Abstract
Brain extraction is a critical preprocessing step in the analysis of neuroimaging studies conducted with magnetic resonance imaging (MRI) and influences the accuracy of downstream analyses. The majority of brain extraction algorithms are, however, optimized for processing healthy brains and thus frequently fail in the presence of pathologically altered brain or when applied to heterogeneous MRI datasets. Here we introduce a new, rigorously validated algorithm (termed HD‐BET) relying on artificial neural networks that aim to overcome these limitations. We demonstrate that HD‐BET outperforms six popular, publicly available brain extraction algorithms in several large‐scale neuroimaging datasets, including one from a prospective multicentric trial in neuro‐oncology, yielding state‐of‐the‐art performance with median improvements of +1.16 to +2.50 points for the Dice coefficient and −0.66 to −2.51 mm for the Hausdorff distance. Importantly, the HD‐BET algorithm, which shows robust performance in the presence of pathology or treatment‐induced tissue alterations, is applicable to a broad range of MRI sequence types and is not influenced by variations in MRI hardware and acquisition parameters encountered in both research and clinical practice. For broader accessibility, the HD‐BET prediction algorithm is made freely available (http://www.neuroAI-HD.org) and may become an essential component for robust, automated, high‐throughput processing of MRI neuroimaging data.
Keywords: artificial neural networks, brain extraction, deep learning, magnetic resonance imaging, neuroimaging, skull stripping
1. INTRODUCTION
Brain extraction, which refers to the process of separating the brain from nonbrain tissues in medical images is a preliminary but critical step in many neuroimaging studies conducted with magnetic resonance imaging (MRI). Consequently, the accuracy of brain extraction may have an essential impact on the quality of the subsequent analyses such as image registration (Kleesiek et al., 2016; Klein et al., 2010; Woods, Mazziotta, & Cherry, 1993), segmentation of brain tumors or lesions (de Boer et al., 2010; Menze et al., 2015; Shattuck, Sandor‐Leahy, Schaper, Rottenberg, & Leahy, 2001; Wang, Chen, Pan, Hong, & Xia, 2010; Zhang, Brady, & Smith, 2001; Zhao, Ruotsalainen, Hirvonen, Hietala, & Tohka, 2010), measurement of global and regional brain volumes (e.g., in neurodegenerative diseases and multiple sclerosis; Frisoni, Fox, Jack Jr., Scheltens, & Thompson, 2010; Radue et al., 2015), estimation of cortical thickness (Haidar & Soul, 2006; MacDonald, Kabani, Avis, & Evans, 2000), cortical surface reconstruction (Dale, Fischl, & Sereno, 1999; Tosun et al., 2006), and for planning of neurosurgical interventions (Leote, Nunes, Cerqueira, Loução, & Ferreira, 2018).
Manual segmentation is currently considered the “gold‐standard” for brain extraction (Smith, 2002; Souza et al., 2018). However, this approach is not only very labor‐intensive and time‐consuming, but also shows a strong interindividual and intraindividual variability (Kleesiek et al., 2016; Smith, 2002; Souza et al., 2018) that could ultimately bias the analysis and consequently hamper the reproducibility of clinical studies. To overcome these shortcomings, several (semi‐) automated brain extraction algorithms have been developed and optimized over the last years (Kalavathi & Prasath, 2016). Their generalizability is, however, limited in the presence of varying acquisition parameters or in the presence of abnormal pathological brain tissue, such as brain tumors. Without additional manual correction, poor brain extraction can introduce errors in downstream analysis (Beers et al., 2018).
Artificial neural networks (ANNs) have recently been successfully applied to a multitude of medical image segmentation tasks. In this context, several approaches based on ANN have been proposed to improve the accuracy of brain extraction. However, these ANN algorithms have focused on learning brain extraction from training datasets either containing a collection of normal (or apparently normal) brain MRI from public datasets (Dey & Hong, 2018; Sadegh Mohseni Salehi, Erdogmus, & Gholipour, 2017), or from a limited number of (single institutional) brain MRI with pathologies (Beers et al., 2018; Kleesiek et al., 2016). Therefore, generalizability of these ANN algorithms to complex multicenter datasets may be limited on unseen data with varying MR hardware and acquisition parameters, pathologies or treatment‐induced tissue alterations. Moreover, most approaches up until now focused on processing precontrast T1‐weighted (T1‐w) MRI sequences, since it provides a good contrast between different brain tissues and is frequently used as standard space for registration of further image sequences (Han et al., 2018; Iglesias, Liu, Thompson, & Tu, 2011; Lutkenhoff et al., 2014). However, they fall short when it comes to processing other types of MRI sequences, which would, however, be desirable for a broad application to research and clinical studies.
To overcome these limitations, we utilize MRI data from a large multicenter clinical trial in neuro‐oncology (EORTC‐26101;Wick et al., 2016 ; Wick et al., 2017) to train and independently validate an ANN for brain extraction (subsequently referred to as HD‐BET). Specifically, we aimed to develop an automated method that (a) performs robustly in the presence of pathological and treatment‐induced tissue alterations, (b) is not influenced by variations in MRI hardware and acquisition parameters, and (c) is applicable to independently process various types of common anatomical MRI sequence.
2. METHODS
2.1. Datasets
Four different datasets including the MRI data from a prospective randomized Phase II and III trials in neuro‐oncology (EORTC‐26101) (Wick et al., 2017; Wick et al., 2016) and three independent public datasets (LONI Probabilistic Brain Atlas [LPBA40], Nathan Kline Institute Enhanced Rockland Sample Neurofeedback Study [NFBS], Calgary‐Campinas‐359 [CC‐359]) (Puccio, Pooley, Pellman, Taverna, & Craddock, 2016; Shattuck et al., 2008; Souza et al., 2018) were used for the present study. The characteristics of the individual datasets were as follows.
2.1.1. EORTC‐26101
The EORTC‐26101 study was a prospective randomized Phase II and III trials in patients with first progression of a glioblastoma after standard chemoradiotherapy. Briefly, Phase II trial evaluated the optimal treatment sequence of bevacizumab and lomustine (four treatment arms with single agent vs. sequential vs. combination) (Wick et al., 2016) whereas the subsequent Phase III trial (two treatment arms) compared patients treated with lomustine alone with those receiving a combination of lomustine and bevacizumab (Wick et al., 2017). Overall, the EORTC‐26101 study included n = 596 patients (n = 159 from Phase II and n = 437 from Phase III) with n = 2,593 individual MRI exams acquired at 37 institutions within Europe. The study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by local ethics committees and patients provided written informed consent (EudraCT# 2010‐023218‐30 and NCT01290939). Full study design and outcomes have been published previously (Wick et al., 2016; Wick et al., 2017). MRI exams were acquired at baseline and every 6 weeks until Week 24, afterward every 3 months. For the present analysis, we included T1‐w, contrast‐enhanced T1‐w (cT1‐w), fluid attenuated inversion recovery (FLAIR), and T2‐weighted (T2‐w) sequences (either acquired 3D and/or with axial orientation) and excluded those with heavy motion artifacts or corrupt data. These criteria were fulfilled by n = 10,005 individual sequences (including n = 2,401 T1‐w, n = 2,248 T2‐w, n = 2,835 FLAIR and n = 2,521 cT1‐w sequences from n = 2,401 exams, and n = 583 patients) which were included for the present analysis. The EORTC‐26101 dataset was divided into a training and test set using a random split of the dataset (∼2:1 ratio) with the constraint that all patients from each of the 37 institution were either assigned to the training or test set (to limit the potential of overfitting the HD‐BET algorithm). By applying this split, the EORTC‐26101 training set included data from n = 25 institutions (n = 6,586 individual MRI sequences from n = 1,568 exams, n = 372 patients) whereas the EORTC‐26101 test set included data from the remaining n = 12 institutions (n = 3,419 individual MRI sequences from n = 833 exams, n = 211 patients). In this context, it is important to emphasize that the EORTC‐26101 test set was entirely independent from the training set, as it is comprised of acquisitions from different institutions (and thus different MRI scanners/field strengths, see Table 1 for the T1 detailed information on the individual MRI sequences, scanner types, field strengths) that are disjunct from the institutions in the training set.
2.1.2. Public datasets
We used three public datasets for independent testing. Specifically, we collected and analyzed data from (a) the single‐institutional Laboratory of Neuro Imaging (LONI) (LPBA40) dataset of the LONI consisting of n = 40 MRI scans from individual healthy human subjects (Shattuck et al., 2008), (b) the single‐institutional NFBS dataset consisting of n = 125 MRI scans from individual patients with a variety of clinical and subclinical psychiatric symptoms (Puccio et al., 2016), and (c) the CC‐359 dataset consisting of n = 359 MRI scans from healthy adults (Souza et al., 2018). For each subject, the repositories contains an anonymized (defaced) T1‐w MRI sequence and a manually corrected ground‐truth (GT) brain mask.
2.2. Brain extraction using competing algorithms
All MRI sequences from each of the datasets were preprocessed identically. First, all images were reoriented to the standard orientation (fslreorient2std, FMRIB software library, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL), followed by the application of reference brain extraction algorithms. We compare HD‐BET to six publicly available and frequently used brain extraction algorithms, namely, BET (Smith, 2002), 3dSkullStrip (Cox, 1996), BSE (Shattuck & Leahy, 2002), ROBEX (Iglesias et al., 2011), BEaST (Eskildsen et al., 2012), and MONSTR (Roy, et al., 2017) (see Methods S1, Supporting Information for detailed description). As we intend HD‐BET to be used out of the box, we also apply the reference methods as they are provided with no dataset‐specific adaptations. For all competing brain extraction algorithms (except MONSTR) the maximum allowed processing time was set to 60 min (to keep processing within an acceptable time frame and execution of the brain extraction process was aborted if an algorithm exceeded this time limit for processing a single MRI sequence). Since BET, 3dSkullStrip, BSE, ROBEX, and BEaST have primarily been developed for processing of T1‐w sequences, we did not perform brain extraction with these algorithms on any other sequence type (i.e., cT1‐w, FLAIR, or T2‐w) that were available in the EORTC‐26101 test set. MONSTR is capable of also processing cT1‐w, FLAIR, and T2‐w sequences and we therefore used it to perform brain extraction on all available sequences of the EORTC‐26101 test set. In summary, this setup resulted in a comparison against six competing algorithms for brain extraction on T1‐w sequences in all four test sets (EORTC‐26101 test set, LPBA40, NFBS, CC‐359) and additional comparison against MONSTR on the remaining MRI sequences (T2‐w, cT1‐w, FLAIR) in the EORTC‐26101 test set.
2.3. Defining a GT (reference) brain mask
A GT reference brain mask is required to evaluate the accuracy of brain extraction algorithms. Moreover, for the purpose of the present study with development of the HD‐BET algorithm for automated brain extraction, these masks are required to train the algorithm (i.e., to learn this specific task), as well as for subsequent evaluation of its accuracy. A GT reference brain mask for the T1‐w sequences was already provided within the three public datasets (LPBA40, NFBS, CC‐359), whereas for the EORTC‐26101 dataset, we generated a radiologist‐annotated GT reference brain mask for T1‐w sequences as follows: The brain mask generated by BET algorithm was selected as a starting point. For each brain mask, visual inspection and corrections were performed using ITK‐SNAP (by applying the different capabilities of this tool, including region‐growing segmentation and manual corrections (http://www.itksnap.org; Yushkevich et al., 2006)). The manual correction took on average about 15 min per brain mask. Given the amount of data, only one rater per GT reference mask was used. Similar to the provided brain masks, we defined the following criteria: (a) including all cerebral and cerebellar gray and white matter as well as the brainstem, (b) including the cerebrospinal fluid in the ventricles and the cerebellar cistern, and (c) excluding the chiasma. In a second step, to enable the use of the HD‐BET algorithm independently of the input MRI sequence type (i.e., not limited to T1‐w sequences) we transferred the GT reference brain masks within the EORTC‐26101 dataset from T1‐w to the remaining anatomical sequences, that is, cT1‐w, FLAIR, and T2‐w sequences. First, all sequences were spatially aligned to the respective T1‐w sequence by rigid registration with 6 degrees of freedom (Greve & Fischl, 2009; Jenkinson & Smith, 2001), resulting in a transformation matrix for each of them. Next, the transformation matrix was inversely back transformed to the individual sequence space of the c T1‐w, FLAIR, and T2‐w sequences and applied to the GT reference brain mask (within the space of the T1‐w sequence) using nearest neighbor interpolation. Thereby a GT brain mask was generated for the remaining sequences (i.e., c T1‐w, FLAIR, and T2‐w) within the individual sequence space. Finally, visual inspection was performed for all brain masks to exclude registration errors.
2.4. Artificial neural network
The topology of the ANN underlying the HD‐BET algorithm was inspired by the U‐Net image segmentation architecture (Ronneberger, Fischer, & Brox, 2015) and its 3D derivatives (Çiçek, Abdulkadir, Lienkamp, Brox, & Ronneberger, 2016; Kayalibay, Jensen, & van der Smagt, 2017; Milletari, Navab, & Ahmadi, 2016) and has recently been shown to have excellent performance in brain tumor segmentation both in an international competition (Isensee, Kickingereder, Wick, Bendszus, & Maier‐Hein, 2018) as well as in the context of a large‐scale multi‐institutional study (Kickingereder et al., 2019). Methods S2, Supporting Information, contain an extended description of the architecture, as well as the training and evaluation procedure. All MRI sequences from the EORTC‐26101 training set (i.e., T1‐w, cT1‐w, FLAIR, and T2‐w) were used to train and validate the HD‐BET algorithm (with fivefold cross‐validation). For independent large‐scale testing and application of the HD‐BET algorithm (done by using the five models from cross‐validation as an ensemble), all MRI sequences from the EORTC‐26101 test set (i.e., T1‐w, cT1‐w, FLAIR, and T2‐w) as well as the T1‐w sequences of the LPBA40, NFBS, and CC‐359 datasets were used. For both training and testing, the HD‐BET algorithm was blinded to the type of MRI sequence used as input (i.e., T1‐w, cT1‐w, FLAIR, or T2‐w) which allowed to develop an algorithm that is capable to perform brain extraction irrespective of the type of anatomical MRI sequence.
2.5. Evaluation metrics
To evaluate the performance of the different brain extraction algorithms, we compared the segmentation results of the different brain extraction methods with the GT reference brain mask from each individual sequence. Among the numerous different metrics for measuring the similarity of two segmentation masks, we calculated a volumetric measure, the Dice similarity coefficient (Dice, 1945) and a distance measure, the Hausdorff distance. The Dice coefficient is a standard metric for reporting the performance of segmentation and measures the extent of spatial overlap between two binary images, GT and predicted brain mask. It is defined as twice the size of the intersection between two masks normalized by the sum of their volumes.
Its values range between 0 (no overlap) and 100 (perfect agreement). However, volumetric measures can be insensitive to differences in edges, especially if this difference leads to an overall small volume effect relative to the total volume. Therefore, we used the Hausdorff distance (Taha & Hanbury, 2015) to measure the maximal contour distance (mm) between the two masks.
The smaller the Hausdorff distance, the more similar the images. Here, we took the 95th percentile of the Hausdorff distance, which is widely used; for example, in the evaluation of brain tumor segmentation (Menze et al., 2015), as it allows to overcome the high sensitivity of the Hausdorff distance to outliers.
2.6. Statistical analysis
The Shapiro–Wilk test was performed to compare all evaluation metrics (Dice coefficient, Hausdorff distance) obtained from the T1‐w sequences among the different brain extraction algorithms for normality. We report descriptive statistics (median, interquartile range [IQR]) for Dice coefficient and Hausdorff distance for all brain extraction algorithms in each of the datasets. To test the general differences of the different brain extraction algorithms in terms of their Dice coefficient and Hausdorff distance, we used a nonparametric Friedman or Skillings–Mack test. The latter was used in the presence of missing data that would prevent a listwise comparison (missing data resulted from those instances where the brain mask from one of the six competing brain extraction algorithms was not generated after exceeding the predefined time limit of 60 min for processing a single T1‐w sequence, no time limit was used for MONSTR. For post hoc comparisons, one‐tailed Wilcoxon matched‐pairs signed‐rank tests were used to assess the performance of the HD‐BET algorithm in comparison to the six competing brain extraction methods. The p‐values from all post hoc tests within each of the dataset were corrected for multiple comparisons using the Bonferroni adjustment. The effect sizes of the post hoc comparisons were interpreted using the Cohen classification (≥0.1 for small effects, ≥0.3 for medium effects, and ≥ 0.5 for large effects; Cohen, 1988).
For all other imaging sequences (i.e., cT1‐w, FLAIR, and T2‐w) analyzed within the EORTC‐26101 dataset using HD‐BET and MONSTR, we report descriptive statistics (median, IQR) for Dice coefficient and Hausdorff distance. Moreover, one‐tailed Wilcoxon matched pairs signed‐rank tests were used to assess the performance of the HD‐BET algorithm in comparison to MONSTR on these imaging sequences.
All statistical analyses were performed with R version 3.4.0 (R Foundation for Statistical Computing, Vienna, Austria). p‐Values <.05 were considered significant.
3. RESULTS
Within the EORTC‐26101 training set (consisting of n = 6,586 individual MRI sequences with precontrast and postcontrast T1‐weighted [T1‐w, cT1‐w], FLAIR, and T2‐weighted [T2‐w] sequences from 1,568 MRI exams in 372 patients acquired across 25 institutions; Table 1), the HD‐BET algorithm acquired the relevant knowledge to generate a brain mask irrespective of the type of MRI sequence and in the presence of pathologies. Independent application and testing of the HD‐BET algorithm in the EORTC‐26101 test set (consisting of n = 3,419 individual MRI sequences from 833 exams in 211 patients acquired across 12 institutions; Table 1) yielded a median Dice coefficient of 97.6 (IQR, 97.0–98.0) on T1‐w, 96.9 (IQR, 96.1–97.4) on cT1‐w, 96.4 (IQR, 95.2–97.0) on FLAIR, and 96.1 (IQR, 95.2–96.7) on T2‐w sequences. Corresponding median Hausdorff distances (95th percentile) were 2.7 mm (IQR, 2.2–3.3 mm) on T1‐w, 3.2 mm (IQR, 2.8–4.1 mm) on cT1‐w, 4.2 mm (IQR, 3.4–5.0 mm) on FLAIR, and 4.4 mm (IQR, 3.9–5.0 mm) on T2‐w (Figure 1 and Table 2). The performance was confirmed upon testing the HD‐BET algorithm in three independent public datasets (LPBA40, NFBS, and CC‐359) which are specifically designed to evaluate the performance of brain extraction algorithms. In contrast to the EORTC‐26101 dataset, application of the HD‐BET algorithm in these public datasets was restricted to T1‐w sequences since no other type of MRI sequence was provided. Specifically, we yielded median Dice coefficients of 97.5 (IQR, 97.4–97.7) for LPBA40, 98.2 (IQR, 98.0–98.4) for NFBS, and 96.9 (IQR, 96.7–97.1) for the CC‐359 datasets with corresponding median Hausdorff distances (95th percentile) of 2.9 mm (IQR, 2.5–3.0 mm), 2.8 mm (IQR, 2.4–2.8 mm), and 1.7 mm (IQR, 1.4–2.0 mm) confirming both reproducibility and generalizability of the performance of the HD‐BET algorithm (Table S1, Supporting Information).
Table 1.
Characteristics of the datasets analyzed within the present study
| EORTC‐26101 | LPBA40 | NFBS | CC‐359 | ||
|---|---|---|---|---|---|
| Training set | Test set | ||||
| Patients (n) | 372 | 211 | 40 | 125 | 359 |
| MRI exams (n) | 1,568 | 833 | 40 | 125 | 359 |
| MRI exams per patient (median, IQR) | 4 (3–6) | 4 (3–6) | 1 | 1 | 1 |
| Institutes (n) | 25 | 12 | 1 | 1 | 2 |
| Patients per institute (median, IQR) | 7 (4–15) | 11 (3–20) | 1 | 1 | 60/299 |
| MRI sequence (n)* | |||||
| T1‐w | 1,568 | 833 | 40 | 125 | 359 |
| cT1‐w | 1,623 | 898 | – | – | – |
| FLAIR | 1,940 | 895 | – | – | – |
| T2‐w | 1,455 | 793 | – | – | – |
| MR vendors (n) | |||||
| Siemens | 535 | 395 | – | 125 | 120 |
| Philips | 350 | 157 | – | – | 119 |
| General electric | 640 | 267 | 40 | – | 120 |
| Toshiba | 12 | – | – | – | – |
| Unknown | 31 | 14 | – | – | – |
| MR field strength (n) | |||||
| 1.0 T | – | 9 | –– | – | – |
| 1.5 T | 631 | 78 | 40 | – | 179 |
| 3.0 T | 216 | 317 | – | 125 | 180 |
| 1.5 or 3 T | 619 | 415 | – | – | – |
| Unknown | 104 | 14 | – | – | – |
Abbreviations: IQR, interquartile range; LPBA40, LONI Probabilistic Brain Atlas; MRI, magnetic resonance imaging; NFBS, Nathan Kline Institute Enhanced Rockland Sample Neurofeedback Study; CC‐359, Calgary‐Campinas‐359.
higher number of MRI sequences (as compared to total number of MRI exams) due to inclusion of both 2D and 3D acquisition (if available).
Figure 1.
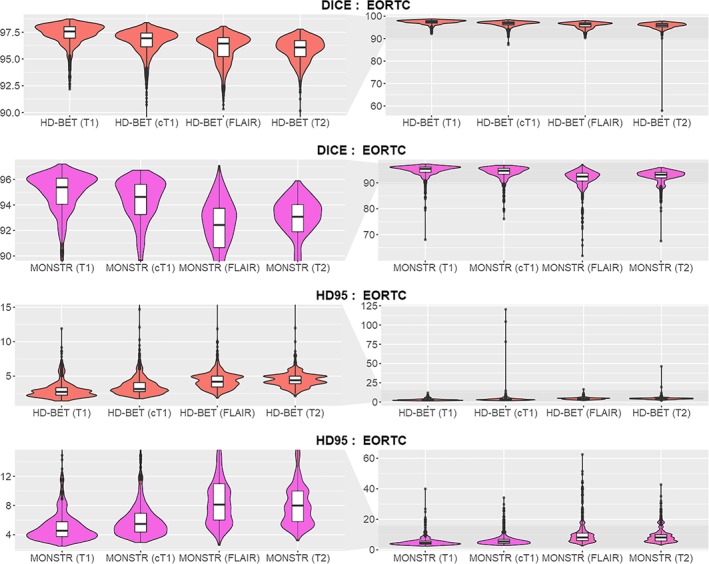
Dice coefficient and Hausdorff distance (95th percentile) obtained from the individual sequences T1‐w, cT1‐w, FLAIR, and T2‐w with the HD‐BET algorithm and for MONSTR in the EORTC‐26101 test set using violin charts (and superimposed box plots). Obtained median Dice coefficients were > 0.95 for all sequences. The performance of brain extraction on cT1‐w, FLAIR, or T2‐w in terms of Dice coefficient (higher values indicate better performance) and Hausdorff distance (lower values indicate better performance) closely replicated the performance seen on T1‐w (left column zoomed to the relevant range of Dice values ≥0.9 and Hausdorff distance [HD95] ≤15 mm; right column depicting the full range of the data) [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Descriptive statistics on brain extraction performance (median and interquartile range (IQR) for Dice coefficient and Hausdorff distance) in the EORTC test set for the different MRI sequences (T1‐w, cT1‐w, FLAIR, T2‐w)
| MRI sequence type | DICE coefficient | Hausdorff distance (95th percentile) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HD‐BET | MONSTR | Statistics | HD‐BET | MONSTR | Statistics | |||||||
| median | IQR | median | IQR | abs(Z) | p | median | IRQ | median | IRQ | abs(Z) | p | |
| T1‐w | 97.6 | (97.0–98.0) | 95.4 | (94.0–96.1) | 30.62 | <.001 | 3.3 | (2.2–3.3) | 4.43 | (3.71–5.79) | 26.72 | <.001 |
| cT1‐w | 96.9 | (96.1–97.4) | 94.6 | (93.2–95.6) | 26.48 | <.001 | 3.9 | (2.8–4.1) | 5.48 | (4.36–6.96) | 26.92 | <.001 |
| FLAIR | 96.4 | (95.2–97.0) | 92.4 | (91.0–93.7) | 32.16 | <.001 | 5.0 | (3.4–5.0) | 8.15 | (6.00–11.0) | 31.30 | <.001 |
| T2‐w | 96.1 | (95.2–96.7) | 93.1 | (92.0–94.0) | 30.64 | <.001 | 5.0 | (3.9–5.0) | 8.0 | (5.78–10.0) | 29.47 | <.001 |
Next, we compared the performance of the HD‐BET algorithm with six publicly available and frequently used brain extraction algorithms on each dataset (EORTC‐26101 test set as well as the public LPBA40, NFBS, and CC‐359 datasets). For all competing brain extraction algorithms (except MONSTR), comparison was restricted to T1‐w sequences since they have primarily been developed for processing of T1‐w sequences and not optimized for independent processing of other sequence types (i.e., cT1‐w, FLAIR, or T2‐w). MONSTR was applied to all available MRI sequences. We applied uniform nonparametric testing due to the evidence of non‐normal data distribution for the majority of measurements (p < .05 on Shapiro–Wilk test for 49/56 measurements—Table S2, Supporting Information). The obtained first‐level statistics showed a significant difference between the investigated brain extraction methods for both evaluation metrics (Dice coefficient, Hausdorff distance) in each dataset (p < .001 for all comparisons—Table S3, Supporting Information).
Specifically, within the EORTC‐26101 test set post hoc Wilcoxon matched‐pairs signed‐rank test revealed significantly higher performance of the HD‐BET algorithm (for both Dice coefficient and Hausdorff distance) as compared to each of the six competing brain extraction algorithms (Bonferroni‐adjusted p < .001 for all comparisons) maintaining a large effect size in 83% of the tests (10/12 comparisons) and medium effect size in the remaining 17% (2/12 comparisons) (Figures 2 and 3 and Table 3). Similarly, within the three public datasets, post hoc Wilcoxon matched‐pairs signed‐rank tests again demonstrated significantly higher performance of the HD‐BET algorithm (for both Dice coefficient and Hausdorff distance) or all but two comparisons Bonferroni‐adjusted p < .001; only the Hausdorff distance of the FSL‐BET algorithm in the LPBA40 dataset and the MONSTR algorithm in the NFBS dataset were not significantly different from the HD‐BET algorithm with an Bonferroni‐adjusted p = .221 and p = 1). Moreover, 91% of the tests (31/34 comparisons) revealed a high effect size and 9% (3/31 comparisons) a medium effect (Figures 2 and 3 and Table 3). The improvement yielded with the HD‐BET algorithm as compared to all competing algorithms within the different datasets ranged from +1.16 to +2.50 for Dice and −0.66 to −2.51 mm for the Hausdorff distance (95th percentile) and was most pronounced in the EORTC‐26101 dataset (Table 4). Figures 4 and 5 depict representative cases for the brain algorithms and sequences at different Dice values (5th percentile and median) from the EORTC‐26101 test set and highlights the challenges associated with brain extraction in the presence of pathology and treatment‐induced tissue alterations.
Figure 2.
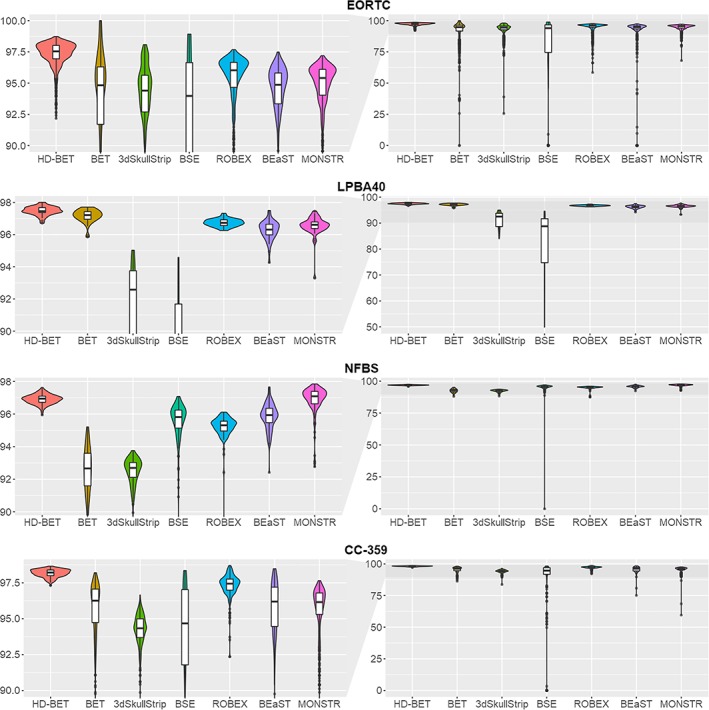
Comparison of Dice coefficients between the HD‐BET brain extraction algorithm and the six public brain extraction methods for each of the test datasets using violin charts (and superimposed box plots) [higher values indicate better performance]. Obtained median Dice coefficients were highest for the HD‐BET algorithm across all datasets (see left column visualizing the relevant range of Dice values ≥0.9). Note the spread of the Dice coefficients, which is consistently lower for the HD‐BET algorithm (right column visualizing the whole range of Dice values) [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
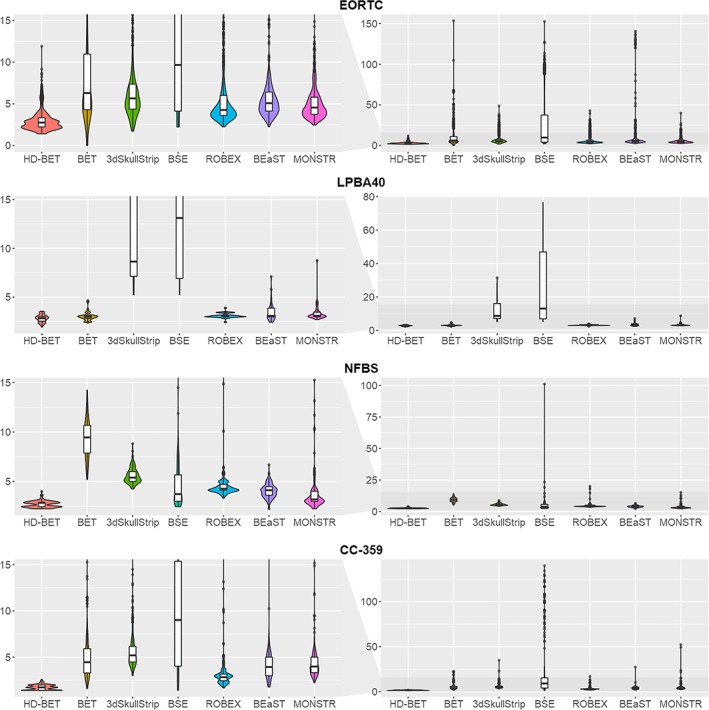
Comparison of Hausdorff distance (95th percentile) between the HD‐BET algorithm and the six public brain extraction methods for each of the test datasets using violin charts (and superimposed box plots; lower values indicate better performance). The median Hausdorff distance was lowest for the HD‐BET algorithm across all datasets (see left column visualizing the relevant range of Hausdorff distance ≤15 mm). Note the spread of the Hausdorff distance, which is consistently lower for the HD‐BET algorithm (right column visualizing the whole range of values) [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 3.
Wilcoxon matched‐pairs signed‐rank tests comparing the performance (Dice coefficient, Hausdorff distance) of the HD‐BET algorithm with six competing brain extraction algorithms. For every test, we reported the absolute value of the Z‐statistics [abs(Z)], the Bonferroni‐adjusted p‐value and the effect size [r] (with r values >.1 corresponding to a small effect, .3 to a medium effect, and .5 to a large effect size; Cohen, 1988)
| Dataset | Variable | BET | 3DSkullStrip | BSE | Robex | BEaST | MONSTR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| abs(Z) | p | r | abs(Z) | p | r | abs(Z) | p | r | abs(Z) | p | r | abs(Z) | p | r | abs(Z) | p | r | ||
| EORTC‐26101 test set | Dice | 24.31 | <.001 | .60 | 29.39 | <.001 | .72 | 27.69 | <.001 | .68 | 26.96 | <.001 | .48 | 3.89 | <.001 | .78 | 30.62 | <.001 | .75 |
| Hausdorffa | 27.14 | <.001 | .66 | 27.88 | <.001 | .68 | 29.18 | <.001 | .72 | 25.69 | <.001 | .46 | 28.16 | <.001 | .71 | 26.72 | <.001 | .66 | |
| LPBA40 | Dice | 3.95 | <.001 | .44 | 7.7 | <.001 | .86 | 7.7 | <.001 | .86 | 7.26 | <.001 | .81 | 7.33 | <.001 | .82 | 7.12 | <.001 | .81 |
| Hausdorffa | 2.03 | .221 | – | 7.7 | <.001 | .86 | 7.7 | <.001 | .86 | 3.94 | <.001 | .44 | 3.69 | .001 | .41 | 4.73 | <.001 | .54 | |
| NFBS | Dice | 13.67 | <.001 | .86 | 13.67 | <.001 | .86 | 12.5 | <.001 | .79 | 13.65 | <.001 | .86 | 11.22 | <.001 | .71 | 2.87 | 1 | – |
| Hausdorffa | 13.68 | <.001 | .87 | 13.68 | <.001 | .87 | 11.08 | <.001 | .70 | 13.63 | <.001 | .86 | 12.79 | <.001 | .81 | 9.53 | <.001 | .61 | |
| CC‐359 | Dice | 22.72 | <.001 | .85 | 23.02 | <.001 | .86 | 21.69 | <.001 | .81 | 17.82 | <.001 | .67 | 21.05 | <.001 | .79 | 23.17 | <.001 | .87 |
| Hausdorffa | 22.97 | <.001 | .86 | 23.05 | <.001 | .86 | 21.57 | <.001 | .80 | 21.77 | <.001 | .81 | 22.64 | <.001 | .84 | 23.20 | <.001 | .87 | |
Using the 95th percentile of the Hausdorff distance (mm).
Abbreviations: LPBA40, LONI Probabilistic Brain Atlas; NFBS, Nathan Kline Institute Enhanced Rockland Sample Neurofeedback Study; CC‐359, Calgary‐Campinas‐359.
Table 4.
Improvement of the performance for brain extraction with the HD‐BET algorithm on T1‐w sequences. The difference for each of the competing algorithms (as compared to HD‐BET) was calculated on a case‐by‐case basis and summarized for all algorithms for each dataset by calculating the median and IQR. Positive values for the change in Dice coefficient (i.e., higher values with HD‐BET), and negative values for the change in the Hausdorff distance (i.e., lower values with HD‐BET) indicate better performance
| Dice coefficient | Hausdorff distancea | |||
|---|---|---|---|---|
| Median | IQR | Median | IQR | |
| EORTC‐26101 test set | +2.50 | +1.47, + 4.26 | −2.46 | −4.82, −1.41 |
| LPBA40 | +1.16 | +0.62, +4.30 | −0.66 | −4.28, −0.14 |
| NFBS | +1,67 | +0.67, +3.85 | −1.91 | −3.39, −0.92 |
| CC‐359 | +2.11 | +1.02, +3.88 | −2.51 | −3.86, −1.43 |
Using the 95th percentile of the Hausdorff distance (mm).
Abbreviations: IQR, interquartile range; LPBA40, LONI Probabilistic Brain Atlas; MRI, magnetic resonance imaging; NFBS, Nathan Kline Institute Enhanced Rockland Sample Neurofeedback Study; CC‐359, Calgary‐Campinas‐359.
Figure 4.
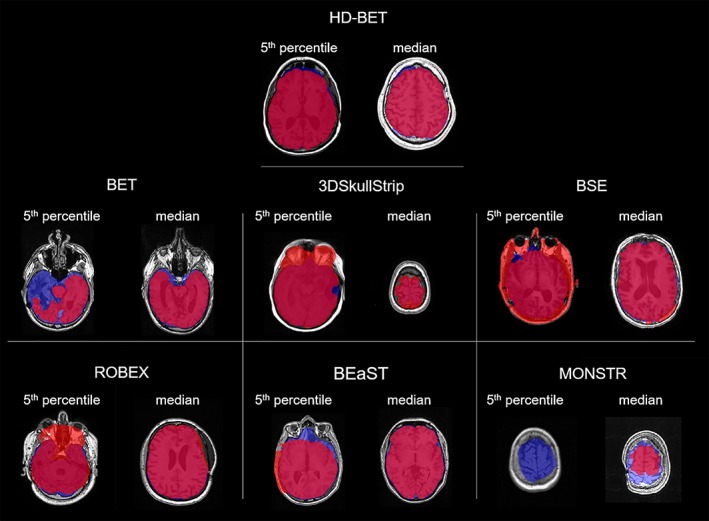
Representative cases showing the performance for T1‐w images of the different brain extraction algorithms at the 5th percentile and the median Dice coefficients in the EORTC‐26101 test set. Depicted in red the calculated brain masks from different brain extraction methods, in blue the ground‐truth brain masks (for illustrative purposes only) and in pink their intersection. While BET, BEaST, and MONSTR tend to underestimate the brain mask in these cases by removing brain tissue from the mask, 3DSkullStrip, BSE, and ROBEX tend to overestimate by including nonbrain tissue (e.g., skull, fat, nasal, and orbital cavity) in the mask [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 5.
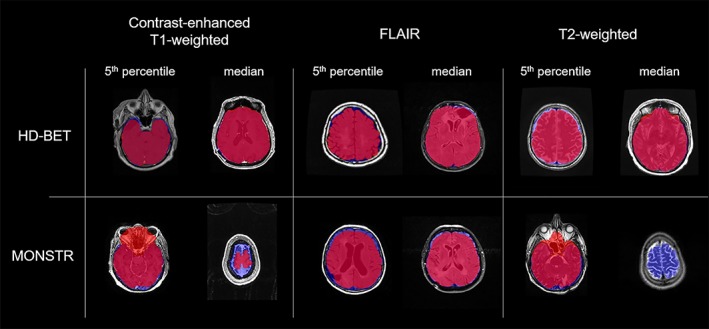
Representative cases showing the performances of HD‐BET and MONSTR for cT1‐w, FLAIR, and T2‐w images at 5th percentiles and medians of the Dice coefficients in the EORTC test set. Depicted in red the calculated brain masks (HD BET or MONSTR), in blue the ground‐truth brain masks (for illustrative purposes only) and in pink their intersection. Similar to T1‐w images MONSTR tends to underestimate in the brain mask in these cases by removing brain tissue from the masks and additionally for the 5th percentile in cT1‐w and T2‐w images tends to overestimate by including nonbrain tissue around the nasal cavities [Color figure can be viewed at http://wileyonlinelibrary.com]
Average processing time for brain extraction of a single MRI sequence required 32 s of processing with the HD‐BET algorithm (NVIDIA TITAN Xp GPU). In contrast, average processing time of a single T1‐w sequence with one of the six competing public brain extraction algorithms ranged from 3 s to 34.6 min (specifically, averages were 3 s for BSE, 17 s for BET, 1.4 min for ROBEX, 4.0 min for 3dSkullstrip, 10.7 min for BEaST, and 34.5 min for MONSTR) on a 8‐core Intel Xeon E5‐2640 v3 CPU.
For broader accessibility, we provide a fully functional version of the presented HD‐BET prediction algorithm for download via http://www.neuroAI-HD.org.
4. DISCUSSION
Here, we present a method (HD‐BET) that enables rapid, automated, and robust brain extraction in the presence of pathology or treatment‐induced tissue alterations, is applicable to a broad range of MRI sequence types, and is not influenced by variations in MRI hardware and acquisition parameters encountered in both research and clinical practice. We demonstrate generalizability of the HD‐BET algorithm on the EORTC‐26101 test set with MRI sequences originating from 12 different institutions covering all major MRI vendors with a broad variety of scanner types and field strengths as well as within three independent public datasets. Importantly the EORTC test set is independent from the EORTC training set, since the institutions from which the imaging data originate differ. The HD‐BET algorithm yields state‐of‐the‐art performance in both the EORTC‐26101 test set as well as three publicly available reference datasets (LPBA40, NFBS, CC‐359). This finding reflects the limitations of many existing brain extraction algorithms which are usually not optimized for processing heterogeneous imaging data with pathological tissue alterations or varying hardware and acquisition parameters (Fennema‐Notestine et al., 2006) and consequently may introduce errors in downstream analysis of MRI neuroimaging data (Beers et al., 2018). We addressed this within our study by training (and independent testing) the HD‐BET algorithm with data from a large multicentric clinical trial in neuro‐oncology which allowed to design a robust and broadly applicable brain extraction algorithm that enables high‐throughput processing of neuroimaging data. Moreover, the improvement in the brain extraction performance yielded by the HD‐BET algorithm was most pronounced in the EORTC‐26101 dataset, again reflecting the limitations of the competing brain extraction algorithms when processing heterogeneous imaging data with abnormal pathologies or varying acquisition parameters.
The HD‐BET algorithm is able to perform brain extraction on various types of common anatomical MRI sequence without prior knowledge of the sequence type. From a practical point of view, this is of particular importance since imaging protocols (and the types of sequences acquired) may vary substantially. The majority of brain extraction algorithms are optimized to process T1‐w MRI sequences (Han et al., 2018; Iglesias et al., 2011; Lutkenhoff et al., 2014) and fall short during processing of other types of MRI sequences (e.g., T2‐w, FLAIR, or cT1‐w images). We addressed this shortcoming and demonstrate that the HD‐BET algorithm also performs well on cT1‐w, FLAIR, or T2‐w MRI and closely replicates the performance observed for brain extraction on T1‐w sequences. Our algorithm also outperformed MONSTR, which is explicitly designed to do brain extraction in the presence of pathologies and on other than T1‐w MRI sequences in the EORTC‐26101 test set as well as the public LPBA40 and CC‐359 test sets.
The runtime of the HD‐BET algorithm for processing a single MRI sequence is in the order of half a minute with modern hardware, including all preprocessing and postprocessing steps. More advanced hardware would allow to further improve processing time, although the existing setup already performed well in comparison to the runtime of the other competing brain extraction algorithms. For example, the second best performing algorithm in the EORTC‐26101 test set (MONSTR) required on average more than 30 min for processing of a single MRI sequence.
We acknowledge that although many different brain extraction algorithms have been proposed and published, we essentially focused on the most commonly used algorithms. Moreover, a case‐specific tuning of parameters from these brain extraction algorithms may have allowed to improve their performance to some extent (Iglesias et al., 2011; Popescu, et al., 2012). This is particularly the case for BEaST, where a mismatch between source and target domain can result in a significant drop in performance (Eskildsen et al., 2012; Novosad & Collins, 2018). Dataset‐specific adaptations are, however, not a practical approach, especially in the context of high‐throughput processing. Moreover, we acknowledge that manually correcting brain masks in a single case can take hours (Puccio et al., 2016). Although our approach with generating a GT brain mask in a large‐scale dataset was more focused on correcting major errors (e.g., around pathologies, resection cavities or due to varying hardware or acquisition parameters), even imperfect GT labels can lead to high quality deep‐learning segmentation algorithms when using the UNET architecture that was employed in our study (Heller, Dean, & Papanikolopoulos, 2018). Moreover, the competitiveness of our approach was rendered by testing on the public datasets (NFBS, CC‐359, and LPBA40) where we confirmed the performance of the HD‐BET algorithm against an independent high‐quality GT. In addition, future studies will need to evaluate the performance of the HD‐BET algorithm in a broader range of diseases in neuroradiology since our evaluation was essentially limited to cases with brain tumors (EORTC‐26101 dataset) or cases with only mild or no structural abnormalities (LPBA40, NFBS, CC‐359 dataset). However, given the broad phenotypic appearance (and associated posttreatment alterations) of brain tumors which were used for training the algorithm we are confident that HD‐BET is equally applicable to the broad disease spectrum encountered in neuroradiology.
In conclusion, the developed and rigorously validated HD‐BET algorithm enables rapid, automated, and robust brain extraction in the presence of pathology or treatment‐induced tissue alterations, is applicable to a broad range of MRI sequence types, and is not influenced by variations in MRI hardware and/or acquisition parameters encountered in both research and clinical practice. Taken together, HD‐BET is made publicly available via http://www.neuroAI-HD.org and may become an essential component for robust, automated, high‐throughput processing of MRI neuroimaging data.
CONFLICT OF INTERESTS
F.I.: none; M.S.: none; I.P.: none, G.B.: none; D.B.: Activities not related to the present article: received payment for lectures, including service on speakers' bureaus, from Profound Medical Inc.; U.N.: none; A.W.: none; H.P.S.: Activities not related to the present article: received payment from Curagita for consultancy and payment from Bayer and Curagita for lectures, including service on speakers' bureaus; S.H.: none; W.W.: Activities not related to the present article: received research grants from Apogenix, Boehringer Ingelheim, MSD, Pfizer, and Roche, as well as honoraria for lectures or advisory board participation or consulting from BMS, Celldex, MSD, and Roche; M.B.: Activities not related to the present article: received grant support from Siemens, Stryker, and Medtronic, consulting fees from Vascular Dynamics, Boehringer Ingelheim, and B. Braun, lecture fees from Teva, grant support and lecture fees from Novartis and Bayer, and grant support, consulting fees, and lecture fees from Codman Neuro and Guerbet; K.H.M.H.: none; and P.K.: none.
AUTHOR CONTRIBUTIONS
P.K., M.S., F.I., I.P. designed the study; P.K., M.S., I.P., G.B., D.B., U.N. performed quality control and preprocessing of the MRI data from the EORTC‐26101 dataset; I.P., M.S. generated and P.K. visually inspected the GT reference brain masks in the EORTC‐26101 dataset; F.I. and P.K. applied the competing brain extraction algorithms to all datasets; F.I. performed development, training, and application of the ANN; F.I. calculated the evaluation metrics; M.S. and P.K. performed statistical analysis; P.K., M.S., F.I. interpreted the findings with essential input from all coauthors; P.K., M.S., F.I., I.P. prepared the first draft of the manuscript; all authors critically revised the manuscript for important intellectual content; all authors approved the final version of the manuscript.
Supporting information
Appendix S1 Supporting Information
ACKNOWLEDGMENT
P.K. was supported by the Medical Faculty Heidelberg Postdoc‐Program and the Else Kröner‐Fresenius Foundation (Else‐Kröner Memorial Scholarship).
Isensee F, Schell M, Pflueger I, et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum Brain Mapp. 2019;40:4952–4964. 10.1002/hbm.24750
Fabian Isensee, Marianne Schell, and Irada Pflueger shared the first authorship.
Funding information Else Kröner‐Fresenius Foundation; Medical Faculty Heidelberg Postdoc‐Program
DATA AVAILABILITY
The MRI data from the EORTC‐26101 trial that were used for training and independent large‐scale testing of the HD‐BET algorithm are not publicly available and restrictions apply to their use. The MRI data from the LPBA40, NFBS, and CC‐359 datasets are publically available and information on download is provided within the respective references cited in Section 2. For broader accessibility, we provide a fully functional version of the presented HD‐BET prediction algorithm for download via http://www.neuroAI-HD.org.
REFERENCES
- Beers, A. , Brown, J. , Chang, K. , Hoebel, K. , Gerstner, E. , Rosen, B. , Kalpathy‐Cramer, J. (2018). DeepNeuro: an open‐source deep learning toolbox for neuroimaging. ArXiv e‐prints. [DOI] [PMC free article] [PubMed]
- Çiçek, Ö. , Abdulkadir, A. , Lienkamp, S. S. , Brox, T. , & Ronneberger, O. (2016). 3D U‐Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin S., Joskowicz L., Sabuncu M., Unal G., Wells W. (eds) Medical Image Computing and Computer‐Assisted Intervention ‐ MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science, vol 9901. Springer, Cham.
- Cohen, J. (1988). Statistical power analysis for the behavioral sciences. Hillsdale, NJ: Erlbaum Associates. [Google Scholar]
- Cox, R. W. (1996). AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research, 29, 162–173. [DOI] [PubMed] [Google Scholar]
- Dale, A. M. , Fischl, B. , & Sereno, M. I. (1999). Cortical surface‐based analysis. I. Segmentation and surface reconstruction. NeuroImage, 9, 179–194. [DOI] [PubMed] [Google Scholar]
- de Boer, R. , Vrooman, H. A. , Ikram, M. A. , Vernooij, M. W. , Breteler, M. M. B. , van der Lugt, A. , & Niessen, W. J. (2010). Accuracy and reproducibility study of automatic MRI brain tissue segmentation methods. NeuroImage, 51, 1047–1056. [DOI] [PubMed] [Google Scholar]
- Dey, R. , Hong, Y. (2018). CompNet: Complementary segmentation network for brain MRI extraction. ArXiv e‐prints.
- Dice, L. R. (1945). Measures of the amount of ecologic association between species. Ecology, 26, 297–302. [Google Scholar]
- Eskildsen, S. F. , Coupe, P. , Fonov, V. , Manjon, J. V. , Leung, K. K. , Guizard, N. , … Collins, D. L. (2012). BEaST: Brain extraction based on nonlocal segmentation technique. NeuroImage, 59, 2362–2373. [DOI] [PubMed] [Google Scholar]
- Fennema‐Notestine, C. , Ozyurt, I. B. , Clark, C. P. , Morris, S. , Bischoff‐Grethe, A. , Bondi, M. W. , … Brown, G. G. (2006). Quantitative evaluation of automated skull‐stripping methods applied to contemporary and legacy images: Effects of diagnosis, bias correction, and slice location. Human Brain Mapping, 27, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni, G. B. , Fox, N. C. , Jack, C. R., Jr. , Scheltens, P. , & Thompson, P. M. (2010). The clinical use of structural MRI in Alzheimer disease. Nature Reviews. Neurology, 6, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve, D. N. , & Fischl, B. (2009). Accurate and robust brain image alignment using boundary‐based registration. NeuroImage, 48, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar, H. , & Soul, J. S. (2006). Measurement of cortical thickness in 3D brain MRI data: Validation of the Laplacian method. Journal of Neuroimaging, 16, 146–153. [DOI] [PubMed] [Google Scholar]
- Han, X. , Kwitt, R. , Aylward, S. , Bakas, S. , Menze, B. , Asturias, A. , … Niethammer, M. (2018). Brain extraction from normal and pathological images: A joint PCA/image‐reconstruction approach. NeuroImage, 176, 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, N. , Dean, J. , Papanikolopoulos, N. (2018). Imperfect segmentation labels: How much do they matter? ArXiv e‐prints.
- Iglesias, J. E. , Liu, C. Y. , Thompson, P. M. , & Tu, Z. (2011). Robust brain extraction across datasets and comparison with publicly available methods. IEEE Transactions on Medical Imaging, 30, 1617–1634. [DOI] [PubMed] [Google Scholar]
- Isensee, F. , Kickingereder, P. , Wick, W. , Bendszus, M. , Maier‐Hein, K.H. (2018). Brain tumor segmentation and radiomics survival prediction: Contribution to the BRATS 2017 challenge. ArXiv e‐prints.
- Jenkinson, M. , & Smith, S. (2001). A global optimisation method for robust affine registration of brain images. Medical Image Analysis, 5, 143–156. [DOI] [PubMed] [Google Scholar]
- Kalavathi, P. , & Prasath, V. B. S. (2016). Methods on skull stripping of MRI head scan images—A review. Journal of Digital Imaging, 29, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayalibay, B. , Jensen, G. , van der Smagt, P. (2017). CNN‐based segmentation of medical imaging data. ArXiv preprint arXiv:1701.03056.
- Kickingereder, P. , Isensee, F. , Tursunova, I. , Petersen, J. , Neuberger, U. , Bonekamp, D. , … Maier‐Hein, K. H. (2019). Automated quantitative tumour response assessment of MRI in neuro‐oncology with artificial neural networks: A multicentre, retrospective study. The Lancet Oncology, 20, 728–740. [DOI] [PubMed] [Google Scholar]
- Kleesiek, J. , Urban, G. , Hubert, A. , Schwarz, D. , Maier‐Hein, K. , Bendszus, M. , & Biller, A. (2016). Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. NeuroImage, 129, 460–469. [DOI] [PubMed] [Google Scholar]
- Klein, A. , Ghosh, S. S. , Avants, B. , Yeo, B. T. T. , Fischl, B. , Ardekani, B. , … Parsey, R. V. (2010). Evaluation of volume‐based and surface‐based brain image registration methods. NeuroImage, 51, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leote, J. , Nunes, R. G. , Cerqueira, L. , Loução, R. , & Ferreira, H. A. (2018). Reconstruction of white matter fibre tracts using diffusion kurtosis tensor imaging at 1.5T: Pre‐surgical planning in patients with gliomas. European Journal of Radiology Open, 5, 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhoff, E. S. , Rosenberg, M. , Chiang, J. , Zhang, K. , Pickard, J. D. , Owen, A. M. , & Monti, M. M. (2014). Optimized brain extraction for pathological brains (optiBET). PLoS One, 9, e115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, D. , Kabani, N. , Avis, D. , & Evans, A. C. (2000). Automated 3‐D extraction of inner and outer surfaces of cerebral cortex from MRI. NeuroImage, 12, 340–356. [DOI] [PubMed] [Google Scholar]
- Menze, B. H. , Jakab, A. , Bauer, S. , Kalpathy‐Cramer, J. , Farahani, K. , Kirby, J. , … Van Leemput, K. (2015). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Transactions on Medical Imaging, 34, 1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milletari, F. , Navab, N. , Ahmadi, S.‐A. (2016) V‐net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, 2016, pp. 565–571. 10.1109/3DV.2016.79. [DOI]
- Novosad, P. , & Collins, D. L. (2018). Alzheimer's disease neuroimaging, I. an efficient and accurate method for robust inter‐dataset brain extraction and comparisons with 9 other methods. Human Brain Mapping, 39, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu, V. , Battaglini, M. , Hoogstrate, W. S. , Verfaillie, S. C. , Sluimer, I. C. , van Schijndel, R. A. , … MAGNIMS Study Group . (2012). Optimizing parameter choice for FSL‐brain extraction tool (BET) on 3D T1 images in multiple sclerosis. NeuroImage, 61, 1484–1494. [DOI] [PubMed] [Google Scholar]
- Puccio, B. , Pooley, J. P. , Pellman, J. S. , Taverna, E. C. , & Craddock, R. C. (2016). The preprocessed connectomes project repository of manually corrected skull‐stripped T1‐weighted anatomical MRI data. GigaScience, 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radue, E. W. , Barkhof, F. , Kappos, L. , Sprenger, T. , Haring, D. A. , de Vera, A. , … Cohen, J. A. (2015). Correlation between brain volume loss and clinical and MRI outcomes in multiple sclerosis. Neurology, 84, 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger, O. , Fischer, P. , & Brox, T. (2015). U‐Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N., Hornegger J., Wells W., Frangi A. (eds) Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham.
- Roy, S. , Butman, J. A. , Pham, D. L. , & Alzheimers Disease Neuroimaging Initiative . (2017). Robust skull stripping using multiple MR image contrasts insensitive to pathology. NeuroImage, 146, 132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh Mohseni Salehi, S. , Erdogmus, D. , Gholipour, A. (2017). Auto‐context convolutional neural network (auto‐net) for brain extraction in magnetic resonance imaging. ArXiv e‐prints. [DOI] [PMC free article] [PubMed]
- Shattuck, D. W. , & Leahy, R. M. (2002). BrainSuite: An automated cortical surface identification tool. Medical Image Analysis, 6, 129–142. [DOI] [PubMed] [Google Scholar]
- Shattuck, D. W. , Mirza, M. , Adisetiyo, V. , Hojatkashani, C. , Salamon, G. , Narr, K. L. , … Toga, A. W. (2008). Construction of a 3D probabilistic atlas of human cortical structures. NeuroImage, 39, 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck, D. W. , Sandor‐Leahy, S. R. , Schaper, K. A. , Rottenberg, D. A. , & Leahy, R. M. (2001). Magnetic resonance image tissue classification using a partial volume model. NeuroImage, 13, 856–876. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. (2002). Fast robust automated brain extraction. Human Brain Mapping, 17, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, R. , Lucena, O. , Garrafa, J. , Gobbi, D. , Saluzzi, M. , Appenzeller, S. , … Lotufo, R. (2018). An open, multi‐vendor, multi‐field‐strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage, 170, 482–494. [DOI] [PubMed] [Google Scholar]
- Taha, A. A. , & Hanbury, A. (2015). An efficient algorithm for calculating the exact Hausdorff distance. IEEE Transactions on Pattern Analysis and Machine Intelligence, 37, 2153–2163. [DOI] [PubMed] [Google Scholar]
- Tosun, D. , Rettmann, M. E. , Naiman, D. Q. , Resnick, S. M. , Kraut, M. A. , & Prince, J. L. (2006). Cortical reconstruction using implicit surface evolution: Accuracy and precision analysis. NeuroImage, 29, 838–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Chen, Y. , Pan, X. , Hong, X. , & Xia, D. (2010). Level set segmentation of brain magnetic resonance images based on local Gaussian distribution fitting energy. Journal of Neuroscience Methods, 188, 316–325. [DOI] [PubMed] [Google Scholar]
- Wick, W. , Gorlia, T. , Bendszus, M. , Taphoorn, M. , Sahm, F. , Harting, I. , … van den Bent, M. J. (2017). Lomustine and Bevacizumab in progressive Glioblastoma. The New England Journal of Medicine, 377, 1954–1963. [DOI] [PubMed] [Google Scholar]
- Wick, W. , Stupp, R. , Gorlia, T. , Bendszus, M. , Sahm, F. , Bromberg, J. E. , … Bent, M. J. V. D. (2016). Phase II part of EORTC study 26101: The sequence of bevacizumab and lomustine in patients with first recurrence of a glioblastoma. Journal of Clinical Oncology, 34, 2019–2019. [Google Scholar]
- Woods, R. P. , Mazziotta, J. C. , & Cherry, R. S. (1993). MRI‐PET registration with automated algorithm. Journal of Computer Assisted Tomography, 17, 536–546. [DOI] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Piven, J. , Hazlett, H. C. , Smith, R. G. , Ho, S. , Gee, J. C. , & Gerig, G. (2006). User‐guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage, 31, 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Brady, M. , & Smith, S. (2001). Segmentation of brain MR images through a hidden Markov random field model and the expectation‐maximization algorithm. IEEE Transactions on Medical Imaging, 20, 45–57. [DOI] [PubMed] [Google Scholar]
- Zhao, L. , Ruotsalainen, U. , Hirvonen, J. , Hietala, J. , & Tohka, J. (2010). Automatic cerebral and cerebellar hemisphere segmentation in 3D MRI: Adaptive disconnection algorithm. Medical Image Analysis, 14, 360–372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information
Data Availability Statement
The MRI data from the EORTC‐26101 trial that were used for training and independent large‐scale testing of the HD‐BET algorithm are not publicly available and restrictions apply to their use. The MRI data from the LPBA40, NFBS, and CC‐359 datasets are publically available and information on download is provided within the respective references cited in Section 2. For broader accessibility, we provide a fully functional version of the presented HD‐BET prediction algorithm for download via http://www.neuroAI-HD.org.


