Abstract
That metabolic syndrome (MetS) is associated with age‐related cognitive decline is well established. The neurobiological changes underlying these cognitive deficits, however, are not well understood. The goal of this study was to determine whether MetS is associated with regional differences in gray‐matter volume (GMV) using a cross‐sectional, between‐group contrast design in a large, ethnically homogenous sample. T1‐weighted MRIs were sampled from the genetics of brain structure (GOBS) data archive for 208 Mexican‐American participants: 104 participants met or exceeded standard criteria for MetS and 104 participants were age‐ and sex‐matched metabolically healthy controls. Participants ranged in age from 18 to 74 years (37.3 ± 13.2 years, 56.7% female). Images were analyzed in a whole‐brain, voxel‐wise manner using voxel‐based morphometry (VBM). Three contrast analyses were performed, a whole sample analysis of all 208 participants, and two post hoc half‐sample analyses split by age along the median (35.5 years). Significant associations between MetS and decreased GMV were observed in multiple, spatially discrete brain regions including the posterior cerebellum, brainstem, orbitofrontal cortex, bilateral caudate nuclei, right parahippocampus, right amygdala, right insula, lingual gyrus, and right superior temporal gyrus. Age, as shown in the post hoc analyses, was demonstrated to be a significant covariate. A further functional interpretation of the structures exhibiting lower GMV in MetS reflected a significant involvement in reward perception, emotional valence, and reasoning. Additional studies are needed to characterize the influence of MetS's individual clinical components on brain structure and to explore the bidirectional association between GMV and MetS.
Keywords: genetics of brain structure, GOBS, gray matter volume, hypercholesterolemia, hyperglycemia, hypertension, hypertriglyceridemia, insulin resistance, insulin resistance syndrome, metabolic syndrome, MetS, Mexican‐American, neuroimaging, obesity, T2DM, Type II diabetes mellitus, VBM, voxel‐based morphometry, waist circumference
1. INTRODUCTION
Metabolic syndrome (MetS), also commonly known as the insulin resistance syndrome, is a cluster of physiological abnormalities associated with the development of cardiovascular disease (CVD), Type 2 diabetes mellitus (T2DM) and early death (Reaven, 1988). It typically comprises a combination of the following: central obesity measured as waist circumference (WC), elevated triglycerides (TG), reduced levels of high‐density lipoprotein (HDL) cholesterol, increased fasting plasma glucose (FPG), and elevated blood pressure (BP; Grundy et al., 2005). Epidemiological data suggest that 34.7% of Americans suffer from MetS with a high prevalence (36.8%) in the Hispanic‐American population (Aguilar, Bhuket, Torres, Liu, & Wong, 2015). Age also plays a role in the prevalence of MetS, such that more than 50% of all adults over the age of 60 are afflicted. In addition to T2DM and CVD, MetS is associated with nonalcoholic fatty liver disease, polycystic ovarian syndrome, several types of cancer (e.g., colon cancer and breast cancer), arthritis, and fibromyalgia (Byrne & Wild, 2011). Most striking, T2DM, and MetS have shown to be associated with dementia, with Mexican Americans exhibiting greater risk than those of European ancestry (Haan et al., 2003). Indeed, individuals with MetS commonly display cognitive impairments in learning and memory, executive function, and generalized processing speed (Cavalieri et al., 2010; Yates, Sweat, Yau, Turchiano, & Convit, 2012). The genetic underpinnings of this observation remain unknown, but are suggested to be partly due to differences in insulin metabolism associated with a genetic admixture (Lee, Zabolotny, Huang, Lee, & Kim, 2016). Additionally, cognitive decline following longstanding MetS has been linked to the development of vascular dementia and Alzheimer's disease (AD; Exalto, Whitmer, Kappele, & Biessels, 2012; Frisardi et al., 2010; Vanhanen et al., 2006).
Neurodegenerative diseases are found to correlate with comorbidities of MetS, such as obesity. These include frontotemporal dementia, cerebrovascular disease, and AD, among others (Lee & Mattson, 2014). Although there are numerous mechanistic hypotheses as to how these degenerative changes occur, several have gained particular notoriety. One such hypothesis suggests that dysregulated insulin receptor binding/activation plays a role in neuronal atrophy of the reward networks (nucleus accumbens, amygdala, VTA, striatum) and learning and memory networks (hippocampus, OFC, temporal lobe; Byrne & Wild, 2011). Impaired insulin receptor binding in the hippocampus, a structure known to express high levels of insulin receptors, is presumed to influence the development of AD (De Felice, Lourenco, & Ferreira, 2014). The hypothesis of AD‐related central nervous system (CNS) insulin resistance, coupled with observations that T2DM is associated with an increased risk of developing AD (Ott et al., 1996; Stranahan, 2015), has led researchers ask whether peripheral metabolic biomarkers could be used to predict neurological decline (Chatterjee & Mudher, 2018; Pruzin, Nelson, Abner, & Arvanitakis, 2018). Besides brain insulin resistance, other mechanisms such as neuroinflammation, oxidative stress, and abnormal brain lipid metabolism have been proposed as pathophysiological contributors to neurocognitive decline in the MetS brain (Yates et al., 2012).
Structural abnormalities in T2DM patients were first observed in a cohort of genetically homogenous Dutch subjects and found that individuals with T2DM had significantly decreased hippocampal and amygdalar gray‐matter volumes (GMVs) compared to normal controls (Den Heijer et al., 2003). Their conclusions gave credence to an earlier study conducted in the Netherlands (The Rotterdam Study), which found a significant correlation between insulin‐dependent T2DM and AD (Ott et al., 1996).
Neuroimaging is an effective method for investigating the neurobiological correlates of cognitive decline in a human population with MetS. Comorbidities of MetS, namely obesity, insulin resistance, and T2DM, have been studied extensively using functional imaging (Byrne & Wild, 2011). Early positron emission tomography (PET) studies comparing insulin resistant subjects with normal controls, reported decreased gray matter glucose uptake in the dopaminergic reward circuit (amygdalae, hippocampi, and orbitofrontal cortex) and increased uptake in the striatum (nucleus accumbens, caudate, and putamen), insula and anterior cingulate (Anthony et al., 2006). Task‐activation fMRI studies (food picture, smell, and taste paradigms) have also shown that glucose infusion is associated with discrete brain activation patterns and prandial satiety, correlating with the brain's reward network (Huerta, Sarkar, Duong, Laird, & Fox, 2014; Michaud, Vainik, Garcia‐Garcia, & Dagher, 2017).
Voxel‐based morphometry (VBM), is a widely‐accepted research technique employed to identify subtle, disease‐related structural changes that cannot be easily inferred through region‐specific or global gray matter volumetric analyses. VBM achieves this via group‐averaging and registration to a standard brain space comparing gray matter densities between cases and controls. This method is optimal for identifying disease‐specific atrophy patterns computed in a univariate, voxel‐wise manner (Ashburner & Friston, 2000). In the literature, we know of only one other study that has looked at cortical thickness and subcortical volume changes (using ANCOVA instead of VBM with n = 86) explicitly in individuals with MetS (Song et al., 2014). However, only five VBM studies (with n ranging from 32 to 54) have investigated the neuroanatomical effects of T2DM (Wu, Lin, Zhang, & Wu, 2017). Other VBM studies have emphasized the GMV effects of MetS's comorbidities, such as the combined effects of T2DM and hypertension (Tchistiakova et al., 2014) and obesity (Masouleh et al., 2016). Allostatic load, the accumulated multisystem physiological response to chronic stress that strongly correlates with MetS has also been studied using VBM as a predictor of stroke, diabetes, and neuroanatomical integrity (Zsoldos et al., 2018).
For this retrospective study, we utilized previously acquired neuroimaging data from a homogenous Mexican‐American cohort comprising an extended‐pedigree imaging dataset known as the Genetics of Brain Structure (GOBS) image archive of San Antonio, TX. In this dataset, participants were recruited at random from the San Antonio community as a measure to minimize selection bias. This archive presented a unique opportunity to study the neurobiological effects of MetS because of the population's increased propensity, both biological and environmental, to develop MetS and its comorbidities. The sample is large enough to conduct a statistically powerful analysis, and includes a wide range of relevant data.
Participants were divided into two groups, randomly matched MetS and metabolically healthy controls (hereafter also referred to as “controls”), who differed only in metabolic status while controlling for age and sex. VBM was subsequently used to identify GMV differences between groups. After initial analysis, the large group was further subdivided in two post hoc analyses of “young” and “old” half samples to assess age‐related effects. Lastly, BrainMap®, an imaging archive designed for meta‐analysis and meta‐analytic data interpretation, was used to interpret behavioral, paradigm class, and disease covariates from our results (Lancaster et al., 2012).
We hypothesized: (a) that a group contrast VBM analysis will reveal discrete MetS‐specific GMV differences; (b) that the patterns observed would recapitulate regions implicated in AD pathology (per the CNS insulin resistance hypothesis); and, (c) that applying meta‐analytic interpretive tools (http://BrainMap.org) to the VBM‐derived regional deficits would confirm a cognitive signature corresponding to cognitive deficits previously reported in MetS.
2. MATERIALS AND METHODS
2.1. Participants
The goal of the GOBS dataset is to localize, identify, and characterize genes/quantitative trait loci associated with variations in brain structure and function (Winkler et al., 2010). The GOBS dataset has been acquired in three blocks of acquisitions, corresponding to the original grant and two renewals. In the original block of acquisitions, the emphasis was almost entirely on imaging and cognitive/behavioral phenotypes with BMI and health history being the only phenotypes referable to metabolic syndrome. In the second block of acquisitions, blood chemistries (including fasting plasma glucose), a lipid panel, and waist circumference (WC) were added. In the third block of acquisitions, BP was added. Participants in the present analysis were drawn only from acquisition Blocks 2 and 3. In all three blocks of acquisition, participants were drawn from the same extended‐pedigree, Mexican‐American families. At the time this study was conducted, a total of 1,911 individuals were enlisted in the GOBS dataset.
In acquisition Blocks 2 and 3, neuroanatomic, neurocognitive, and biometric phenotypes were obtained on each participant. Participants were subjected to the standardized Composite International Diagnostic Interview (CIDI; Kessler, Andrews, Mroczek, Ustun, & Wittchen, 1998) and the Mini‐International Neuropsychiatric Interview (MINI‐Plus) for DSM‐IV and ICD‐10 psychiatric disorders (Sheehan et al., 1998). Blood samples were collected after a 12‐hr fast and processed in order to obtain blood chemistry and blood lipid data, such as triglycerides, cholesterol, and fasting plasma glucose.
Acquiring BP values did not become standard protocol until acquisition Block 3. This left ~70% of the eligible participants from the GOBS cohort without measured BP. However, from the 835 participants with all pertinent imaging and lipid data (Figure 1), a total of 356 (or 42.6%) had available BP data. Excluding participants with no BP values, but who nonetheless had all other pertinent data, would have left us with a small and underpowered cohort. We overcame this obstacle by diagnosing our data as missing at random (MAR), indicating that there is a systematic relationship between observed data (the 42.6% of BP data available) and missing values (the 57.4% of missing BP data). BP can therefore be imputed from available observed BP data and other observed variables known to correlate with BP (e.g., age, sex, height, weight, WC, fasting plasma glucose, triglycerides, blood urea nitrogen, creatinine, sodium, potassium, chloride, calcium, smoking status, total cholesterol, high density, low density, and very low‐density lipoprotein cholesterol, and previous diagnosis of hypertension; Little & Rubin, 2002). Missing BP values were thus imputed using a well‐validated model‐based full information maximum likelihood estimation from the aforementioned observed variables (Enders, 2010).
Figure 1.
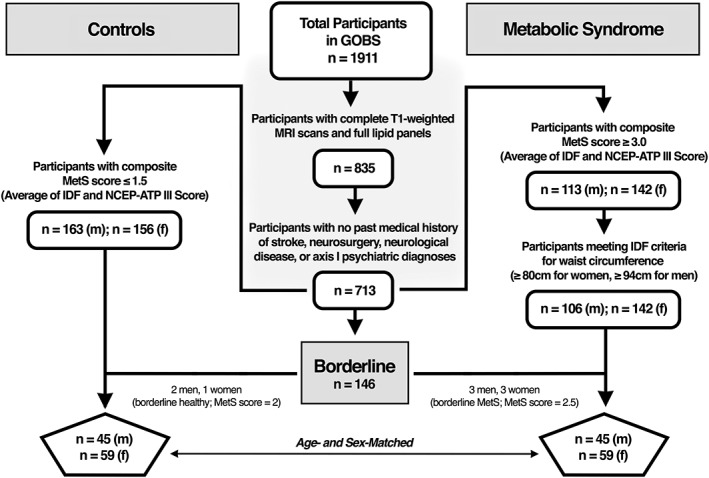
Participant selection pipeline from the original GOBS dataset applying MetS criteria scores based on a composite of the International Diabetes Federation (IDF) and National Cholesterol Education Program‐Adult Treatment Plan III (NCEP‐ATP III). Criteria for metabolically healthy control group was determined by including participants with a composite MetS score of ≤1.5. Criteria for MetS case group was determined by including participants fulfilling the WC criteria of the IDF and achieving a MetS score of ≥3.0. A few borderline individuals (3 controls and 6 MetS) were added to provide age‐ and sex‐matched symmetry in the final analysis
As a cross‐check we evaluated whether the MAR assumption was tenable and performed sensitivity analysis under the missing not at random (MNAR) assumption. We employed the fully conditional specification for multiple imputation to impute BP values for participants in the study group based on observed values of participants not included in the study group (Ratitch & O'Kelly, 2011). Using this test of model sensitivity, we were able to discern whether differences existed between groups on BP values assuming the MAR versus MNAR assumption. The results of the sensitivity analysis yielded the same conclusions as those observed under MAR.
2.2. Image acquisition
MRI acquisition was carried out on a 3T Siemens Tim MRI scanner at the University of Texas Health Science Center at San Antonio's Research Imaging Institute. An 8‐element high‐resolution phase array head coil equipped with foam padding was employed to comfortably restrict head motion. A standard localizer image was obtained for each participant for determining head placement, followed by a standard shim sequence. Seven high quality T1‐weighted 3D structural images were collected per participant via a TurboFLASH sequence with an adiabatic inversion recovery pulse (TE = 3.04 ms, TR = 2000 ms, TI = 795 ms, flip angle = 8°, NEX = 6) optimized to achieve a gray/white contrast of ~25% with signal to noise ratio of 25. Each image contained 0.8 mm3 isotropic voxels and a 20 cm field of view. Scan time per participant totaled 60 min. Each of the seven MPRAGE T1‐weighted images was motion corrected and all were subsequently averaged to generate a single high‐resolution anatomical image per participant (Kochunov et al., 2006).
2.3. Study sample
Of the 1,911 total participants in the GOBS cohort, we excluded 519 participants who were lacking successfully acquired T1‐weighted MRI scans, and 557 participants from the first acquisition phase who were lacking full biometric data such as blood lipids and WC necessary to establish a MetS diagnosis. Upon reviewing data obtained from CIDI and MINI‐Plus evaluations, we then excluded 122 participants with past medical histories of stroke, neurosurgery, neurological diseases, or Axis I psychiatric diagnoses (e.g., schizophrenia, bipolar disorder, major depressive disorder, substance use disorder, and so on). Our final sample of neurologically and psychiatrically healthy individuals who met basic imaging, psychiatric, biometric, and metabolic criteria totaled 713 participants.
We applied both the National Cholesterol Education Program Adult Treatment Plan III (NCEP‐ATP III) and the International Diabetes Federation (IDF) criteria (Figure 1) to the selection of metabolically healthy and MetS participants (Eckel & Cornier, 2014). The NCEP‐ATP III defines MetS as patients meeting at least three of the following five criteria: 1. central obesity measured by WC (≥102 cm in men, ≥ 88 cm in women), 2. raised TGs (≥ 150 mg/dL), 3. reduced HDL cholesterol (<40 mg/dL in men, < 50 mg/dL in women), 4. raised FPG (> 110 mg/dL), and 5. elevated BP (≥ 130 mmHg systolic or ≥ 85 mmHg diastolic) and/or diagnosis of HTN (Eckel & Cornier, 2014). Similarly, the IDF defines MetS as a disease in which patients must exhibit central obesity (WC of ≥94 cm for men, ≥ 80 cm for women), along with at least two of the following four criteria: 1. raised TGs (>150 mg/dL) and/or taking medication for hypertriglyceridemia, 2. reduced HDL cholesterol (<40 mg/dL in men, < 50 mg/dL in women) and/or taking medication for hypercholesterolemia, 3. raised BP (>130 mmHg systolic or >85 mmHg diastolic) and/or taking medication for hypertension, and 4. raised FPG (>100 mg/dL) and or having a diagnosis of T2DM (Grundy et al., 2005).
A score of 0–5 was generated for each of the 713 eligible participants based on how many components (WC, TG, HDL, FPG, and BP) met the MetS criteria for NCEP‐ATP III and IDF respectively (Tables 1 and 2). For example, a male participant with a WC of 98 cm would receive a score of 1 for WC under the IDF criteria, but receive a score of 0 for WC under the NCEP‐ATP III criteria. Both the IDF and NCEP‐ATP III scores were then averaged to give each participant a single composite MetS score based on both criteria. A score of 3 or greater would be considered indicative of MetS under the standards for both criteria.
Table 1.
Demographics, MetS components and neuroanatomical characteristics of participants
| [Female] | Total control (SD) n = 104 [59] | Total MetS (SD) n = 104 [59] | p | Young control (SD) n = 52 [23] | Young MetS (SD) n = 52 [23] | p | Old control (SD) n = 52 [22] | Old MetS (SD) n = 52 [22] | p |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 37.3 (13.2) | 37.3 (13.2) | .996 | 26.4 (4.6) | 26.5 (4.6) | .932 | 48.1 (9.6) | 48.1 (9.8) | .976 |
| Education (years) | 12.2 (2.7) | 12.3 (2.6) | .872 | 12.7 (1.8) | 12.4 (2.0) | .486 | 11.7 (3.4) | 12.1 (3.2) | .550 |
| MetS components | |||||||||
| Waist circumference (cm) | 84.1 (11.2) | 110.0 (11.2) | <.001* | 80.3 (9.1) | 110.4 (12.1) | <.001* | 87.9 (11.9) | 109.6 (10.4) | <.001* |
| Triglycerides (mg/dL)a | 87.8 (31.5) | 223.2 (95.6) | <.001* | 85.5 (35.1) | 220.0 (87.7) | <.001* | 90.0 (27.7) | 226.5 (103.8) | <.001* |
| HDL cholesterol (mg/dL) | 61.8 (15.4) | 38.8 (8.8) | <.001* | 60.1 (14.5) | 38.3 (8.7) | <.001* | 63.5 (16.2) | 39.3 (9.0) | <.001* |
| Fasting plasma glucose (mg/dL) | 86.3 (7.5) | 119.8 (55.1) | <.001* | 86.3 (6.8) | 104.7 (49.2) | .010* | 86.3 (8.3) | 134.9 (56.9) | <.001* |
| Systolic blood pressure (mmHg)b | 113.4 (15.2) | 126.7 (17.3) | <.001* | 113.1 (15.17) | 123.8 (14.1) | <.001* | 113.8 (15.4) | 129.5 (19.8) | <.001* |
| Diastolic blood pressure (mmHg)b | 69.3 (10.9) | 77.4 (11.3) | <.001* | 68.9 (10.6) | 76.7 (10.3) | <.001* | 69.8 (11.2) | 78.0 (12.4) | .001* |
| IDF score | 0.57 (0.63) | 4.11 (0.72) | <.001* | 0.37 (0.60) | 3.75 (0.65) | <.001* | 0.77 (0.61) | 4.46 (0.61) | <.001* |
| NCEP‐ATP III score | 0.24 (0.49) | 3.60 (0.90) | <.001* | 0.15 (0.41) | 3.40 (0.72) | <.001* | 0.33 (0.55) | 3.79 (1.02) | <.001* |
| Composite MetS score | 0.40 (0.51) | 3.85 (0.74) | <.001* | 0.23 (0.46) | 3.58 (0.64) | <.001* | 0.55 (0.53) | 4.13 (0.74) | <.001* |
| Other lipid measures of interest | |||||||||
| Total cholesterol (mg/dl) | 174.4 (31.7) | 202.5 (45.3) | <.001* | 167.9 (30.8) | 197.0 (42.1) | <.001* | 180.8 (31.6) | 208.0 (48.0) | .001* |
| LDL cholesterol (mg/dL) | 95.0 (28.3) | 119.2 (40.3) | <.001* | 90.7 (28.0) | 114.5 (37.8) | <.001* | 99.3 (28.2) | 123.9 (42.6) | .001* |
| BMI (kg/m2) | 24.3 (3.8) | 34.5 (5.5) | <.001* | 23.3 (3.5) | 34.7 (5.6) | <.001* | 25.4 (3.9) | 34.2 (5.5) | <.001* |
| Whole brain gray and white matter measures derived from VBM | |||||||||
| Average gray matter density | 0.482 (0.028) | 0.474 (0.032) | .072 | 0.495 (0.018) | 0.493 (0.019) | .445 | 0.468 (0.030) | 0.456 (0.032) | .045* |
| Average white matter density | 0.378 (0.013) | 0.373 (0.014) | .008* | 0.370 (0.010) | 0.365 (0.016) | .413 | 0.372 (0.007) | 0.374 (0.019) | .012* |
Abbreviations: BMI, body mass index; HDL, high density lipoprotein cholesterol; IDF, International Diabetes Federation; LDL, low density lipoprotein; NCEP‐ATP III, National Cholesterol Education Program Adult Treatment Plan III; SD, standard deviation; VBM, voxel‐based morphometry. *Two‐sample t‐test of group differences reported as significant p‐value ≤.05.
Excludes outliers with TG values >500 mg/dL.
BP values obtained were generated in part by full‐information maximum likelihood statistical modeling.
Table 2.
Percentage of participants meeting MetS criteria by group
| Metabolic syndrome components | Total control | Total MetS | Young control | Young MetS | Old control | Old MetS |
|---|---|---|---|---|---|---|
|
International diabetes federation | ||||||
|
Waist circumference: ≥96 cm in men, ≥80 cm in women |
41.3% | 100% | 23.1% | 100% | 59.6% | 100% |
|
Triglycerides: ≥150 mg/dL and/or HTG Rx |
1.0% | 93.3% | 1.9% | 94.2% | 0% | 92.3% |
|
HDL cholesterol: <40 mg/dL in men, <50 mg/dL in women and/or HCL Rx |
3.8% | 94.2% | 3.8% | 90.4% | 3.8% | 98.1% |
|
Fasting plasma glucose: >100 mg/dL and/or T2DM dx |
1.0% | 60.6% | 0% | 38.5% | 1.9% | 78.8% |
|
Blood pressure a: ≥130 mmHg systolic or ≥85 mmHg diastolic and/or HTN dx |
13.5% | 61.5% | 11.5% | 48.1% | 15.4% | 75.0% |
| Meet IDF criteria for MetS | 0% | 100% | 0% | 100% | 0% | 100% |
| National cholesterol education program adult treatment plan III | ||||||
|
Waist circumference: ≥102 cm in men, ≥88 cm in women |
9.6% | 92.3% | 1.9% | 90.4% | 17.3% | 94.2% |
|
Triglycerides: ≥150 mg/dL |
1.0% | 87.5% | 1.9% | 94.2% | 0% | 80.8% |
|
HDL cholesterol: <40 mg/dL in men, <50 mg/dL in women |
2.9% | 83.7% | 1.9% | 88.5% | 3.8% | 78.8% |
|
Fasting plasma glucose: >110 mg/dL |
0% | 33.7% | 0% | 14.8% | 0% | 51.9% |
|
Blood pressure a: ≥130 mmHg systolic or ≥ 85 mmHg diastolic and/or HTN dx |
13.5% | 61.5% | 11.5% | 48.1% | 15.4% | 75.0% |
| Meet NCEP‐ATP III criteria for MetS | 0% | 87.5% | 0% | 90.4% | 0% | 84.6% |
| Previously diagnosed comorbidities | ||||||
| Hypertriglyceridemia | 0% | 21.2% | 0% | 3.8% | 0% | 38.5% |
| Hypercholesterolemia | 1.9% | 33.7% | 1.9% | 11.5% | 1.9% | 55.8% |
| Type II diabetes mellitus | 0% | 31.7% | 0% | 13.5% | 0% | 50% |
| Hypertension | 3.8% | 35.6% | 0% | 17.3% | 7.7% | 53.8% |
Abbreviations: Dx, previously diagnosed; HCL, hypercholesterolemia; HDL, high density lipoprotein cholesterol; HTG, hypertriglyceridemia; HTN, hypertension; IDF, International Diabetes Federation; NCEP‐ATP III, National Cholesterol Education Program Adult Treatment Plan III; Rx, currently receiving treatment; T2DM, Type 2 diabetes mellitus.
BP values obtained were generated in part by full‐information maximum likelihood statistical modeling.
Metabolically healthy controls were selected from the remaining 713 eligible participants if they had a composite MetS score of 1.5 or lower (n = 319) and MetS participants were selected if they had a composite MetS score of 3 or higher (n = 255). From the latter group, we further excluded MetS subjects who did not meet the IDF criteria for WC (n = 7). We then employed an algorithm that optimally and randomly age‐ and sex‐matched the participants who met the study‐specific criteria for controls and for MetS. A total of 101 controls (n = 44 males, n = 57 females; plus 3 borderline) and 98 MetS participants (n = 42 males, n = 56 females; plus 6 borderline) were matched successfully (Figure 1). We included borderline individuals to optimally configure the group‐matching framework between age and sex that is required for a two‐group difference general linear model analysis. Borderline control individuals had a composite score of 2. They did not meet the ≤1.5 cut‐off, nor the IDF or NCEP‐ATP III criteria for MetS. Similarly, borderline MetS individuals had a composite score of 2.5, meeting the IDF criteria for MetS, but not for NCEP‐ATP III.
These criteria were derived in consultation with Dr. Ralph DeFronzo, director of the Texas Diabetes Institute and were considered the best accepted estimates to define two groups that are metabolically distinct as “healthy” and “MetS” (Miranda, DeFronzo, Califf, & Guyton, 2005). Particular emphasis was made on defining the MetS group as meeting at least the IDF criteria for WC. This emphasis is important because central obesity as measured by WC is an easily measurable biometric variable that closely correlates with insulin resistance (Simonson & Kendall, 2005).
2.4. VBM and univariate analysis
T1 structural images were acquired retrospectively from the GOBS dataset. Freesurfer was used for initial processing with autorecon1 (motion correction, nonuniform intensity normalization, Talairach transform computation, intensity normalization, and skull stripping; Fischl, 2012). Brain‐extracted data were then analyzed with FSL‐VBM (Douaud et al., 2007), an optimized VBM protocol carried out with FSL tools (FMRIB Software Library; Good et al., 2001; Smith et al., 2004). Structural images were gray matter‐segmented and nonlinearly registered to MNI‐152 standard space (Andersson, Jenkinson, & Smith, 2007). The resulting images were averaged, flipped along the x‐axis, and re‐averaged to create a left‐right symmetric, study‐specific gray matter template. All native gray matter images were nonlinearly registered to this study‐specific template and “modulated” to correct for local expansion (or contraction) due to the nonlinear component of the spatial transformation. The modulated gray matter images were smoothed with an isotropic Gaussian kernel (sigma = 3 mm).
To compare MetS versus metabolically healthy control group differences in GMV, a two‐group difference (two‐sampled unpaired t‐test) analysis using voxel‐wise general linear modeling (GLM) was applied in a control vs. MetS contrast analysis involving all participants (n = 208) with “years of education” as a nuisance covariate. Two separate post hoc contrast analyses were subsequently conducted along age‐defined half‐samples. We ran the GLM analysis with FSL's Randomize, a nonparametric permutation‐testing tool (10,000 permutations), that corrects for multiple comparisons across voxels. Significance was determined at the voxel level using a family‐wise error (FWE) correction (p < .05). In VBM, the null hypothesis states that there is no difference in GMV between the groups being studied. The null hypothesis is refuted when statistical maps are generated exhibiting voxels, often clustered together, that overcome a predetermined and study‐specific statistical threshold (e.g., FWE p05, in the case of the present study; Whitwell, 2009). We did not further correct for the total number of VBMs performed (i.e., 3) since the post hoc age‐defined half samples were independent of the large primary analysis.
The output comprised of threshold‐free cluster enhancement (TFCE) based statistical parametric maps depicting locations of gray matter differences between MetS and metabolically healthy control groups. TFCE avoids the arbitrary definition of an initial cluster‐forming threshold, as in cluster‐based thresholding, by enhancing cluster‐like structures while maintaining a voxel‐wise approach (Smith & Nichols, 2009). The final results were overlaid onto the MNI‐152 standard template. Maxima locations for the large 208‐subject contrast were derived from the Talairach Daemon (Table 3; Lancaster et al., 2000). A summary of the VBM image analysis pipeline is shown in Figure 2.
Table 3.
Results of the whole‐brain VBM analysis comparing GMV between groups (MetS: n = 104 vs. metabolically healthy control: n = 104
| Metabolic syndrome VBM contrast analysis (n = 208) | ||||||
|---|---|---|---|---|---|---|
| Cluster | Cluster size (voxels) | Foci: MNI coordinates | Max t‐score within cluster | Cluster foci: MNI Daemon label | ||
| x | y | z | ||||
| Brainstem | 13,187 | 2 | −28 | −48 | 4.98 | Pons |
| L. Cerebellum | 28,818 | −24 | −62 | −46 | 5.73 | L. Crus I |
| −44 | −46 | −30 | 3.62 | L. Culmen | ||
| R. Cerebellum | 24,419 | 20 | −66 | −46 | 5.44 | R. Cerebellar tonsil |
| Orbitofrontal cortex | 9,175 | 4 | 14 | −24 | 5.66 | R. Rectal gyrus (BA11) |
| 12 | 62 | −22 | 3.90 | R. Superior Frontal Gyrus (BA10) | ||
| 12 | 44 | −28 | 4.14 | R. Medial frontal gyrus (BA11) | ||
| −6 | 14 | −28 | 4.68 | L. Rectal gyrus | ||
| −12 | 34 | −32 | 3.64 | L. Orbital gyrus (BA47) | ||
| −2 | 28 | −30 | 4.49 | L. Rectal gyrus (BA11) | ||
| L. Limbic lobe | 891 | −16 | −28 | −12 | 3.00 | L. Parahippocampal gyrus |
| R. Limbic lobe | 3,071 | 18 | 2 | −18 | 4.25 | Subcallosal gyrus (BA34) |
| 27 | −2 | −16 | 3.60 | R. Amygdala | ||
| R. insula | 4,026 | 34 | 0 | −12 | 3.73 | R. Claustrum |
| 42 | −2 | −8 | 3.41 | R. Insula | ||
| L. Caudate | 2,752 | −12 | 16 | 10 | 4.70 | L. Caudate body |
| R. Caudate | 4,054 | 8 | 22 | 8 | 4.59 | R. Caudate body |
| 6 | 14 | 4 | 4.00 | R. Caudate body | ||
| Cuneus | 2,787 | −4 | −64 | 8 | 3.51 | L. Cuneus |
| −6 | −84 | 8 | 3.05 | L. Lingual gyrus | ||
| R. Temporal lobe | 2,444 | 70 | −24 | 0 | 4.15 | R. Superior temporal gyrus (BA22) |
| 68 | −12 | −6 | 3.33 | R. Middle temporal gyrus | ||
| Lingual gyrus | 637 | 8 | −98 | 4 | 3.19 | R. Lingual gyrus |
Results are significant at FWE p < .05, corrected for multiple comparisons at the voxel level.
Abbreviations: BA, Brodmann area; L., left; R., right.
Figure 2.
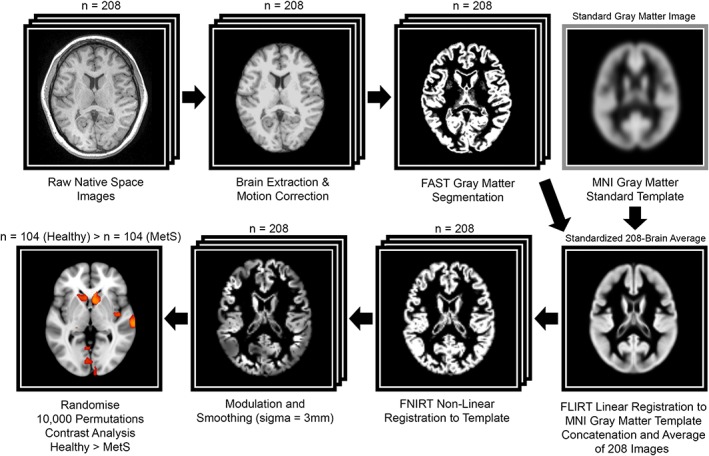
Depiction of the VBM image analysis pipeline as applied in this study. Acronym disambiguation: FAST, FMRIB automated segmentation tool; FLIRT, FMRIB linear image registration tool; FNIRT, FMRIB nonlinear image registration tool; MNI, montreal Neurological nstitute (standard brain space)
2.5. Post hoc analysis on age effects
Two additional post hoc analyses involving the subdivided large group of 208 participants were performed using identical thresholds and permutations. In this analysis we divided participants by the median age (35.5 years) and grouped them into half samples termed “young” and “old.” The “young” group included all participants with age ≤ 35 years (mean = 26.4 ± 4.6 years; range = 18–35 years); 52 metabolically healthy controls and 52 MetS participants. The “old” group included all participants with age ≥36 (mean = 48.1 ± 9.6 years; range = 36–74 years); 52 metabolically healthy controls and 52 MetS participants (Tables 1 and 2, Figure 3).
Figure 3.
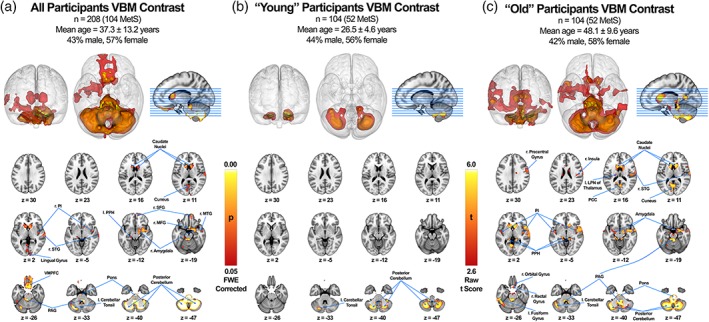
VBM cluster analysis of statistically significant gray matter structural changes associated with MetS across all subjects (a), subjects with age range below total subject median (“young”) (b), and subjects with age range above total subject median (“old”) (c). Regional GMV was reduced significantly across all subjects in the caudate nuclei bilaterally, right posterior insula (r. PI), right superior temporal gyrus (r. STG), lingual gyrus, left posterior parahippocampus (l. PPH), right superior frontal gyrus (r. SFG), right medial frontal gyrus (r. MFG), right amygdala, right middle temporal gyrus (r. MTG), ventromedial prefrontal cortex (VMPFC), periaqueductal gray (PAG), pons, left cerebellar tonsil and posterior cerebellum (a). Regional GMV was reduced significantly among “young” subjects in the left cerebellar tonsil and posterior cerebellum (b). Regional GMV was reduced significantly among “old” subjects in the right precentral gyrus, right insula, left lateral posterior nucleus of the thalamus (l. LPN), caudate nuclei bilaterally, r. STG, cuneus, posterior insula bilaterally, PPH bilaterally, amygdala bilaterally, right orbital gyrus, right rectal gyrus, left fusiform gyrus, PAG, pons, left cerebellar tonsil and posterior cerebellum (c). All values are FWE‐corrected (p < .05) and were not further correct for the total number of VBMs performed (i.e., 3) since the post hoc age‐defined half samples were independent of the large primary analysis
2.6. Behavior, paradigm class, and disease analyses
BrainMap is a neuroimaging database which, at the time of this study, contained 15,243 published functional imaging experiments (125,588 locations) from 3,261 peer‐reviewed publications and 3,151 published VBM, or structural imaging experiments, (21,827 coordinates) from 994 peer‐reviewed publications. BrainMap reports statistically significant results from the published, peer‐reviewed neuroimaging literature in the form of standardized coordinates that report functional and structural effects (Fox, Lancaster, Laird, & Eickhoff, 2014). It allows investigators to perform robust meta‐analyses from thousands of subjects, experiments, and paradigms using the standardized, coordinate‐based (x, y, z) mapping system. The results have been coupled to experimental and behavioral conditions in the functional task‐driven database (Lancaster et al., 2012), and disease diagnoses in the structural VBM database (Vanasse et al., 2018).
BrainMap's® Behavior and Paradigm Class analysis plugins are invaluable meta‐data inference function tools that quantitatively characterize regions that are anatomically altered in MetS and show how these regions relate to healthy brain functions (Crossley et al., 2014; Smith et al., 2009). Using this method, we can test the hypothesis that negative behavioral effects of MetS—specifically concerning impairment of executive function, cognitive reasoning, and reward perception— can be at least partially explained by structural changes in the form of GMV loss (Crossley, Scott, Ellison‐Wright, & Mechelli, 2015; Fox et al., 2014; Glahn et al., 2008). Similarly, BrainMap's Disease Inference Function also uses meta‐data inferencing to correlate GMV changes in MetS to neurodegenerative diseases present in the BrainMap VBM structural database (Kotkowski, Price, Fox, Vanasse, & Fox, 2018; Vanasse et al., 2018). We can use this function to independently quantify the degree of structural similarity between neurodegenerative diseases like AD and MetS using a z‐score.
BrainMap's software allows for behavior, paradigm class, and disease comparisons by imputing a 3D spatial region of interest known as a “mask.” Masks in this study were generated by the VBM analyses and represent the regions of significant GMV reduction. The software allows us to compare regions derived in our study to behaviors and paradigm classes ascribed to a number of functional studies, or diseases in VBM studies, from across the BrainMap database. This is achieved by comparing our mask to a null reference of random spatial distribution. Z‐scores are generated for observed‐minus‐expected values for each behavior sub‐domain, or disease sub‐domain in the case of VBM, with the operative threshold z‐score of 3.0 as comparable to a group p‐value of .05. This analysis has been previously validated in both the behavior domain and paradigm class analyses (Lancaster et al., 2012) and recently in disease class analyses (Kotkowski et al., 2018). Due to a lack of inclusion of the cerebellum and brainstem in the vast majority of task activation studies involving PET and fMRI within the BrainMap database, the cerebellum and brainstem regions were excluded from the ROI masks in this analysis.
3. RESULTS
3.1. Mass univariate VBM contrast analysis
A VBM contrast analysis was performed to investigate regional GMV differences between age‐ and sex‐matched metabolically healthy controls (n = 104) and individuals meeting the criteria for MetS (n = 104; see Tables 1 and 2, Figure 1). The most notable between‐group differences were found in the following regions: the posterior cerebellum, brainstem, orbitofrontal cortex (aka ventromedial prefrontal cortex), bilateral caudate nuclei, right posterior insula, right amygdala, lingual gyrus, and superior temporal gyrus (Figure 3, Table 3).
3.2. Structural age differences
Age differences were observed when contrasting participants in the post hoc age‐defined half‐samples. The “young” participants contrast analysis (≤35 years) reported significantly lower GMV in the posterior cerebellum only (Figure 3, Table S1). Furthermore, the “old” participants contrast analysis (≥36 years) reported significantly lower GMV in a more diffuse pattern than the whole group analysis and involved a greater number of affected regions. The notable regions in the “old” group analysis included: the posterior cerebellum, brainstem, orbitofrontal cortex, caudate nuclei, bilateral posterior insula, bilateral amygdalae, superior temporal gyrus, posterior cingulate cortex, bilateral posterior parahippocampi, and left fusiform gyrus (Figure 3, Table S2).
3.3. BrainMap behavior, paradigm class, and disease meta‐analysis
MetS‐associated regions exhibiting decreased GMV in the cortex (not including the cerebellum and brainstem) from the whole sample (n = 208) were associated with cognitive reasoning (z = 5.4), emotional valence (z = 5.4), and reward paradigms (z = 6.7). These regions were also found to be similar in gray matter pathology to Huntington's disease (HD; z = 3.6) and AD (z = 3.0; Figure 4). A z‐score of ≥3.0 is considered statistically significant using this method (Lancaster et al., 2012).
Figure 4.
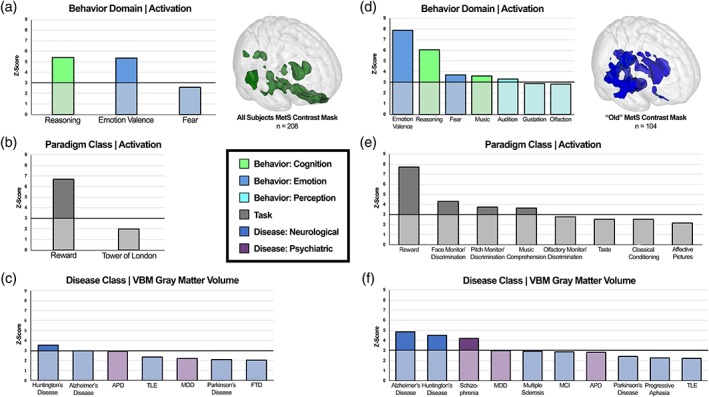
Specific behavioral (a), paradigm class (b), and disease weightings (c) on the MetS atrophy mask in all subjects and in “old” subjects (d–f). Mask regions of interest are FWE‐corrected (p < .05) as coordinate‐based search function. Functional characterization of MetS volume loss mask is based on behavior domain (a,d) and paradigm class (b,e) meta‐data of the BrainMap functional database representing peak functional domains related to mask regions. Structural characterization of MetS volume loss mask is based on disease classification meta‐data of the BrainMap VBM structural database representing peak structural atrophy domains related to mask regions (c,f). Disease class disambiguation: Antisocial personality disorder (APD), temporal lobe epilepsy (TLE), major depressive disorder (MDD), frontotemporal dementia (FTD), mild cognitive impairment (MCI). Only paradigms fulfilling a z‐score threshold ≥2.0 are reported, with z‐scores of ≥3.0 deemed statistically significant
No statistically significant findings were reported for the “young” half‐sample subgroup (n = 104). Cortical brain regions did not exhibit any significant GMV differences in this analysis. However, findings of significantly lower GMV in the posterior cerebellum were reported.
MetS‐associated regions exhibiting decreased GMV in the cortex from the “old” half‐sample subgroup (n = 104) were associated with cognitive reasoning (z = 6.0), fear modulation (z = 3.7), music composition (z = 3.6), audition perception (z = 3.3), reward paradigms (z = 7.7), face monitor/discrimination paradigms (z = 4.3), pitch monitor/discrimination paradigms (z = 3.7), and music comprehension paradigms (z = 3.6). These regions were also found to be similar in gray matter pathology to AD (z = 4.9), HD (z = 4.5), and schizophrenia (z = 4.2; Figure 4). Definitions for these terms can be found in Table S3.
4. DISCUSSION
Two of the three hypotheses put forward in the present study were supported by our findings. The first hypothesis, that a discrete pattern of GMV differences would be identified in individuals with MetS, was strongly confirmed. Specific GMV reductions were found in the cerebellum, brainstem, OFC, caudate, amygdala, insula, and superior temporal gyrus, among others. Additionally, age‐related effects were observed in two separate post hoc young versus old analyses from the larger group sample, suggesting increased age‐related effects influencing GMV between MetS participants and metabolically healthy controls. The second hypothesis—that patterns of GMV loss would recapitulate AD pathology—was weakly confirmed in the overall analysis. BrainMap's disease inference function identified AD's pattern of GMV changes as the neurodegenerative disease pattern most closely resembling our MetS VBM findings. This was especially noticeable in the “old” half‐sample analysis. HD and schizophrenia were the other diseases exhibiting significant similarities in GMV loss with MetS. Importantly, the hippocampus, a region prominently affected by AD, demonstrated no GMV reductions in our study. Although other regions implicated in AD pathology did exhibit GMV reductions, such as the insula, amygdala, and caudate nuclei. Finally, a BrainMap meta‐analysis of task‐activation functional studies of the neural signature of MetS gray matter atrophy mask revealed significant associations with regions involved in cognitive reasoning, emotional valence, and reward perception.
4.1. Cerebellum
Decreased GMV in the posterior cerebellum was the most significant and consistent regional difference in MetS across all age groups. The effects of decreased cerebellar GMV tend to vary depending on the cerebellar‐subregion where differences are seen. For instance, a commonly cited cause of cerebellar damage is alcohol dependence. Previous VBM studies have identified GMV in the vermis and anterior cerebellar regions—regions known to be involved in motor control and coordination—as significantly lower in alcoholics versus nonalcoholics (Mechtcheriakov et al., 2007). In contrast, atrophy in the posterior region of the cerebellum is attributed to “slowness,” defined as slow reaction time and walking speed (Chen et al., 2015). The effects of posterior cerebellar degeneration have been further supported by functional studies demonstrating that cortical regions significantly co‐activating with the anterior cerebellum are the association motor areas and those most significantly co‐activating with posterior cerebellum are regions implicated in cognition (Riedel et al., 2015). Furthermore, lesion studies investigating the effects of regional damage to the posterior cerebellum have identified deficits in executive function, visual spatial processing, linguistic skills, and regulation of affect (Schmahmann, 2004). This collection of symptoms termed “dysmetria of thought” have since been given the name of Cerebellar Cognitive Affective Syndrome, also known by its eponym as Schmahmann's Syndrome (Schmahmann, 2019).
4.2. Brainstem
Our study demonstrated significant MetS‐associated decreased GMV in the pons and periaqueductal gray (PAG) regions of the brainstem. Human studies have shown that increased hemoglobin A1c, another blood glucose measure, is independently associated with the severity and prognosis of brainstem infarcts, primarily involving the pons (Li et al., 2012). Decreased activity in the PAG is associated with increased levels of pain (migraine‐like headaches, fibromyalgia, nonspecific back pain) and anxiety. Animal studies have also found that diabetes‐related neuropathic pain is associated with a decrease in functional activity of the PAG (Paulson, Wiley, & Morrow, 2007). Of note, forebrain projections to the PAG arise predominantly from the prefrontal cortex, insular cortex, and amygdala (Linnman, Moulton, Barmettler, Becerra, & Borsook, 2012), structures implicated in our study's neural signature of MetS. PAG atrophy can thus serve as a potential neuroimaging biomarker responsible for increased levels of generalized pain symptoms in MetS patients.
4.3. The appetitive network
Regions in the appetitive network are involved in appetite‐related behaviors such as craving, feeding, and satiety. It consists of the lateral hypothalamus and the reward circuit, which includes the OFC, caudate, insula, amygdala, nucleus accumbens, substantia nigra, and ventral tegmentum (Kenny, 2011). Hyperactivation of the appetitive network has been hypothesized to lead to overeating and obesity. Individuals with high trait reward sensitivity have demonstrated increased neural activity in the appetitive network when exposed to highly palatable foods such as pizza and chocolate cake (Beaver et al., 2006), including obese patients (Rothemund et al., 2007). As weight increases, deficits in the brain's appetitive network emerge. For example, deficits in the caudate nuclei have led researchers to speculate that partial reward hyposensitivity may perpetuate the overconsumption of palatable foods as a compensatory mechanism for reward deficits (Kenny, 2011). As the motivation to consume palatable food increases, the hedonic value from the consumption of such foods decreases. These observations could have broader implications for neurocognitive functioning in other behavioral‐cognitive domains involving overlapping brain structures (Cornier, 2011).
The OFC is an integral part of the cognitive processes related to decision‐making, emotion valuation, and reward perception. This region also comprises the secondary taste cortex, a region responsible for the representation of the reward value of taste (Price, 2007). Furthermore, dysfunction in the OFC has been attributed to the overconsumption of food in obese patients (Cameron, Chaput, Sjödin, & Goldfield, 2017).
The caudate nuclei are involved in the modulation of inhibitory control, goal‐directed actions, and procedural learning. Reduced activity in the caudate nuclei is associated with obesity in older adults (Green, Jacobson, Haase, & Murphy, 2011). Additionally, adolescents with T2DM have been found to exhibit reduced GMV in the caudate nuclei compared to metabolically healthy age‐ and weight‐matched controls (Nouwen et al., 2017).
The insular cortex is predominantly associated with emotion, empathy, self‐awareness, and interoception. It also processes information related to the hedonic valuation and taste of food (Small, 2010). The right insula in particular, has previously been reported to show GMV loss in MetS after correcting for allostatic load (Zsoldos et al., 2018).
The amygdala lends affective significance to perceived stimuli and enhances memory for emotionally‐relevant stimuli. Its activity increases in response to hunger, particularly when exposed to high‐calorie palatable food (Goldstone et al., 2009). FDG‐PET studies show that increased levels of circulating insulin are correlated with decreased activity in the right amygdala and cerebellar vermis relative to whole brain, possibly explaining an individual's impaired emotional response to high‐calorie food (Anthony et al., 2006).
4.4. Relation to AD, HD, and schizophrenia
The incidence and progression of AD is thought to be influenced by metabolic diseases, dysregulations, and comorbidities (Stranahan, 2015). In the Rotterdam Study (the first study to propose this link) T2DM was associated with a twofold increased risk of developing AD and all‐type dementia (Ott et al., 1996). An association that has since been replicated in more recent studies (Arvanitakis, Wilson, Bienias, Evans, & Bennet, 2004; Chatterjee & Mudher, 2018). In our study we found a modest relationship between the MetS neural signature and that of AD, as shown in the BrainMap VBM meta‐analysis. This is a relationship that conspicuously does not include the hippocampus. MetS, obesity, HTN, dyslipidemia and T2DM all are associated with an increased risk of developing AD and other dementias. However, the reverse could also be true; for instance, that neurodegeneration contributes to the development of metabolic disease (Biessels & Reagan, 2015). Therefore, although AD may moderately correlate with MetS, we consider our findings to only weakly support the relationship. We speculate that it is possible that AD‐like symptoms are exhibited by certain individuals with MetS because of overlapping structural involvement, yet do not reflect the same underlying pathophysiological mechanism.
HD is a heritable neurodegenerative disease implicating the basal ganglia and cerebral cortex. Patients in advanced states of the disease are known to exhibit poor glycemic control and progressive weight loss (Aziz, Swaab, Pijl, & Roos, 2007). However, research within the past decade has shed light on the role that obesity‐induced inflammation can play in the pathogenesis of HD. For instance, it has been demonstrated that insulin resistance and decreased circulating insulin‐like growth factor 1 (IGF‐1) can accelerate HD onset in individuals with the genetic predisposition (Lalic et al., 2008). Furthermore, mouse models have also indicated that mutations in the huntingtin gene expressed in the hypothalamus may be the causal factor for metabolic abnormalities seen in the disease, such as: impaired glucose metabolism, insulin resistance, and leptin resistance (Hult et al., 2011; Procaccini et al., 2016).
Schizophrenia is a neuropsychiatric disorder in which patients exhibit dysfunctions in language, behavior and affect, often in the form of delusions, hallucinations, and disorganized speech that tend to present in early adulthood. Among neuropsychiatric disorders, the association between schizophrenia and obesity is well documented (Saha, Chant, & McGrath, 2007). Moreover, a commonly cited association between schizophrenia and metabolic syndrome is thought to be treatment‐emergent. A number of second‐generation anti‐psychotic drugs, most notably olanzapine, have been shown to cause weight gain, metabolic syndrome, and T2DM. It is thought that olanzapine's appetite‐stimulating effects via its antagonistic actions on the serotonin 5‐HT2C and dopamine D2 receptors contribute to its metabolic disease side‐effects (Patel et al., 2009). One key limitation for the BrainMap VBM database is its absence of flags indicating whether patient populations in VBM analyses suffer from metabolic comorbidities such as metabolic syndrome or obesity. We believe that schizophrenic patients on antipsychotics such as olanzapine might be weighing VBM effects related to MetS. However, more evidence is needed to support this speculation.
4.5. Mechanisms of atrophy
Trophic effects associated with gray matter have been attributed to four primary mechanisms: transneuronal spread, nodal stress, trophic failure, and shared vulnerability (Zhou, Gennatas, Kramer, Miller, & Seeley, 2012). We believe that trophic failure, involving the subcellular dysfunction of trophic factors such as insulin, insulin‐like growth factor, and leptin, is a likely mechanism for explaining our findings that is worth exploring (Verdile, Fuller, & Martins, 2015). When trophic factor expression and regulation is impaired, trophic failure in the form of poor cellular and synaptic maintenance hinders gray matter structural integrity (Fornito, Zalesky, & Breakspear, 2015). Shared vulnerability is also a likely candidate since similar neuronal cell types can be the targets of disease‐specific changes due to shared genetic and metabolic profiles (Cioli, Abdi, Beaton, Burnod, & Mesmoudi, 2014). For example, animal models of obesity and wild‐type mice fed high fat diets have found associations with dopamine‐dependent mesocorticolimbic‐prefrontal alterations which could impact reward learning, motivation, and executive functions (Stoeckel et al., 2016). In summary, a number of potential mechanisms emerging in the periphery that disturb the normal neurophysiological processes in gray matter can, collectively, be used to explain our reported findings.
4.6. Limitations
The criteria used to describe the metabolically healthy and MetS groups were a mixture of the IDF and NCEP‐ATP III classification systems with the notable absence of BP values in 57.4% of our samples. Although this limitation was addressed using sophisticated statistical methods involving available observations and related biometric data, the absence of directly measured BP values stands as a limitation. The nature of our choice for defining metabolically healthy controls and MetS participants as meeting a composite score of IDF and NCEP‐ATP III criteria may differ from criteria used in other studies, especially in different ethnic populations. Importantly, the criteria chosen to define our samples was optimized for the dataset available.
This study focused on identifying GMV differences associated with MetS. The pattern identified moderately correlates to neurodegenerative diseases involving gray matter as HD, AD, and schizophrenia. Microinfarcts and other vascular‐related events are also known to play a role in the pathophysiology of neurodegeneration (e.g., blood‐brain barrier breakdown, white matter lesions, and inflammation; Stranahan, 2015). Given the strong evidence for small vessel disease in the comorbidities of MetS, it could be argued that white matter integrity is also an important parameter to investigate (Van Bloemendaal et al., 2016), and we agree. However, an important part of the present study was to establish a link to cognition. White matter integrity as assessed by various imaging modalities does not predict cognitive status as well as gray matter (Lansley, Mataix‐Cols, Grau, Radua, & Sastre‐Garriga, 2013). Nevertheless, a subsequent investigative report addressing white matter involvement in MetS is underway.
Sex‐based differences in BMI‐related responses to food cues, identified using functional MRI, indicate the presence of significant regional differences in brain activity between males and females (Atalayer et al., 2014; Cornier, Salzberg, Endly, Bessesen, & Tregellas, 2010). Although studies have shown that individuals with MetS exhibit worse cognitive performance with increasing number and severity of MetS components (Yaffe et al., 2004; Cavalieri et al., 2010), no study to our knowledge has demonstrated that gender differences are related to reduced GMV in MetS or T2DM.
The association between MetS, T2DM, obesity, and cognition has been well‐characterized using psychometric evaluations (Yates et al., 2012). In this study, we demonstrated that regions associated with MetS also are related to cognition, supporting reports from the literature that link peripheral metabolic dysfunction to impaired emotion salience and cognition (Cameron et al., 2017). Nevertheless, we did not probe psychometric scores available to us in in the GOBS dataset on a per‐subject level. For the purposes of this study, we ensured that participants were nondemented and did not differ in level of educational attainment.
Finally, we intentionally chose to use a cohort of geographically and ethno‐culturally homogenous Mexican‐Americans from an extended pedigree due to the genetic and environmental predisposition for developing MetS in this population. However, in selecting these participants, we did not account for family or household effects. We encourage others to reproduce our findings in other ethno‐cultural populations.
5. CONCLUSION
We report that MetS is associated with reduced GMV, a finding that is amplified with age. Gray matter regions associated with MetS include the posterior cerebellum, brainstem, and regions involved in the appetitive network such as the orbitofrontal cortex, caudate, amygdala and insula. Functionally, these regions were identified to correlate with reward perception, reasoning and emotional valence. Patterns of atrophy also indicated similarities to those seen in AD, but failed to recapitulate its most important features, namely hippocampal involvement.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Eithan Kotkowski and Peter Fox devised the project and its main conceptual ideas with help from Ralph DeFronzo. Eithan Kotkowski wrote the manuscript with support from Peter Fox, Ralph DeFronzo, Crystal Franklin, and David Glahn. Crystal Franklin was instrumental in the design of the methods involving data curation and image processing with help from Maximino Salazar. Larry Price assisted in the design of statistical methods and analyses used in the study. Mary Woolsey, John Blangero, David Glahn, and Peter Fox obtained the individual subject data and MRI scans used in this study.
Supporting information
Supplementary Table 1 Anatomical coordinates in MNI space showing reduced GMV in “young” participants (n = 52 healthy vs. 52 MetS, FWE‐corrected p < 0.05) and their regional labels.
Supplementary Table 2: Anatomical coordinates in MNI space showing reduced GMV in “old” participants (n = 52 healthy vs. 52 MetS, FWE‐corrected p < 0.05) and their regional labels.
Supplementary Table 3: BrainMap® labels, definitions, and meta‐data attributed to MetS VBM contrast regional findings
Supplementary Table 4 Metabolically healthy control participants
Supplementary Table 5: Participants with metabolic syndrome
ACKNOWLEDGMENTS
The authors thank Drs. Amy Garret, Tom Vanasse, and Cheasequah Blevins for their valuable edits and insight, and the UT Health San Antonio Medical Scientist Training Program for their institutional and academic support. This study was supported by funds from the National Institutes of Health (NIH South Texas Medical Scientist Training Program: T32GM113898, NIMH: R01MH074457‐11S1, NCATS Translational Scientist Training Program: TL1TR002647‐01.) The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Kotkowski E, Price LR, Franklin C, et al. A neural signature of metabolic syndrome. Hum Brain Mapp. 2019;40:3575–3588. 10.1002/hbm.24617
Data Availability Statement:The data that support the findings of this study are available from Drs. David Glahn and John Blangero. Restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors with the permission of Drs. David Glahn and John Blangero.
Funding information National Center for Advancing Translational Sciences, Grant/Award Number: TL1TR002647‐01; National Institute of Mental Health, Grant/Award Number: R01MH074457‐11S1; National Institutes of Health, Grant/Award Number: T32GM113898
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from Drs. David Glahn and John Blangero. Restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors with the permission of Drs. David Glahn and John Blangero.
REFERENCES
- Aguilar, M. , Bhuket, T. , Torres, S. , Liu, B. , & Wong, R. J. (2015). Prevalence of the metabolic syndrome in the United States, 2003‐2012. JAMA, 313(19), 1973–1974. [DOI] [PubMed] [Google Scholar]
- Andersson, J. L. R. , Jenkinson, M. , Smith, S. (2007) Non‐linear registration, aka spatial normalization. FMRIB technical report TR07JA2 Retrieved from http://www.fmrib.ox.ac.uk/analysis/techrep
- Anthony, K. , Reed, L. J. , Dunn, J. T. , Bingham, E. , Hopkins, D. , Marsden, P. K. , & Amiel, S. A. (2006). Attenuation of insulin‐evoked responses in brain networks controlling appetite and reward in insulin resistance: The cerebral basis for impaired control of food intake in metabolic syndrome? Diabetes, 55, 2986–2992. [DOI] [PubMed] [Google Scholar]
- Arvanitakis, Z. , Wilson, R. S. , Bienias, J. L. , Evans, D. A. , & Bennet, D. A. (2004). Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Archives of Neurology, 61, 661–666. [DOI] [PubMed] [Google Scholar]
- Ashburner, J. , & Friston, K. J. (2000). Voxel‐based morphometry—The methods. NeuroImage, 11(6), 805–821. [DOI] [PubMed] [Google Scholar]
- Atalayer, D. , Pantazatos, S. P. , Gibson, C. D. , McOuatt, H. , Puma, L. , Astbury, N. M. , & Geliebter, A. (2014). Sexually dimorphic functional connectivity in response to high vs. low energy‐dense food cues in obese humans: An fMRI study. NeuroImage, 100, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, N. A. , Swaab, D. F. , Pijl, H. , & Roos, R. A. (2007). Hypothalamic dysfunction and neuroendocrine and metabolic alterations in Huntington's disease: Clinical consequences and therapeutic implications. Reviews in the Neurosciences, 18, 223–251. [DOI] [PubMed] [Google Scholar]
- Beaver, J. D. , Lawrence, A. D. , van Ditzhuijzen, J. , Davis, M. H. , Woods, A. , & Calder, A. J. (2006). Individual differences in reward drive predict neural responses to images of foods. The Journal of Neuroscience, 26, 5160–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessels, G. J. , & Reagan, L. P. (2015). Hippocampal insulin resistance and cognitive dysfunction. Nature Reviews Neuroscience, 16(11), 660–671. [DOI] [PubMed] [Google Scholar]
- Byrne, C. D. , & Wild, S. H. (2011). Brain insulin resistance In Byrne C. D. & Wild S. H. (Eds.), The metabolic syndrome (2nd ed.). Hoboken, NJ: Wiley‐Blackwell; ISBN: 978–1–4443‐3658‐0. [Google Scholar]
- Cameron, J. D. , Chaput, J.‐P. , Sjödin, A. M. , & Goldfield, G. S. (2017). Brain on fire: Incentive salience, hedonic hot spots, dopamine, obesity, and other hunger games. Annual Review of Nutrition, 37(1), 183–205. [DOI] [PubMed] [Google Scholar]
- Cavalieri, M. , Ropele, S. , Petrovic, K. , Pluta‐Fuerst, A. , Homayoon, N. , Enzinger, C. , … Schmidt, R. (2010). Metabolic syndrome, brain magnetic resonance imaging, and cognition. Diabetes Care, 33(12), 2489–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. , & Mudher, A. (2018). Alzheimer's disease and type 2 diabetes: A critical assessment of the shared pathological traits. Frontiers in Neuroscience, 12, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.‐T. , Chou, K.‐H. , Liu, L.‐K. , Lee, P.‐L. , Lee, W.‐J. , Chen, L.‐K. , … Lin, C. P. (2015). Reduced cerebellar gray matter is a neural signature of physical frailty. Human Brain Mapping, 36(9), 3666–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioli, C. , Abdi, H. , Beaton, D. , Burnod, Y. , & Mesmoudi, S. (2014). Differences in human cortical gene expression match the temporal properties of large‐scale functional networks. PLoS One, 9, e115913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornier, M. A. , Salzberg, A. K. , Endly, D. C. , Bessesen, D. H. , & Tregellas, J. R. (2010). Sex‐based differences in the behavioral and neuronal responses to food. Physiology & Behavior, 99(4), 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornier, M. A. (2011). Is your brain to blame for weight regain? Physiology & Behavior, 104(4), 608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley, N. A. , Mechelli, A. , Scott, J. , Carletti, F. , Fox, P. T. , McGuire, P. , & Bullmore, E. T. (2014). The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain, 137(8), 2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley, N. A. , Scott, J. , Ellison‐Wright, I. , & Mechelli, A. (2015). Neuroimaging distinction between neurological and psychiatric disorders. The British Journal of Psychiatry, 207(5), 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Heijer, T. , Vermeer, S. E. , van Dijk, E. J. , Prins, N. D. , Koudstaal, P. J. , Hofman, A. , & Breteler, M. M. (2003). Type 2 diabetes and atrophy of medial temporal lobe structures on brain MRI. Diabetologia, 46, 1604–1610. [DOI] [PubMed] [Google Scholar]
- De Felice, F. G. , Lourenco, M. V. , & Ferreira, S. T. (2014). How does brain insulin resistance develop in Alzheimer's disease? Jalz, 10(Supplement, S26–S32. [DOI] [PubMed] [Google Scholar]
- Douaud, G. , Smith, S. , Jenkinson, M. , Behrens, T. , Johansen‐Berg, H. , Vickers, J. , … James, A. (2007). Anatomically related grey and white matter abnormalities in adolescent‐onset schizophrenia. Brain, 130, 2375–2386. [DOI] [PubMed] [Google Scholar]
- Eckel, R. H. , & Cornier, M. A. (2014). Update on the NCEP ATP‐III emerging cardiometabolic risk factors. BMC Medicine, 12(115), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders, C. K. (2010). Applied missing data analysis. New York, NY: The Guilford Press. [Google Scholar]
- Exalto, L. G. , Whitmer, R. A. , Kappele, L. J. , & Biessels, G. J. (2012). An update on type 2 diabetes, vascular dementia and Alzheimer's disease. Experimental Gerontology, 47, 858–864. [DOI] [PubMed] [Google Scholar]
- Fischl, B. (2012). FreeSurfer. NeuroImage, 62(2), 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito, A. , Zalesky, A. , & Breakspear, M. (2015). The connectomics of brain disorders. Nature Reviews. Neuroscience, 16, 159–172. [DOI] [PubMed] [Google Scholar]
- Fox, P. T. , Lancaster, J. L. , Laird, A. R. , & Eickhoff, S. B. (2014). Meta‐analysis in human neuroimaging: Computational modeling of large‐scale databases. Annual Review of Neuroscience, 37(1), 409–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisardi, V. , Solfrizzi, V. , Seripa, D. , Capurso, C. , Santamato, A. , Sancarlo, D. , … Panza, F. (2010). Metabolic‐cognitive syndrome: A cross‐talk between metabolic syndrome and Alzheimer's disease. Ageing Research Reviews, 9(4), 399–417. [DOI] [PubMed] [Google Scholar]
- Glahn, D. C. , Laird, A. R. , Ellison‐Wright, I. , Thelen, S. M. , Robinson, J. L. , Lancaster, J. L. , … Fox, P. T. (2008). Meta‐analysis of gray matter anomalies in schizophrenia: Application of anatomic likelihood estimation and network analysis. Biological Psychiatry, 64(9), 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone, A. P. , Prechtl de Hernandez, C. G. , Beaver, J. D. , Muhammed, K. , Croese, C. , Bell, G. , … Bell, J. D. (2009). Fasting biases brain reward systems towards high‐calorie foods. European Journal of Neuroscience, 30(8), 1625–1635. [DOI] [PubMed] [Google Scholar]
- Good, C. D. , Johnarude, I. B. , Ashburner, J. , Hensop, R. N. , Riston, K. J. , & Frackowiak, R. B. (2001). Voxel‐based morphometric study. NeuroImage, 14, 21–36. [DOI] [PubMed] [Google Scholar]
- Green, E. , Jacobson, A. , Haase, L. , & Murphy, C. (2011). Reduced nucleus accumbens and caudate nucleus activation to a pleasant taste is associated with obesity in older adults. Brain Research, 1386, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, S. M. , Cleeman, J. I. , Daniels, S. R. , Donato, K. A. , Eckel, R. H. , Franklin, B. A. , … Costa, F. (2005). Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute scientific statement: Executive summary. Circulation, 112(17), 285–290. [DOI] [PubMed] [Google Scholar]
- Haan, M. N. , Mungas, D. M. , Gonzalez, H. M. , Ortiz, T. A. , Acharya, A. , & Jagust, W. J. (2003). Prevalence of dementia in older Latinos: The influence of type 2 diabetes mellitus, stroke and genetic factors. Journal of the American Geriatrics Society, 51, 169–177. [DOI] [PubMed] [Google Scholar]
- Huerta, C. I. , Sarkar, P. R. , Duong, T. Q. , Laird, A. R. , & Fox, P. T. (2014). Neural bases of food perception: Coordinate‐based meta‐analyses of neuroimaging studies in multiple modalities. Obesity, 22(6), 1439–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hult, S. , Soylu, R. , Björklund, T. , Belgardt, B. F. , Mauer, J. , Brüning, J. C. , & a. (2011). Mutant huntingtin causes metabolic imbalance by disruption of hypothalamic neurocircuits. Cell Metabolism, 13, 428–439. [DOI] [PubMed] [Google Scholar]
- Kenny, P. J. (2011). Reward mechanisms in obesity: New insights and future directions. Neuron, 69(4), 664–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, R. C. , Andrews, G. , Mroczek, D. , Ustun, T. B. , & Wittchen, H. U. (1998). The World Health Organization composite international diagnostic interview short form (CIDI‐SF). International Journal of Methods in Psychiatric Research, 7, 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov, P. , Lancaster, J. L. , Glahn, D. C. , Purdy, D. , Laird, A. R. , Gao, F. , & Fox, P. (2006). Retrospective motion correction protocol for high‐resolution anatomical MRI. Human Brain Mapping, 27(12), 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotkowski, E. , Price, L. R. , Fox, P. M. , Vanasse, T. J. , & Fox, P. T. (2018). The hippocampal network model_ a transdiagnostic metaconnectomic approach. Neuroimage Clinical, 18, 115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalic, N. M. , Maric, J. , Svetel, M. , Jotic, A. , Stefanova, E. , Lalic, K. , et al. (2008). Glucose homeostasis in Huntington disease: Abnormalities in insulin sensitivity and early‐phase insulin secretion. Archives of Neurology, 65, 476–480. [DOI] [PubMed] [Google Scholar]
- Lancaster, J. L. , Laird, A. R. , Eickhoff, S. B. , Martinez, M. J. , Fox, M. P. , & Fox, P. T. (2012). Automated regional behavioral analysis for human brain images. Frontiers in Neuroinformatics, 6(23), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, J. L. , Woldorff, M. G. , Parsons, L. M. , Liotti, M. , Freitas, C. S. , Rainey, L. , … Fox, P. T. (2000). Automated Talairach atlas labels for functional brain mapping. Human Brain Mapping, 10, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansley, J. , Mataix‐Cols, D. , Grau, M. , Radua, J. , & Sastre‐Garriga, J. (2013). Localized grey matter atrophy in multiple sclerosis: A meta‐analysis of voxel‐based morphometry studies and associations with functional disability. Neuroscience and Biobehavioral Reviews, 37(5), 819–830. [DOI] [PubMed] [Google Scholar]
- Lee, E. B. , & Mattson, M. P. (2014). The neuropathology of obesity: Insights from human disease. Acta Neuropathologica, 127(1), 3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. H. , Zabolotny, J. M. , Huang, H. , Lee, H. , & Kim, Y.‐B. (2016). Insulin in the nervous system and the mind: Functions in metabolism, memory, and mood. Molecular Metabolism, 5(8), 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Kang, Z. , Qiu, W. , Hu, B. , Wu, A.‐M. , Dai, Y. , … Lu, Z. (2012). Hemoglobin A1C is independently associated with severity and prognosis of brainstem infarctions. Journal of the Neurological Sciences, 317(1–2), 87–91. [DOI] [PubMed] [Google Scholar]
- Linnman, C. , Moulton, E. A. , Barmettler, G. , Becerra, L. , & Borsook, D. (2012). Neuroimaging of the periaqueductal gray: State of the field. NeuroImage, 60(1), 505–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, R. J. A. , & Rubin, D. B. (2002). Statistical analysis with missing data (2nd ed.). Hoboken, NJ: Wiley. [Google Scholar]
- Masouleh, S. K. , Arélin, K. , Horstmann, A. , Lampe, L. , Kipping, J. A. , Luck, T. , et al. (2016). Higher body mass index in older adults is associated with lower gray matter volume: Implications for memory performance. Neurobiology of Aging, 40(C), 360–372. [DOI] [PubMed] [Google Scholar]
- Mechtcheriakov, S. , Brenneis, C. , Egger, K. , Koppelstaetter, F. , Schocke, M. , & Marksteiner, J. (2007). A widespread distinct pattern of cerebral atrophy in patients with alcohol addiction revealed by voxel‐based morphometry. Journal of Neurology, Neurosurgery & Psychiatry, 78(6), 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, A. , Vainik, U. , Garcia‐Garcia, I. , & Dagher, A. (2017). Overlapping neural Endophenotypes in addiction and obesity. Frontiers in Endocrinology, 8, 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, P. J. , DeFronzo, R. A. , Califf, R. M. , & Guyton, J. R. (2005). Metabolic syndrome: Definition, pathophysiology, and mechanisms. American Heart Journal, 149(1), 33–45. [DOI] [PubMed] [Google Scholar]
- Nouwen, A. , Chambers, A. , Chechlacz, M. , Higgs, S. , Blissett, J. , Barrett, T. G. , & Allen, H. A. (2017). Microstructural abnormalities in white and gray matter in obese adolescents with and without type 2 diabetes. Neuroimage Clinical, 16, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, A. , Stolk, R. P. , Hofman, A. , van Harskamp, F. , Grobbee, D. E. , & Breteler, M. M. (1996). Association of diabetes mellitus and dementia: The Rotterdam study. Diabetologia, 39, 1392–1397. [DOI] [PubMed] [Google Scholar]
- Patel, J. K. , Buckley, P. F. , Woolson, S. , Hamer, R. M. , McEvoy, J. P. , Perkins, D. O. , Lieberman, J. A. , & CAFE Investigators . (2009). Metabolic profiles of second‐generation antipsychotics in early psychosis: findings from the CAFE study. Schizophrenia Research, 111(3), 9–16. [DOI] [PubMed] [Google Scholar]
- Paulson, P. E. , Wiley, J. W. , & Morrow, T. J. (2007). Concurrent activation of the somatosensory forebrain and deactivation of periaqueductal grey associated with diabetes‐induced neuropathic pain. Experimental Neurology, 208(2), 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, J. L. (2007). Definition of the orbital cortex in relation to specific connections with limbic and visceral structures and other cortical regions. Annals of the New York Academy of Sciences, 1121(1), 54–71. [DOI] [PubMed] [Google Scholar]
- Procaccini, C. , Santopaolo, M. , Faicchia, D. , Colamatteo, A. , Formisano, L. , de Candia, P. , … Matarese, G. (2016). Role of metabolism in neurodegenerative disorders. Metabolism, 65(9), 1376–1390. [DOI] [PubMed] [Google Scholar]
- Pruzin, J. J. , Nelson, P. T. , Abner, E. L. , & Arvanitakis, Z. (2018). Invited review: Relationship of type 2 diabetes to human brain pathology. Neuropathology and Applied Neurobiology, 92(53), 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratitch, B. & O'Kelly, M. (2011). Implementation of Pattern‐Mixture Models Using Standard SAS/STAT Procedures. Proceedings of PharmaSUG 2011 (Pharmaceutical Industry SAS Users Group), SP04, Nashville.
- Reaven, G. M. (1988). Banting lecture 1988: Role of insulin resistance in human disease. Diabetes, 37, 1595–1607. [DOI] [PubMed] [Google Scholar]
- Riedel, M. C. , Ray, K. L. , Dick, A. S. , Sutherland, M. T. , Hernandez, Z. , Fox, P. M. , … Laird, A. R. (2015). Meta‐analytic connectivity and behavioral parcellation of the human cerebellum. NeuroImage, 117(C), 327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund, Y. , Preuschhof, C. , Bohner, G. , Bauknecht, H. C. , Klingebiel, R. , Flor, H. , & Klapp, B. F. (2007). Differential activation of the dorsal striatum by high‐calorie visual food stimuli in obese individuals. NeuroImage, 37, 410–421. [DOI] [PubMed] [Google Scholar]
- Saha, S. , Chant, D. , & McGrath, J. (2007). A systematic review of mortality in schizophrenia: Is the differential mortality gap worsening over time? Archives of General Psychiatry, 64, 1123–1131. [DOI] [PubMed] [Google Scholar]
- Schmahmann, J. D. (2004). Disorders of the cerebellum: Ataxia, dysmetria of thought, and the cerebellar cognitive affective syndrome. Neuropsychiatric Practice and Opinion, 16(3), 367–378. [DOI] [PubMed] [Google Scholar]
- Schmahmann, J. D. (2019). The cerebellum and cognition. Neuroscience Letters, 688, 62–75. [DOI] [PubMed] [Google Scholar]
- Sheehan, D. V. , Lecrubier, Y. , Sheehan, K. H. , Amorim, P. , Janavs, J. , Weiller, E. , … Dunbar, G. C. (1998). The Mini‐international neuropsychiatric interview (M.I.N.I.): The development and validation of a structured diagnostic psychiatric interview for DSM‐IV and ICD‐10. The Journal of Clinical Psychiatry, 59(Suppl 20), 22–33 quiz 34–57. [PubMed] [Google Scholar]
- Simonson, G. D. , & Kendall, D. M. (2005). Diagnosis of insulin resistance and associated syndromes: The spectrum from the metabolic syndrome to type 2 diabetes mellitus. Coronary Artery Disease, 16, 465–472. [DOI] [PubMed] [Google Scholar]
- Small, D. M. (2010). Taste representation in the human insula. Brain Structure and Function, 214(5–6), 551–561. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , & Nichols, T. E. (2009). Threshold‐free cluster enhancement: Addressing problems of smoothing, threshold dependence and localization in cluster inference. NeuroImage, 44(1), 83–98. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , Jenkinson, M. , Woolrich, M. W. , Beckmann, C. F. , Behrens, T. E. , Johansen‐Berg, H. , … Matthews, P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23(Suppl 1), S208–S219. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , Fox, P. T. , Miller, K. L. , Glahn, D. C. , Fox, P. M. , Mackay, C. E. , … Beckmann, C. F. (2009). Correspondence of the brain's functional architecture during activation and rest. Proceedings of the National Academy of Sciences, 106(31), 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. W. , Chung, J. H. , Rho, J. S. , Lee, Y. A. , Lim, H. K. , Kang, S. G. , et al. (2014). Regional cortical thickness and subcortical volume changes in patients with metabolic syndrome. Brain Imaging and Behavior, 9(3), 588–596. [DOI] [PubMed] [Google Scholar]
- Stoeckel, L. E. , Arvanitakis, Z. , Gandy, S. , Small, D. , Kahn, C. R. , Pascual‐Leone, A. , et al. (2016). “White paper” meeting summary and catalyst for future inquiry: Complex mechanisms linking neurocognitive dysfunction to insulin resistance and other metabolic dysfunction. F1000Research, 5, 353–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranahan, A. M. (2015). Models and mechanisms for hippocampal dysfunction in obesity and diabetes. Neuroscience, 309, 125–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchistiakova, E. , Anderson, N. D. , Greenwood, C. E. , & MacIntosh, B. J. (2014). Combined effects of type 2 diabetes and hypertension associated with cortical thinning and impaired cerebrovascular reactivity relative to hypertension alone in older adults. Neuroimage Clinical, 5, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bloemendaal, L. , Ijzerman, R. G. , ten Kulve, J. S. , Barkhof, F. , Diamant, M. , Veltman, D. J. , & van Duinkerken, E. (2016). Alterations in white matter volume and integrity in obesity and type 2 diabetes. Metabolic Brain Disease, 31(3), 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanasse, T. J. , Fox, P. M. , Barron, D. S. , Robertson, M. , Eickhoff, S. B. , Lancaster, J. L. , & Fox, P. T. (2018). BrainMap VBM: An environment for structural meta‐analysis. Human Brain Mapping, 39(8), 3308–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhanen, M. , Koivisto, K. , Moilanen, L. , Helkala, E. L. , Hänninen, T. , Soininen, H. , … Kuusisto, J. (2006). Association of metabolic syndrome with Alzheimer disease. Neurology, 67(5), 843–847. [DOI] [PubMed] [Google Scholar]
- Verdile, G. , Fuller, S. J. , & Martins, R. N. (2015). The role of type 2 diabetes in neurodegeneration. Neurobiology of Disease, 84(C), 22–38. [DOI] [PubMed] [Google Scholar]
- Whitwell, J. L. (2009). Voxel‐based morphometry: An automated technique for assessing structural changes in the brain. Journal of Neuroscience, 29(31), 9661–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, A. M. , Kochunov, P. , Blangero, J. , Almasy, L. , Zilles, K. , Fox, P. T. , … Glahn, D. C. (2010). Cortical thickness or gray matter volume? The importance of selecting the phenotype for imaging genetics studies. NeuroImage, 15(3), 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , Lin, L. , Zhang, Q. , & Wu, J. (2017). Brain gray matter changes in type 2 diabetes mellitus: A meta‐analysis of whole‐brain voxel‐based morphometry study. Journal of Diabetes and its Complications, 31, 1698–1703. [DOI] [PubMed] [Google Scholar]
- Yaffe, K. , Blackwell, T. , Kanaya, A. M. , Davidowitz, N. , Barrett‐Connor, E. , & Krueger, K. (2004). Diabetes, impaired fasting glucose, and development of cognitive impairment in older women. Neurology, 63, 658–663. [DOI] [PubMed] [Google Scholar]
- Yates, K. F. , Sweat, V. , Yau, P. L. , Turchiano, M. M. , & Convit, A. (2012). Impact of metabolic syndrome on cognition and brain: A selected review of the literature. Arteriosclerosis, Thrombosis, and Vascular Biology, 32(9), 2060–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Gennatas, E. D. , Kramer, J. H. , Miller, B. L. , & Seeley, W. W. (2012). Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron, 73, 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsoldos, E. , Filippini, N. , Mahmood, A. , Mackay, C. E. , Singh‐Manoux, A. , Kivimamp, M. , et al. (2018). Allostatic load as a predictor of grey matter volume and white matter integrity in old age: The Whitehall II MRI study. Scientific Reports, 8(6411), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Anatomical coordinates in MNI space showing reduced GMV in “young” participants (n = 52 healthy vs. 52 MetS, FWE‐corrected p < 0.05) and their regional labels.
Supplementary Table 2: Anatomical coordinates in MNI space showing reduced GMV in “old” participants (n = 52 healthy vs. 52 MetS, FWE‐corrected p < 0.05) and their regional labels.
Supplementary Table 3: BrainMap® labels, definitions, and meta‐data attributed to MetS VBM contrast regional findings
Supplementary Table 4 Metabolically healthy control participants
Supplementary Table 5: Participants with metabolic syndrome
Data Availability Statement
The data that support the findings of this study are available from Drs. David Glahn and John Blangero. Restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors with the permission of Drs. David Glahn and John Blangero.


