Abstract
Programmed death 1 (PD-1)-targeted therapy has benefited patients with microsatellite instability-high metastatic colorectal cancer (mCRC). However, the efficacy of PD-1-targeted therapy is poor in patients with microsatellite-stable (MSS) mCRC. Therefore, it is imperative to explore additional co-inhibitory molecular signalling pathways to improve the efficacy of immunotherapy in MSS mCRC treatment. In the present study, the association between cyclin-dependent kinase 9 (CDK9) expression and the survival of patients with CRC was analysed using RNA sequencing data from 605 patients, including 121 cases of mortality, from human cancer datasets. Furthermore, 35 clinical MSS stage III–IV CRC specimens were collected to assess CDK9 protein expression by immunohistochemistry, and the frequency of tumor-infiltrating CD8+ T cells was assessed by flow cytometry. The human cancer datasets demonstrated that upregulation CDK9 significantly shortened the survival of patients with stage II–IV colon cancer. Additionally, CDK9 mRNA expression was positively correlated with the expression levels of genes associated with immune evasion in the tumor. Notably, CDK9 was expression was upregulated in stage IV CRC compared with para-cancerous tissues and early-stage tumors. Interestingly, CDK9 expression was negatively associated with the infiltration of CD8+ T cells at the tumor site. In addition, the expression levels of T-cell immunoglobulin mucin family member 3 and CD39, proteins associated with exhaustion, on tumor-infiltrating CD8+ T cells were significantly elevated in patients with abnormal CDK9 expression levels. The present study demonstrated that CDK9 expression was negatively associated with CD8+ T cell infiltration and positively associated with CD8+ T cell exhaustion in MSS mCRC. In conclusion, CDK9 may be utilized to evaluate the prognosis and the immune-type of the tumor microenvironment in patients with MSS mCRC.
Keywords: CDK9, immunotherapy, CD8+ T cell exhaustion, microsatellite stable, colorectal cancer
Introduction
Colorectal cancer (CRC) has become the third most common type of cancer and the second leading cause of cancer-associated mortality worldwide (1). In the United States, the 5-year relative survival rate is 90.1 or 69.2% for patients with CRC with localized (stage I–II) or regional metastasis (stage III), respectively. However, the 5-year relative survival rate is only 11.7% for patients with distant metastasis (stage IV) (2). Therefore, improving the diagnosis and treatment efficacy of patients with stage IV CRC is the key to ameliorating the overall survival of CRC.
The American Society of Clinical Oncology (ASCO) Annual Meeting in 2016 named ‘Immunotherapy’ as the ‘Primary Progress’ in Cancer Research in 2015 (3). The Federal Drug Agency and China Food and Drug Administration (CFDA) have approved pembrolizumab, an anti-PD-1 inhibitor, for the treatment of microsatellite instability-high (MSI-H) or mismatch repair-deficient (dMMR) metastatic CRC (mCRC) (4). Numerous studies have reported that 44.8–89% of patients with CRC express PD-L1, thus suggesting that abnormal expression of PD-L1 is an independent poor prognostic indicator in CRC (5,6). However, data from clinical trials revealed that PD-1/PD-L1 blockades are only effective in 2.5–5% of patients with mCRC (MSI-H or dMMR), and the vast majority of patients [microsatellite stable (MSS)] does not benefit from this treatment (7–9). This is a great obstacle to the application of immunotherapy in CRC.
Chen and Mellman (10) analysed data from patients undergoing immunotherapy. According to the clinical efficacy and the immunophenotype of the patients, the tumor immuno-microenvironment was divided into three basic types: The immune-desert phenotype, immune-excluded tumor and inflamed tumor (10). Based on this classification, the effectiveness of PD-L1 inhibitors depends not only on the expression of intra-tumor PD-L1 but also on the existence of sufficient immune effector cells, particularly CD8+ T cells, in the tumor microenvironment (TME). Without the infiltration of CD8+ T cells, even if elevated expression of PD-L1 is detected in tumor tissues, a therapeutic effect could not be achieved; this is the case for patients with the immune-desert phenotype (10). It has previously been reported that the number of tumor-infiltrating CD4+ and CD8+ T cells in patients with MSI-H CRC who benefit from pembrolizumab immunotherapy is significantly higher than that in MSS CRC (11). This may be due to patients with MSI-H CRC producing a large number of mutant proteins in tumor tissues, which can stimulate the immune response and promote the local lymphocyte infiltration of tumors (7,12). Therefore, promoting the infiltration of immune effector cells, particularly CD8+ T cells, into tumor tissues and improving their immune function are effective measures to increase the efficacy of immunotherapy in the majority of patients with CRC (MSS phenotype).
Cyclin-dependent kinase 9 (CDK9) is a key regulator of transcriptional elongation, which is a promising therapeutic target in cancer, particularly for types of cancer driven by transcriptional dysregulation (13,14). However, the mechanism and clinical transformation of CDK9 in CRC have been rarely reported (15). In addition, it has been demonstrated that APC/BRAF/SMAD4 gene mutations lead to upregulation of transcription, which leads to the occurrence and development of CRC (16–18). Positive transcription elongation factor (P-TEFb)/CDK9 triggers the release and nuclear export of β-catenin by the α-catenin:APC complex (19). CDK8 and CDK9 provide the coordinated regulation of SMAD transcriptional activators in the bone morphogenetic protein and transforming growth factor β1 signalling pathways (20). Therefore, CDK9 serves an important role in the development of CRC. In addition, studies have demonstrated that CDK9 promotes the proliferation and differentiation of immune cells, including neutrophils, macrophages and lymphocytes, alters the ratio of CD4+CD25+ forkhead box P3+ Tregs, and regulates the expression of chemokines, thereby affecting the infiltration of effector T cells in an inflammatory environment (21–23). The present study utilized human cancer datasets and clinically resected CRC specimens to assess CDK9 expression and CD8+ T cell infiltration, and examined their alterations in mCRC. The results of the present study suggested that CDK9 may be a promising therapeutic target for patients with MSS mCRC.
Materials and methods
Analysis of public datasets
RNA sequencing-based gene expression data in CRC was obtained from the Gene Expression Profiling Interactive Analysis (GEPIA) database for cancer genomics (24). The association between CDK9 expression and the survival of 605 patients with CRC, including 121 patients who died, was analysed using the Tumor Immune Estimation Resource (TIMER) database (25). The co-expression of proteins was analysed by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (26). For gene expression, P<0.01 was used as the cut-off. The Kaplan-Meier method with a log-rank test was used for estimating patient survival rates. P<0.05 was considered to indicate a statistically significant difference.
Patient tissue samples
A total of 35 patients with CRC who were admitted and treated at Tianjin Medical University Cancer Institute and Hospital between May 2018 and September 2018 were enrolled randomly. The patients comprised 16 men and 19 women, aged 27–79 years. During surgical resection, specimen (both tumor and paired control tissues) were obtained from the treatment-naïve patients with CRC. The gene stability of BAT-25, BAT-26, NR-21, NR-24 and Mono-27 in the specimens from all patients was examined using next-generation sequencing (Hongzhong Precision Medicine). All patients had MSS CRC according to the definition of the National Cancer Institute (there was no ‘instability’ in the results of the five aforementioned loci) (27). All samples and clinical data were collected after ethical approval was granted by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Written informed consent was obtained from all patients for participation in the present study. The clinical features of the patients are presented in Table I. The records of all of the patients contained basic information, including Tumor-Node-Metastasis (TNM) stage, the degree of differentiation, lymph node metastasis and distant metastasis, according to the 2002 International Cancer Alliance TNM staging criteria (28).
Table I.
Clinical and pathological characteristics of the patients with colorectal cancer.
| Patient no. | Sex | Age, years | Primary tumor location | TNM stage | Tumor stage (28) |
|---|---|---|---|---|---|
| P#1 | Male | 61 | Ascending colon | T4aN1bM0 | IIIB |
| P#2 | Female | 77 | Rectum | T3N1bM0 | IIIB |
| P#3 | Female | 64 | Descending colon | T4bN2bM0 | IIIC |
| P#4 | Male | 79 | Ascending colon | T3N2aM0 | IIIB |
| P#5 | Female | 60 | Rectum | T3N0M1c | IVC |
| P#6 | Female | 60 | Ascending colon | T3N2bM1a | IVA |
| P#7 | Female | 49 | Descending colon | T3N2bM0 | IIIB |
| P#8 | Female | 41 | Rectum | T3N2bM0 | IIIB |
| P#9 | Male | 67 | Sigmoid | T3N2bM1b | IVB |
| P#10 | Male | 65 | Ascending colon | T2N1bM0 | IIIA |
| P#11 | Female | 27 | Rectum | T4aN1bM0 | IIIB |
| P#12 | Female | 54 | Rectum | T2N1cM0 | IIIA |
| P#13 | Female | 70 | Rectum | T3N1cM0 | IIIB |
| P#14 | Male | 56 | Descending colon | T3N2aM0 | IIIB |
| P#15 | Female | 68 | Sigmoid | T3N1bM0 | IIIB |
| P#16 | Male | 73 | Rectum | T3N2aM1a | IVA |
| P#17 | Male | 50 | Descending colon | T3N2aM1a | IVA |
| P#18 | Female | 65 | Ascending colon | T4bN0M1a | IVA |
| P#19 | Female | 53 | Ascending colon | T4aN2aM1a | IVA |
| P#20 | Male | 76 | Ascending colon | T4bN2bM1a | IVA |
| P#21 | Female | 48 | Ileocecus | T4bN0M1b | IVB |
| P#22 | Male | 48 | Ascending colon | T4bN2aM1b | IVB |
| P#23 | Male | 70 | Descending colon | T4aN2M1a | IVA |
| P#24 | Female | 65 | Ascending colon | T3N2M1a | IVA |
| P#25 | Male | 65 | Ascending colon | T4aN0M1b | IVB |
| P#26 | Female | 60 | Transverse colon | T4bN0M1b | IVB |
| P#27 | Male | 77 | Transverse colon | T3N1aM1b | IVB |
| P#28 | Female | 29 | Ascending colon | T3N1M1b | IVB |
| P#29 | Male | 59 | Ascending colon | T3N1bM0 | IIIB |
| P#30 | Female | 79 | Rectum | T2N2bM0 | IIIB |
| P#31 | Male | 60 | Sigmoid | T4aN2bM0 | IIIC |
| P#32 | Male | 52 | Ascending colon | T4aN2bM0 | IIIC |
| P#33 | Female | 68 | Ascending colon | T3N1cM0 | IIIB |
| P#34 | Male | 62 | Sigmoid | T4aN1bM0 | IIIB |
| P#35 | Female | 65 | Ascending colon | T3N1aM0 | IIIB |
Isolation of tissue-infiltrating cells
Fresh colorectal tumor tissues and paired control tissues from patients with CRC were prepared by mechanical disruption, followed by digestion with 0.5 mg/ml collagenase type IV (cat. no. C5138; Sigma-Aldrich; Merck KGaA) in 10% FBS with 10 U/ml DNase I in RPMI-1640 medium (both from Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 30 min. Digested tissues were incubated for 5 min at 37°C with EDTA (0.5 M) to prevent dendritic cell/T-cell aggregation and filtered through a 100-µm and a 40-µm filter. The isolation and culture of tissue/tumor-infiltrating cells were performed as previously described (29).
Immunohistochemistry (IHC)
Colon and rectal tumor tissues, and paired normal tissues were were fixed with 4% formaldehyde for 24 h at room temperature (RT), embedded in paraffin and cut into 4-µm sections. The sections were dewaxed with xylene then hydrated through a graded series of ethanol (100, 95, 90, 80 and 70%) for 5 min in each percentage at RT. Slides were boiled in 10 mM sodium citrate buffer pH 6.0 at 95°C for 10 min and subsequently incubated in 3% hydrogen peroxide for 10 min. Sections were blocked with 100–400 µl 5% normal goat serum in TBS-Tween (cat. no. 5425; Cell Signaling Technology, Inc.) for 1 h at RT. To assess CDK9 expression in the MSS phenotype mCRC specimens, sections were incubated with a CDK9 antibody (dilution, 1:100; cat. no. 2316; Cell Signaling Technology, Inc.) overnight at 4°C, and were then incubated with a horseradish peroxidase-conjugated secondary antibody (dilution, 1:1,000; cat. no. ab6721; Abcam) at room temperature for 1 h. Following washing with PBS, the sections were incubated for 30 min at RT with streptavidin-biotin conjugated with horseradish peroxidase [dilution, 1:100; cat. no. KIT-0305; UltraSensitive™ SP (Mouse/Rabbit) IHC kit]. Subsequently, the slides were stained with 3,3′-diaminobenzidine (Fuzhou Maixin Biotech Co., Ltd.), and the nuclei were counterstained with haematoxylin for 5 min at RT. Morphometric analyses of the tumor and paired normal tissues were performed using an Olympus BX51 light microscope (magnification, ×20; Olympus Corporation). Images were obtained from 10 randomly selected areas.
Staining intensity was scored as follows: 0, Negative; 1, weakly positive (<25% of cells stained); and 2, positive (>25% of cells stained). Samples were subsequently grouped into 2 categories: Low expression (0 and 1) and high expression (2).
Flow cytometric analysis
Tumor/paired control tissue-infiltrating cells (1×106 cells) were incubated with human FITC anti-CD3 monoclonal antibody (mAb) (1:20; cat. no. 300452; BioLegend, Inc.), APC anti-CD8 mAb (1:20; cat. no. 344722; BioLegend, Inc.), phycoerythrin (PE) anti-CD39 mAb (1:20; cat. no. 328208; BioLegend, Inc.), APC/Cy7 anti-PD-1 mAb (1:20; cat. no. 329921; BioLegend, Inc.), and PE/Cy7 anti-T-cell immunoglobulin mucin family member 3 (Tim-3) mAb (1:20; cat. no. 345013; BioLegend, Inc.) in cell staining buffer (cat. no. 420201; BioLegend, Inc.) for 15 min at RT. After washing with PBS and centrifugation at 400 × g for 5 min at 4°C, 1×106 cells were suspended in 300 µl cell staining buffer (cat. no. 420201; BioLegend, Inc.) and analysed on a BD FACSCalibur (BD Biosciences) flow cytometer. The data were analysed using FlowJo v10 software (FlowJo, LLC).
Statistical analysis
Statistical analysis was performed using GraphPad Prism v5 software (GraphPad Software, Inc.). Data are presented as the mean ± standard error of the mean of three repeats. A χ2 test was used to evaluate the association between CDK9 expression and the clinicopathological parameters. A log-rank test was used to compare the survival curves of two groups. Cox regression analysis was performed for multivariate analysis of prognostic variables. A correlation between transcripts per million (TPM) of CDK9 and other genes was calculated for statistical significance, correlation co-efficient, and is represented using a scatter plot. One-way ANOVA followed by Tukey's multiple comparison test was used for multiple-group analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
CDK9 significantly shortens the survival of patients with colon cancer
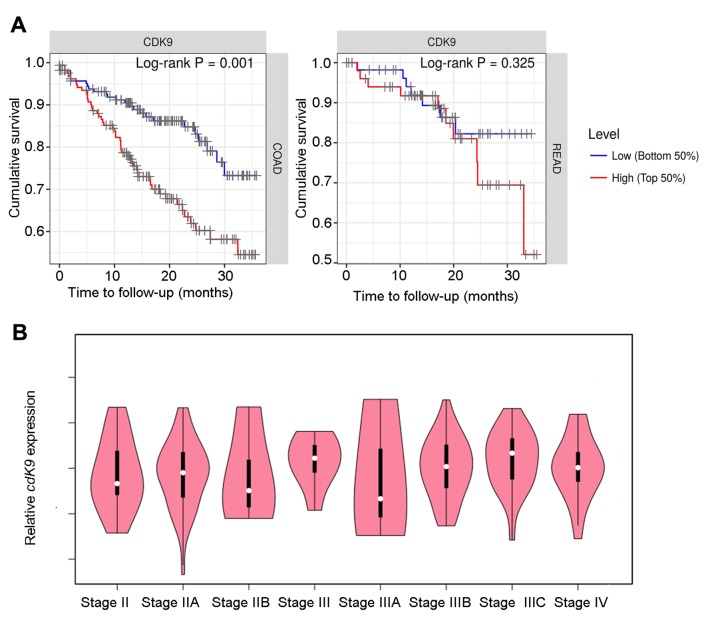
To examine the association between CDK9 and prognosis in patients with colon and rectal cancer, the present study analysed the TIMER database, and found that high CDK9 expression was significantly associated with a shortened survival of patients with colon cancer (P=0.001; 445 cases total; 98 cases of mortality). The same trend was observed in patients with rectal cancer; however, this was not statistically significant (P=0.325; 160 cases total; 23 cases of mortality; Fig. 1A). As shown in Table II, CDK9 was a risk factor for survival in patients with colon cancer based on a Cox proportional hazard model analysis. However, Cox model analysis demonstrated that CDK9 did not affect rectal cancer progression (Table III). In addition, CDK9 expression had no significant effect on prognosis when the survival time was >3 years (data not shown). Therefore, CDK9 may serve an important role in the progression of advanced colon cancer. The GEPIA database was searched and CDK9 mRNA expression in stage III–IV patients was upregulated compared with that in stage II patients, and the expression level was positively associated with the clinical tumor stage in stage IIIA-IIIC patients although the association was not significant (Fig. 1B). These data suggested that CDK9 may promote lymph node metastasis in colon cancer.
Figure 1.
CDK9 significantly shortens the survival of patients with colon cancer. (A) Association between CDK9 expression and the survival of patients with colon and rectal cancer. Kaplan-Meier plots were used to visualize the survival differences. Levels were divided into low and high levels (cut-off, 50%). P-values calculated using a log-rank test are shown in each plot. (B) CDK9 mRNA expression in different stages of colorectal cancer based on data btained from the GEPIA database. CDK9, cyclin-dependent kinase 9; COAD, colon adenocarcinoma; READ, rectum adenocarcinoma.
Table II.
Multivariate survival analyses of stage and CDK9 with overall survival of patients with COAD using the Cox proportional hazards model.
| Factor | Coefficient | HR | 95% CI_l | 95% CI_u | P-value |
|---|---|---|---|---|---|
| Stage II | 0.646 | 1.908 | 0.737 | 4.941 | 0.183 |
| Stage III | 1.166 | 3.208 | 1.243 | 8.277 | 0.016a |
| Stage IV | 2.241 | 9.406 | 3.629 | 24.381 | 0.000b |
| CDK9 | 0.625 | 1.868 | 1.151 | 3.033 | 0.011a |
Model: Survival (COAD) ~ Stage + CDK9. Data from a total of 445 patients were analysed, including 98 cases of mortality. Data was obtained from the TIMER database. The coefficient reads as a regression coefficient. HR, and its lower and upper 95% CI are shown.
P<0.05
P<0.001. 95% CI-l, lower 95% CI; 95% CI-u, upper 95% CI; CDK9, cyclin dependent kinase 9; COAD, colon adenocarcinoma; HR, hazard ratio.
Table III.
Multivariate survival analyses of stage and CDK9 with overall survival of patients with READ using the Cox proportional hazards model.
| Factor | Coefficient | HR | 95% CI_l | 95% CI_u | P-value |
|---|---|---|---|---|---|
| Stage II | 0.156 | 1.169 | 0.225 | 6.090 | 0.853 |
| Stage III | 0.726 | 2.066 | 0.426 | 10.026 | 0.368 |
| Stage IV | 1.657 | 5.244 | 1.104 | 24.907 | 0.037a |
| CDK9 | 0.823 | 2.277 | 0.775 | 6.696 | 0.135 |
Model: Survival (READ) ~ Stage + CDK9. Data from a total of 160 patients were analysed, including 23 cases of mortality. Data was obtained from TIMER database. The coefficient reads as a regression coefficient. HR, and its lower and upper 95% confidential interval are shown.
P<0.05. 95% CI_l, lower 95% CI; 95% CI_u, upper 95% CI; CDK9, cyclin dependent kinase 9; HR, hazard ratio; READ, rectum adenocarcinoma.
CDK9 is negatively associated with the infiltration of CD8+ T cells in CRC
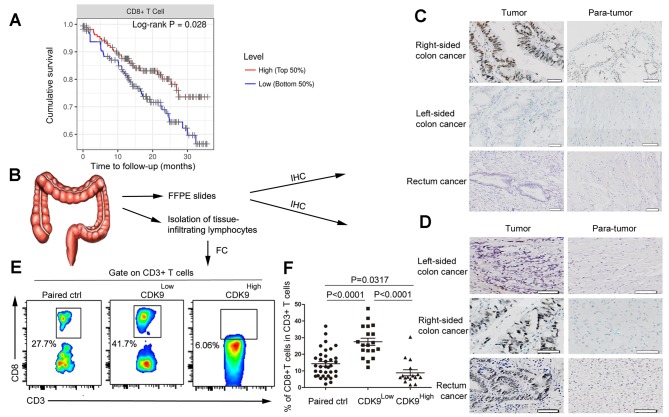
By analysing RNA sequencing (RNA-seq) data obtained from GEPIA, and the pathological estimation data from human CRC tumors, the present study revealed that patients with high expression levels of CDK9 and fewer CD8+ T cells in the TME exhibited poorer prognoses (Figs. 1A and 2A). Next it was determined whether CDK9 affected infiltration of CD8+ T cells in CRC. Specimens were collected from 35 patients with stage III–IV MSS CRC, and the clinical and pathological characteristics of the patients are shown in Table III. CDK9 expression in these patients was detected by immunohistochemical staining. The results demonstrated that the CDK9-positive expression rate (a score of 2) in stage III right-sided colon cancer was 85.7% (6/7), which was significantly higher than that in paracancerous tissues, with a positivity rate of 28.6% (2/7; χ2=4.667; P=0.03; Fig. 2B and C). However, there was no significant difference identified between CDK9 expression in all cases of left-sided colon cancer and rectal cancer (3/12) compared with the corresponding right-sided cancer (4/12; Fig. 2C). CDK9 expression in stage IV colon cancer was much higher than that in the paracancerous region, regardless of the primary tumor site (Fig. 2D). Clinically, the survival of patients with stage III–IV right-sided colon cancer was significantly shorter than that of patients with left-sided colon cancer (30,31). Therefore, these results suggested that abnormally high CDK9 expression predicted poor prognosis, and this may be a significant factor in the prognosis of patients with left/right-sided colon cancer.
Figure 2.
CDK9 is negatively associated with the infiltration of CD8+ T cells in colorectal cancer. (A) Kaplan-Meier curves for CD8+ T cell infiltration in colorectal cancer. Levels were divided into low and high levels (cut-off, 50%). The P-value was calculated using a log-rank test. (B) Experimental scheme. (C) Immunohistochemistry of CDK9 expression in stage III colorectal cancer tissues and adjacent non-cancerous tissues. Scale bar, 50 µm. (D) Immunohistochemistry of CDK9 expression in stage IV colorectal cancer tissues and adjacent non-cancerous tissues. Scale bar, 50 nm. (E) Flow cytometric analysis of tumor-infiltrating CD8+ T cells in stage III–IV colorectal cancer tissues and paired normal tissues. (F) Frequency of tumor-infiltrating CD8+ T cells in stage III–IV colorectal cancer tissues and paired normal tissues. Each dot represents data generated from one patient (n=35). CDK9, cyclin-dependent kinase 9; IHC, immunohistochemistry; FFPE, formalin-fixed and paraffin-embedded; FC, flow cytometry; Paired ctrl, paired normal control tissues; CDK9Low T, tumor tissues with low expression of cyclin-dependent kinase 9; CDK9High T, tumor tissues with high expression of cyclin-dependent kinase 9.
Additionally, cells were isolated from the fresh tumor samples and matched normal tissues of these patients. Flow cytometry was utilized to detect tumor infiltration by CD8+ T cells. According to the immunohistochemical staining results, patients with the MSS-phenotype CRC were divided into the CDK9 high- and low-expression groups, and the results of flow cytometry were analysed. The frequency of tumor-infiltrating CD8+ T cells in the CDK9 low-expression group was markedly higher than that in the CDK9 high-expression group (Fig. 2E and F). This result demonstrated that CDK9 inhibited the recruitment and infiltration of CD8+ T cells in the TME.
CDK9 is associated with genes responsible for immune cell migration and exhaustion in CRC
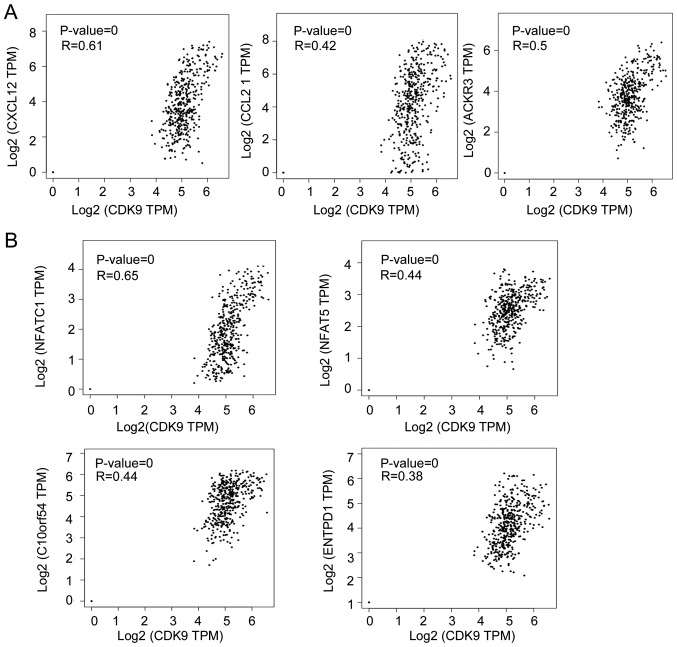
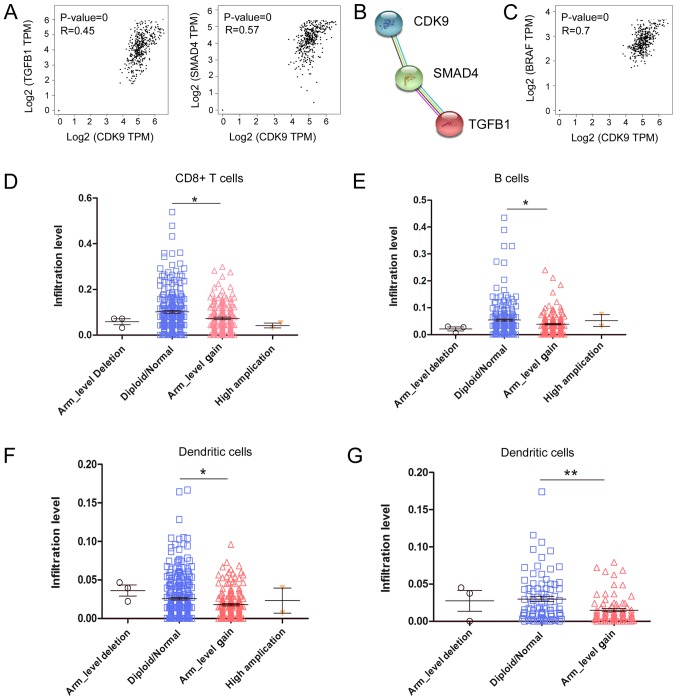
Subsequently, it was explored how CDK9 inhibited the recruitment and migration of CD8+ T cells to the TME by analysing RNA-seq data from CRC samples. As shown in Fig. 3A, CDK9 mRNA expression was positively correlated with the expression of C-X-C motif chemokine ligand 12 (CXCL12), C-C motif chemokine ligand 21 (CCL21) and atypical chemokine receptor 3 (ACKR3; also referred to as CXCR7), which are associated with lymphocyte migration. It has been reported that CXCL12 decreases the number of tumor-infiltrating natural killer (NK) cells and CD8+ T cells (32,33). These data indicated that CDK9 may be involved in CXCL12/CCL21/CXCR7-axis-mediated negative immune regulation in the TME. Since CDK9 was associated with the expression of numerous chemokines in the TME and affected the migration of immune cells, the present study investigated whether CDK9 mediated ‘editing’ of the TME, and the function and phenotype of immune cells. To explore this, the present study analysed the association between CDK9 and CD8+ T cell exhaustion-associated genes by analysing RNA-seq data from CRC. Notably, there was a significant positive correlation between the transcription of CDK9 and several genes that induce T lymphocyte exhaustion, including nuclear factor of activated T cells 1 (NFATC1), V-set immunoregulatory receptor, nuclear factor of activated T cells 5 (NFAT5) and ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1; also referred to as CD39; Fig. 3B). Additionally, the anti-inflammatory molecules transforming growth factor β1 (TGFB1) and SMAD4 were correlated with CDK9 expression in CRC tumors (Fig. 4A). Furthermore, CDK9 was co-expressed with TGFB1 and SMAD4 according to STRING database analysis (Fig. 4B). Interestingly, CDK9 was significantly positively correlated with BRAF mRNA expression, which serves an important role in the development of CRC (Fig. 4C). It was revealed that 26.2% (96/367) of patients with CRC had BRAF mutations, and their antitumor immunity was more inhibited than that of wild-type patients, based on analysis of the TIMER database. The decrease in tumor-infiltrating immune cells caused by the BRAF mutation was significantly different between colon and rectal cancer (Fig. 4D and E). Compared with wild-type rectal cancer (diploid/normal), the arm-level gain (affecting ≥50% of the chromosome) of BRAF led to decreased tumor-infiltrating CD8+ T cells (P<0.001), B cells (P<0.05) and dendritic cells (P<0.05) in colon cancer (Fig. 4D-F), whereas only tumor-infiltrating dendritic cells (P<0.01) were decreased in rectal cancer (Fig. 4G). These data indicated that CDK9 was involved in BRAF-mediated immunosuppression, which was affected by the site of the primary tumor. Overall, these data implied that CDK9 may contribute to CRC immune escape by promoting CD8+ T cell exhaustion and inhibiting the infiltration of numerous immune cell populations.
Figure 3.
CDK9 is correlated with genes responsible for immune cell migration and exhaustion in colorectal cancer. (A) Correlations of CDK9 expression with CXCL12, CCL21 and ACKR3 mRNA expression in CRC. (B) Correlations of CDK9 with NFATC1, NFAT5, C10orf54 and ENTPD1 mRNA expression in CRC. CRC, colorectal cancer; CDK9, cyclin-dependent kinase 9; CXCL12, C-X-C motif chemokine ligand 12; CCL21, C-C motif chemokine ligand 21; ACKR3, atypical chemokine receptor 3; NFATC1, nuclear factor of activated T cells 1; NFAT5, nuclear factor of activated T cells 5; C10orf54 V-set immunoregulatory receptor; ENTPD1, ectonucleoside triphosphate diphosphohydrolase 1; TPM, transcripts per million.
Figure 4.
CDK9 is associated with anti-inflammatory molecules in CRC. (A) Correlations of CDK9 with TGFB1 and SMAD4 mRNA expression in CRC. (B) Co-expression analysis of CDK9, TGFB1 and SMAD4 proteins. (C) Correlation of CDK9 with BRAF mRNA expression in CRC. (D) Influence of the BRAF mutation on CD8+ T cell infiltration in colon cancer. (E) Influence of the BRAF mutation on B cell infiltration in colon cancer. (F) Influence of the BRAF mutation on dentritic cell infiltration in colon cancer. (G) Influence of the BRAF mutation on dendritic cells infiltration in rectal cancer. Scatter plots are presented to show the distributions of each immune subset at each somatic copy number status for BRAF in colorectal cancer. The infiltration level for each category was compared with the diploid/normal group using one-way ANOVA followed by Tukey's multiple comparison test. *P<0.05, **P<0.001 vs. normal. CRC, colorectal cancer; CDK9, cyclin-dependent kinase 9; TGFB1, transforming growth factor ß1; SMAD4, SMAD family member 4; TPM, transcripts per million; COAD, colon adenocarcinoma; READ, rectum adenocarcinoma.
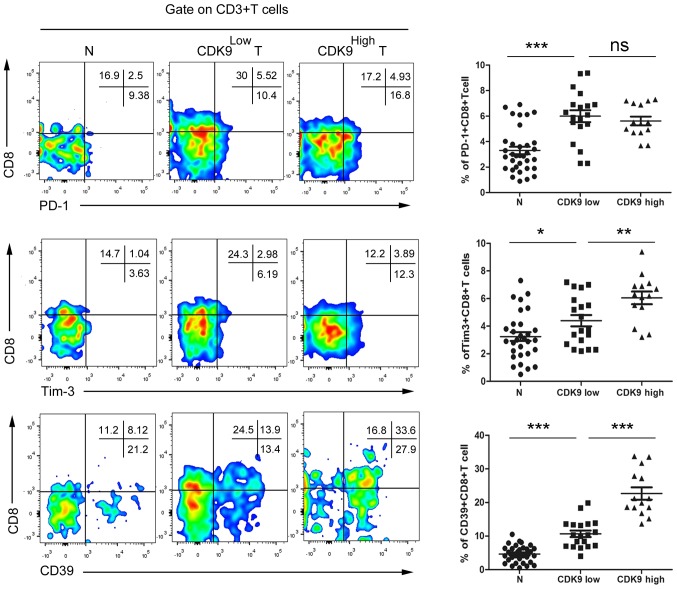
Association of CDK9 expression with tumor-infiltrating CD8+ T cell exhaustion in CRC
To confirm the association between CDK9 and tumor-infiltrating CD8+ T cell exhaustion, tumor-infiltrating cells were isolated from fresh CRC tissues and paired normal tissues from advanced CRC tumors, and the cells were co-cultured with antibodies and analysed by flow cytometry. According to the aforementioned results of the CDK9 immunohistochemistry, patients with MSS phenotype CRC were divided into two groups, a CDK9 high- and a CDK9 low-expression group, for statistical analysis. The results revealed that there were no significant differences identified in the CD8+PD-1+ T cell frequency between the CDK9high group and the CDK9low group (Fig. 5). Compared with the CDK9low group, the frequency of CD8+Tim-3+ T cells was increased by 37.5% in the CDK9high group (Fig. 5). Notably, the frequency of infiltrating CD8+CD39+ T cells in CDK9high CRC tumors was increased 1.12-fold compared with that in CDK9low tumors (Fig. 5). These data indicated that CDK9 was positively associated with tumor-infiltrating CD8+ T cell exhaustion, independent of the PD-1/PD-L1 signal, and may be used to evaluate the immune-type of the TME in patients with MSS mCRC.
Figure 5.
Association of CDK9 expression with tumor-infiltrating CD8+ T cell exhaustion in colorectal cancer. The frequencies of PD-1+CD8+ T cells, Tim-3+CD8+ T cells and CD39+CD8+ T cells were determined by flow cytometry. Each dot presents data generated from one patient (n=35). *P<0.05; **P<0.001; ***P<0.0001. CDK9, cyclin-dependent kinase 9; PD-1, programmed cell death 1; Tim-3, T-cell immunoglobulin mucin family member 3; N, paired normal tissue; CDK9Low, low expression of cyclin-dependent kinase 9; CDK9High, high expression of cyclin-dependent kinase 9; ns, not significant; T, tumor tissue.
Discussion
CDK9 has been widely used in the research and development of antitumor drugs due to its key regulatory role in transcription, and its contribution to the progression and maintenance of several types of cancer (34–37). A preclinical in vivo study demonstrated that treatment with a small-molecule CDK9 inhibitor impairs the growth of human melanoma xenografts (38). BAY 1143572, a novel and highly selective CDK9/P-TEFb inhibitor that is currently being investigated in phase I studies, decreases c-Myc and MCL1 apoptosis regulator levels in ATL-derived or HTLV-1-transformed lines, which inhibits their growth (39). It has been reported that CDK9 functions in immune responses and that its inhibition effectively suppresses the inflammatory response in chondrocytes (40). However, the regulatory effect of CDK9 on the antitumor immune response in CRC remains to be elucidated. The present study investigated CDK9 expression in different primary tumor sites in CRC and provided evidence that CDK9 expression was negatively associated with CD8+ T cell antitumor function in MSS mCRC.
With the development of precision medicine, it has become gradually understood that the prognoses of left- and right-sided CRC are different. Venook's CALGB/SWOG 80405 clinical data were published in ASCO in 2016 and revealed that the median survival of KRas wild-type right-sided and left-sided mCRCs was 19.4 and 34.2 months, respectively, and that of KRas mutant right-sided and left-sided mCRCs was 23.1 and 30.3 months, respectively (3). Therefore, the difference in primary tumor location has become a novel focus in CRC research. The results of the present study demonstrated that CDK9 expression in advanced left-sided CRC was significantly higher than that in early-stage left-sided CRC, and CDK9 expression in stage III–IV right-sided CRC was significantly higher than that in stage III–IV left-sided CRC. Notably, the prognostic difference between left-sided and right-sided CRC is only seen in tumors that are at least at stage III (30). Therefore, CDK9 may be a prognostic factor for left/right-sided colon cancer, which broadens the knowledge for future research.
Since the 5-year relative survival of patients with stage IV CRC is only 11.7%, regardless of primary tumor location, improving the diagnosis and treatment efficacy of stage IV patients is the key to ameliorating the survival of CRC (41). Anti-PD-1 immunotherapy has been demonstrated to be effective only in patients with MSI-H phenotype stage IV CRC, since they have more tumor-infiltrating immune cells (11). However, only 3.5–5% of all stage IV CRC cases are MSI-H (9). The present study determined CDK9 expression and the frequency of CD8+ T cells in tumor and paired normal control specimens of 35 patients with MSS-phenotype CRC, which accounts for >95% of all advanced CRC (9), and identified that CDK9 was negatively associated with tumor-infiltrating CD8+ T cells. This result suggested that CDK9 inhibition may benefit patients with MSS phenotype CRC in combination with anti-PD-1 immunotherapy.
The recruitment and migration of immune cells upon stimulation are regulated by numerous factors, of which the chemokine family serves an important role. It has been reported that fibroblasts help pancreatic cancer to develop chemotherapy resistance, and the chemokine CXCL12 is secreted by fibroblasts and prevents the infiltration of CD8+ T cells into the tumor (42). Zboralski et al (33) demonstrated that inhibition of CXCL12 by ‘NOX-A12’ can promote the infiltration of T cells and NK cells, thereby increasing the efficacy of anti-PD-1 drugs in colon cancer models. CXCR7/C-X-C motif chemokine receptor 4 heterodimer-induced histone demethylation promotes colorectal tumorigenesis (43). In addition, CCL21 is involved in T-cell migration and trafficking to secondary lymphoid organs (44). The present study revealed that CDK9 expression was positively correlated with CXCL12, ACKR3 (also referred to as CXCR7) and CCL21 expression, suggesting that CDK9 may inhibit CD8+ T cell infiltration via the CXCL12/CXCR7 axis.
The present study investigated how the TME ‘edits’ T cells in CRC and whether CDK9 serves a role in this process. tumor antigens are weakly immunogenic self-molecules, and the majority of tumor-specific T cells have a low T-cell receptor (TCR) affinity, since tumor-specific T cells with high avidity are cleared during the thymic selection process (45). Therefore, the process of antigen presentation is impaired in the TME, leading to insufficient priming and boosting of T cells (45). Consequently, the complex components in the TME drive T cells to terminally differentiate into ‘exhausted’ T cells (46). ‘Exhausted’ CD8+ T cells overexpress a large number of cell surface inhibitory receptors, including PD-1 and Tim-3. NFAT activates the nuclear factor protein of T cells, which results in inability of the NFATC1 gene to synergize with AP-1 during gene modification, resulting in loss of the TCR signal and upregulation of the expression of inhibitory receptors on the cell surface, thereby weakening the antitumor ability of CD8+ T cells (47). In addition, CD39 may be a functional surface marker for identifying exhausted CD8+ T cell subsets and regulating purine signalling pathways, and the combined blockade of the Tim-3 and PD-1 signalling pathways reverses the exhaustion of T cells that is induced by rectal cancer (48). The present study revealed that CDK9 expression was positively correlated with NFATC1, ENTPD1 (also referred to as CD39) and NFAT5 expression, suggesting that CDK9 may be involved in the ‘cancer immunoediting’ of T cells by the TME. Furthermore, CDK9 promoted CD39 and Tim-3 expression in tumor-infiltrating CD8+ T cells in MSS CRC tumors, whereas CDK9 did not significantly affect PD-1 expression.
To the best of our knowledge, the present study was the first to evaluate the association of CDK9 and tumor-infiltrating CD8+ T cells in MSS CRC. A number of studies have demonstrated that CDK9/P-TEFb is involved in the cell growth and survival of several types of cancer, including CRC (15,49,50). However, these studies have focused on the cancer cells themselves, ignoring the effect of CDK9 on the TME. The present study explored the association between CDK9 expression and CD8+ T cells in the TME of MSS CRC. Our previous study demonstrated that PHA767491, a selective CDK9 inhibitor, could impair the activation and proliferation of effector T cells but preserve the function of Treg cells in a mouse inflammatory model (51). To the best of our knowledge, no study has reported that a CDK9 inhibitor could alter anti-inflammatory molecules or T cells in CRC. The present study focused on the immune microenvironment of MSS CRC. In future studies, the effect of CDK9 inhibitors on tumor-infiltrating lymphocytes in CRC will be explored.
In conclusion, the present study demonstrated that CDK9 expression was positively associated with poor survival among patients with colon cancer and was negatively associated with CD8+ T cell antitumor function in MSS mCRC. These findings suggested that CDK9 may be utilized to evaluate the prognosis and the immune-type of the TME in patients with MSS mCRC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- mCRC
metastatic colorectal cancer
- CDK9
cyclin-dependent kinase 9
- ASCO
American Society of Clinical Oncology
- CFDA
China Food and Drug Administration
- MSI-H
microsatellite instability-high
- MSS
microsatellite stable
- dMMR
mismatch repair deficient
Funding
The present study was supported by grants from The Science and Technology Development Fund of Tianjin Education Commission for Higher Education (grant no. 2017KJ204) and Scientific Research Foundation of Tianjin Medical University Cancer Institute and Hospital (grant no. B1704), awarded to YZ.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the GEPIA repository, http://gepia.cancer-pku.cn/detail.php?gene=CDK9.
Authors' contributions
YZ and JL performed the majority of the experiments. FT collected clinical specimens. JW performed data analysis and interpretation. YZ and DK designed and supervised the study. DK wrote the manuscript. YZ revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Patient samples were collected at the Tianjin Medical University Cancer Institute and Hospital, and ethical approval was granted by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Written informed consent was obtained from all patients for participation in the present study.
Patient consent for publication
Written consent was obtained from all participants for the publication of data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Kibble A, Al-Shamahi A, Kuennemann K, Marqués F, Tremosa L, Cole P. Highlights from the 52nd Annual Meeting of the American Society of Clinical Oncology (ASCO) (June 3–7, 2016-Chicago, Illinois, USA) Drugs Today (Barc) 2016;52:407–423. doi: 10.1358/dot.2016.52.7.2533873. [DOI] [PubMed] [Google Scholar]
- 4.Xie S, Huang J, Qiao Q, Zang W, Hong S, Tan H, Dong C, Yang Z, Ni L. Expression of the inhibitory B7 family molecule VISTA in human colorectal carcinoma tumors. Cancer Immunol Immunother. 2018;67:1685–1694. doi: 10.1007/s00262-018-2227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Chen L, Zou L, Yang P, Wu R, Mao Y, Zhou H, Li R, Wang K, Wang W, et al. MiR-20b, −21, and −130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463–1473. doi: 10.1136/gutjnl-2016-311421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koopman M, Kortman GA, Mekenkamp L, Ligtenberg MJ, Hoogerbrugge N, Antonini NF, Punt CJ, van Krieken JH. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, Kaplan R, Quirke P, Seymour MT, Richman SD, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: A pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20:5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 11.De Smedt L, Lemahieu J, Palmans S, Govaere O, Tousseyn T, Van Cutsem E, Prenen H, Tejpar S, Spaepen M, Matthijs G, et al. Microsatellite instable vs. stable colon carcinomas: Analysis of tumor heterogeneity, inflammation and angiogenesis. Br J Cancer. 2015;113:500–509. doi: 10.1038/bjc.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beauchamp EM, Abedin SM, Radecki SG, Fischietti M, Arslan AD, Blyth GT, Yang A, Lantz C, Nelson A, Goo YA, et al. Identification and targeting of novel CDK9 complexes in acute myeloid leukemia. Blood. 2019;133:1171–1185. doi: 10.1182/blood-2018-08-870089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H, Seebacher NA, Hornicek FJ, Duan Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in osteosarcoma. EBioMedicine. 2019;39:182–193. doi: 10.1016/j.ebiom.2018.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahaman MH, Lam F, Zhong L, Teo T, Adams J, Yu M, Milne RW, Pepper C, Lokman NA, Ricciardelli C, et al. Mol Oncol; 2019. Targeting CDK9 for treatment of colorectal cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaiyapan W, Duangpakdee P, Boonpipattanapong T, Kanngern S, Sangkhathat S. Somatic mutations of K-ras and BRAF in Thai colorectal cancer and their prognostic value. Asian Pac J Cancer Prev. 2013;14:329–332. doi: 10.7314/APJCP.2013.14.1.329. [DOI] [PubMed] [Google Scholar]
- 17.Novellasdemunt L, Antas P, Li VS. Targeting Wnt signaling in colorectal cancer. A Review in the Theme: Cell Signaling: Proteins, Pathways and Mechanisms. Am J Physiol Cell Physiol. 2015;309:C511–C521. doi: 10.1152/ajpcell.00117.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C, Lipton L, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73:725–735. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 19.Choi SH, Estarás C, Moresco JJ, Yates JR, III, Jones KA. α-Catenin interacts with APC to regulate β-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev. 2013;27:2473–2488. doi: 10.1101/gad.229062.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Falco G, Leucci E, Onnis A, Bellan C, Tigli C, Wirths S, Cerino G, Cocco M, Crupi D, De Luca A, et al. Cdk9/Cyclin T1 complex: A key player during the activation/differentiation process of normal lymphoid B cells. J Cell Physiol. 2008;215:276–282. doi: 10.1002/jcp.21311. [DOI] [PubMed] [Google Scholar]
- 22.Kanazawa S, Okamoto T, Peterlin BM. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/S1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 23.Marshall RM, Salerno D, Garriga J, Graña X. Cyclin T1 expression is regulated by multiple signaling pathways and mechanisms during activation of human peripheral blood lymphocytes. J Immunol. 2005;175:6402–6411. doi: 10.4049/jimmunol.175.10.6402. [DOI] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta S, Provenzale D, Regenbogen SE, Hampel H, Slavin TP, Hall MJ, Llor X, Chung DC, Ahnen DJ, Bray T, et al. NCCN guidelines insights: Genetic/Familial high-risk assessment: Colorectal, version 3.2017. J Natl Compr Canc Netw. 2017;15:1465–1475. doi: 10.6004/jnccn.2017.0176. [DOI] [PubMed] [Google Scholar]
- 28.Hu H, Krasinskas A, Willis J. Perspectives on current tumor-node-metastasis (TNM) staging of cancers of the colon and rectum. Semin Oncol. 2011;38:500–510. doi: 10.1053/j.seminoncol.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Lee YH, Martin-Orozco N, Zheng P, Li J, Zhang P, Tan H, Park HJ, Jeong M, Chang SH, Kim BS, et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 2017;27:1034–1045. doi: 10.1038/cr.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park JH, Kim MJ, Park SC, Kim MJ, Hong CW, Sohn DK, Han KS, Oh JH. Difference in time to locoregional recurrence between patients with right-sided and left-sided colon cancers. Dis Colon Rectum. 2015;58:831–837. doi: 10.1097/DCR.0000000000000426. [DOI] [PubMed] [Google Scholar]
- 31.Meguid RA, Slidell MB, Wolfgang CL, Chang DC, Ahuja N. Is there a difference in survival between right- versus left-sided colon cancers? Ann Surg Oncol. 2008;15:2388–2394. doi: 10.1245/s10434-008-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zboralski D, Hoehlig K, Eulberg D, Frömming A, Vater A. Increasing tumor-infiltrating T cells through inhibition of CXCL12 with NOX-A12 synergizes with PD-1 blockade. Cancer Immunol Res. 2017;5:950–956. doi: 10.1158/2326-6066.CIR-16-0303. [DOI] [PubMed] [Google Scholar]
- 34.Minzel W, Venkatachalam A, Fink A, Hung E, Brachya G, Burstain I, Shaham M, Rivlin A, Omer I, Zinger A, et al. Small molecules Co-targeting CKIα and the transcriptional kinases CDK7/9 control AML in preclinical models. Cell. 2018;175:171.e25–185.e25. doi: 10.1016/j.cell.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahaman MH, Yu Y, Zhong L, Adams J, Lam F, Li P, Noll B, Milne R, Peng J, Wang S. CDKI-73: An orally bioavailable and highly efficacious CDK9 inhibitor against acute myeloid leukemia. Invest New Drugs. 2019;37:625–635. doi: 10.1007/s10637-018-0661-2. [DOI] [PubMed] [Google Scholar]
- 36.Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15:519–527. doi: 10.1080/15384101.2016.1138186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin T, Lallena MJ, Kreklau EL, Fales KR, Carballares S, Torrres R, Wishart GN, Ajamie RT, Cronier DM, Iversen PW, et al. A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol Cancer Ther. 2014;13:1442–1456. doi: 10.1158/1535-7163.MCT-13-0849. [DOI] [PubMed] [Google Scholar]
- 38.Abdullah C, Wang X, Becker D. Expression analysis and molecular targeting of cyclin-dependent kinases in advanced melanoma. Cell Cycle. 2011;10:977–988. doi: 10.4161/cc.10.6.15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narita T, Ishida T, Ito A, Masaki A, Kinoshita S, Suzuki S, Takino H, Yoshida T, Ri M, Kusumoto S, et al. Cyclin-dependent kinase 9 is a novel specific molecular target in adult T-cell leukemia/lymphoma. Blood. 2017;130:1114–1124. doi: 10.1182/blood-2016-09-741983. [DOI] [PubMed] [Google Scholar]
- 40.Yik JH, Hu Z, Kumari R, Christiansen BA, Haudenschild DR. Cyclin-dependent kinase 9 inhibition protects cartilage from the catabolic effects of proinflammatory cytokines. Arthritis Rheumatol. 2014;66:1537–1546. doi: 10.1002/art.38378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 42.Sleightholm RL, Neilsen BK, Li J, Steele MM, Singh RK, Hollingsworth MA, Oupicky D. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol Ther. 2017;179:158–170. doi: 10.1016/j.pharmthera.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Song ZY, Wang F, Cui SX, Gao ZH, Qu XJ. CXCR7/CXCR4 heterodimer-induced histone demethylation: A new mechanism of colorectal tumorigenesis. Oncogene. 2019;38:1560–1575. doi: 10.1038/s41388-019-0836-0. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Zhao L, Yang Y, Gao J, Hu C, Guo B, Zhu B. Cytotoxic chemotherapy reduces T cell trafficking to the spleen by downregulating the expression of C-C motif chemokine ligand 21 and C-C motif chemokine ligand 19. Oncol Lett. 2018;16:5013–5019. doi: 10.3892/ol.2018.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim PS, Ahmed R. Features of responding T cells in cancer and chronic infection. Curr Opin Immunol. 2010;22:223–230. doi: 10.1016/j.coi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baitsch L, Fuertes-Marraco SA, Legat A, Meyer C, Speiser DE. The three main stumbling blocks for anticancer T cells. Trends Immunol. 2012;33:364–372. doi: 10.1016/j.it.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Martinez GJ, Pereira RM, Äijö T, Kim EY, Marangoni F, Pipkin ME, Togher S, Heissmeyer V, Zhang YC, Crotty S, et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity. 2015;42:265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Zhang S, Hu Y, Yang Z, Li J, Liu X, Deng L, Wang Y, Zhang X, Jiang T, Lu X. Targeting PD-1 and Tim-3 pathways to reverse CD8 T-cell exhaustion and enhance ex vivo T-cell responses to autologous dendritic/tumor vaccines. J Immunother. 2016;39:171–180. doi: 10.1097/CJI.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 49.Franco LC, Morales F, Boffo S, Giordano A. CDK9: A key player in cancer and other diseases. J Cell Biochem. 2018;119:1273–1284. doi: 10.1002/jcb.26293. [DOI] [PubMed] [Google Scholar]
- 50.Boquoi A, Chen T, Enders GH. Chemoprevention of mouse intestinal tumorigenesis by the cyclin-dependent kinase inhibitor SNS-032. Cancer Prev Res (Phila) 2009;2:800–806. doi: 10.1158/1940-6207.CAPR-09-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan Y, Han Y, Sun H, Liang T, Zhang C, Song J, Hou G. Down-regulating cyclin-dependent kinase 9 of alloreactive CD4+ T cells prolongs allograft survival. Oncotarget. 2016;7:24983–24994. doi: 10.18632/oncotarget.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the GEPIA repository, http://gepia.cancer-pku.cn/detail.php?gene=CDK9.