soh KP, Szczurek E, Sakoparnig T, Beerenwinkel N. Predicting cancer type from tumour DNA signatures. Genome Med. 2017;9(1):104. doi: 10.1186/s13073-017-0493-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferracin M, Pedriali M, Veronese A, et al. . MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43-53. doi: 10.1002/path.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang S, Li Q, Chen Q, et al. . CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 2017;18(1):53. doi: 10.1186/s13059-017-1191-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao X, Luo H, Krawczyk M, et al. . DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A. 2017;114(28):7414-7419. doi: 10.1073/pnas.1703577114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1-2):57-68. doi: 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman JS, Asthana S, Cade L, et al. . Clinical sequencing contributes to a BRCA-associated cancer rediagnosis that guides an effective therapeutic course. J Natl Compr Canc Netw. 2015;13(7):835-845. doi: 10.6004/jnccn.2015.0101 [DOI] [PubMed] [Google Scholar]
- 13.Zehir A, Benayed R, Shah RH, et al. . Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703-713. doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A, et al. . Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251-264. doi: 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breiman L. Random forests. Mach Learn. 2001;45:5-32. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 16.Bella A, Ferri C, Hernández-Orallo J, Ramírez-Quintana MJ. On the effect of calibration in classifier combination. Appl Intell. 2013;38(4):566-585. doi: 10.1007/s10489-012-0388-2 [DOI] [Google Scholar]
- 17.Chang MT, Asthana S, Gao SP, et al. . Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34(2):155-163. doi: 10.1038/nbt.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adalsteinsson VA, Ha G, Freeman SS, et al. . Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. doi: 10.1038/s41467-017-00965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flaherty KT, Infante JR, Daud A, et al. . Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694-1703. doi: 10.1056/NEJMoa1210093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long GV, Stroyakovskiy D, Gogas H, et al. . Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877-1888. doi: 10.1056/NEJMoa1406037 [DOI] [PubMed] [Google Scholar]
- 21.Pentsova EI, Shah RH, Tang J, et al. . evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34(20):2404-2415. doi: 10.1200/JCO.2016.66.6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Ahmadie HA, Iyer G, Lee BH, et al. . Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat Genet. 2016;48(4):356-358. doi: 10.1038/ng.3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltran H, Eng K, Mosquera JM, et al. . Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1(4):466-474. doi: 10.1001/jamaoncol.2015.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sholl LM, Do K, Shivdasani P, et al. . Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19):e87062. doi: 10.1172/jci.insight.87062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirshfield KM, Tolkunov D, Zhong H, et al. . Clinical actionability of comprehensive genomic profiling for management of rare or refractory cancers. Oncologist. 2016;21(11):1315-1325. doi: 10.1634/theoncologist.2016-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frampton GM, Fichtenholtz A, Otto GA, et al. . Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. doi: 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roychowdhury S, Iyer MK, Robinson DR, et al. . Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3(111):111ra121. doi: 10.1126/scitranslmed.3003161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh RR, Patel KP, Routbort MJ, et al. . Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15(5):607-622. doi: 10.1016/j.jmoldx.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 29.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44-56. doi: 10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 30.Shortliffe EH, Sepúlveda MJ. Clinical decision support in the era of artificial intelligence. JAMA. 2018;320(21):2199-2200. doi: 10.1001/jama.2018.17163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Detailed Methods
eFigure 1. Most Informative Features for Each Tumor Type
eFigure 2. Calibration of Probability Scores
eFigure 3. Number of Correct and Total Predictions Made Within Each Probability Range
eFigure 4. Classification Performance for Cancers of Unknown Primary
eFigure 5. Prediction of Colorectal Cancer for a Cancer of Unknown Primary
eFigure 6. Molecular Re-Diagnosis Changes Therapeutic Intervention
eReferences
eTable 1. Distinct Tumor Types Considered for Classification
eTable 2. Clinical and Technical Characteristics of the Training and Validation Cohorts
eTable 3. Tumor Type Predictions From Cross Validation for All Tumors Included in the Training Cohort
eTable 4. Sensitivity and Specificity of Predictions for Each Tumor Type
eTable 5. Prediction Accuracy for Detailed Histological Subtypes
eTable 6. Individual Molecular Features Selected by the Classifier