Abstract
Neurobiological models to explain vulnerability of major depressive disorder (MDD) are scarce and previous functional magnetic resonance imaging studies mostly examined “static” functional connectivity (FC). Knowing that FC constantly evolves over time, it becomes important to assess how FC dynamically differs in remitted‐MDD patients vulnerable for new depressive episodes. Using a recently developed method to examine dynamic FC, we characterized re‐emerging FC states during rest in 51 antidepressant‐free MDD patients at high risk of recurrence (≥2 previous episodes), and 35 healthy controls. We examined differences in occurrence, duration, and switching profiles of FC states after neutral and sad mood induction. Remitted MDD patients showed a decreased probability of an FC state (p < 0.005) consisting of an extensive network connecting frontal areas—important for cognitive control—with default mode network, striatum, and salience areas, involved in emotional and self‐referential processing. Even when this FC state was observed in patients, it lasted shorter (p < 0.005) and was less likely to switch to a smaller prefrontal–striatum network (p < 0.005). Differences between patients and controls decreased after sad mood induction. Further, the duration of this FC state increased in remitted patients after sad mood induction but not in controls (p < 0.05). Our findings suggest reduced ability of remitted‐MDD patients, in neutral mood, to access a clinically relevant control network involved in the interplay between externally and internally oriented attention. When recovering from sad mood, remitted recurrent MDD appears to employ a compensatory mechanism to access this FC state. This study provides a novel neurobiological profile of MDD vulnerability.
Keywords: cognitive control, dynamic FC, functional networks, major depressive disorder, resting‐state fMRI
1. INTRODUCTION
Major depressive disorder (MDD) is a severe psychiatric disease, which globally accounts for the greatest loss of years due to disability (Smith, 2014). This high impact of MDD is related to its high incidence and recurrence rates, particularly in patients with multiple previous episodes (Bockting, Spinhoven, Wouters, Koeter, & Schene, 2009). However, this vulnerability during MDD remission has scarcely been studied from a level of intrinsic brain connectivity. Elucidating neural vulnerability factors in remitted recurrent MDD (rrMDD) could facilitate the development of effective prediction tools or improve preventive treatments against MDD recurrence (Fischer, Keller, & Etkin, 2016).
Previous studies have linked MDD to disrupted resting‐state functional connectivity (FC) in several resting‐state networks (RSNs) and systems, including frontal networks (FNs), regulating cognitive control and attention, the default mode network (DMN), involved in internal attention, the salience network (SN) and frontostriatal pathways, both involved in salience detection and emotion (Furman, Hamilton, & Gotlib, 2011; Kaiser, Andrews‐Hanna, Wager, & Pizzagalli, 2015; Menon, 2011; Mulders, van Eijndhoven, Schene, Beckmann, & Tendolkar, 2015). During remission between episodes, MDD vulnerability might specifically relate to a failure of control systems to downregulate DMN activity, with the SN as switching hub between the networks (Marchetti, Koster, Sonuga‐Barke, & De Raedt, 2012; Servaas et al., 2017). However, the scarce research that has been conducted in remitted‐MDD previously examined “static” FC, representing mean connectivity over a period of scanning. Instead, growing evidence shows that brain activity at rest is not stable during the scan, but slowly wanders through a repertoire of time‐varying, but reoccurring, states of coupling among brain regions (Cabral, Kringelbach, & Deco, 2017; Deco, Jirsa, & McIntosh, 2011; Hansen, Battaglia, Spiegler, Deco, & Jirsa, 2015). Dynamic‐FC (dFC) analysis allows characterizing these reoccurring FC states.
dFC changes during tasks (Sakoglu et al., 2010) and resting‐state (Damaraju et al., 2014; Jin et al., 2017; Kaiser et al., 2016) have been associated with psychiatric disorders and with reduced behavioral/cognitive performance in healthy subjects (Cabral, Vidaurre, et al., 2017; Jia, Hu, & Deshpande, 2014; Madhyastha, Askren, Boord, & Grabowski, 2015). For MDD specifically, findings from recent studies suggest aberrations in dFC involving DMN, (Wise et al., 2017) frontal, and SN areas (Demirtaş et al., 2016; Kaiser et al., 2015). However, it remains unclear whether this persists during MDD remission.
The best way to characterize dFC remains under debate (Hutchison et al., 2013). Although the sliding‐window analysis is most commonly used to calculate successive dFC matrices (Sakoglu et al., 2010), the window size affects the temporal resolution, challenging its validity (Deco et al., 2017; Hindriks et al., 2016; Hutchison et al., 2013; Lindquist, Xu, Nebel, & Caffo, 2014; Preti, Bolton, & Ville, 2016; Shine et al., 2015). In the current study, we instead use a recently developed method, the Leading Eigenvector Dynamics Analysis (LEiDA), which calculates dFC at the instantaneous level (for each recorded frame), and allows identifying patterns of blood oxygen level dependent (BOLD) phase coherence, or FC states, that reoccur over time both within and across scanning sessions (Cabral, Vidaurre, et al., 2017). Operating in the temporal domain, this method allows characterizing recurrent FC states in terms of probabilities of occurrence, duration and transition profiles on a subject‐by‐subject level, which allows statistical comparisons between groups. Previously, the dynamical properties of recurrent FC states have been shown to relate with cognitive performance in healthy participants (Cabral, Vidaurre, et al., 2017). Here, we used the same LEiDA method to investigate whether there are specific configurations of dFC that differentiate between antidepressant‐free rrMDD and controls without personal and familial MDD history. We compared differences in occurrence, duration, and switching profiles of FC states both after neutral and sad mood induction. We hypothesized to find alterations in dFC, particularly involving the frontoparietal network, the DMN, and the SN, which would be influenced by sad mood (Cohen, 2017; Harrison et al., 2008).
2. METHODS
2.1. Participants
After approval by the local Medical Ethical Committee and written informed consent, 62 rrMDD patients with ≥2 depressive episodes as defined by the Structured Clinical Interview for DSM disorders (SCID), in stable remission for ≥2 months according to DSM IV criteria, and 41 healthy controls were scanned. Hamilton Depressive Rating Scale (HDRS‐17) scores, an observer rated MDD‐symptom scale to assess depression severity, were ≤7 (Rush et al., 1986). Patients were antidepressant free for ≥8 weeks. Controls did not have a personal or familial history for psychiatric disease (assessed by SCID). All participants were aged 35–65 years. We excluded participants with alcohol/drug dependency; psychotic or bipolar disorder; predominant anxiety disorder; severe personality disorder; electroconvulsive therapy ≤2 months before scanning; history of severe head trauma; neurological disease; severe general physical illness; and no Dutch/English proficiency. rrMDD patients and controls were matched for age, sex, educational level, and working class. Participants were recruited through identical advertisements in freely available online and house‐to‐house papers, posters in public spaces and from previous studies in our and affiliated research centers (Mocking et al., 2016). See Appendix S1 (Supporting Information) for information about psychiatric comorbidity of rrMDD patients.
2.2. Mood‐induction paradigm
As described in more detail in previous work regarding this mood induction (Figueroa et al., 2017; Mocking et al., 2016), before the scan, participants described with as much detail as possible a memory which they regarded as neutral, (e.g., doing the dishes) and one which they regarded as being among the saddest in their life (e.g., losing a job and death of a relative). In addition, participants chose one neutral and one sad fragment of music from 10 different fragments. Memories were scripted in key sentences for display on the screen in the MRI scanner. During memory display, we played the chosen neutral or sad music. Participants were asked to rate their current mood on a scale of 0 to 10 (0 being extremely sad; 10 extremely happy) after the neutral mood induction, after the neutral resting‐state scan and before and after the sad mood induction. After the sad resting‐state scan, the most extreme sadness was rated. The gap between the neutral and sad mood induction, in which participants completed other fMRI tasks was ±125 min, including a 30 min break (Mocking et al., 2016). We designed the sad mood induction to be at the end of all fMRI scanning, as it might have been too stressful for participants to continue fMRI scanning and tasks after the sad mood induction (Figure S1, Supporting Information).
2.3. Image acquisition and analyses
A 3 Tesla Philips Achieva XT scanner (Philips Medical Systems, Best, the Netherlands), equipped with a 32‐channel SENSE head coil, was used to obtain the images. A high‐resolution T1‐weighted 3D structural image was acquired using fast‐field echo for anatomical reference (220 slices; repetition time (TR): 8.3 ms; TE: 3.8 ms; field of view (FOV): 240 × 188; 240 × 240 matrix; voxel size: 1 × 1 × 1 mm3). Functional images were acquired with T2*‐weighted gradient echo planar imaging (EPI) sequences. Participants were instructed to close their eyes without falling asleep. The scans comprised 210 volumes of 37 axial slices (TR: 2,000 ms; TE: 27.6 ms; FOV: 240 × 240; 80 × 80 matrix; voxel size = 3 × 3 × 3 mm3). Slices were oriented parallel to the anterior commissure‐posterior commissure (AC–PC) transverse plane and acquired in ascending order with a gap of 0.3 mm.
2.4. Preprocessing
We preprocessed functional MRI data with FMRIB's Software Library (FSL, https://www.fmrib.ox.ac.uk/fsl). We used the default parameters of an imaging preprocessing pipeline on all participants: Multivariate Exploratory Linear Optimized Decomposition into Independent Components (MELODIC 3.14). MELODIC is usually used to conduct an independent component analysis but here we only used it for motion correction and high‐pass filtering of the data. This pipeline further consisted of motion correction using FMRIB's linear image registration tool (MCFLIRT) (Jenkinson, Bannister, Brady, & Smith, 2002); nonbrain removal using brain extraction tool (BET) (Smith, 2002); spatial smoothing using a Gaussian kernel of full width half maximum (FWHM) 5 mm; grand‐mean intensity normalization of the entire 4D dataset by a single multiplicative factor applying a standard high‐pass temporal filtering (Gaussian‐weighted least‐squares straight line fitting, with σt = 50.0 s and σf = 0.02 Hz). The EPI images were coregistered to the T1‐ weighted structural images, and the T1‐weighted images were coregistered to standard MNI space.
We used the Anatomical Automatic Labeling (AAL) atlas (Tzourio‐Mazoyer et al., 2002) to parcelate the MNI brain into N = 90 cortical and subcortical noncerebellar brain areas and the BOLD signals were then averaged over all voxels belonging to each brain area. The BOLD signals in each of the 90 brain areas were subsequently band‐pass filtered between 0.02 and 0.1 Hz (using a second‐order Butterworth filter), partially discarding the high frequency components associated with cardiac and respiratory signals (>0.1 Hz), and focusing on the most meaningful frequency range of resting‐state fluctuations (Biswal, Yetkin, Haughton, & Hyde, 1995; Cabral, Vidaurre, et al., 2017; Glerean, Salmi, Lahnakoski, Jaaskelainen, & Sams, 2012).
2.5. Dynamic functional connectivity
We used BOLD phase coherence connectivity (Deco et al., 2017; Deco & Kringelbach, 2016; Glerean et al., 2012; Ponce‐Alvarez et al., 2015) to obtain a time‐resolved dFC tensor, with size N × N × T, where N = 90 is the number of brain areas considered in the current parcellation scheme (see Section 2.4), and T = 210 is the number of recording frames in each scan. We first estimated the phase of the BOLD signals in all N = 90 areas over time, θ(n,t), using the Hilbert transform, which expresses a signal X as X(t) = A(t)cos(θ(t)), where A(t) is the instantaneous amplitude, and θ(t) is the instantaneous phase. In Figure 1a, we represent the portrait of all N = 90 BOLD phases at time t, which can be represented in Cartesian coordinates with cos(θ(n,t)) in the horizontal axis and sin(θ(n,t)) in the vertical axis. Each entry dFC(n,p,t) contains to the BOLD phase coherence between brain areas n and p at time t, obtained using the following equation:
Figure 1.
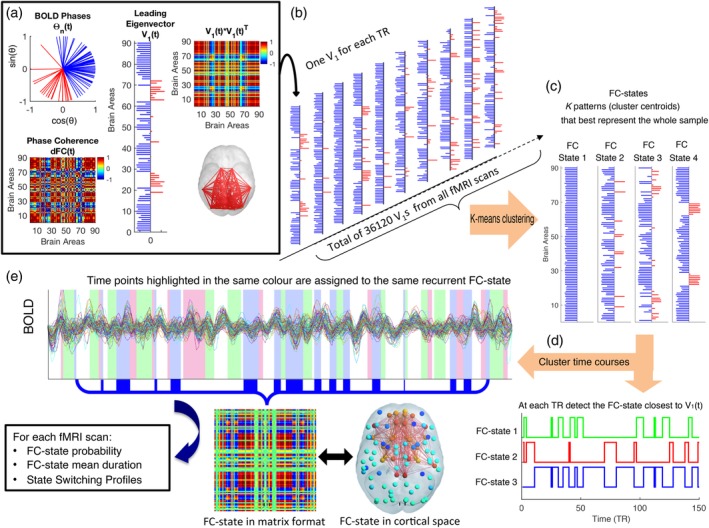
Time courses of recurrent functional connectivity (FC) states obtained with Leading Eigenvector Dynamics Analysis (LEiDA). (a) Leading eigenvector of BOLD phase coherence (top left) phase portrait of BOLD signal phases at a given time point t in all N = 90 brain areas (bottom left). The N × N phase coherence matrix, dynamic‐FC (dFC(t)) indicates for each pair of areas how coherent they are, where 1 (red) means full synchrony and −1 (blue) indicates a phase difference of 180° (middle). The leading eigenvector V 1(t) of this matrix is a N × 1 vector that, when multiplied by its transpose V 1(t) × V 1(t) reveals the dominant pattern of the dFC(t) matrix (top right). Note that the product of elements with the same sign (be they negative or positive) is always positive, so negative values in the matrix are between pairs with different signs. The signs of the elements in V 1 (red/blue) are used to divide brain areas into communities according to their BOLD phase, which can be visualized in cortical space (here links between the areas with positive elements in V 1 are plotted in red). (b) The leading eigenvectors V 1 are obtained for each time point from all fMRI scans in all subjects, resulting in a large sample of 36,120 leading eigenvectors. (c) This sample is partitioned into a reduced number of K clusters (here we varied K between 2 and 20). Each cluster is represented by a central vector, which may not necessarily be a member of the data set. We take these cluster centroid vectors as representing recurrent patterns of BOLD phase coherence, or FC states. (d) To obtain the FC‐state time courses, we select, at each TR, the cluster number to which V 1(t) is the most similar. The cluster time courses (illustrated as color‐shaded bars, over a single fMRI session) are then used to calculate, for each scan, the probability, mean duration, and the state‐switching probabilities of each FC state (bottom). For illustration, the FC state assigned to the blue‐shaded time points are displayed as an N × N matrix (outer product) and as a network in cortical space (here lighter and darker colors show stronger and weaker coherence, respectively, within the positive [yellow‐red] versus the negative [cyan‐blue] communities) [Color figure can be viewed at http://wileyonlinelibrary.com]
where cos() is the cosine function. dFC(n,p,t) is 1 if two areas n and p have synchronized BOLD signals at time t, and dFC(n,p,t) is 0 if the BOLD signals are orthogonal (with a phase difference of 90°).
2.6. FC states
To identify recurrent patterns in the dFC, LEiDA considers, at each time t, only the leading eigenvector V 1(t) of each dFC(t) (i.e., the one associated with the largest magnitude eigenvalue), which captures only the dominant pattern of FC, instead of considering the whole matrices (Cabral, Vidaurre, et al., 2017). As can be seen in Figure 1a, this vector contains N elements (each representing one brain area) and their sign (positive or negative) can be used to separate brain areas into two communities according to their BOLD‐phase relationship (Newman, 2006). When all elements of the largest magnitude eigenvector, V 1(t), have the same sign, it means all BOLD phases are following in the same direction with respect to the orientation determined by V 1(t), which is indicative of a global mode governing all BOLD signals. If instead the first eigenvector V 1(t) has elements of different signs (i.e., positive and negative), the BOLD signals follow different directions with respect to the leading eigenvector, which we use to divide brain areas into two “communities” according to their BOLD phase relationship (see Figure 1a). Moreover, the magnitude of each element in V 1(t) indicates the “strength” with which brain areas belong to the communities in which they are placed (Newman, 2006). The dominant FC state can also be represented back into matrix format (N × N) by computing the outer product V 1 V 1 T, which is visually similar to the original dFC(t) matrix, despite being a matrix of Rank 1 (i.e., a matrix that is obtained from a single vector). Since V and –V represent the same vector orientation, we use a convention ensuring that most of the elements have negative values.
Conveniently, eigenvectors can be represented in cortical space by representing each element as a sphere placed at the center of gravity of the corresponding brain area, and scaling the color of each sphere according to the value of the corresponding eigenvector element. As such, areas with coherent BOLD signals are colored alike (yellow‐to‐red for the smallest community and cyan‐to‐blue for the largest community), where lighter colors (cyan/yellow) indicate stronger contributions and darker colors (blue/red) weaker contributions. To highlight the network formed by the smallest community of brain areas, we plot links between the corresponding areas. For example, Figure 1 (bottom right) shows an FC state represented in cortical space, where the BOLD signals can be divided into two modes: a larger set of brain areas (cyan areas) and a smaller functional network (orange/red areas) formed by areas whose BOLD signal is coherent but phase shifted with respect to the other community. In the matrix format, all links between pairs of areas with the same sign (be it positive or negative) have a positive value (red). Negative values in the matrix format (blue) correspond to links between areas with different signs in the eigenvector.
In this work, we aimed to explore whether there are specific FC configurations that differentiate rrMDD patients from controls. To do so, we first cluster all the samples of FC states into a reduced number of recurrent patterns, applying a k‐means clustering algorithm to all leading eigenvectors V 1(t) across all subjects (rrMDD and controls in neutral and sad mood, resulting in 210 × 86 × 2 = 36,120 leading eigenvectors) (see Figure 1b). The clustering divides the samples into k clusters (each representing a recurrent FC configuration), with higher k revealing more rare and more fine‐grained network configurations. Although there is not a consensus regarding the number of FC states revealed by fMRI (and whether FC states can be discretized in the first place), the number of RSNs reported in the literature generally falls between 7 and 17, depending on the selected criteria (Damoiseaux et al., 2006; Yeo et al., 2011).
Here, we do not aim to determine the optimal number of FC states, but rather to explore if there is an FC state that significantly differs (consistently) between remitted patients and controls, even if that FC state is different from previously reported networks and/or occurs only rarely in time. As such, we varied k (number of clusters) over a wide range between 2 and 20 and for each k, examined how each FC state changed between groups. Importantly, the clustering assigns a single FC state to each fMRI time frame, as highlighted by the shaded bars in Figure 1d. The clustering obtained for each value of k is obtained independently of each other and thus represent independent models of FC configuration space.
2.7. Between‐group comparisons
To assess how the repertoire of k FC states explored during rest varied between groups, we first calculated, for each subject and condition, the probability of occurrence of each FC state (fraction of epochs it occurred throughout the scan duration), the mean duration of each FC state (mean number of consecutive epochs in the same state), the switching frequency (number of transitions per second [Hz]) and the switching profiles (probabilities of switching from a given FC state to another). All values were compared between rrMDD patients and controls, after neutral and sad mood induction using (nonparametric) permutation‐based t tests (10,000 permutations). For each FC repertoire obtained by k‐means clustering, k hypotheses are tested. To correct for multiple comparison, we adjusted the significance threshold to 0.05/k, using a Bonferroni correction (green dashed line in Figure 2). We then evaluated the consistency of the FC states that were found to be significantly different between groups across the range of k explored (Figures 2 and 3, see Section 3.2). We also report within‐group differences for neutral versus sad mood (rrMDD patients and controls separately) in the Results, Supporting Information. For statistical testing involving clinical and demographic characteristics, we used SPSS version 25. For the LEiDA analysis, we used MATLAB version R‐2017b.
Figure 2.
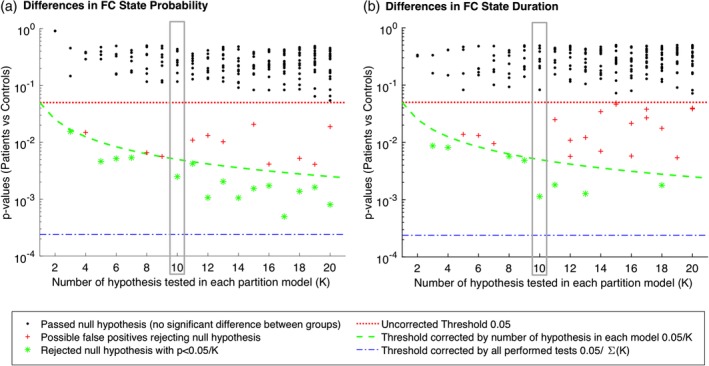
Significance of between‐group differences in functional connectivity (FC)‐state probability and lifetime as a function of k. For each partition of the sample into k = 2 to 20 FC states, we plot the p‐values associated with the between‐group comparison between patients and controls in both FC‐state probability (a) and lifetimes (b). We find that, although most FC states do not show significant differences between groups (black dots falling above the 0.05 threshold, in red), for all k > 2, there is one FC state that consistently falls below (or very near) the corrected threshold by the number of clusters (<0.05/k, green dashed line). The p‐values marked as red crosses pass the standard threshold (<0.05) but do not survive the correction for multiple comparisons within each partition model (>0.05/k) and are hence considered false positives. The blue dashed line refers to the threshold correction if all hypothesis were independent across models, losing statistical power (i.e., increased probability of false negatives) when the hypothesis are not independent, as it is the case when considering the whole sample of tests performed [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.

Consistency of the functional connectivity (FC) state with the most significant differences in probability of occurrence between remitted patients and controls over partition models. After clustering all eigenvectors into k = 2 to 20 clusters, we consistently find, for all k > 2, that the FC state most different between groups is a functional network consisting of areas of the frontal cortex coupled (in terms of BOLD phase alignment) with basal ganglia (i.e., caudate, putamen, and pallidum) and the angular gyrus (default mode network [DMN]). Note that the involvement of basal ganglia in the relevant network becomes more pronounced for more fine‐grained partitions (higher k). For all partition models with k > 2, the probability of occurrence of these FC states between groups passed the standard significance threshold of 0.05, but only 15 (highlighted with a green title) passed the significance threshold when corrected by the number of states, or independent hypothesis, compared within each partition model [Color figure can be viewed at http://wileyonlinelibrary.com]
2.8. Code availability statement
The LEiDA codes are publicly available at http://github.com/juanitacabral/LEiDA in the folder: “Remission from Major Depression.”
3. RESULTS
3.1. Sample characteristics
Here, 72 rrMDD patients and 46 controls were initially eligible, of which 62 and 41 were scanned, respectively. Of these participants, we excluded nine rrMDD and six controls because of abnormal brain anatomy and two rrMDD due to technical difficulties (corrupted scans). Then, 51 rrMDD patients and 35 controls were included in the final analysis (Figure S2, Supporting Information). No significant differences were observed between rrMDD patients and controls for sex, age, education, IQ, living situation, employment status, and handedness. rrMDD patients showed higher levels of residual depressive symptoms (HDRS) p < 0.001 (Table 1). Comparisons between rrMDD and controls did not change when restricted to the sample selected for the present fMRI analyses.
Table 1.
Sample characteristics
| Between‐group statistics | |||||||
|---|---|---|---|---|---|---|---|
| rrMDD | HC | χ2 | t | U | p | ||
| (n = 62) | (n = 41) | ||||||
| Female | N (%) | 43 (69.3%) | 28 (68.3%) | 0.01 | 0.91 | ||
| Age | Years; mean (SD) | 53.7 (7.9) | 51.8 (8.1) | 1.17 | 0.25 | ||
| Education | Levelsa | 0/0/0/4/21/23/14 | 0/0/0/1/16/17/7 | 1.49 | 0.69 | ||
| IQ | Mean (SD) | 108 (8.5) | 106 (9.9) | 878.5 | 0.14 | ||
| Living situation | Levelsb | 26/0/18/14/2/0/2 | 10/0/16/11/4/0/0 | 6.23 | 0.18 | ||
| Employment status | Levelsc | 24/23/15/0 | 21/16/4/0 | 3.7 | 0.16 | ||
| Handedness | Levelsd | 4/50/4 | 2/33/4 | 0.44 | 0.8 | ||
| Age of onset | Years; mean (SD) | 27.2 (11.2) | – | – | |||
| Episodes | Median (IQR) | 4.0 (2/4/7) | – | – | |||
| HDRS | Median (IQR) | 2.0 (1/2/5) | 1.0 (0/1/1) | 644 | <0.001 | ||
HC = healthy control; HDRS = Hamilton Depression Rating Scale; IQR: interquartile range; LEIDS‐R = Leiden Index Depression Sensitivity‐Revised; rrMDD = remitted recurrent major depressive disorder; RRS = Ruminative Response Scale.
Level of educational attainment (Verhage, 1964): levels range from 1 to 7 (1 = primary school not finished; 7 = preuniversity/university degree).
Living situation: alone/living with parents/cohabiting/cohabiting with children/single living with children/other/unknown.
Employment status: low/middle/high/never worked.
Handedness = left/right/ambidexter; IQR = interquartile range; χ2 = chi‐square test statistic; p = p‐value; U = Mann–Whitney U nonparametric test statistic; t = independent‐samples t‐test statistic. This table was published before (Figueroa et al., 2017).
Both groups reported comparable neutral to positive mood after neutral mood induction (p = 0.095). Mood scores (±SD) decreased only slightly during neutral resting state (rrMDD: −0.41 ± 0.93, controls: −0.022 ± 0.65), without a group × mood interaction (p = 0.36). The sad mood induction significantly decreased mood scores in both groups (rrMDD: −2.13 ± 1.50, controls: −2.15 ± 1.17; p < 0.001), also without a group × mood‐induction interaction (p = 0.95). The lowest mood reported during the second resting‐state was significantly lower in rrMDD (rrMDD: 4.99 ± 1.88 vs. controls: 5.94 ± 1.39; p = 0.027). See Table S1 (Supporting Information) for all mood ratings.
3.2. Detection of the most different FC states
The repertoire of FC states obtained depends on the number of clusters determined in the k‐means clustering algorithm, with generally a higher number of clusters resulting in more fine‐grained, less frequent and often less symmetric networks. Importantly, here we do not aim to determine the optimal number of FC states explored during rest but instead to search for FC configurations that most significantly and consistently differentiate patients remitted from recurrent MDD from controls. In Figure 2, we show, for each clustering model of the FC state samples into k FC state categories, the k p‐values obtained from between‐group comparisons in terms of probability and lifetimes. In each model, k hypotheses are tested. Thus, to account for the increased probability of false positives, we Bonferroni corrected the significance threshold to 0.05/k (green dashed line in Figure 2). We find that, irrespective of the number of clusters selected (as long as k > 2), the clustering consistently returns an FC state that significantly differs in probability between patients in remission and controls, falling below (or very near) the threshold corrected by the number of clusters within each partition model (green dashed line in Figure 2 ).
In detail, of the 19 partition models considered (i.e., with k ranging from 2 to 20), 15 solutions revealed an FC state that occurred significantly less in rrMDD compared to controls after correcting for the number of clusters in each repertoire of FC states (p < 0.05/k). The most significantly different FC states of each partition model are reported in Figure 3 in vector format, where areas with the same color are assigned to the same mode of phase coherence. The 15 FC states that passed the corrected significance threshold, were highly correlated (Pearson's r > 0.84 for all pairs of FC states), which indicate that they refer to variant forms of the same underlying FC state, with differences arising from the number of output states constrained by k. In terms of duration, this FC state also lasted significantly shorter in seven partition models (p < 0.05/k), of which four had overlap with a significant lower probability (see Figure 2). Of note, for k = 18, the FC state that lasted significantly shorter in rrMDD patients includes only the subcortical areas (caudate, putamen, pallidus, thalamus, and amygdala) (2.74 ± 0.26 vs. 4.55 ± 0.63 s, p = 0.0018, uncorrected). This reveals that more detailed partitions (higher k) can further subdivide the FC states, revealing finer grained structures that differentiate between groups (see Figure S3, Supporting Information for dominant FC states from k = 2 to 20 and Figure S4, Supporting Information for the variance in each k‐means derived FC state for each partition model).
The consistency of our findings for a range of partition models reinforces the existence of a specific pattern of BOLD phase coherence that differentiates patients in remission from controls. For the subsequent analysis, we selected the partition into k = 10 FC states, since it returned a repertoire of 10 FC states where only one FC state significantly differed both in terms of probability and lifetime (see Figure 2, blue dots in the gray rectangle in both [a] and [b]), whereas all other nine FC states in this partition model did not show significant differences (p > 0.05 for all black dots within the gray rectangles). Moreover, the partition into 10 states is aligned with previous studies in the resting‐state literature (Damoiseaux et al., 2006).
3.3. Relevant FC state
Our analysis revealed an FC state, which consistently appeared less and lasted shorter in rrMDD compared to controls in neutral mood (Figure 4 ; FCstate.mp4; Figure S3, Supporting Information). This underrepresented FC state consists of an extensive network including frontal (dorsolateral prefrontal cortex [DLPFC] and fronto‐orbital cortex), DMN (posterior cingulate cortex, angular gyrus, and medial prefrontal cortex [MPFC]), subcortical (the dorsal striatum [Str] [caudate, putamen] and pallidus), and SN areas (anterior cingulate cortex, frontal operculum). We further indicate this FC state as the “FN‐DMN‐Str‐SN state.” See Figure S5, Supporting Information for the overlap of this FN‐DMN‐Str‐SN state with RSNs defined by previous whole brain parcellations (Choi, Yeo, & Buckner, 2012; Yeo et al., 2011).
Figure 4.
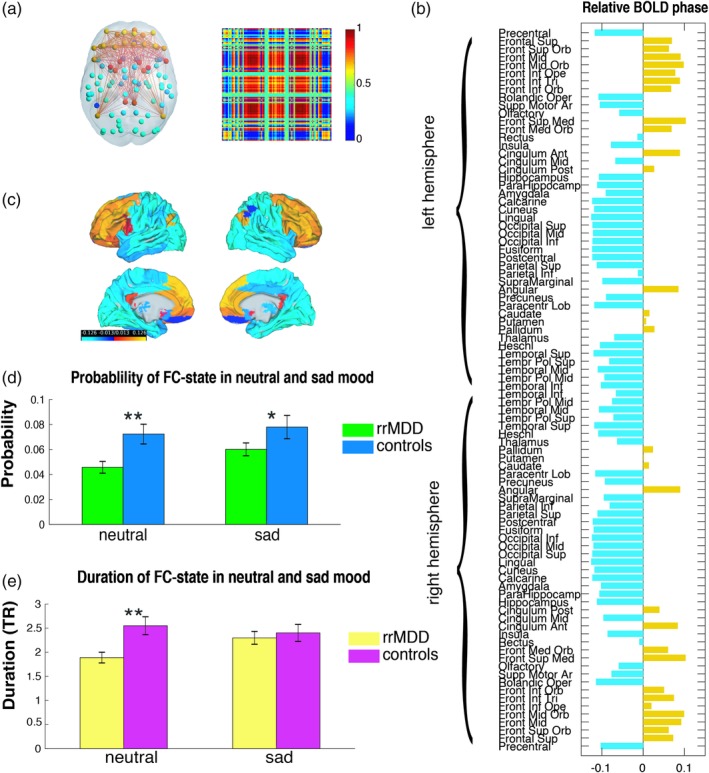
Functional connectivity (FC) state with a significant difference between rrMDD patients and controls (k = 10) in probability of occurrence and lifetime during neutral mood. (a) The dominant connectivity state is represented in the cortical space, where functionally connected brain areas (represented as spheres) are colored alike. The spheres colored in yellow, orange, and red, represent areas in the FN, default mode network (DMN), striatum, and SN, which are all positively correlated between each other, but negatively correlated with the rest of the brain (cyan/blue colored spheres). The dominant state is also represented as the eigenvector's outer product, which is a 90 × 90 matrix representing the number of brain areas and red or blue indicate positive or negative BOLD phase synchronization between them. (b) Contribution of different brain areas to the dominant FC state. Bars in yellow represent areas in the FN, DMN, striatum, and SN and bars in light blue represent the rest of the brain. The magnitude of values indicates the “strength” with which brain areas belong to the FC state. (c) The significant FC state rendered on the cortex. (d) Differences in probability of occurrence of this state between rrMDD patients and controls (4.58 ± 0.47% vs. 7.24 ± 0.79%, respectively, p = 0.0022 in neutral mood and 6.0 ± 0.51% vs. 7.80 ± 0.93%, p = 0.049 in sad mood). (e) Differences in lifetime of this state between rrMDD patients and controls (3.78 ± 0.11 vs. 5.10 ± 0.18 s, p = 0.0020 and 4.60 ± 0.26 vs. 4.80 ± 0.36, p = 0.32 in sad mood). ** Significant group difference after correcting for multiple comparisons. * Significant group difference before correcting for multiple comparisons. Abbreviations: MDD = major depressive disorder; rrMDD = remitted recurrent MDD; TR = repetition time [Color figure can be viewed at http://wileyonlinelibrary.com]
For the selected clustering model (k = 10), the FN‐DMN‐Str‐SN state occurred less frequently (4.58 ± 0.47% compared to 7.24 ± 0.79% of the time, p = 0.0025, uncorrected, Hedge's g = 0.67; Hedge's g = 0.67; medium to large effect size) and lasted shorter when dominant (3.78 ± 0.11 vs. 5.10 ± 0.18 s, p = 0.0011, Hedge's g = 0.71; medium to large effect size) (Figure 4; FCstate.mp4).
In Figure 5 , we show the full repertoire of FC states that are returned by LEiDA when choosing k = 10. This reveals different network configurations that appear, dissolve, and reoccur in all subjects during the scan, with the state of global coherence showing not only the highest probability but also the highest variability. Notably, these networks overlap with previously described RSNs (Choi et al., 2012; Yeo et al., 2011) and with cognitive states based on a large‐scale automatic synthesis of human functional neuroimaging data (Yarkoni, Poldrack, Nichols, Van Essen, & Wager, 2011).
Figure 5.
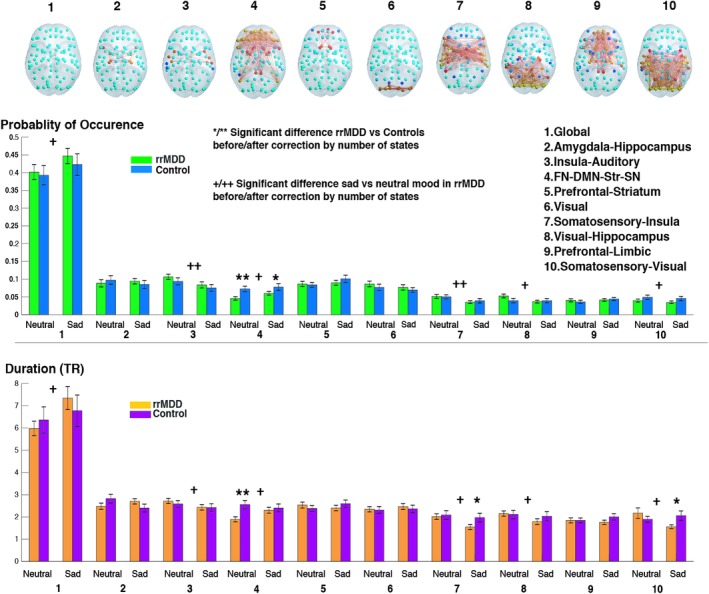
Differences in probability and duration of each functional connectivity (FC) state between and within groups for neutral and sad mood conditions for k = 10. FC states are represented in the cortical space, where functionally connected brain areas (represented as spheres) are colored alike. The spheres colored in yellow/red represent areas that are all positively correlated between them, but negatively correlated with the rest of the brain (cyan/blue colored spheres). Names of FC states were defined by loading network maps into neurosynth, http://neurosynth.org/decode/, a platform for large‐scale, automated synthesis of functional magnetic resonance imaging (fMRI) data. In the case of the state that was significantly different between groups, (FC state 4), we additionally compared the FC state with resting‐state networks (RSNs) defined by a whole‐brain parcellation scheme (Choi et al., 2012; Yeo et al., 2011) (Figure S5, Supporting Information). Of note, when considering other clustering solutions other FC states may be identified. */** Significant difference between rrMDD and controls before/after correcting for number of states (*p < 0.05, **p < 0.05/k), +/++ Significant within‐group difference for rrMDD, neutral versus sad before/after correcting for number of states (+p < 0.05, ++p < 0.05/k). Abbreviations: DMN = default mode network; FN = frontal network; MDD = major depressive disorder; rrMDD = remitted recurrent MDD; SN = salience network; Str = striatum; TR = repetition time [Color figure can be viewed at http://wileyonlinelibrary.com]
3.4. Correction for residual symptoms
For the FN‐DMN‐Str‐SN state, we examined whether the differences between rrMDD and controls in duration and lifetime remained significant after correcting for residual depressive symptoms (HDRS scores). In a regression analysis with group (rrMDD or control) and HDRS as independent variables and probability/lifetime as the dependent variable, the group differences remained significant (p = 0.007 and p = 0.006, respectively). This indicates that current level of depressive symptoms likely does not explain the group differences.
3.5. Effect of sad mood induction on FC states
During sad mood, of the 19 partition models considered, the FN‐DMN‐Str‐SN state had a lower probability of occurrence in three clustering solutions of a higher order (k = 17, 19, and 20), that is, when more subdivisions in FC states are made (Figure S3, Supporting Information), after correction for multiple comparisons (p < 0.05/k). Furthermore, significantly shorter lifetimes were found for this FC state in partition models of a lower order (k = 5 and 6) (Figure S3, Supporting Information).
As can be seen in Figure 4d,e, for k = 10, the probability of being in the FN‐DMN‐Str‐SN state in sad mood was only significantly different between groups before correcting by k (6.0 ± 0.51% compared to 7.80 ± 0.93% of the time, respectively, p = 0.048, uncorrected, Hedge's g = 0.41, indicating a small to medium effect size). Further, no differences were detected in terms of duration of FC states (4.60 ± 0.26 vs. 4.80 ± 0.36 s, respectively, p = 0.32, uncorrected, Hedge's g = 0.10, indicating a small effect size).
3.6. Group × mood interactions
We examined if there were significant group × mood interaction effects for the probability of occurrence and mean lifetime of the FN‐DMN‐Str‐SN state for k = 10 by means of a repeated measures ANOVA. We found no significant interaction effects for this FC pattern for probability, F = 0.634 p = 0.425, but there was a significant interaction effect for lifetime, F = 4.32, p = 0.041. These interaction effects did not significantly change when correcting for residual symptoms in a repeated measures ANCOVA, p = 0.259 and p = 0.024, respectively.
3.7. Within‐group differences in FC states for neutral versus sad mood
Remitted‐MDD showed lower probabilities and lifetimes for sad versus neutral mood in multiple FC states (Results/Figure S6, Supporting Information). For controls, there were no within‐group differences for neutral versus sad mood.
3.8. Switching frequencies
Overall, mean switching frequencies (number of switches/second) did not differ between rrMDD and controls for all clustering solutions (from k = 2 to 20) in neutral and in sad mood (all p‐values >0.05).
3.9. Switching probabilities
We examined the transition patterns between FC states in detail for the selected partition model (k = 10) by calculating the probability of, being in a given FC state, switching to any of the other FC states. In Figure 6, we illustrate the general switching pattern for the whole group, with blue arrows indicating the switches that exceed a probability of 20% of occurring (numeric values reported in the switching matrix on the right). On a whole‐group level, the most common switches were observed from FC states 2, 3, 5, and 6 toward the global FC state (first column in the switching matrix), suggesting that after being coherent in BOLD phase for some time, the areas involved in these functional networks realign their BOLD phases with the global signal, returning to a state of global BOLD phase coherence, which is the most prevalent FC state.
Figure 6.
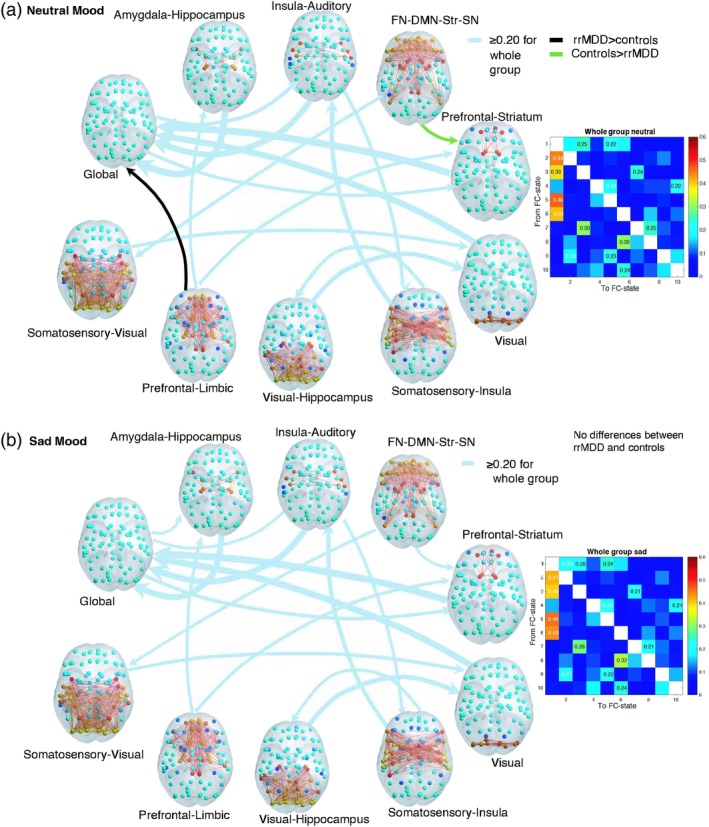
(a,b) Switching probabilities for the whole group and differences between rrMDD and controls, for (a) neutral and (b) sad mood. Switching probabilities for the whole group are shown above a threshold of 20% probability of switching to show more frequent switches. The whole group switching matrices (titled “whole group neutral” and “whole group sad”) indicate the probability of, being in a given functional connectivity (FC) state (rows), transitioning to any of the other states (columns) for the whole group. FC states are represented in the cortical space, where functionally connected brain areas (represented as spheres) are colored alike. The spheres colored in yellow/red represent areas that are all positively correlated between them, but negatively correlated with the rest of the brain (cyan/blue colored spheres). The light blue arrows from and to the FC states indicate the whole group switching probabilities, scaled to the magnitude of probability of switching. Significantly different transitions after correcting for k = 10 (p < 0.05/10) for rrMDD compared to controls are illustrated in this figure, with the black arrow representing the transition that occurs with higher probability in rrMDD and in green the one that occurs with higher probability in controls (note these arrows have not been scaled according to magnitude of probability of switching). Transitioning differences between groups were calculated using a permutation‐based two sample t test with 10,000 permutations. rrMDD patients showed a lower probability of switching from the FN‐DMN‐Str‐SN state to the prefrontal–striatum state (16.5 vs. 23%, p = 0.004), and a higher probability to switch from the prefrontal–limbic state to the global state (6 vs. 1%, p = 0.003). We identified no significant between group differences in sad mood after correction for k = 10 (p > 0.05/k). Abbreviations: DMN = default mode network; FN = frontal network; MDD = major depressive disorder; rrMDD = remitted recurrent MDD; SN = salience network; Str = striatum [Color figure can be viewed at http://wileyonlinelibrary.com]
Comparing the switching patterns between rrMDD and controls, we found that, even when rrMDD patients displayed the FN‐DMN‐Str‐SN state, they showed a significantly lower probability of switching from this state to the prefrontal–striatum state compared to controls (16.5 vs. 23%, p = 0.004, uncorrected, p = 0.04 corrected by k; Figure 6a, green arrow). In addition, we also detected a higher probability to switch from the prefrontal–limbic state to the state of global BOLD coherence (6% rrMDD vs. 1% controls, p = 0.002, uncorrected, p = 0.02 corrected by k; Figure 6a, black arrow). Conducting a deeper analysis of the transition profiles shown in Figure 6, we can see that the trajectory from the FN‐DMN‐Str‐SN state before returning to the state of global BOLD coherence occurs preferentially via the prefrontal–striatum state in controls. In rrMDD, this trajectory appears disrupted, suggesting that rrMDD patients not only have decreased ability to access the FN‐DMN‐Str‐SN network but also to switch from this to the prefrontal–striatum state.
3.10. Effect of sad mood on switching profiles
The overall switching pattern after sad mood induction for the whole group was similar to the switching pattern during neutral mood (Figure 6b). During sad mood, for k = 10, we no longer identified significant differences in switching probabilities between groups (all p > 0.05/k, k = 10). See Figure S7, Supporting Information for differences with p < 0.05.
3.11. Within‐group differences for neutral versus sad mood
Differences in switching probabilities were observed both for rrMDD and controls for neutral versus sad mood, which we describe in the Results and Figures S7 and 8a,b (Supporting Information).
4. DISCUSSION
This study investigated differences in FC states reoccurring over time during resting‐state in rrMDD patients not taking antidepressants compared with never depressed controls. We identified decreased ability in rrMDD patients to access an FN‐DMN‐Str‐SN state, consisting of frontal, DMN, striatum, and SN areas during neutral mood. Our study provides a new framework for detecting network abnormalities associated with vulnerability during MDD remission.
The FN‐DMN‐Str‐SN state, which consistently differed between rrMDD and controls, with a medium to high effect size, reveals an extensive network of clinically relevant areas including key areas from the DMN (posterior cingulate cortex (PCC) and MPFC), executive network (dorsolateral prefrontal cortex (DLFPC)), and SN (anterior cingulate cortex (ACC)) (Figure S4, Supporting Information). These networks have been identified as affected in MDD before, and together form the “triple network,” a model employed for understanding affective and neurocognitive dysfunctions across multiple disorders (Menon, 2011). The extensive FN included in this FC state consists of areas activated during cognitive control and flexible switching of attention from internal thought processes to the external environment, with the DLPFC being particularly relevant (Japee, Holiday, Satyshur, Mukai, & Ungerleider, 2015).
Additionally, the FC state includes a large DMN component. Dominance of the DMN over networks involved in cognitive control has been associated with depressive symptoms (Knyazev, Savostyanov, Bocharov, Tamozhnikov, & Saprigyn, 2016), rumination (Hamilton et al., 2011), and might be related to depressive recurrence, though this has not been empirically tested (Marchetti et al., 2012). Furthermore, the state included areas of the SN, a network that has been proposed to play a key role in switching brain activity between introspective, ruminative DMN functions and task‐based executive networks functions (Menon & Uddin, 2010). Finally, the state included the dorsal striatum (caudate and putamen) and globus pallidus, structures involved in the focusing (and filtering) of cortical input (Grillner, Hellgren, Menard, Saitoh, & Wikstrom, 2005). Abnormal functioning of frontostriatal pathways might additionally lead to maladaptive regulation of emotions (Drevets, 2007), contributing to anhedonia and rumination in MDD (Furman et al., 2011).
Taken together, our results suggest that, patients remitted from recurrent MDD, show decreased ability to access an FC state in neutral mood regulating cognitive control (FN/SN) to diminish negative self‐referential processes (DMN) and effectively regulate emotions (SN/corticostriatal pathways) (Marchetti et al., 2012).
Additionally, we observed a lower probability in rrMDD to switch from the FN‐DMN‐Str‐SN state to a smaller prefrontal–striatum state. Interestingly, on a group level, the prefrontal–striatum state has a high probability of switching to the state of global BOLD coherence. It has been suggested that the more frequently occurring global brain states allow for a greater range of either integration or segregation between neural networks and brain areas, that is, more flexible switching to different brain states (Nomi et al., 2017). This greater neural flexibility might facilitate cognitive flexibility (Kringelbach, McIntosh, Ritter, Jirsa, & Deco, 2015; Nomi et al., 2017). As an additional measure of neural flexibility, in a post hoc supplementary analysis, we calculated the entropy (Shannon, 1948) for k = 10 and the FN‐DMN‐Str‐SN state separately (Discussion, Supporting Information). We found that the entropy associated with the FN‐DMN‐Str‐SN state was significantly decreased in patients in neutral mood. This indicates a higher predictability of occurrence (Wang, 2008) of this FC state in rrMDD patients, supporting the idea that this brain state occurs in a less flexible manner.
After the sad mood induction, the duration of the FN‐DMN‐Str‐SN state increased in rrMDD but stayed similar in controls (significant group × mood interaction). This suggests that rrMDD have a decreased ability to access this state in neutral mood but manage to recruit this state during sad mood (albeit slightly less than controls). This differential increase supports our hypothesis that this FC state is relevant for processes associated with affect and emotion in rrMDD.
We speculate that this increase might reflect a compensatory mechanism in rrMDD to regulate brain activity. This could be an attempt (which might or might not be successful) to regulate negative self‐referential and emotional processing when sad mood is induced. In controls, this FC state is already more easily activated in neutral mood and therefore regulation of brain activity during sad mood and in daily life might be more automatic and less effortful for never depressed subjects. This is corroborated by the fact that this FC state occurs more flexibly (as measured by the entropy) in controls during neutral mood than in remitted‐MDD (but not during sad mood, see Results, Supporting Information). Therefore, we hypothesize that an inability to access this state during neutral mood might reflect increased difficulty for rrMDD patients to effectively and flexibly handle daily life stressors and changes in affect (Van der Lande et al., 2016). This hypothesis needs to be tested by future research. Of note, we additionally observed within‐group differences only in rrMDD for sad versus neutral mood (Results, Supporting Information), indicating that the induction of sad mood (i.e., recall of the saddest life experience) affects brain network dynamics more strongly in patients in remission compared to controls.
Of note, though rrMDD reported higher severity of residual depressive symptoms, this did not explain group differences in probability or lifetime of the FN‐DMN‐Str‐SN state. Additionally, rrMDD reported higher levels of sadness than controls before and after the sad mood induction (although both groups' mood decreased equally during the sad mood induction). Overall, it thus seems that these symptoms did not contribute to group differences in the ability to access the FN‐DMN‐Str‐SN state in neutral or sad mood. This could indicate that these dynamic FC differences represent more of a trait effect of depression than a state effect associated with depressive symptomatology.
Interestingly, previous studies in MDD have also found increased dynamic FC in areas overlapping with the FN‐DMN‐Str‐SN state: between the frontoparietal network and DMN (Kaiser et al., 2016) within the DMN (Wise et al., 2017) and between DMN (MPFC) and the insula (Kaiser et al., 2016); related to levels of self‐report rumination (Kaiser et al., 2016). This suggests that dynamic changes in FC in these areas might be particularly important for depression and a ruminative thinking style. However, these studies included acute MDD patients and did not use a mood‐induction procedure, which makes it difficult to compare results. In remitted‐MDD, Foland‐Ross, Cooney, Joormann, Henry, & Gotlib, 2014 found that during both a sad mood induction and automatic mood regulation by positive autobiographical recall, remitted participants exhibited a decrease in activation in the left ventrolateral prefrontal cortex and cuneus, which are both involved in autobiographical memory processing (Foland‐Ross et al., 2014). However, since this study examined brain activation this is also challenging to compare. Further, we examined resting‐state dynamic FC during the recovery of sad mood, and not during the actual mood induction. It should be examined further how induction of sad mood or stress changes dynamic FC.
Results of this study are novel because our approach differs from common static‐FC analyses, which previously identified aberrations in MDD in networks assumed to be temporally stable over the whole recording time, whereas we here show differences in networks that reoccur and dissolve over time. Of note, the FN‐DMN‐Str‐SN state is only dominant during a small proportion of time. However, it consists of clinically relevant areas implicated in cognitive control and emotional/self‐referential processing and occurred less, especially during neutral mood, in rrMDD patients at high risk for recurrence. Importantly, we found that this FC state consistently differed between groups after multiple comparisons correction, largely independent of which clustering solution was chosen (Figure S3, Supporting Information). Further, using this approach, we found that an FC state of areas traditionally belonging to spatially defined RSNs derived from static‐FC analysis forms a separate network of increased coherence over time. This emphasizes transient dysfunctional interactions between multiple networks involved in psychological functioning, as opposed to unistructural or single network abnormalities in the pathophysiology of MDD.
Although this is still unclear, the more a functional network is accessed, the more stable it might become during rest because it reinforces underlying structural connections, perhaps trough Hebbian learning mechanisms (Martin & Morris, 2002). For instance, it has been shown that cognitive training over time does not only alter FC but occurs alongside changes in the structural connectome (Caeyenberghs, Metzler‐Baddeley, Foley, & Jones, 2016). Here, healthy controls might have accessed the FN‐DMN‐Str‐SN state more throughout life, which might be associated with a lower risk of occurrence of a depressive episode. If this hypothesis is true, it could be examined whether interventions such as cognitive control training (Hoorelbeke & Koster, 2017) or neurofeedback (Enriquez‐Geppert, Huster, & Herrmann, 2017), which allows participants to regulate brain processes/states in real time (Zilverstand, Sorger, Zimmermann, Kaas, & Goebel, 2014), could increase the occurrence and duration of this FC state.
Interestingly, two previous studies on dFC in acute MDD also found alterations in similar networks involving DMN/frontoparietal and SN areas, albeit using different methods (Demirtaş et al., 2016; Kaiser et al., 2016). Kaiser et al. (2016) used a sliding‐window analysis, which has limitations related to window size, and Demirtaş et al. (2016) used instantaneous FC, more comparable to our study. Our approach of focusing on the dominant FC state has the advantage of being more robust to high‐frequency noise, as recurrences of the same pattern are more clearly detected (Cabral et al., 2017; Cabral, Vidaurre, et al., 2017). Merit for future studies lies in examining whether FC states are altered when MDD patients change from a remitted to a depressed state, and whether FC alterations predict short‐ and long‐term MDD recurrence.
The state that occurred most in both groups was a state of global coherence of BOLD phases. This global state shows the greatest variability of all FC states and might therefore allow for a greater range of correlations between areas to form, thereby functioning as a baseline state from which other FC states are organized (Nomi et al., 2017). However, this FC state might relate to what is commonly described as the “global signal,” defined as the time series of signal intensity averaged across all brain voxels, composed of both neural and nonneural signals (Murphy & Fox, 2017). In this study, movement was corrected using MCFLIRT but no further artifact rejection was carried out. In particular, we chose not to regress out motion parameters or use global signal regression, a procedure which has been suggested to reduce motion‐based signals (Power et al., 2014), though this remains a topic of debate. Furthermore, we did not correct for direct physiological measures nor did we use COMPCOR (Behzadi, Restom, Liau, & Liu, 2007). Not removing these factors could affect our results. It has recently been argued that including/removing this global signal might produce different complementary insights into the brain's functional organization (Murphy & Fox, 2017). The functional properties of this global state of coherence merit further examination.
4.1. Strengths and limitations
The main strength of our study is that we have a large sample of antidepressant‐free patients in remission from at least two episodes of MDD, which allows investigating biomarkers for patients at risk of MDD excluding possible medication effects. Further, using a method to characterize FC at the instantaneous level, we identify FC states that, despite occurring with relatively low probability and duration, govern the pattern of BOLD phase coherence recurrently and consistently across subjects, which could be otherwise missed with analysis over longer time windows. Importantly, instead of focusing on single pairwise connections between regions, the identification of recurrent brain‐wide FC patterns allows characterizing FC states on a subject‐by‐subject level, whose properties can subsequently be statistically compared between groups. Although we only examined partition models from k = 2 to 20 clusters, we show that results can be robust across a range of partition models, with differences arising from the granularity inherent to the number of selected clusters. The FC states identified in the current work are strongly constrained by the selected parcellation atlas (AAL). Due to the anatomical basis of this atlas and since all areas have different and relatively large sizes, it might have relatively low BOLD signal homogeneity within its regions compared to other fMRI‐based parcellations, due to mixing of functional signals (Craddock et al., 2013). Therefore, extending this methodological approach to parcellations based on functional homogeneity and with similar size (Craddock et al., 2013; Glasser et al., 2016; Shen, Tokoglu, Papademetris, & Constable, 2013), and/or focusing on more fine‐grained FC states is likely to reveal subdivisions within the FN‐DMN‐Str‐SN state. This would allow to potentially identify specific subsystems driving the activation of the FN‐DMN‐Str‐SN state.
Further, we selected a solution of k = 10 for further analysis in detail, because this solution yielded the strongest group difference in dynamic properties. This approach could be critiqued as circular and could thus be considered a limitation of our analyses. However, relying on standard algorithms for evaluating cluster performance (which consistently have a penalty for higher dimensionalities) may not necessarily be the appropriate approach for detecting altered patterns in neuroimaging data. Importantly, independent validation experiments should be conducted to confirm that the core of our findings is reproducible and potentially extendable to patients with MDD (see COBIDAS for guidelines on reproducible research; Nichols et al., 2017).
The unclear functional meaning of FC states remains a limitation in studies of dynamic FC. Moreover, the dynamic nature of FC is likely to occur at a much faster timescale than captured by the BOLD signal (here with a TR of 2 s), with magnetoencephalography (MEG) studies pointing to timescales in the order of 200 ms (Baker et al., 2014; Vidaurre et al., 2016). As such, measures like “FC state duration” must be interpreted with caution, having in mind that the BOLD signals were smoothed below 0.1 Hz. Therefore, long durations of a specific FC state (~10 s) most likely reflect periods of high probability occurring at fast frequencies, rather than expressing a prolonged sustained period of a fixed FC configuration. Analyzing the results from a statistical perspective and comparing between groups, as we do here, allows overcoming this limitation in order to capture the most meaningful between‐group differences in connectivity patterns with inherently faster dynamics. Nevertheless, being able to detect meaningful dFC characteristics differentiating people at risk for MDD from controls reinforces the potential of dFC measures to provide biomarkers for psychiatry.
Further, even though we selected remitted participants that were free of current diagnoses of alcohol/drug dependence, psychotic or bipolar, predominant anxiety or severe personality disorder (all assessed by the SCID), a proportion of participants (41%) had other past or current psychiatric comorbidities. Although we performed a sensitivity analysis correcting for the presence of comorbidity in our group comparison for probability and lifetime for the FN‐DMN‐str‐SN state (see Results, Supporting Information), we cannot rule out that these comorbidities did not partly influence the group differences. An additional limitation is that the sad mood induction was at the end of our scanning paradigm. We took this approach because it might have been too emotionally straining for participants to continue with tasks afterward. However, we cannot rule out order effects and/or additional effects of fatigue after other scanning components.
5. CONCLUSION
Using the novel LEiDA approach to examine instantaneous dynamic FC, this study provides new insights on aberrations in dynamic brain network connectivity in remitted MDD patients. This new framework for exploring dynamic FC could potentially be extended to other diseases that have been related to pathological resting‐state connectivity. Overall, our findings suggest reduced ability and flexibility of patients remitted from MDD, but at high risk for recurrence, to access a clinically relevant control network involved in the interplay between emotional and attentional processing and self‐referential attention.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Appendix S1 Supplementary information
ACKNOWLEDGMENTS
This study is supported by unrestricted personal grants from the Academic Medical Centre to C.A.F. (AMC MD‐PhD Scholarship) and Dr. R.J.M. (AMC PhD Scholarship) and a dedicated grant from the Dutch Brain Foundation (Hersenstichting The Netherlands: 2009(2)‐72). Dr. J.C. was supported under the project NORTE‐01‐0145‐FEDER‐000023, by the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER). Dr. G.D. is supported by the Spanish Research Project PSI2016‐75688‐P (AEI/FEDER) and by the European Union's Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 (Human Brain Project SGA2). Dr. M.L.K. is supported by the ERC Consolidator Grant: CAREGIVING (no. 615539) and Center for Music in the Brain, funded by the Danish National Research Foundation (DNRF117). Dr. H.G.R. is supported by a NWO/ZonMW VENI‐Grant #016.126.059. Dr. PE acknowledges support from the EPSRC award EP/N014529/1 funding the EPSRC Centre for Mathematics of Precision Healthcare at Imperial. First, the authors would like to thank the study subjects that participated in this research. Second, the authors acknowledge the thoughtful comments of Prof. Z. Segal and Prof. C.L. Bockting regarding their in‐scanner mood‐induction procedure. Third, the authors would like to thank Eline Meijer, Lisa Bouma, and Gelera Mahmoud who helped with collection of data; and Henk Hallie helped with input and checking of questionnaire data. No one who helped collect/process data received financial compensation for their contributions. These persons are affiliated with: University of Toronto, Toronto, Canada; Academic Medical Centre, University of Amsterdam, The Netherlands; and University Medical Center Groningen, University of Groningen, Groningen, The Netherlands.
Figueroa CA, Cabral J, Mocking RJT, et al. Altered ability to access a clinically relevant control network in patients remitted from major depressive disorder. Hum Brain Mapp. 2019;40:2771–2786. 10.1002/hbm.24559
Caroline A. Figueroa and Joana Cabral share first authorship.
Morten L. Kringelbach and Henricus G. Ruhé share senior authorship.
Funding information Danish National Research Foundation, Grant/Award Number: (DNRF117); Dutch Brain Foundation, Grant/Award Number: 2009(2)‐72; ERC Consolidator Grant, Grant/Award Number: CAREGIVING (n. 615539); European Regional Development Fund, Grant/Award Number: NORTE‐01‐0145‐FEDER‐000023; Horizon 2020 Framework Programme, Grant/Award Number: No. 785907 (Human Brain Project SGA2); Spanish Research Project, Grant/Award Number: PSI2016‐75688‐P (AEI/FEDER); ZonMw, Grant/Award Number: VENI‐Grant #016.126.059
Contributor Information
Caroline A. Figueroa, Email: carolinefigueroa8@gmail.com.
Henricus G. Ruhé, Email: H.G.Ruhe@gmail.com.
REFERENCES
- Baker, A. P. , Brookes, M. J. , Rezek, I. A. , Smith, S. M. , Behrens, T. , Probert Smith, P. J. , & Woolrich, M. (2014). Fast transient networks in spontaneous human brain activity. eLife, 3, e01867 10.7554/eLife.01867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi, Y. , Restom, K. , Liau, J. , & Liu, T. T. (2007). A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage, 37(1), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal, B. , Yetkin, F. Z. , Haughton, V. M. , & Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magnetic Resonance in Medicine, 34(4), 537–541. [DOI] [PubMed] [Google Scholar]
- Bockting, C. L. , Spinhoven, P. , Wouters, L. F. , Koeter, M. W. , & Schene, A. H. (2009). Long‐term effects of preventive cognitive therapy in recurrent depression: A 5.5‐year follow‐up study. The Journal of Clinical Psychiatry, 70(12), 1621–1628. 10.4088/JCP.08m04784blu [DOI] [PubMed] [Google Scholar]
- Cabral, J. , Kringelbach, M. , & Deco, G. (2017). Functional connectivity dynamically evolves on multiple time‐scales over a static structural connectome: Models and mechanisms. NeuroImage, 160, 84–96. 10.1016/j.neuroimage.2017.03.045 [DOI] [PubMed] [Google Scholar]
- Cabral, J. , Vidaurre, D. , Marques, P. , Magalhães, R. , Moreira, P. S. , Soares, J. M. , … Kringelbach, M. L. (2017). Cognitive performance in healthy older adults relates to spontaneous switching between states of functional connectivity during rest. Scientific Reports, 7(1), 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caeyenberghs, K. , Metzler‐Baddeley, C. , Foley, S. , & Jones, D. K. (2016). Dynamics of the human structural connectome underlying working memory training. The Journal of Neuroscience, 36(14), 4056–4066. 10.1523/JNEUROSCI.1973-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, E. Y. , Yeo, B. T. , & Buckner, R. L. (2012). The organization of the human striatum estimated by intrinsic functional connectivity. Journal of Neurophysiology, 108(8), 2242–2263. 10.1152/jn.00270.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. R. (2017). The behavioral and cognitive relevance of time‐varying, dynamic changes in functional connectivity. NeuroImage, 180, 515–525. 10.1016/j.neuroimage.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock, R. C. , Jbabdi, S. , Yan, C.‐G. , Vogelstein, J. , Castellanos, F. X. , Di Martino, A. , … Milham, M. P. (2013). Imaging human connectomes at the macroscale. Nature Methods, 10(6), 524–539. 10.1038/nmeth.2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju, E. , Allen, E. A. , Belger, A. , Ford, J. M. , McEwen, S. , Mathalon, D. H. , … Calhoun, V. D. (2014). Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage: Clinical, 5, 298–308. 10.1016/j.nicl.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux, J. S. , Rombouts, S. A. , Barkhof, F. , Scheltens, P. , Stam, C. J. , Smith, S. M. , & Beckmann, C. F. (2006). Consistent resting‐state networks across healthy subjects. Proceedings of the National Academy of Sciences of the United States of America, 103(37), 13848–13853. 10.1073/pnas.0601417103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco, G. , Cabral, J. , Woolrich, M. W. , Stevner, A. B. , van Hartevelt, T. J. , & Kringelbach, M. L. (2017). Single or multiple frequency generators in on‐going brain activity: A mechanistic whole‐brain model of empirical MEG data. NeuroImage, 152, 538–550. 10.1016/j.neuroimage.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco, G. , Jirsa, V. K. , & McIntosh, A. R. (2011). Emerging concepts for the dynamical organization of resting‐state activity in the brain. Nature Reviews. Neuroscience, 12(1), 43–56. 10.1038/nrn2961 [DOI] [PubMed] [Google Scholar]
- Deco, G. , & Kringelbach, M. (2016). Metastability and coherence: Extending the communication through coherence hypothesis using a whole‐brain computational perspective. Trends in Neurosciences, 39(6), 432 10.1016/j.tins.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Demirtaş, M. , Tornador, C. , Falcón, C. , López‐Solà, M. , Hernández‐Ribas, R. , Pujol, J. , … Deco, G. (2016). Dynamic functional connectivity reveals altered variability in functional connectivity among patients with major depressive disorder. Human Brain Mapping, 37(8), 2918–2930. 10.1002/hbm.23215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets, W. C. (2007). Orbitofrontal cortex function and structure in depression. Annals of the New York Academy of Sciences, 1121(1), 499–527. 10.1196/annals.1401.029 [DOI] [PubMed] [Google Scholar]
- Enriquez‐Geppert, S. , Huster, R. J. , & Herrmann, C. S. (2017). EEG‐neurofeedback as a tool to modulate cognition and behavior: A review tutorial. Frontiers in Human Neuroscience, 11, 51 10.3389/fnhum.2017.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, C. A. , Mocking, R. J. T. , van Wingen, G. , Martens, S. , Ruhe, H. G. , & Schene, A. H. (2017). Aberrant default‐mode network‐hippocampus connectivity after sad memory‐recall in remitted‐depression. Social Cognitive and Affective Neuroscience, 12(11), 1803–1813. 10.1093/scan/nsx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, A. S. , Keller, C. J. , & Etkin, A. (2016). The clinical applicability of functional connectivity in depression: Pathways toward more targeted intervention. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 1(3), 262–270. 10.1016/j.bpsc.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Foland‐Ross, L. C. , Cooney, R. E. , Joormann, J. , Henry, M. L. , & Gotlib, I. H. (2014). Recalling happy memories in remitted depression: A neuroimaging investigation of the repair of sad mood. Cognitive, Affective, & Behavioral Neuroscience, 14(2), 818–826. 10.3758/s13415-013-0216-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman, D. J. , Hamilton, J. P. , & Gotlib, I. H. (2011). Frontostriatal functional connectivity in major depressive disorder. Biology of Mood & Anxiety Disorders, 1(1), 11 10.1186/2045-5380-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F. , Coalson, T. S. , Robinson, E. C. , Hacker, C. D. , Harwell, J. , Yacoub, E. , … Van Essen, D. C. (2016). A multi‐modal parcellation of human cerebral cortex. Nature, 536, 171–178. 10.1038/nature18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerean, E. , Salmi, J. , Lahnakoski, J. M. , Jaaskelainen, I. P. , & Sams, M. (2012). Functional magnetic resonance imaging phase synchronization as a measure of dynamic functional connectivity. Brain Connectivity, 2(2), 91–101. 10.1089/brain.2011.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner, S. , Hellgren, J. , Menard, A. , Saitoh, K. , & Wikstrom, M. A. (2005). Mechanisms for selection of basic motor programs‐‐roles for the striatum and pallidum. Trends in Neurosciences, 28(7), 364–370. 10.1016/j.tins.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Hamilton, J. P. , Furman, D. J. , Chang, C. , Thomason, M. E. , Dennis, E. , & Gotlib, I. H. (2011). Default‐mode and task‐positive network activity in major depressive disorder: Implications for adaptive and maladaptive rumination. Biological Psychiatry, 70(4), 327–333. 10.1016/j.biopsych.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, E. C. , Battaglia, D. , Spiegler, A. , Deco, G. , & Jirsa, V. K. (2015). Functional connectivity dynamics: Modeling the switching behavior of the resting state. NeuroImage, 105, 525–535. 10.1016/j.neuroimage.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Harrison, B. J. , Pujol, J. , Ortiz, H. , Fornito, A. , Pantelis, C. , & Yücel, M. (2008). Modulation of brain resting‐state networks by sad mood induction. PLoS One, 3(3), e1794 10.1371/journal.pone.0001794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriks, R. , Adhikari, M. H. , Murayama, Y. , Ganzetti, M. , Mantini, D. , Logothetis, N. K. , & Deco, G. (2016). Can sliding‐window correlations reveal dynamic functional connectivity in resting‐state fMRI? NeuroImage, 127, 242–256. 10.1016/j.neuroimage.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoorelbeke, K. , & Koster, E. H. (2017). Internet‐delivered cognitive control training as a preventive intervention for remitted depressed patients: Evidence from a double‐blind randomized controlled trial study. Journal of Consulting and Clinical Psychology, 85(2), 135–146. 10.1037/ccp0000128 [DOI] [PubMed] [Google Scholar]
- Hutchison, R. M. , Womelsdorf, T. , Allen, E. A. , Bandettini, P. A. , Calhoun, V. D. , Corbetta, M. , … Chang, C. (2013). Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage, 80, 360–378. 10.1016/j.neuroimage.2013.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japee, S. , Holiday, K. , Satyshur, M. D. , Mukai, I. , & Ungerleider, L. G. (2015). A role of right middle frontal gyrus in reorienting of attention: A case study. Frontiers in Systems Neuroscience, 9, 23 10.3389/fnsys.2015.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson, M. , Bannister, P. , Brady, M. , & Smith, S. (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage, 17(2), 825–841. [DOI] [PubMed] [Google Scholar]
- Jia, H. , Hu, X. , & Deshpande, G. (2014). Behavioral relevance of the dynamics of the functional brain connectome. Brain Connectivity, 4(9), 741–759. 10.1089/brain.2014.0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C. , Jia, H. , Lanka, P. , Rangaprakash, D. , Li, L. , Liu, T. , … Deshpande, G. (2017). Dynamic brain connectivity is a better predictor of PTSD than static connectivity. Human Brain Mapping, 38(9), 4479–4496. 10.1002/hbm.23676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, R. H. , Andrews‐Hanna, J. R. , Wager, T. D. , & Pizzagalli, D. A. (2015). Large‐scale network dysfunction in major depressive disorder: A meta‐analysis of resting‐state functional connectivity. JAMA Psychiatry, 72(6), 603–611. 10.1001/jamapsychiatry.2015.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, R. H. , Whitfield‐Gabrieli, S. , Dillon, D. G. , Goer, F. , Beltzer, M. , Minkel, J. , … Pizzagalli, D. A. (2016). Dynamic resting‐state functional connectivity in major depression. Neuropsychopharmacology, 41(7), 1822–1830. 10.1038/npp.2015.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knyazev, G. G. , Savostyanov, A. N. , Bocharov, A. V. , Tamozhnikov, S. S. , & Saprigyn, A. E. (2016). Task‐positive and task‐negative networks and their relation to depression: EEG beamformer analysis. Behavioural Brain Research, 306, 160–169. 10.1016/j.bbr.2016.03.033 [DOI] [PubMed] [Google Scholar]
- Kringelbach, M. L. , McIntosh, A. R. , Ritter, P. , Jirsa, V. K. , & Deco, G. (2015). The rediscovery of slowness: Exploring the timing of cognition. Trends in Cognitive Sciences, 19(10), 616–628. 10.1016/j.tics.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Lindquist, M. A. , Xu, Y. , Nebel, M. B. , & Caffo, B. S. (2014). Evaluating dynamic bivariate correlations in resting‐state fMRI: A comparison study and a new approach. NeuroImage, 101, 531–546. 10.1016/j.neuroimage.2014.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhyastha, T. M. , Askren, M. K. , Boord, P. , & Grabowski, T. J. (2015). Dynamic connectivity at rest predicts attention task performance. Brain Connectivity, 5(1), 45–59. 10.1089/brain.2014.0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti, I. , Koster, E. H. , Sonuga‐Barke, E. J. , & De Raedt, R. (2012). The default mode network and recurrent depression: A neurobiological model of cognitive risk factors. Neuropsychology Review, 22(3), 229–251. 10.1007/s11065-012-9199-9 [DOI] [PubMed] [Google Scholar]
- Martin, S. J. , & Morris, R. G. (2002). New life in an old idea: The synaptic plasticity and memory hypothesis revisited. Hippocampus, 12(5), 609–636. 10.1002/hipo.10107 [DOI] [PubMed] [Google Scholar]
- Menon, V. (2011). Large‐scale brain networks and psychopathology: A unifying triple network model. Trends in Cognitive Sciences, 15(10), 483–506. 10.1016/j.tics.2011.08.003 [DOI] [PubMed] [Google Scholar]
- Menon, V. , & Uddin, L. Q. (2010). Saliency, switching, attention and control: A network model of insula function. Brain Structure & Function, 214(5–6), 655–667. 10.1007/s00429-010-0262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocking, R. J. T. , Figueroa, C. A. , Rive, M. M. , Geugies, H. , Servaas, M. N. , Assies, J. , … Ruhé, H. G. (2016). Vulnerability for new episodes in recurrent major depressive disorder: Protocol for the longitudinal DELTA‐neuroimaging cohort study. BMJ Open, 6(3), e009510 10.1136/bmjopen-2015-009510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders, P. C. , van Eijndhoven, P. F. , Schene, A. H. , Beckmann, C. F. , & Tendolkar, I. (2015). Resting‐state functional connectivity in major depressive disorder: A review. Neuroscience and Biobehavioral Reviews, 56, 330–344. 10.1016/j.neubiorev.2015.07.014 [DOI] [PubMed] [Google Scholar]
- Murphy, K. , & Fox, M. D. (2017). Towards a consensus regarding global signal regression for resting state functional connectivity MRI. NeuroImage, 154, 169–173. 10.1016/j.neuroimage.2016.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M. E. (2006). Finding community structure in networks using the eigenvectors of matrices. Physical Review. E, Statistical, Nonlinear, and Soft Matter Physics, 74(3 Pt. 2), 036104 10.1103/PhysRevE.74.036104 [DOI] [PubMed] [Google Scholar]
- Nichols, T. E. , Das, S. , Eickhoff, S. B. , Evans, A. C. , Glatard, T. , Hanke, M. , … Yeo, B. T. (2017). Best practices in data analysis and sharing in neuroimaging using MRI. Nature Neuroscience, 20(3), 299–303. 10.1038/nn.4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi, J. S. , Vij, S. G. , Dajani, D. R. , Steimke, R. , Damaraju, E. , Rachakonda, S. , … Uddin, L. Q. (2017). Chronnectomic patterns and neural flexibility underlie executive function. NeuroImage, 147, 861–871. 10.1016/j.neuroimage.2016.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce‐Alvarez, A. , Deco, G. , Hagmann, P. , Romani, G. L. , Mantini, D. , & Corbetta, M. (2015). Resting‐state temporal synchronization networks emerge from connectivity topology and heterogeneity. PLoS Computational Biology, 11(2), e1004100 10.1371/journal.pcbi.1004100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Mitra, A. , Laumann, T. O. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2014). Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage, 84, 320–341. 10.1016/j.neuroimage.2013.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti, M. G. , Bolton, T. A. , & Ville, D. V. (2016). The dynamic functional connectome: State‐of‐the‐art and perspectives. NeuroImage, 160, 41–54. 10.1016/j.neuroimage.2016.12.061 [DOI] [PubMed] [Google Scholar]
- Rush, A. J. , Giles, D. E. , Schlesser, M. A. , Fulton, C. L. , Weissenburger, J. , & Burns, C. (1986). The inventory for depressive symptomatology (IDS): Preliminary findings. Psychiatry Research, 18(1), 65–87. [DOI] [PubMed] [Google Scholar]
- Sakoglu, U. , Pearlson, G. D. , Kiehl, K. A. , Wang, Y. M. , Michael, A. M. , & Calhoun, V. D. (2010). A method for evaluating dynamic functional network connectivity and task‐modulation: Application to schizophrenia. Magma, 23(5–6), 351–366. 10.1007/s10334-010-0197-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servaas, M. N. , Riese, H. , Renken, R. J. , Wichers, M. , Bastiaansen, J. A. , Figueroa, C. A. , … Ruhe, H. G. (2017). Associations between daily affective instability and connectomics in functional subnetworks in remitted patients with recurrent major depressive disorder. Neuropsychopharmacology, 42(13), 2583–2592. 10.1038/npp.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, C. E. (1948). A mathematical theory of communication (parts I and II). Bell System Technical Journal, 27, 379–423. [Google Scholar]
- Shen, X. , Tokoglu, F. , Papademetris, X. , & Constable, R. T. (2013). Groupwise whole‐brain parcellation from resting‐state fMRI data for network node identification. NeuroImage, 82, 403–415. 10.1016/j.neuroimage.2013.05.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine, J. M. , Koyejo, O. , Bell, P. T. , Gorgolewski, K. J. , Gilat, M. , & Poldrack, R. A. (2015). Estimation of dynamic functional connectivity using multiplication of temporal derivatives. NeuroImage, 122, 399–407. 10.1016/j.neuroimage.2015.07.064 [DOI] [PubMed] [Google Scholar]
- Smith, K. (2014). Mental health: A world of depression. Nature, 515(7526), 181 10.1038/515180a [DOI] [PubMed] [Google Scholar]
- Smith, S. M. (2002). Fast robust automated brain extraction. Human Brain Mapping, 17(3), 143–155. 10.1002/hbm.10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer, N. , Landeau, B. , Papathanassiou, D. , Crivello, F. , Etard, O. , Delcroix, N. , … Joliot, M. (2002). Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. NeuroImage, 15(1), 273–289. 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- Van der Lande, H. , Mocking, R. J. , Servaas, M. , Figueroa, C. , Maaskant, J. , Schene, A. , & Ruhe, H. (2016). Associations of positive and negative affect with physical activity in drug‐free patients in remission of recurrent major depressive disorder: P‐291. Bipolar Disorders, 18, 160–161. [Google Scholar]
- Verhage, F. (1964). Intelligentie en leeftijd bij volwassenen en bejaarden. Groningen: Koninklijke Van Gorcum, 98. [Google Scholar]
- Vidaurre, D. , Quinn, A. J. , Baker, A. P. , Dupret, D. , Tejero‐Cantero, A. , & Woolrich, M. W. (2016). Spectrally resolved fast transient brain states in electrophysiological data. NeuroImage, 126, 81–95. 10.1016/j.neuroimage.2015.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. A. (2008). Probability distribution and entropy as a measure of uncertainty. Journal of Physics A: Mathematical and Theoretical, 41(6), 065004. [Google Scholar]
- Wise, T. , Marwood, L. , Perkins, A. M. , Herane‐Vives, A. , Joules, R. , Lythgoe, D. J. , … Arnone, D. (2017). Instability of default mode network connectivity in major depression: A two‐sample confirmation study. Translational Psychiatry, 7(4), e1105 10.1038/tp.2017.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni, T. , Poldrack, R. A. , Nichols, T. E. , Van Essen, D. C. , & Wager, T. D. (2011). Large‐scale automated synthesis of human functional neuroimaging data. Nature Methods, 8(8), 665–670. 10.1038/nmeth.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, B. T. , Krienen, F. M. , Sepulcre, J. , Sabuncu, M. R. , Lashkari, D. , Hollinshead, M. , … Buckner, R. L. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology, 106(3), 1125–1165. 10.1152/jn.00338.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilverstand, A. , Sorger, B. , Zimmermann, J. , Kaas, A. , & Goebel, R. (2014). Windowed correlation: A suitable tool for providing dynamic fMRI‐based functional connectivity neurofeedback on task difficulty. PLoS One, 9(1), e85929 10.1371/journal.pone.0085929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary information


