Abstract
Histone H3K27 demethylase Jumonji domain-containing protein 3 (JMJD3) is involved in somatic cell differentiation and tumor progression; however, the underlying mechanisms of JMJD3 in cancer progression are yet to be fully explored. To improve understanding regarding the function of JMJD3 in brain tumor cells, the present study investigated the effects of JMJD3 on the epithelial-mesenchymal transition (EMT) and migration in glioma cells, and the underlying mechanisms involving the C-X-C motif chemokine ligand 12 (CXCL12)/C-X-C motif chemokine receptor 4 (CXCR4) axis. Immunohistochemical staining of a tissue microarray of glioma samples confirmed that JMJD3 overexpression could stratify highly metastatic glioma. The overexpression of JMJD3 induced a spindle-shaped morphology, promoted N-cadherin expression, inhibited E-cadherin expression and enhanced the migration ability of U-251MG and U-87MG American Type Culture Collection cells. The expression of E-cadherin and N-cadherin were assessed by western blotting and reverse transcription-quantitative polymerase chain reaction, and cell migration was evaluated using a Transwell migration assay and wound-healing. The overexpression of JMJD3 upregulated CXCL12 expression in a demethylase activity-dependent manner as ChIP assays revealed a decrease in H3K27 trimethylation at the CXCL12 promoter following overexpression of JMJD3 in U-87MG ATCC cells. Accordingly, CXCL12 overexpression was sufficient to rescue the suppressive effects of JMJD3 inhibition on the EMT and migration in glioma cells. In addition, CXCR4 expression was not regulated by JMJD3, but the interruption of CXCR4 caused by the CXCR4 inhibitor AMD3100 abolished the promotional effect of JMJD3 on EMT and migration in glioma cells. Collectively, these results suggested that JMJD3 promoted EMT and migration in glioma cells via the CXCL12/CXCR4 axis. The present study described a novel epigenetic mechanism regulating tumor cell EMT and migration, and provided a novel direction for glioma diagnosis and treatment.
Keywords: Jumonji domain-containing protein 3, C-X-C motif chemokine ligand 12, epithelial-mesenchymal transition, migration, glioma
Introduction
Transcriptional repression or activation of critical genes, as regulated by epigenetic regulation via histone methylation, serves critical roles in various biological processes, including embryonic development, tissue differentiation and tumorigenesis (1). The histone H3 lysine 27 (H3K27) demethylase Jumonji domain-containing protein 3 (JMJD3; also known as KDM6B) has been reported to affect the processes of tumorigenesis, invasion and metastasis (2–4). JMJD3 binds to, and activates, the cyclin dependent kinase inhibitor 2A locus in human fibroblasts (5,6). In glioma cells, JMJD3 serves as an oncogene and is essential for proliferation and migration (7). Numerous studies have suggested that another important aspect of JMJD3 function is its regulatory effect on cytokines (8–11). In lipopolysaccharide-activated microphages, JMJD3 directly transcriptionally promotes the expression of inflammatory cytokine genes by demethylating the promoter region H3K27 of these genes (8). In microglia, STAT1 and STAT3 regulate JMJD3 expression and activate a transcriptional network of inflammation-associated genes, which are required for inflammatory activation (9). Similar aberrant stimulation of cytokines controlled by JMJD3 has been observed in cancer (10). JMJD3 mediates the effects of STAT3 on the maintenance of the inhibition of differentiation-specific genes, and the self-renewal of normal neural and glioblastoma stem cells (10). Perrigue et al (11) reported that JMJD3 stimulates the secretion of inflammatory cytokines and senescence-associated secretory phenotype proteases in glioma cells, and promotes tumor cell migration.
Numerous chemokines/cytokines and receptors affect invasion and metastasis in the tumor microenvironment. The C-X-C motif chemokine ligand 12 (CXCL12)/C-X-C motif chemokine receptor 4 (CXCR4) pathway serves a critical role in the process of tumor invasion and metastasis (12,13). CXCL12, a ligand of CXCR4, is expressed in a number of organs and modulates multiple physiological processes, such as endocytosis, inflammatory processes and neovascularization (14–17). Previous studies have highlighted that the CXCR4/CXCL12 axis could mediate metastasis in various types of cancer (18–23). Epithelial-mesenchymal transition (EMT) is a dynamic process in which epithelial cells lose epithelial proteins and polarize to become mesenchymal cells, enhancing their metastatic ability; this process is associated with the invasion, dissemination and metastasis processes of tumor development (24,25). Previous studies have demonstrated that the CXCL12/CXCR4 signaling pathway promotes the EMT process in breast and gastric cancers, and glioblastoma cells (26–28).
The present study first demonstrated that the overexpression of JMJD3 stimulated EMT and significantly promoted migration in glioma cells. Additionally, the data demonstrated that JMJD3 promoted CXCL12 expression through the demethylation of H3K27 at its promoter region, and that the CXCL12/CXCR4 axis mediated the metastatic effect of JMJD3 in glioma cells.
Materials and methods
Tissue microarray
JMJD3 expression was examined on tissue microarrays (cat. no. HBra-Gli65PG-01; Shanghai Outdo Biotech Co., Ltd.) with samples from a cohort of 60 patients with glioma, which included 30 males and 30 females with an age range of 27–75 years (mean age, 54.7 years). The quantification of JMJD3 expression in glioma tissues was carried out according to a previously established method (29). Basic information of the patients with glioma is listed in Table SI.
Cell lines and reagents
Human glioma U-251MG cells and U-87MG American Type Culture Collection (ATCC; probable glioblastoma of unknown origin) cells were obtained from the Institute for Biological Sciences of the Chinese Academy of Sciences. Cells were grown in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences) and 100 U/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) with 5% CO2 at 37°C. The JMJD3-specific inhibitor GSK-J4 and CXCR4 antagonist plerixafor (AMD3100) were purchased from Selleck Chemicals and ApexBio Technology LLC, respectively.
Vector construction
The pCMV-JMJD3-HA vector was purchased from Addgene, Inc. The CXCL12 cDNA fragment from U87 ATCC cells was cloned into the Bam HI and XbaI restriction sites of the multiple cloning site of the pLV-EF1α-MCS-IRES-Bsd plasmid (BioSettia, Inc.) to generate the human CXCL12 expression plasmid. The short hairpin RNA (shRNA) sequence of JMJD3 (5′-GCATCTATCTGGAGAGCAAAC-3′) was searched and blasted using the BLOCK-iT™ RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/), and a scrambled sequence (5′-GCTACACTATCGAGCAA-3′) was used as a control for knockdown analysis. The shRNA was cloned into the pLV-H1-EF1α-puro vector (BioSettia, Inc., San Diego, CA, USA).
Protein and shRNA expression
Human glioma cells were transfected with 2 µg JMJD3/CXCL12/JMJD3shRNA/MCS/scrambled shRNA plasmids per well. A total of 1×105 cells per well were seeded into a 6-well plate and then transfected using Lipofectamine® 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The effects of the overexpression or suppression of JMJD3 or CXCL12 were investigated in cells that were either harvested for western blotting and quantitative (q)PCR, or were used for wound healing, Transwell and chromatin immunoprecipitation (ChIP) assays 48 h following transfection. The cell morphology of U-251MG and U-87MG ATCC cells after transfection were imaged with a Nikon light microscope (magnification, ×100).
RNA isolation, reverse transcription and qPCR
Total RNA was isolated from U-251MG and U-87MG ATCC cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was generated with an All-in-One First-Strand cDNA Synthesis kit (GeneCopoeia, Inc.). The reverse transcription were as follows: 25°C for 5 min, 42°C for 20 min and 85°C for 5 min. The experiment was performed according to the manufacturer's protocol. qPCR was performed on a StepOnePlus Real-Time PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) using an UltraSYBR Mixture kit (CoWin Biosciences, Inc. Beijing, China). The quantification cycle (Cq) value of the examined genes was normalized to the GAPDH level, and the relative fold expression was computed using the 2−∆∆Cq method (30). The reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers used for qPCR are listed in Table I.
Table I.
Primers used for reverse transcription-quantitative PCR and ChIP assays.
| Primer sequences (5′-3′) | ||
|---|---|---|
| Primer name | Forward primer | Reverse primer |
| Homo JMJD3 | CCACAGCGCCCTTCGATAC | CCATAGTTCCGTTTGTGCTCAAG |
| Homo CXCL12 | TCCAAACTGTGCCCTTCA | CTCTTCTTCTGTCGCTTCTT |
| Homo CXCR4 | CCCCATCCTCTATGCTTT | GAATGTCCACCTCGCTTT |
| Homo E-cadherin | ACAGCCCCGCCTTATGATT | TCGGAACCGCTTCCTTCA |
| Homo N-cadherin | CCGGTTTCATTTGAGGGCACATGC | GCCGTGGCTGTGTTTGAAAGGC |
| Homo Fibronectin | TCCAAGTTGATGCCGTTCCA | GAGAGAGCTTCTTGTCCTGTCT |
| Homo GAPDH | GCTGGCGCTGAGTACGTCGT | ACGTTGGCAGTGGGGACACG |
| ChIP-CXCL12-1 | CCTGAATCCCTGGTTCGC | GCCCTGGAGCTCGGAGAT |
| ChIP-CXCL12-2 | GGATCTCCGAGCTCCAGG | AGGCAGTGAGGTCCAGTGAA |
| ChIP-CXCL12-3 | CGACGGCTGCCTTCTTCA | GGTGGGAGTAGGTGTCTTCTGT |
| ChIP-CXCL12-4 | CAGAAGACACCTACTCCCACCA | CTTTGGGCCTCGCTTTGT |
| ChIP-CXCL12-5 | ACAAAGCGAGGCCCAAAG | TCCGTGCGACCCAAAGAG |
| ChIP-CXCL12-6 | TTCTCAGCTCTTTGGGTCG | AATGTAGCCTTTGTACTTCC |
| ChIP-CXCL12-7 | GGTCGGAAGTACAAAGGCTACA | GCTCGATCTGCGGGAATG |
| ChIP-CXCL12-8 | GCCACGCTGACTGCAAAGA | GCCACGCTGACTGCAAAGA |
| ChIP-CXCL12-9 | ATTCAGTTCCCGCCATCGA | GCTCCGGCCTTTGACCTTCT |
| ChIP-CXCL12-10 | CCAGCGGAGCCTGAGAAG | CCAGCGGAGCCTGAGAAG |
Primers with names starting with ‘homo’ were used for quantitative PCR, and primers with names starting with ‘ChIP’ were used for ChIP-PCR assay. ChIP, chromatin immunoprecipitation; CXCL12, C-X-C motif chemokine ligand 12; CXCR4, C-X-C motif chemokine receptor 4; JMJD3, Jumonji domain-containing protein 3.
Western blotting
To collect the whole cell lysates, 5×106 U-251MG and U-87MG cells were washed twice with cold PBS at 48 h after transfection and lysed in RIPA lysis buffer (CoWin Biosciences, Inc., Cambridge, MA, USA) containing protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The lysates were kept on ice for 30 min and centrifuged at 13,000 × g for 15 min at 4°C. The supernatant was collected and protein concentration was detected using a bicinchoninic acid assay. Target proteins (30 µg) were separated via 8–12% SDS-PAGE and then transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk (BioRad Laboratories, Inc.) in PBS for 1.5 h at room temperature and incubated at 4°C overnight with antibodies against JMJD3 (cat. no. ab38113; 1:500; Abcam), E-cadherin (cat. no. ab152102; 1:1,000; Abcam), N-cadherin (cat. no. ab19348; 1:1,000; Abcam), CXCL12 (cat. no. ab9797; 1:500; Abcam), CXCR4 (cat. no. ab77909; 1:500; Abcam), H3K27me3 (cat. no. ab6002; 1:1,000; Abcam) and β-actin (cat. no. A00702; 1:5,000; GenScript). Subsequently, the membranes were incubated at room temperature for 1 h with the secondary horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G (IgG; cat. nos. 7074 and 7076, respectively; 1:5,000; Cell Signaling Technology, Inc.), followed by detection using an enhanced chemiluminescent reagent (EMD Millipore). The grey values of the bands were quantified using Scion Image software (version 4.0.3.2; Scion Corporation, Frederick, MD, USA) and were normalized against β-actin (E-cadherin and N-cadherin protein expression was performed only in U-251MG cells transfected with shRNA).
Immunohistochemical (IHC) staining
The glioma tumor specimens were fixed in 4% paraformaldehyde for 24 h at room temperature, embedded in paraffin and then sliced into sections (3-µm thick). For IHC, the staining was performed using an SPlink Detection kit (cat. no. SP-9001; OriGene Technologies, Inc.) according to the manufacturer's protocol. Paraffin sections of glioma were dewaxed, hydrated in 100, 95, 90, 85 and 75% alcohol solutions, and washed with PBS three times for 5 min each. For antigen retrieval, the sections were boiled in 10 mM sodium citrate buffer (pH 6.0) and maintained at a sub-boiling temperature for 10 min. Slides were cooled on the bench top to room temperature and sections were incubated in 3% hydrogen peroxide for 10 min, then the sections were blocked with goat serum working solution (cat. no. SP-9001) for 1 h at room temperature. Sections were incubated at 4°C overnight with the JMJD3 antibody (cat. no. ab38113; 1:50; Abcam), incubated with a biotinylated secondary antibody (cat. no. SP-9001; OriGene Technologies, Inc.) for 1.5 h at room temperature and avidin-peroxidase for 30 min at room temperature, stained with DAB for 1.5 min at room temperature and counterstained with hematoxylin for 1 min at room temperature. The slides were dehydrated and mounted with neutral resin. Images were captured at three different fields of each sample with a Nikon light microscope (magnification, ×100).
ChIP
For the ChIP experiments, 1×107 U-87MG ATCC cells were transfected with the JMJD3 vectors for 48 h at 37°C. Cross-linking with 1% formaldehyde for 10 min at room temperature, collection, sonication (non-contact sonication: time on 30 sec, time off 30 sec, 15 cycle at 4°C) and immunoprecipitation were performed using an EZ-ChIP™ kit (cat. no. 17-371; EMD Millipore) according to the manufacturer's protocol. Antibodies against H3K27me3 (cat. no. ab6002; 1:50; Abcam) were used for immunoprecipitation. Antibodies against RNA polymerase (cat. no. 17-371; 1:50; EMD Millipore) and normal mouse IgG (cat. no. 17-371; 1:50; EMD Millipore) were used as the positive and negative controls, respectively. ChIP-PCR was performed using a 2×Es Taq MasterMix (Dye) kit (CoWin Biosciences, Inc.) to detect the immunoprecipitated DNA fragments of the CXCL12 promoter. The thermocycling conditions were as follows: 94°C For 2 min, followed by 30 cycles of 94°C for 30 sec, 56°C for 30 sec and 72°C for 30 sec, 72°C for 2 min. A 1% agarose gel and ethidium bromide were used for agarose gel electrophoresis. The primer sequences used for ChIP-PCR are listed in Table I.
Wound healing and Transwell migration assays
For each test group, 1×105 U-251MG or U-87MG ATCC cells were treated with GSK-J4 (5 µM) or AMD3100 (500 nM) for 24 h and subsequently transfected with pCMV-JMJD3-HA, pLV-EF1α-CXCL12-IRES-Bsd, pLV-shJMJD3-H1-EF1α-puro or a control vector for 48 h at 37°C. The cells were incubated in serum-free medium at 37°C in a humidified atmosphere of 5% CO2, the wound healing (31) and Transwell assays (30) were performed according to established protocols. Wound healing was performed only in U-251MG cells transfected with the shRNA group. For transwell assay, the cells which had migrated to the bottom side of the chamber were fixed with absolute methanol at −20°C for 15 min and then stained with DAPI (1:1,000) for 2 min at room temperature.
Statistical analysis
Statistical analysis of all data was conducted using SPSS software (version 16.0; SPSS, Inc.). A total of three independent experiments were performed for statistical analysis, and the results are presented as the mean ± the standard error of the mean. Statistical significance among different groups was determined with one-way ANOVA followed by a Bonferroni post hoc test or χ2 analysis. An unpaired t-test was used to compare the means of two independent groups. P<0.05 was considered to indicate a statistically significant difference.
Results
JMJD3 expression in glioma is positively associated with the clinical tumor metastatic progress
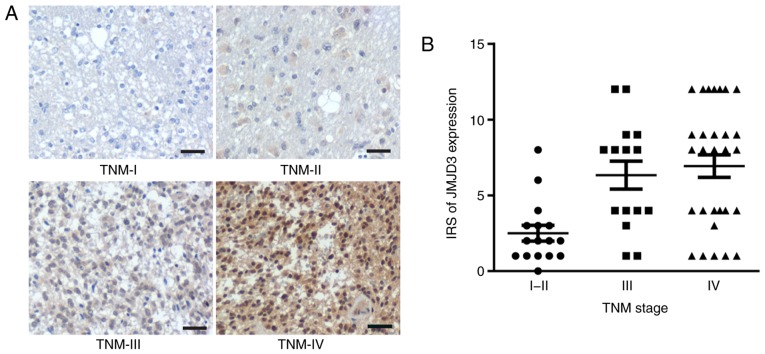
To investigate the association between JMJD3 expression in glioma and the clinical tumor metastatic status, IHC staining of JMJD3 on a tissue microarray containing 60 surgical glioma samples was performed. The results of IHC staining demonstrated that JMJD3 expression in human glioma tissue was associated with stage, JMJD3 expression was lower in glioma tissue of TNM I and II, medium in tissue of TNM III and higher in tissue of TNM IV (Fig. 1). JMJD3 expression was positively associated with TNM stage (Table II). Taken together, these observations suggested a general tumor-facilitating role of JMJD3 in the metastatic progression of glioma.
Figure 1.
JMJD3 expression is positively associated with the clinical progression of glioma. (A) Representative images of the immunohistochemical staining of JMJD3 on a glioma tissue microarray. Scale bar, 100 µm. (B) Distribution of JMJD3 expression in glioma tissues at different TNM stages. IRS, immunoreactivity score; JMJD3, Jumonji domain-containing protein 3.
Table II.
Association between JMJD3 expression and TNM stage of patients with glioma.
| IRS of JMJD3 expression | ||||
|---|---|---|---|---|
| Tissue classification | 0–4 (%) | 6–12 (%) | Total | P-value |
| TNM stage | <0.05 | |||
| I–II | 15 (93.8) | 1 (6.2) | 16 | |
| III | 7 (46.7) | 8 (53.3) | 15 | |
| IV | 12 (41.4) | 17 (58.6) | 29 | |
IRS is a method to quantify the protein expression in tissues. IRS, immunoreactivity score; JMJD3, Jumonji domain-containing protein 3; TNM, Tumor-Node-Metastasis.
Overexpression of JMJD3 promotes EMT in glioma cells
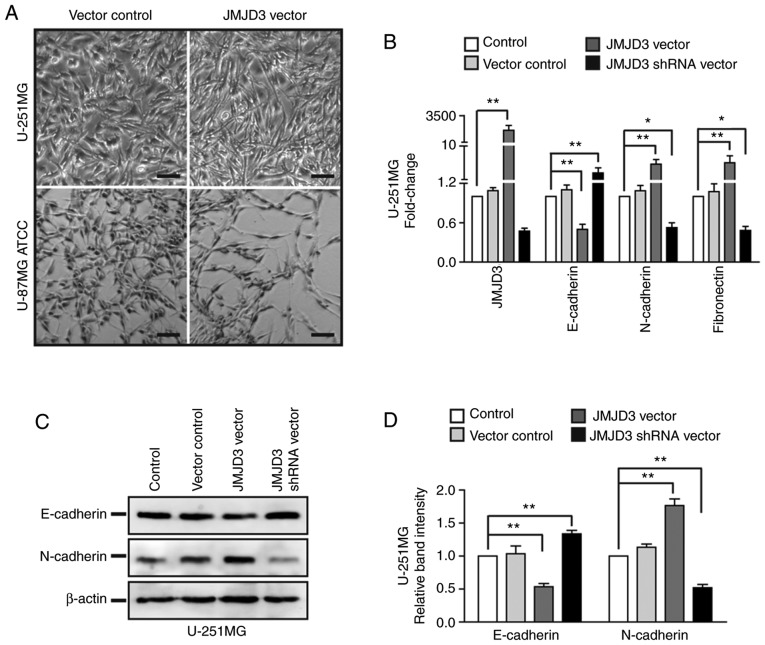
To further explore the role of JMJD3 in glioma metastasis, JMJD3 overexpression or depletion was established in U-251MG and U-87MG ATCC cells by transfection with a JMJD3 expression vector or a JMJD3 shRNA vector for 48 h (Fig. S1A and B). The microscopy results demonstrated that the overexpression of JMJD3 induced a spindle-shaped morphology in U-251MG and U-87MG ATCC cells (Fig. 2A). As EMT is a process in which epithelial cells lose cell-cell adhesion proteins and acquire mesenchymal molecules to enhance their motile ability, and affect cancer progression and migration through multiple mechanisms, the present study examined whether JMJD3 affected EMT-associated gene expression. In U-251MG cells, JMJD3 overexpression markedly decreased E-cadherin expression and increased the expression of N-cadherin and fibronectin. Accordingly, JMJD3 knockdown increased E-cadherin and decreased N-cadherin and fibronectin expression (Fig. 2B-D). These results suggested that JMJD3 promoted the EMT process in glioma cells.
Figure 2.
Upregulation of JMJD3 promotes EMT in glioma cells. (A) Microscopy images of morphological alterations of U-251MG and U-87MG ATCC cells following JMJD3 overexpression. Scale bar, 100 µm. (B) mRNA expression levels of JMJD3 and EMT-associated genes (E-cadherin, N-cadherin and fibronectin) in U-251MG cells in which JMJD3 was overexpressed or depleted. (C) Western blot analysis examining the expression levels of EMT-associated proteins (E-cadherin and N-cadherin) in U-251MG cells in which JMJD3 was overexpressed or depleted. (D) Relative protein expression levels of E-cadherin and N-cadherin was determined in U-251MG cells. β-actin was used as an endogenous control. *P<0.05, **P<0.01. ATCC, American Type Culture Collection; EMT, epithelial-mesenchymal transition; JMJD3, Jumonji domain-containing protein 3; shRNA, short hairpin RNA.
JMJD3 increases migration in glioma cells
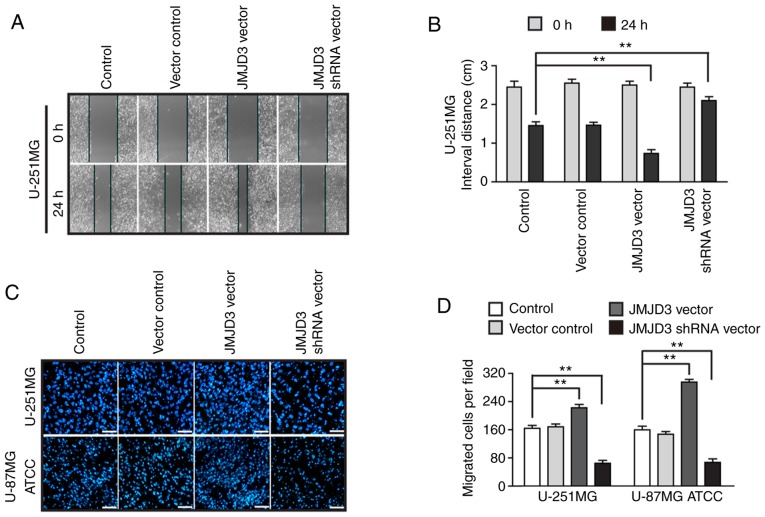
To further assess the function of JMJD3 in tumor metastasis, wound healing assays were carried out using U-251MG cells in which JMJD3 had been overexpressed or silenced (Fig. S1A and B). As shown in Fig. 3A, compared with control cells, the U-251MG cells overexpressing JMJD3 exhibited enhanced migratory capability at 24 h. However, the migratory capability was decreased in cells in which JMJD3 had been silenced compared with that in the control cells (Fig. 3B). The results were further confirmed by a Transwell assay, which is another method to test the migratory properties of cancer cells (Fig. 3C and D). The data revealed that JMJD3 overexpression enhanced the serum-induced migratory abilities of U-251MG and U-87MG ATCC cells compared with that of the control cells.
Figure 3.
Ectopic overexpression of JMJD3 promotes the migration of glioma cells. (A) Wound healing assay to evaluate the migration ability of U-251MG cells in which JMJD3 was overexpressed or depleted. Magnification, ×10. (B) Quantification results of the interval distance in the wound healing assay. (C) Representative DAPI staining images and (D) statistical results for the Transwell assay of wild type control, vector control, JMJD3 overexpressing and JMJD3 depleted U-251MG and U-87MG ATCC cells. Scale bar, 200 µm. **P<0.01. American Type Culture Collection; JMJD3, Jumonji domain-containing protein 3; shRNA, short hairpin RNA.
CXCL12 is a target gene regulated by JMJD3
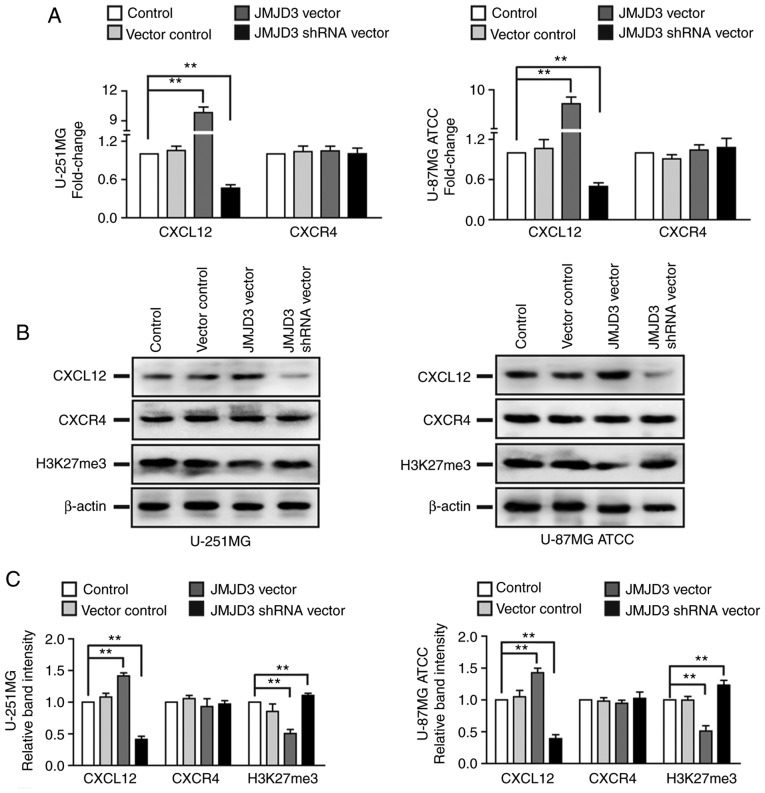
Considering the promoting effects of JMJD3 on the migration of glioma cells and the critical role of the CXCL12/CXCR4 axis in cancer cell metastasis, the present study subsequently assessed whether JMJD3 affected the ligand CXCL12 or its receptor CXCR4 in glioma cells. qPCR and western blotting revealed that JMJD3 overexpression increased the mRNA and protein expression levels of CXCL12 and decreased the protein expression level of H3K27me3 in U-251MG and U-87MG ATCC cells compared with those of control cells. Accordingly, JMJD3 knockdown inhibited CXCL12 and increased H3K27me3 expression compared with that of the control cells; however, the mRNA or protein expression levels of CXCR4 were not affected by JMJD3 knockdown/overexpression (Fig. 4). Therefore, CXCL12 expression was further analyzed.
Figure 4.
JMJD3 regulates the expression levels of CXCL12. (A) mRNA expression levels of CXCL12 and its receptor CXCR4 in U-251MG (left panel) and U-87MG ATCC (right panel) cells in which JMJD3 was overexpressed or depleted. (B) Western blotting results revealing CXCL12, CXCR4 and H3K27me3 protein expression in U-251MG (left panel) and U-87MG ATCC (right panel) cells in which JMJD3 was overexpressed or depleted. (C) Relative protein expression of CXCL12, CXCR4 and H3K27me3 was determined in U-251MG (left panel) and U-87MG ATCC (right panel) cells. β-actin was used as an endogenous control. **P<0.01. ATCC, American Type Culture Collection; CXCL12, C-X-C motif chemokine ligand 12; CXCR4, C-X-C motif chemokine receptor 4; H3K27me3, histone H3 lysine 27 trimethyl; JMJD3, Jumonji domain-containing protein 3; shRNA, short hairpin RNA.
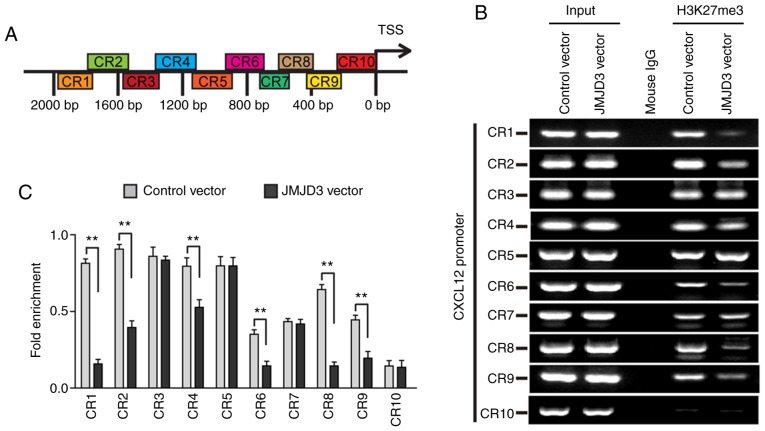
A ChIP assay was performed to evaluate the influence of JMJD3 on the H3K27 trimethylation of the CXCL12 promoter region. The results demonstrated that a remarkable decrease in H3K27 trimethylation was observed at the CXCL12 promoter following JMJD3 overexpression compared with that of the control cells (Fig. 5). Taken together, the data indicated that JMJD3 regulated CXCL12 expression by decreasing the trimethylation of H3K27 at the CXCL12 promoter.
Figure 5.
JMJD3 regulates H3K27 trimethylation at the CXCL12 promoter. (A) Distribution of the tested DNA fragments of the CXCL12 promoter region in ChIP. (B) ChIP-PCR results of the immunoprecipitated DNA fragments of CXCL12 promoter in U-87MG ATCC cells in which JMJD3 was overexpressed or not. Input was the loading control, and mouse IgG was the negative control. (C) Quantification of the ChIP-PCR results. **P<0.01. ChIP, chromatin immunoprecipitation; CR, combine Region; CXCL12, C-X-C motif chemokine ligand 12; H3K27me3, histone H3 lysine 27 trimethyl; IgG, immunoglobulin G; JMJD3, Jumonji domain-containing protein 3; TSS, transcription start site.
CXCL12 mediates the effect of JMJD3 on metastasis in glioma cells
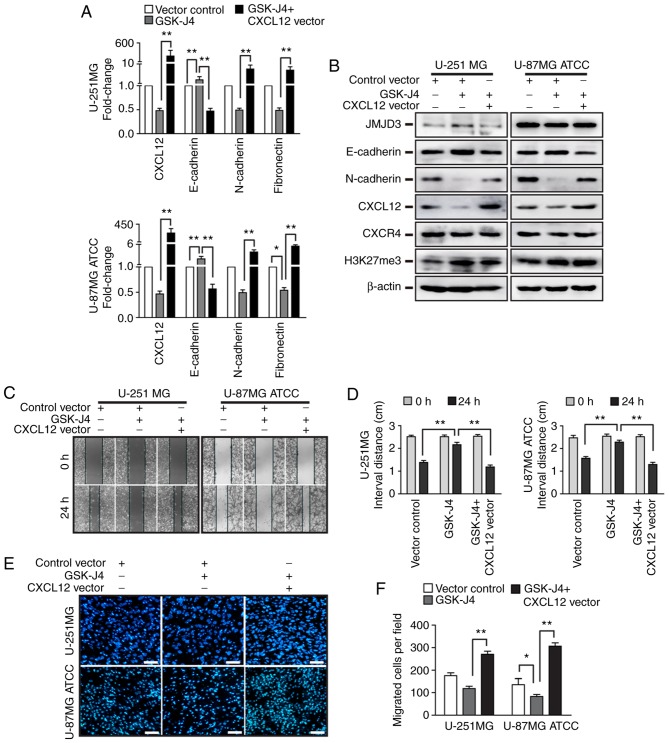
To investigate whether CXCL12 is involved in the regulation of EMT and migration elicited by JMJD3 overexpression in glioma cells, CXCL12 was overexpressed in U-251MG and U-87MG ATCC cells (Fig. S1E and F). Furthermore, CXCL12 was overexpressed in U-251MG and U-87MG ATCC cells that were pretreated with the JMJD3 inhibitor GSK-J4 for 24 h (Fig. S1E and F). Consistent with the aforementioned observations, the inhibition of JMJD3 activity with GSK-J4 decreased CXCL12 expression, affected the expression levels of EMT-associated genes (increased E-cadherin and decreased N-cadherin) and reduced the migratory ability of glioma cells (Fig. 6). In agreement with these results, CXCL12 overexpression was sufficient to affect the expression of EMT-associated genes, induce migration in glioma cells (CXCL12 vector group in Fig. 7) and rescue the inhibitory effect of JMJD3 on EMT and migration in U-251MG and U-87MG ATCC cells. Taken together, these results suggested that CXCL12 is a downstream target gene that mediates the EMT and migration effect of JMJD3 in glioma cells.
Figure 6.
Overexpression of CXCL12 rescues the EMT and migration abilities that were suppressed by GSK-J4. U-251MG and U-87MG ATCC cells were treated with GSK-J4 alone or GSK-J4 following the ectopic expression of CXCL12. (A) mRNA expression levels of CXCL12 and EMT-associated genes (E-cadherin, N-cadherin and fibronectin) in U-251MG (upper panel) and U-87MG ATCC (lower panel) cells. (B) Protein expression levels of JMJD3, E-cadherin, N-cadherin, CXCL12, CXCR4 and H3K27me3 were determined by western blotting in U-251MG and U-87MG ATCC cells treated with GSK-J4 or GSK-J4 and CXCL12 overexpression. (C) Wound healing assay to evaluate the migration ability of U-251MG and U-87MG ATCC cells treated with GSK-J4 or GSK-J4 and CXCL12 overexpression. Magnification, ×10. (D) Statistical results for the interval distance of the wound healing assay. (E) Representative DAPI staining images and (F) statistical results from the Transwell assay of the vector control, GSK-J4 treated and GSK-J4 plus CXCL12 overexpressing U-251MG and U-87MG ATCC cell groups. Scale bar, 200 µm. *P<0.05, **P<0.01. ATCC, American Type Culture Collection; CXCL12, C-X-C motif chemokine ligand 12; EMT, epithelial-mesenchymal transition; H3K27me3, histone H3 lysine 27 trimethyl.
Figure 7.
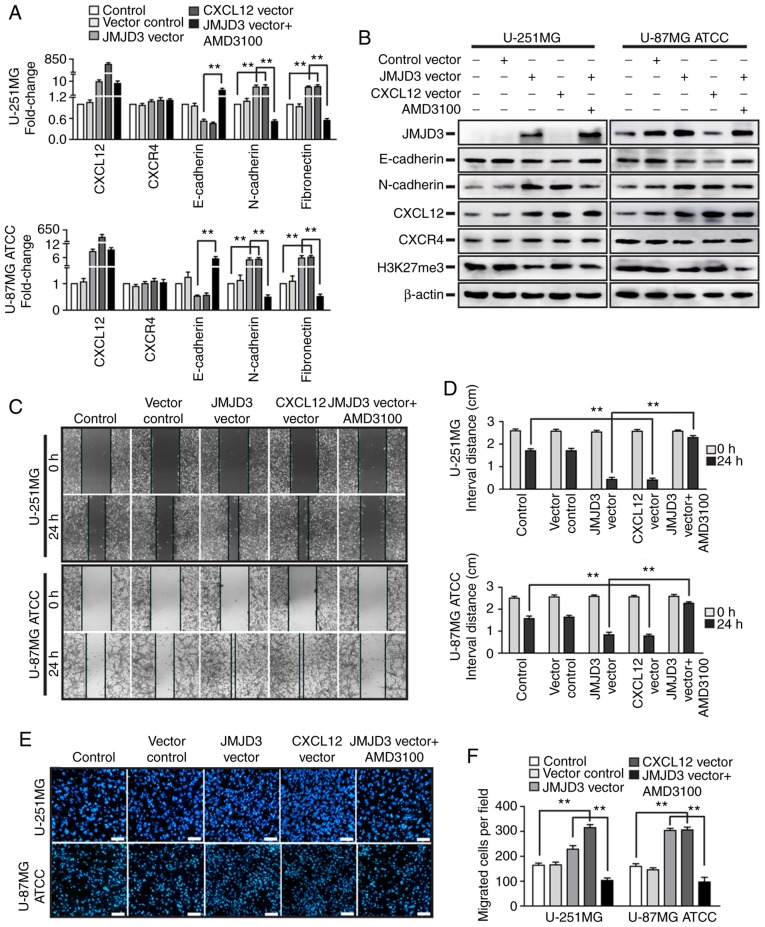
CXCL12-CXCR4 axis mediates the effects of JMJD3 on the epithelial-mesenchymal transition and migration in glioma cells. U-251MG and U-87MG ATCC cells were ectopically expressing JMJD3 or CXCL12, respectively, or JMJD3 plus pretreatment with AMD3100. (A) mRNA expression levels of CXCL12, CXCR4, E-cadherin, N-cadherin and fibronectin in U-251MG (upper panel) and U-87MG ATCC (lower panel) cells. (B) Protein expression levels of JMJD3, E-cadherin, N-cadherin, CXCL12, CXCR4 and H3K27me3 were determined by western blotting in U-251MG and U-87MG ATCC cells. (C) Wound healing assay to evaluate the motility of the wild-type control, vector control, JMJD3 overexpression, CXCL12 overexpression and JMJD3 overexpression plus pretreatment with AMD3100 U-251MG and U-87MG ATCC cells. Magnification, ×10. (D) Statistical results for the interval distance of the wound healing assay. (E) Representative DAPI staining images and (F) quantification of fields in the Transwell assay for the wild-type control, vector control, JMJD3 overexpression, CXCL12 overexpression and JMJD3 overexpression and pretreatment with AMD3100 U-251MG and U-87MG ATCC cell groups. Scale bar, 200 µm. **P<0.01. ATCC, American Type Culture Collection; CXCL12, C-X-C motif chemokine ligand 12; CXCR4, C-X-C motif chemokine receptor 4; H3K27me3, histone H3 lysine 27 trimethyl; JMJD3, Jumonji domain-containing protein 3.
CXCR4 is required for the promotional effects of JMJD3 on the EMT and migration of glioma cells
In the tumor microenvironment, numerous chemokines and receptors affect EMT and metastasis. The CXCL12/CXCR4 pathway-associated proteins were observed to be aberrantly expressed in various human types of cancer and to serve a critical role in tumor metastasis. However, the aforementioned results revealed that the ectopic expression of JMJD3 did not influence CXCR4 expression (Fig. 4).
To further investigate whether CXCR4 is essential for the ability of JMJD3 to enhance metastatic properties, JMJD3 was overexpressed in U-251MG and U-87MG ATCC cells that were pretreated with AMD3100 for 24 h (Fig. S1C and D), and the expression levels of EMT-associated genes and the migratory ability of cells were assessed. Notably, the results of qPCR and western blotting demonstrated that pretreatment with AMD3100 abolished the inhibition of E-cadherin and the promotion of N-cadherin that had been induced by JMJD3 overexpression (Fig. 7A and B). The results of wound healing and Transwell assays revealed that JMJD3 overexpression increased the migration ability of U-251MG and U-87MG ATCC cells compared with that of the control cells. Furthermore, pretreatment with AMD3100 abolished the effect of JMJD3 overexpression (Fig. 7C-F). Collectively, these data suggested that the binding of CXCL12 to CXCR4 mediated the effects of the JMJD3-CXCL12 axis on EMT and metastasis in glioma cells.
Discussion
Previous studies have demonstrated that the process of tumor metastasis is closely associated with EMT (24,25). The crucial feature of the EMT process is that epithelial cells lose cell-cell adhesion molecules, including E-cadherin (32,33), and express mesenchymal proteins, such as N-cadherin, to acquire motility (34), which could enhance the invasion of tumor cells into adjacent tissues and promote metastasis to distant sites (35). Numerous studies have also demonstrated that various signaling pathways are involved in the regulation of EMT in tumor cells or in the microenvironment (21,36–38). It has been well documented that the CXCL12/CXCR4 axis promotes EMT and metastasis in numerous different types of cancer cells. For example, the CXCL12/CXCR4 axis increases microRNA-125b expression and induces EMT in human colorectal cancer (21). Additionally, the overexpression of CXCL12 and CXCR4 in esophageal cancer stem cells accelerates esophageal tumor spread and metastasis (39). Conversely, blocking the CXCL12/CXCR4 axis with a neutralizing antibody or antagonist of CXCR4 interrupts the tumor metastasis process (40). The present study demonstrated that JMJD3 activated the CXCL12/CXCR4 axis by regulating CXCL12 expression, and enhanced EMT and migration in glioma cells.
Based on previous studies, it has been well established that JMJD3 is closely associated with the epigenetic regulation of development, differentiation and senescence (8,10,11,41). JMJD3 induces EMT and increases the motile ability of tumor cells, which conflicts with its suppressive role in the induction of tumor cell senescence (5,6). Several studies have reported that JMJD3 is downregulated in human types of cancer and acts as a tumor suppressor (2,5). In addition, JMJD3 has been demonstrated to be overexpressed in several types of cancer and to be associated with tumor progression (10,42). Consistent with these studies, the present study revealed that JMJD3 was overexpressed in high-grade glioma tissue, and the ectopic overexpression of JMJD3 promoted EMT and metastasis in glioma cells. Conversely, JMJD3 knockdown suppressed this process. In accordance with the results of the present study, JMJD3 has been reported to exhibit similar effects in different types of cancer cells (43–45). These results implied that the regulation of EMT and metastasis is another important mechanism via which JMJD3 contributes to tumor progression.
The present study demonstrated that JMJD3 regulated CXCL12 expression at the mRNA and protein levels. As JMJD3 is a histone H3K27me3 demethylase, accumulating evidence has revealed that JMJD3 regulates gene expression via two distinct mechanisms: A demethylase-dependent mechanism and a demethylase- independent mechanism (46,47). Therefore, the present study further demonstrated that JMJD3 regulated CXCL12 expression via a demethylase-dependent mechanism in glioma cells. Additionally, overexpression of CXCL12 rescued the suppressive effect of GSK-J4 on the EMT and migration processes of glioma cells. The results of the present study indicated that CXCL12 mediated the promoting effects of JMJD3 on EMT and metastasis in glioma cells, and was directly induced by JMJD3. Of note, it has been reported that JMJD3 induces EMT and enhances metastasis by regulating snail family transcriptional repressor 2 (SLUG) expression in clear cell renal cell carcinoma (44). Additionally, SLUG promotes cell invasion and metastasis via the CXCL12/CXCR4 axis in prostate cancer cells (48). These findings imply that JMJD3 may also regulate the CXCL12/CXCR4 axis via SLUG in an indirect manner in cancer, but direct evidence is required to further confirm this hypothesis.
The receptor of CXCL12, CXCR4, has been demonstrated to be widely expressed in a variety of tumors (49,50) and has also been identified to serve crucial roles in the process of cancer growth, invasion and migration (51,52). In the present study, it was examined whether JMJD3 directly upregulated CXCR4 expression in glioma cells. Notably, the data revealed that there was no association between JMJD3 and CXCR4 expression, but blocking CXCR4 with the CXCR4 antagonist AMD3100 abolished the effects of JMJD3 on the EMT and migration of glioma cells. The data indicated that CXCR4 was involved in the regulation of the migration process in glioma cells and was required for the JMJD3-CXCL12-CXCR4 axis. In addition, previous studies have demonstrated that JMJD3 is involved in tumorigenesis, senescence, apoptosis and the maintenance of stemness in cancer by stimulating the secretion of multiple inflammatory cytokines and through other mechanisms (11,53,54). Therefore, JMJD3 is extensively involved in the regulation of various aspects of cancer progression, and targeting JMJD3 may be a feasible novel therapeutic method for the treatment of various types of cancer.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Long Shen (Department of Immunology of the Medical School, Nankai University) for providing the pCMV-JMJD3-HA and pLV-EF1α-CXCL12-IRES-Bsd vectors.
Funding
The present study was supported by the Nature Science Foundation of Tianjin (grant no. 18JCQNJC12200) and Logistics University of People's Armed Police Force Grant (grant no. WHB201502).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YZ designed the experiments, drafted the manuscript and analyzed the data. SZ and DZ performed the experiments. ZX and XW participated in the study design, assisted with the data analysis and helped to revise the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu SH, Zhu KY, Chen J, Liu XZ, Xu PF, Zhang W, Yan L, Guo HZ, Zhu J. JMJD3 facilitates C/EBPβ-centered transcriptional program to exert oncorepressor activity in AML. Nat Commun. 2018;9:3369. doi: 10.1038/s41467-018-05548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams K, Christensen J, Rappsilber J, Nielsen AL, Johansen JV, Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS One. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubner MR, Spector DL. Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation. Cold Spring Harb Symp Quant Biol. 2010;75:43–49. doi: 10.1101/sqb.2010.75.020. [DOI] [PubMed] [Google Scholar]
- 5.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui A, Xu Y, Li Y, Hu Q, Wang Z, Zhang H, Yang J, Guo X, Zhao W. The pharmacological role of histone demethylase JMJD3 inhibitor GSK-J4 on glioma cells. Oncotarget. 2017;8:68591–68598. doi: 10.18632/oncotarget.19793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Przanowski P, Dabrowski M, Ellert-Miklaszewska A, Kloss M, Mieczkowski J, Kaza B, Ronowicz A, Hu F, Piotrowski A, Kettenmann H, et al. The signal transducers Stat1 and Stat3 and their novel target Jmjd3 drive the expression of inflammatory genes in microglia. J Mol Med (Berl) 2014;92:239–254. doi: 10.1007/s00109-013-1090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherry-Lynes MM, Sengupta S, Kulkarni S, Cochran BH. Regulation of the JMJD3 (KDM6B) histone demethylase in glioblastoma stem cells by STAT3. PLoS One. 2017;12:e0174775. doi: 10.1371/journal.pone.0174775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrigue PM, Silva ME, Warden CD, Feng NL, Reid MA, Mota DJ, Joseph LP, Tian YI, Glackin CA, Gutova M, et al. The histone demethylase jumonji coordinates cellular senescence including secretion of neural stem cell-attracting cytokines. Mol Cancer Res. 2015;13:636–650. doi: 10.1158/1541-7786.MCR-13-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki K, Natsugoe S, Ishigami S, Matsumoto M, Okumura H, Setoyama T, Uchikado Y, Kita Y, Tamotsu K, Sakurai T, et al. Expression of CXCL12 and its receptor CXCR4 correlates with lymph node metastasis in submucosal esophageal cancer. J Surg Oncol. 2008;97:433–438. doi: 10.1002/jso.20976. [DOI] [PubMed] [Google Scholar]
- 13.Koishi K, Yoshikawa R, Tsujimura T, Hashimoto-Tamaoki T, Kojima S, Yanagi H, Yamamura T, Fujiwara Y. Persistent CXCR4 expression after preoperative chemoradiotherapy predicts early recurrence and poor prognosis in esophageal cancer. World J Gastroenterol. 2006;12:7585–7590. doi: 10.3748/wjg.v12.i47.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacitto R, Gaeta I, Swanson JA, Yoshida S. CXCL12-induced macropinocytosis modulates two distinct pathways to activate mTORC1 in macrophages. J Leukoc Biol. 2017;101:683–692. doi: 10.1189/jlb.2A0316-141RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patalano S, Rodriguez-Nieves J, Colaneri C, Cotellessa J, Almanza D, Zhilin-Roth A, Riley T, Macoska J. CXCL12/CXCR4-mediated procollagen secretion is coupled to Cullin-RING ubiquitin ligase activation. Sci Rep. 2018;8:3499. doi: 10.1038/s41598-018-21506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du LL, Liu P. CXCL12/CXCR4 axis regulates neovascularization and lymphangiogenesis in sutured corneas in mice. Mol Med Rep. 2016;13:4987–4994. doi: 10.3892/mmr.2016.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Hale JS, Rich JN, Ransohoff RM, Lathia JD. Chemokine CXCL12 in neurodegenerative diseases: An SOS signal for stem cell-based repair. Trends Neurosci. 2012;35:619–628. doi: 10.1016/j.tins.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie S, Zeng W, Fan G, Huang J, Kang G, Geng Q, Cheng B, Wang W, Dong P. Effect of CXCL12/CXCR4 on increasing the metastatic potential of non-small cell lung cancer in vitro is inhibited through the downregulation of CXCR4 chemokine receptor expression. Oncol Lett. 2014;7:941–947. doi: 10.3892/ol.2014.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C, Zhang H, Wang B, He J, Ge G. SDF-1/CXCR4 axis promotes the growth and sphere formation of hypoxic breast cancer SP cells by c-Jun/ABCG2 pathway. Biochem Biophys Res Commun. 2018;505:593–599. doi: 10.1016/j.bbrc.2018.09.130. [DOI] [PubMed] [Google Scholar]
- 20.De-Colle C, Menegakis A, Monnich D, Welz S, Boeke S, Sipos B, Fend F, Mauz PS, Tinhofer I, Budach V, et al. SDF-1/CXCR4 expression is an independent negative prognostic biomarker in patients with head and neck cancer after primary radiochemotherapy. Radiother Oncol. 2018;126:125–131. doi: 10.1016/j.radonc.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Shi W, Zhang Y, Wang X, Sun S, Song Z, Liu M, Zeng Q, Cui S, Qu X. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci Rep. 2017;7:42226. doi: 10.1038/srep42226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Ren CC, Yang L, Xu YM, Chen YN. Role of CXCL12-CXCR4 axis in ovarian cancer metastasis and CXCL12-CXCR4 blockade with AMD3100 suppresses tumor cell migration and invasion in vitro. J Cell Physiol. 2018;234:3897–3909. doi: 10.1002/jcp.27163. [DOI] [PubMed] [Google Scholar]
- 23.Jeng KS, Jeng CJ, Jeng WJ, Chang CF, Sheen IS. Role of C-X-C chemokine ligand 12/C-X-C chemokine receptor 4 in the progression of hepatocellular carcinoma. Oncol Lett. 2017;14:1905–1910. doi: 10.3892/ol.2017.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 25.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ, Feng YM. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br J Cancer. 2014;110:724–732. doi: 10.1038/bjc.2013.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao A, Shi R, Jiang Y, Tian S, Li P, Song F, Qu Y, Li J, Yun H, Yang X. SDF-1/CXCR4 axis regulates cell cycle progression and epithelial-mesenchymal transition via up-regulation of survivin in glioblastoma. Mol Neurobiol. 2016;53:210–215. doi: 10.1007/s12035-014-9006-0. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y, Song Y, Qu J, Che X, Song N, Fan Y, Wen T, Xu L, Gong J, Wang X, et al. The chemokine receptor CXCR4 and c-MET cooperatively promote epithelial-mesenchymal transition in gastric cancer cells. Transl Oncol. 2018;11:487–497. doi: 10.1016/j.tranon.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Guo Q, Zhang Z, Bai N, Liu Z, Xiong M, Wei Y, Xiang R, Tan X. VDR status arbitrates the prometastatic effects of tumor-associated macrophages. Mol Cancer Res. 2014;12:1181–1191. doi: 10.1158/1541-7786.MCR-14-0036. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, Liu Y, Li X, Xiang R, Li N. SOX2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of WNT/beta-catenin signal network. Cancer Lett. 2013;336:379–389. doi: 10.1016/j.canlet.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Karicheva O, Rodriguez-Vargas JM, Wadier N, Martin-Hernandez K, Vauchelles R, Magroun N, Tissier A, Schreiber V, Dantzer F. PARP3 controls TGFβ and ROS driven epithelial-to-mesenchymal transition and stemness by stimulating a TG2-Snail-E-cadherin axis. Oncotarget. 2016;7:64109–64123. doi: 10.18632/oncotarget.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang ZC, Yi MJ, Ran N, Wang C, Fu P, Feng XY, Xu L, Qu ZH. Transforming growth factor-beta1 induces bronchial epithelial cells to mesenchymal transition by activating the Snail pathway and promotes airway remodeling in asthma. Mol Med Rep. 2013;8:1663–1668. doi: 10.3892/mmr.2013.1728. [DOI] [PubMed] [Google Scholar]
- 34.Johnson JR, Nishioka M, Chakir J, Risse PA, Almaghlouth I, Bazarbashi AN, Plante S, Martin JG, Eidelman D, Hamid Q. IL-22 contributes to TGF-β1-mediated epithelial-mesenchymal transition in asthmatic bronchial epithelial cells. Respir Res. 2013;14:118. doi: 10.1186/1465-9921-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendt MK, Tian M, Schiemann WP. Deconstructing the mechanisms and consequences of TGF-β-induced EMT during cancer progression. Cell Tissue Res. 2012;347:85–101. doi: 10.1007/s00441-011-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shien K, Papadimitrakopoulou VA, Ruder D, Behrens C, Shen L, Kalhor N, Song J, Lee JJ, Wang J, Tang X, et al. JAK1/STAT3 activation through a proinflammatory cytokine pathway leads to resistance to molecularly targeted therapy in non-small cell lung cancer. Mol Cancer Ther. 2017;16:2234–2245. doi: 10.1158/1535-7163.MCT-17-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen EN, Gao H, Anfossi S, Mego M, Reddy NG, Debeb B, Giordano A, Tin S, Wu Q, Garza RJ, et al. Inflammation mediated metastasis: Immune induced epithelial-to-mesenchymal transition in inflammatory breast cancer cells. PLoS One. 2015;10:e0132710. doi: 10.1371/journal.pone.0132710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aversa I, Zolea F, Ierano C, Bulotta S, Trotta AM, Faniello MC, De Marco C, Malanga D, Biamonte F, Viglietto G, et al. Epithelial-to-mesenchymal transition in FHC-silenced cells: The role of CXCR4/CXCL12 axis. J Exp Clin Cancer Res. 2017;36:104. doi: 10.1186/s13046-017-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Cao Y, Zhang S, Chen Z, Fan L, Shen X, Zhou S, Chen D. Stem cell autocrine CXCL12/CXCR4 stimulates invasion and metastasis of esophageal cancer. Oncotarget. 2017;8:36149–36160. doi: 10.18632/oncotarget.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramsey DM, McAlpine SR. Halting metastasis through CXCR4 inhibition. Bioorg Med Chem Lett. 2013;23:20–25. doi: 10.1016/j.bmcl.2012.10.138. [DOI] [PubMed] [Google Scholar]
- 41.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Shen L, Stupack DG, Bai N, Xun J, Ren G, Han J, Li L, Luo Y, Xiang R, Tan X. JMJD3 promotes survival of diffuse large B-cell lymphoma subtypes via distinct mechanisms. Oncotarget. 2016;7:29387–29399. doi: 10.18632/oncotarget.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Hou L, Ding G, Li Y, Wang J, Qian B, Sun J, Wang Q. KDM6B induces epithelial-mesenchymal transition and enhances clear cell renal cell carcinoma metastasis through the activation of SLUG. Int J Clin Exp Pathol. 2015;8:6334–6344. [PMC free article] [PubMed] [Google Scholar]
- 45.Xia M, Yao L, Zhang Q, Wang F, Mei H, Guo X, Huang W. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget. 2017;8:19795–19802. doi: 10.18632/oncotarget.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, Lai C, Belova G, Cam M, Walling J, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS One. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, Chen Y, Wang HY, Wang RF. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uygur B, Wu WS. SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol Cancer. 2011;10:139. doi: 10.1186/1476-4598-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue LJ, Mao XB, Ren LL, Chu XY. Inhibition of CXCL12/CXCR4 axis as a potential targeted therapy of advanced gastric carcinoma. Cancer Med. 2017;6:1424–1436. doi: 10.1002/cam4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res. 2014;124:31–82. doi: 10.1016/B978-0-12-411638-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berning P, Schaefer C, Clemens D, Korsching E, Dirksen U, Potratz J. The CXCR4 antagonist plerixafor (AMD3100) promotes proliferation of Ewing sarcoma cell lines in vitro and activates receptor tyrosine kinase signaling. Cell Commun Signal. 2018;16:21. doi: 10.1186/s12964-018-0233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y, Tang L, Zhao S, Sun B, Cheng L, Tang Y, Luo Z, Lin Z, Zhu J, Zhu W, et al. CXCR4-mediated osteosarcoma growth and pulmonary metastasis is suppressed by MicroRNA-613. Cancer Sci. 2018;109:2412–2422. doi: 10.1111/cas.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xun J, Wang D, Shen L, Gong J, Gao R, Du L, Chang A, Song X, Xiang R, Tan X. JMJD3 suppresses stem cell-like characteristics in breast cancer cells by downregulation of Oct4 independently of its demethylase activity. Oncotarget. 2017;8:21918–21929. doi: 10.18632/oncotarget.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Yu D, Qi P, Wang X, Guo X, Zhang A. Calcitriol induces cell senescence of kidney cancer through JMJD3 mediated histone demethylation. Oncotarget. 2017;8:100187–100195. doi: 10.18632/oncotarget.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.