Abstract
Myeloma bone disease (MBD) is one of the clinical features of multiple myeloma, which contributes to the attenuation of osteoblast function. Bone marrow mesenchymal stem cells exhibit a high potential for differentiation into osteoblasts. A number of studies have reported that microRNAs (miRs) serve a vital role in mesenchymal stem cell (MSC) osteogenesis; however, the role of miR-221-5p in the osteogenic differentiation of MBD-MSCs remains unclear. The present study revealed that the osteogenic differentiation capacity of MBD-MSCs was reduced compared with that of normal (N)-MSCs. Further experiments demonstrated that miR-221-5p expression was downregulated in N-MSCs following osteoblast induction while no obvious alterations in expression levels were observed in MBD-MSCs. The inhibition of miR-221-5p promoted the osteogenic differentiation of MBD-MSCs. Bioinformatics, luciferase reporter assays, reverse transcription-quantitative PCR and western blotting assays indicated that smad family member 3 (smad3) was a direct target of miR-221-5p in MBD-MSCs. A negative association was identified between the expression levels of smad3 and miR-221-5p. Investigations of the molecular mechanism indicated that suppressed miR-221-5p could regulate the osteogenic differentiation of MBD-MSCs by upregulating smad3 expression. It was also identified that the PI3K/AKT/mTOR signaling pathway was activated following miR-221-5p inhibition, and this increased the osteogenic differentiation capacity of MBD-MSCs. The present study may improve the understanding regarding the role of miR-221-5p in the regulation of osteogenic differentiation, and may contribute to the development of a novel therapy for MBD.
Keywords: myeloma bone disease, bone marrow mesenchymal stem cell, microRNA-221-5p, smad family member 3, PI3K/AKT/mTOR
Introduction
Multiple myeloma (MM), a common malignancy that accounts for ~1% of all human malignant tumors and 10–15% of all cases of hematological malignancy globally, is characterized by the abnormal proliferation of plasma cells in the bone marrow (1). The majority of patients with MM develop myeloma bone disease (MBD), which is characterized by severe bone pain, fractures, osteoporosis and hypercalcemia (2,3). Bone destruction in MBD is usually attributed to an imbalance between enhanced osteoclast activity and the attenuation of osteoblast function (4). Osteogenesis is mediated by the recruitment of bone marrow mesenchymal stem cells (BMMSCs), which have the potential for osteogenic differentiation (5). Previous study has indicated that the osteogenic differentiation of BMMSCs from MM-mesenchymal stem cells (MSCs) in patients with MM is impaired (6). Therefore, improving the osteogenic differentiation ability of MM-MSCs is a key issue that may promote bone regeneration and repair.
MicroRNAs (miRNAs/miRs) are highly conserved, short non-coding RNA molecules that have been identified as regulators involved in cell proliferation, differentiation, organ development and metabolism (7). A number of studies have reported that numerous miRNAs, including miR-133 (8), miR-141 (9) and miR-34a (10), promote or inhibit the differentiation of preosteoblasts and osteoblasts by regulating key transcription factors and osteogenic markers in the osteoblast signaling pathway (11,12). However, to the best of our knowledge, the role of miR-221-5p in the osteogenic differentiation of BMMSCs from MBD-MSCs in patients with MBD remains unclear.
The present study aimed to examine the role of miR-221-5p in the osteogenic differentiation of MBD-MSCs, and to investigate the molecular mechanism of miR-221-5p-regulated osteogenic differentiation. It was identified that miR-221-5p was downregulated in MBD-MSCs, and participated in osteogenic differentiation. Inhibition of miR-221-5p resulted in an increase in the osteogenic potential of MBD-MSCs via the upregulation of smad family member 3 (smad3). The potential mechanism may involve the excessive phosphorylation of PI3K/AKT/mTOR due to the action of miR-221-5p.
Materials and methods
Patients
The present study was approved by the General Hospital of Western Theater Command (Chengdu, China), and written informed consent was obtained from all participants. Bone marrow (BM) samples were obtained from 5 patients with MBD (age range, 36–57 years; 2 males and 3 females) who underwent surgery at The General Hospital of Western Theater Command (Chengdu, China) between April 2016 and April 2017. Additionally, five healthy controls (age range, 32–46 years; 3 males, 2 females) were included in the present study to obtain N-MSCs. The inclusion criteria were as follows: i) Patients who were diagnosed with MM at first admission and who had one or more osteolytic lesions on imaging examination; ii) patients who were willing to participate in the study; iii) patients without comorbid diseases that may result in osteolytic destruction, including prostate, breast, thyroid and lung cancer. The exclusion criteria were as follows: i) Patients who received chemotherapy prior to the study; and ii) patients with MM who developed plasma cell leukemia.
Isolation and propagation of BMMSC
N-MSCs and MBD-MSCs were isolated from BM samples using Ficoll (Sigma-Aldrich; Merck KGaA). Following centrifugation at 450 × g for 20 min at room temperature, the mononuclear cells were resuspended and seeded in a T25 cell culture bottle at a density of 5,000 cells/cm2 with DMEM (HyClone; GE Healthcare Life Sciences) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and maintained at 37°C in a humidified atmosphere (5% CO2). The medium was replaced twice per week until the cultures attained 80–90% confluence.
MSC identification by flow cytometry
The immunophenotype of the N-MSCs and MBD-MSCs were evaluated at the third passage. The collected cells were resuspended in 100 µl PBS (1×106 cells) with 5 µl of the following antibodies: Mouse anti-human CD44-PE (cat. no. 338807; 1:20), CD90-FITC (cat. no. 328107; 1:20), CD105-PC5.5 (cat. no. 323215; 1:20), CD34-FITC (cat. no. 343603; 1:20) and CD45-PE (cat. no. 368509; 1:20; all BioLegend, Inc.) for 30 min at 4°C. Subsequently, N-MSCs and MBD-MSCs were analyzed using a FACSCalibur flow cytometer and Cell Quest software (version 3.3; BD Biosciences).
MSC differentiation
The third-generation N-MSCs and MBD-MSCs were seeded at a density of 1×105 cells/well in 6-well plates. Once the cultures attained 60–70% confluence, the medium was discarded and 2 ml fresh DMEM containing 10% FBS, 1×10−8 mol/l dexamethasone, 10 mmol/ml sodium β-glycerophosphate and 50 µg/ml vitamin C was added to incubate the N-MSCs and MBD-MSCs for 2 weeks, and the media were replaced twice a week. The osteogenic differentiation of N-MSCs and MBD-MSCs was evaluated by using Alizarin Red S staining (1%; pH 4.2; Novon Scientific) for 5 min at room temperature. Images of the N-MSCs and MBD-MSCs were captured using a light microscope (magnification, ×40).
Cell transfection
N-MSCs were seeded into a 6-well plate at a density of 1×105 cells/well 1 day prior to transfection. Cell transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol with 50 pmol/ml miR-221-5p inhibitor or mimic (Guangzhou RiboBio Co., Ltd.). The medium was changed after 6 h. Following culture for 48 h at 37°C in a humidified atmosphere (5% CO2), the transfected cells were collected for western blotting and reverse transcription-quantitative PCR (RT-qPCR) analyses.
Lentiviruses overexpressing smad3 (lv-smad3; Smad3 mRNA sequence, NM_001145102; vector name, GV358) and the corresponding control lentiviruses (lv-green fluorescent protein) were purchased from Shanghai GeneChem Co., Ltd. N-MSCs were seeded into a 6-well plate at a density of 1×105 cells/well. Lentivirus (1×108 TU/ml) infection was performed when the cells reached 20–40% confluence. The lentiviruses were transfected into N-MSCs with a multiplicity of infection of 30. After transfection for 8–12 h, fresh medium was added for further incubation at 37°C in a humidified atmosphere (5% CO2). Cells were collected for RT-qPCR or western blot analysis 72 h after transfection.
RT-qPCR assay
Total RNA was extracted from N-MSCs and MBD-MSCs (1×105 cells/well) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and converted to cDNA using a PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol at 37°C for 15 min and 85°C for 5 sec. Subsequently, primers and SYBR® Premix Ex Taq II (Takara Biotechnology Co., Ltd.) were used to detect the expression of alkaline phosphatase (ALP), osteopontin (OPN), osteocalcin (OC), and smad3 in an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 3 min at 95°C; 40 cycles of 95°C for 5 sec and 60°C for 30 sec followed by 72°C for 30 sec. For the quantification of miRNA expression, RT was performed at 42°C for 60 min and 70°C for 10 min using Bulge-Loop™ miRNA RT-qPCR Primer and Bulge-Loop™ miRNA RT-qPCR Starter kit (cat. no. C10211-1; Guangzhou RiboBio Co., Ltd.). miR-221-5p expression levels were quantified at 95°C for 10 min, followed by 40 cycles of 95°C for 2 sec, 60°C for 20 sec and 70°C for 10 sec using Bulge-Loop™ miRNA RT-qPCR Starter kit (cat. no. C10211-1). The 2−∆∆Cq method was used for comparative quantitation (13). β-actin and U6 small nuclear RNA genes were used as endogenous normalization controls. The data were analyzed using Bio-Rad CFX Manager software (version 3.0; Bio-Rad Laboratories, Inc.). The primer sequences (Sangon Biotech Co., Ltd.) are presented in Table I. The catalog number of miR-221-5p primer is MQPS0000852-1 and the catalog number of U6 is miRAN0002-1.
Table I.
Primer sequences for reverse transcription-quantitative PCR.
| Primer sequence (5′-3′) | ||
|---|---|---|
| Gene | Forward | Reverse |
| ALP | GACCTCCTCGGAAGACACTCTG | CGCCTGGTAGTTGTTGTGAGC |
| OPN | GCCGACCAAGGAAAACTCACT | GGCACAGGTGATGCCTAGGA |
| OC | CCAGGCGCTACCTGTATCAATG | ATGTGGTCAGCCAACTCGTCA |
| Smad3 | AGGGCTTTGAGGCTGTCTACC | GTCCACGCTGGCATCTTCTG |
| β-actin | TGGCACCCAGCACAATGAA | CTAAGTCATAGTCCGCCTAGAAGCA |
ALP, alkaline phosphatase; OC, osteocalcin; OPN, osteopontin; Smad3, smad family member 3.
Western blotting
Cell specimens were lysed using radioimmunoprecipitation assay lysis buffer (Wuhan Boster Biological Technology, Ltd.) and quantified using a Bicinchoninic Acid Protein assay kit (Wuhan Boster Biological Technology, Ltd.). The cellular lysate (20 µg/lane) was separated by 10% SDS-PAGE and then transferred onto PVDF membranes (EMD Millipore). Membranes were blocked with 5% skimmed milk powder at 37°C for 1 h and incubated with the following primary antibodies: Anti-smad3 (cat. no. 9523; 1:1,000), anti-PI3K (cat. no. 4249; 1:1,000), anti-phosphorylated (p)-PI3K (cat. no. 4228; 1:1,000), anti-AKT (cat. no. 4685; 1:1,000), anti-p-AKT (cat. no. 4060; 1:1,000), anti-mTOR (cat. no. 2983; 1:1,000), anti-p-mTOR (cat. no. 5536; 1:1,000) and anti-β-actin (cat. no. 4970; 1:1,000 dilution; all Cell Signaling Technology, Inc.), anti-ALP (cat. no. ab83259; 1:500), anti-OPN (cat. no. ab8448; 1:1,000) and anti-OC (cat. no. ab93876; 1:500; all Abcam), at 4°C overnight. Subsequently, the membranes were incubated with HRP-conjugated goat anti-rabbit immunoglobulin G secondary antibody (cat. no. BA1054; 1:5,000 dilution; Wuhan Boster Biological Technology, Ltd.) for 1 h at room temperature. Protein bands were detected using an ECL chemiluminescence kit (EMD Millipore) according to the manufacturer's protocol. Protein levels were calculated relative to β-actin. The protein bands were visualized using the ChemiDoc™ MP imaging system (Bio-Rad Laboratories, Inc.). Image-ProPlus software (version 6.0; Media Cybernetics, Inc.) was used for densitometry analysis.
Luciferase reporter assay
miR-221-5p targets were predicted using bioinformatics software, including TargetScan (www.targetscan.org), mirDB (mirdb.org), Diana Tools (diana.imis.athena-innovation.gr) and venny2.1.0 (bioinfogp.cnb.csic.es/tools/venny/index.html). N-MSCs (4×104/well) were seeded into 24-well plates, followed by transfection with the Renilla luciferase pRL-TK plasmid (100 ng/ml; Shanghai GenePharma Co., Ltd.) with the recombinant firefly luciferase pGL3 reporters containing the 3′-untranslated region (3′-UTR) region of human smad3 (2 µg/ml; Shanghai GenePharma Co., Ltd.) in combination with miR-221-5p mimic, miR-221-5p inhibitor and controls using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). At 48 h after transfection, cells were collected and lysed. Luciferase and Renilla signals were measured using a Dual-Luciferase Reporter assay kit (cat. no. E1910; Promega Corporation), according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS version 20.0 (IBM Corp.). Data are presented as the means ± standard error and derived from three independent experiments. The unpaired Student's t-test (for parametric data) or Mann-Whitney U test (for non-parametric data) were used to compare two independent samples. Differences among multiple groups were compared by one-way ANOVA with Dunnett's post hoc test or two-way ANOVA with Bonferroni's post hoc test. P<0.05 was considered to indicate a statistically significant difference. P<0.01 was considered to indicate a statistically highly significant difference.
Results
Osteogenic differentiation capacity is reduced in MBD-MSCs
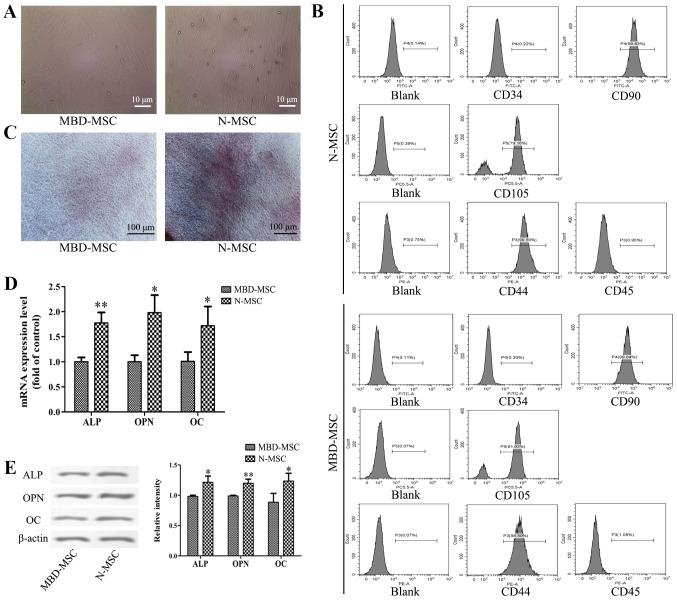
After 7 days of primary culture, partial cell colonies were identified, and the cells were fusiform and pleomorphic. After 14 days of primary culture, the N-MSCs and MBD-MSCs proliferated rapidly and attained 80–90% confluence (Fig. 1A). MBD-MSCs exhibited slower cell proliferation and higher morphological variation compared with normal (N)-MSCs. Flow cytometry analysis revealed that the cells were negative for CD34 and CD45 but positive for CD44, CD90 and CD105 (Fig. 1B). These results suggested that the cultured cells were MSCs. The osteogenic induction of N-MSCs and MBD-MSCs was visualized by staining with Alizarin Red S, and the results revealed that the calcium deposition of N-MSCs was markedly higher compared with MBD-MSCs (Fig. 1C). Furthermore, the RT-qPCR and western blotting results demonstrated that the mRNA and protein expression levels of ALP, OPN and OC were significantly lower in MBD-MSCs compared with in N-MSCs (Fig. 1D and E). These data indicated that the osteogenic differentiation ability of MBD-MSCs was inhibited.
Figure 1.
Osteogenic differentiation ability of MBD-MSCs is inhibited compared with that of N-MSCs. (A) Images captured using a light microscope showing MSCs in primary culture on the 14th day. Scale bar, 10 µm. (B) Surface markers of the third generation MSCs were identified by flow cytometry. (C) Images of Alizarin Red staining were captured using a light microscope following osteogenic induction of N-MSCs and MBD-MSCs. Scale bar, 100 µm. (D) Reverse transcription-quantitative PCR was performed to detect the mRNA expression levels of ALP, OPN and OC following osteogenic induction of N-MSCs and MBD-MSCs. (E) Western blotting was performed to detect the protein expression levels of ALP, OPN and OC following osteogenic induction of N-MSCs and MBD-MSCs. *P<0.05, **P<0.01 vs. MBD-MSC. ALP, alkaline phosphatase; MBD, myeloma bone disease; MSC, mesenchymal stem cell; N, normal; OC, osteocalcin; OPN, osteopontin.
Inhibition of miR-221-5p promotes osteogenic differentiation of MBD-MSCs
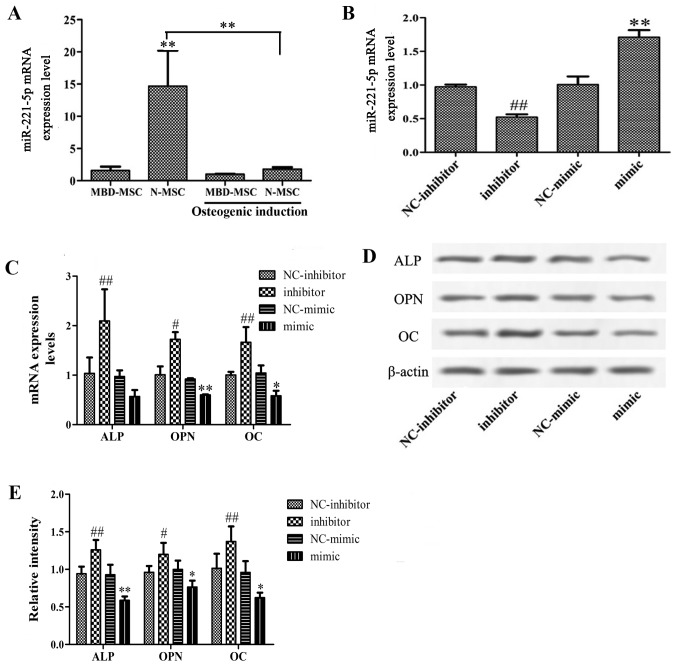
To investigate whether miR-221-5p is dysregulated in the osteogenic differentiation of MSCs, the expression levels of miR-221-5p were detected in N-MSCs and MBD-MSCs prior to and following osteogenic induction. As presented in Fig. 2A, the mRNA expression levels of miR-221-5p in MBD-MSCs were significantly lower compared with those in N-MSCs. Following osteoblast induction, the miR-221-5p mRNA level in N-MSCs was significantly decreased, while no obvious change was detected in MBD-MSCs. These data suggested that decreased expression levels of miR-221-5p may contribute to osteogenic differentiation. Furthermore, to verify the role of miR-221-5p in the osteogenic differentiation of MBD-MSCs, MBD-MSCs were transfected with miR-221-5p mimic or inhibitor. RT-qPCR revealed that the expression levels of miR-221-5p were significantly increased in the mimic group and decreased in the inhibitor group compared with the respective control groups, which indicated a high transfection efficiency in MBD-MSCs (Fig. 2B). Subsequently, the expression levels of marker genes of osteogenic differentiation were detected following transfection. As presented in Fig. 2C-E, the mRNA and protein expression levels of ALP, OPN and OC were significantly increased in the inhibitor group and decreased in the mimic group compared with the respective control groups. These data demonstrated that the inhibition of miR-221-5p can effectively promote the osteogenic differentiation of MBD-MSCs.
Figure 2.
Effects of miR-221-5p inhibition on the osteogenic differentiation of MBD-MSCs. (A) RT-qPCR was used to quantify the expression levels of miR-221-5p in N-MSCs and MBD-MSCs prior to and following osteoblast induction. (B) RT-qPCR was used to quantify the expression levels of miR-221-5p in MBD-MSCs following transfection with miR-221-5p mimic or inhibitor. (C) Expression levels of marker genes of osteogenic differentiation were detected by RT-qPCR following transfection. (D) Western blotting was performed to detect the protein expression levels of ALP, OPN and OC following transfection. (E) Relative band intensity of ALP, OPN and OC protein. *P<0.05, **P<0.01 vs. NC-mimic, unless indicated otherwise. #P<0.05, ##P<0.01 vs. NC-inhibitor. ALP, alkaline phosphatase; MBD, myeloma bone disease; miR-221-5p, microRNA-221-5p; MSC, mesenchymal stem cell; N, normal; NC, negative control; OC, osteocalcin; OPN, osteopontin; RT-qPCR, reverse transcription-quantitative PCR.
Smad3 is a direct target of miR-221-5p
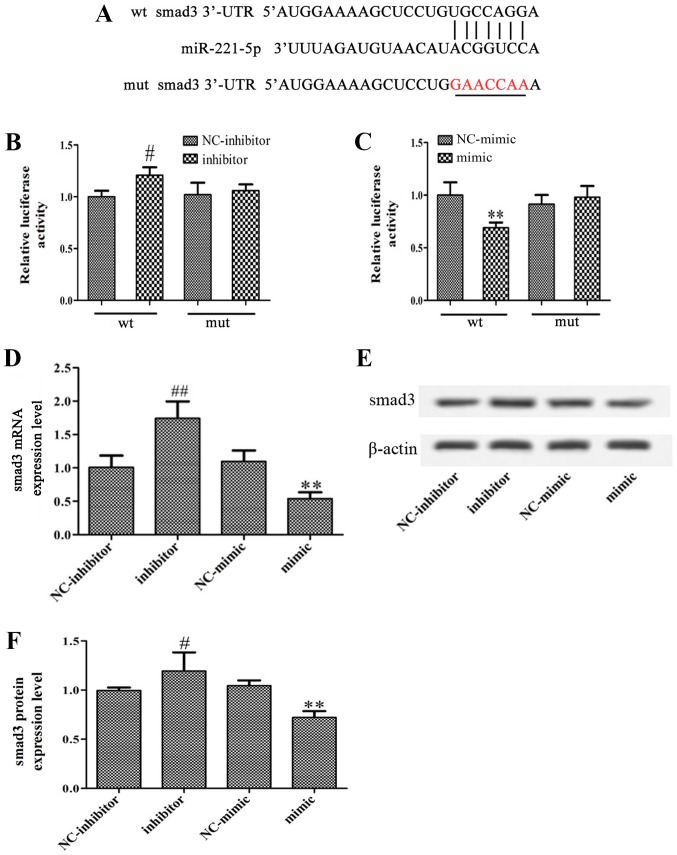
Using bioinformatics analysis, it was identified that the 3′-UTR of smad3 contains a conserved putative target site for miR-221-5p (Fig. 3A). A luciferase reporter assay was performed to validate the association between miR-221-5p and smad3. As presented in Fig. 3B and C, miR-221-5p inhibitor significantly increased luciferase activity, while miR-221-5p mimic significantly inhibited the luciferase activity compared with the controls. Furthermore, the present study revealed that overexpression of miR-221-5p significantly decreased smad3 expression, whereas transfection with miR-221-5p inhibitor significantly increased smad3 expression at the mRNA and protein levels (Fig. 3D-F). In summary, it may be suggested that miR-221-5p targets smad3 and negatively regulate smad3 expression.
Figure 3.
miR-221-5p targets smad3 in MBD-MSCs. (A) Predicted miR-221-5p target site in the 3′-UTR of smad3. (B and C) Luciferase activities were measured following transfection with miR-221-5p mimic or inhibitor. (D) mRNA expression levels of smad3 were measured following transfection by reverse transcription-quantitative PCR. (E) Protein expression levels of smad3 were detected following transfection by western blotting. (F) Relative band intensity of smad3 protein. **P<0.01 vs. NC-mimic. #P<0.05, ##P<0.01 vs. NC-inhibitor. MBD, myeloma bone disease; miR-221-5p, microRNA-221-5p; MSC, mesenchymal stem cell; mut, mutant; NC, negative control; smad3, smad family member 3; 3′-UTR, 3′-untranslated region; wt, wild-type.
Overexpression of smad3 enhances the effect of miR-221-5p inhibitor on the osteogenic differentiation of MBD-MSCs
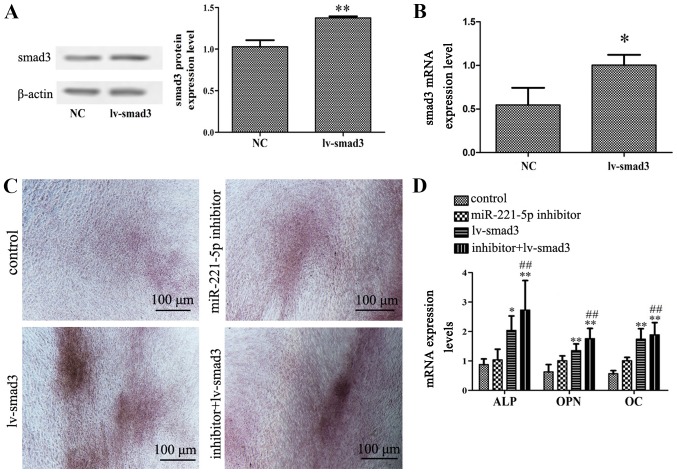
To determine whether smad3 is involved in miR-221-5p-regulated osteogenic differentiation of MBD-MSCs, the present study used a lentiviral vector overexpressing smad3. Western blotting and RT-qPCR results demonstrated that the protein and mRNA expression levels of smad3 were significantly increased following transduction with lentiviral vector, which indicated high transduction efficiency in MBD-MSCs (Fig. 4A and B). Furthermore, it was identified that the overexpression of smad3 could not only promote calcium deposition and the expression of osteogenic markers, but it could also enhance the effect of miR-221-5p inhibitor on osteogenic differentiation (Fig. 4C and D). These results indicated that miR-221-5p regulates osteogenic differentiation by upregulating smad3.
Figure 4.
Effects of smad3 overexpression on miR-221-5p inhibitor-mediated osteogenic differentiation. (A) Protein expression levels of smad3 were detected by western blotting following transduction with lv-smad3 lentiviral vector. **P<0.01 vs. NC. (B) mRNA expression levels of smad3 were detected by RT-qPCR following transfection with lentiviral vector. *P<0.05 vs. NC. (C) N-mesenchymal stem cells transfected with miR-221-5p inhibitors were subsequently infected with the lentiviral vector. Images of Alizarin Red staining were captured using a light microscope. Scale bar, 100 µm. (D) RT-qPCR was performed to detect the mRNA expression levels of ALP, OPN and OC following transfection with miR-221-5p inhibitor and/or infection with lentiviral vector. *P<0.05, **P<0.01 vs. control. ##P<0.01 vs. lv-smad3 group. ALP, alkaline phosphatase; lv-smad3, lentiviral vector overexpressing smad3; miR-221-5p, microRNA-221-5p; NC, negative control; OC, osteocalcin; OPN, osteopontin; smad3, smad family member 3; RT-qPCR, reverse transcription-quantitative PCR.
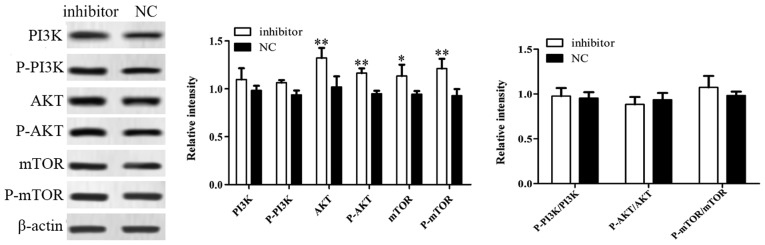
Inhibition of miR-221-5p may promote the osteogenic differentiation of MBD-MSCs by activating the PI3K/AKT/mTOR pathway
Western blotting was used to further investigate the potential signaling pathway associated with miR-221-5p regulated osteogenic differentiation. The results demonstrated that the protein expression levels of AKT, p-AKT, mTOR and p-mTOR were significantly increased following transfection with miR-221-5p inhibitor compared with the control, while no significant changes in the expression levels of PI3K and p-PI3K were observed. In addition, there were no significant difference in the ratios of p-PI3K to PI3K, p-AKT to AKT and p-mTOR to mTOR between groups was identified (Fig. 5). These results suggested that the inhibition of miR-221-5p may increase the expression levels of phosphorylated proteins by inducing total protein expression, thus activating the PI3K/AKT/mTOR signaling pathway and promoting osteogenic differentiation.
Figure 5.
Inhibition of miR-221-5p expression activates the PI3K/AKT/mTOR signaling pathway. The expression levels of key proteins of the PI3K/AKT/mTOR pathway were detected using western blotting following transfection of myeloma bone disease mesenchymal stem cells with microRNA-221-5p inhibitor. *P<0.05, **P<0.01 vs. NC. AKT, protein kinase B; mTOR, mammalian target of rapamycin; NC, negative control; P-, phosphorylated; PI3K, phosphoinositide 3-kinase.
Discussion
A prominent clinical feature of MBD is progressive bone destruction, including osteoporosis, osteolytic lesions, pathological fracture, hypercalcemia and bone pain (14). Increased bone resorption and decreased bone formation result in bone damage in patients with MM. MSCs are ideal seed cells for bone tissue engineering due to their strong renewal ability and multidirectional differentiation potentiality (15). Therefore, osteogenic defects in patients with MBD may be treated by improving the osteogenic capability of MSCs. Osteoblasts act as the major cells that contribute to bone formation by secreting ALP and bone matrix proteins that induce bone matrix mineralization (16). The present study demonstrated that calcium deposition and the expression levels of bone formation indicators were decreased in MBD-MSCs following osteoblast induction compared with N-MSCs, which indicates that the osteogenic differentiation ability of MSCs is reduced in patients with MBD.
miRNAs are essential regulatory molecules that can regulate the expression levels of genes associated with bone homeostasis via transcriptional inhibition or mRNA cleavage (17). A number of studies (11,18,19) have highlighted the biological roles of dysregulated miRNAs in osteoblast differentiation. For example, it has been suggested that miR-21 promotes the osteogenic differentiation of MSCs by regulating the PI3K/β-catenin pathway (20). In another study, miR-208a-3p has been reported to suppress osteogenic differentiation and inhibit bone formation by targeting activin A receptor type 1 (21). Recently, miR-221 was reported to regulate the osteoblast differentiation of MSCs in patients with osteoporosis by targeting runt-related transcription factor 2 (22). Despite these findings, further investigations are required to identify additional mechanisms underlying the effects of miR-221-5p in osteogenic differentiation. The present study identified that the expression levels of miR-221-5p were significantly lower in MBD-MSCs compared with in N-MSCs. Following osteoblast induction, the expression levels of miR-221-5p in N-MSCs were significantly decreased, while no notable alteration in expression was detected in MBD-MSCs. These results suggested that miR-221-5p may act as an inhibitor in the osteogenic differentiation of MSCs. Furthermore, it was identified that knockdown of miR-221-5p could induce osteogenic differentiation in MBD-MSCs, which was indicated by the increased mRNA expression levels of typical osteoblast differentiation markers, including ALP, OPN and OC. In addition, the overexpression of miR-221-5p inhibited the expression of osteogenic differentiation markers. These data suggested that miR-221-5p serves as a negative regulator in the osteogenic differentiation of MBD-MSCs. In future studies, ALP staining and Alizarin Red staining should be used to verify this result.
miRNA regulation in the osteogenic differentiation of MSCs is achieved by miRNAs acting on specific target genes and signal transduction pathways. Smad3 is an important member of the transforming growth factor-β1/smad signaling pathway, as it can form complexes with smad4 and translocate into the nucleus, which results in the activation or suppression of downstream target genes, and the regulation of cell function and metabolism (23). A number of studies have indicated that smad3 serves an important role in bone formation, remodeling and maintenance (24–26). Loss of smad3 decreases bone formation and induces osteopenia via the dysregulation of osteoblast differentiation and apoptosis (27). Additionally, in a previous study, smad3 knockout mice developed skeletal abnormalities shortly after weaning and this effect worsened with age (28). The present study used bioinformatics analysis to identify that the 3′-UTR of smad3 contained a conserved putative target site for miR-221-5p in MBD-MSCs. In the present study, a luciferase reporter assay, RT-qPCR and western blotting demonstrated that smad3 is a direct target of miR-221-5p and a negative association was revealed between the expression levels of smad3 and miR-221-5p. Furthermore, we demonstrated that overexpression of smad3 could enhance the effect of miR-221-5p inhibitor on the osteogenic differentiation of MBD-MSCs, indicating that miR-221-5p may be a negative regulator of the osteogenic differentiation of MBD-MSCs by upregulating smad3. In future studies, additional candidate targets of miR-221-5p should be investigated to support the use of miR-221-5p as a potential therapeutic target for MBD.
The PI3K/AKT/mTOR signaling pathway is closely associated with the osteogenic differentiation of MSCs (29,30). Under physiological conditions, the PI3K/AKT signaling pathway is an important signaling pathway that serves a key role in inhibiting apoptosis and inducing osteogenic differentiation (31). Zhang et al (32) reported that all-trans retinoic acid induces the osteogenic differentiation of MSCs by activating the PI3K/AKT/glycogen synthase kinase-3β signaling pathway. In addition, miR-378 reduces glucose-suppressed osteogenic differentiation by targeting caspase-3 and activating the PI3K/AKT signaling pathway (33). The present study revealed that inhibition of miR-221-5p increased the expression levels of osteogenic differentiation markers and significantly increased the levels of total proteins and phosphorylated proteins associated with the PI3K/AKT/mTOR pathway.
In conclusion, the present study demonstrated that miR-221-5p was expressed at a low level in MBD-MSCs and may be a negative regulator that participates in osteogenic differentiation. Additionally, the inhibition of miR-221-5p increased osteoblast differentiation by directly targeting smad3. The potential mechanism may involve increased phosphorylation of PI3K/AKT/mTOR via the activity of miR-221-5p. These results suggest that miR-221-5p may be used as a promising therapeutic against MBD in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key Research Program ‘Stem Cell and Transformation Research’ Key Project (grant. no. 2017YFA0105502).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FYF, RD, YS and XZ conceived and designed the experiments. FYF, RD, SHL, QW, YZ and LG performed the experiments. YL, PK and JZ analyzed the data and assisted with the experiments. FYF, RD, YS and XZ wrote the paper. YS and XZ revised the manuscript and supervised the study. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of General Hospital of Western Theater Command (Chengdu, China) and written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kyle RA, Rajkumar SV. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2014;23:3–9. doi: 10.1038/leu.2008.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson KC. Progress and paradigms in multiple myeloma. Clin Cancer Res. 2016;22:5419–5427. doi: 10.1158/1078-0432.CCR-16-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaccoby S. Advances in the understanding of myeloma bone disease and tumour growth. Br J Haematol. 2010;149:311–321. doi: 10.1111/j.1365-2141.2010.08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heider U, Hofbauer LC, Zavrski I, Kaiser M, Jakob C, Sezer O. Novel aspects of osteoclast activation and osteoblast inhibition in myeloma bone disease. Biochem Biophys Res Commun. 2005;338:687–693. doi: 10.1016/j.bbrc.2005.09.146. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Li T, Fan J, Li T, Fan L, Wang S, Weng X, Han Q, Zhao RC. miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death Differ. 2015;22:1935–1945. doi: 10.1038/cdd.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raje N, Roodman GD. Advances in the biology and treatment of bone disease in multiple myeloma. Clin Cancer Res. 2011;17:1278–1286. doi: 10.1158/1078-0432.CCR-10-1804. [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 8.Lv H, Sun Y, Zhang Y. miR-133 is involved in estrogen Deficiency-Induced osteoporosis through modulating osteogenic differentiation of mesenchymal stem cells. Med Sci Monit. 2015;21:1527–1534. doi: 10.12659/MSM.894323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Liu W, Sinha KM, Yasuda H, de Crombrugghe B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor osterix. PLoS One. 2013;8:e58104. doi: 10.1371/journal.pone.0058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vimalraj S, Selvamurugan N. MicroRNAs: Synthesis, gene regulation and osteoblast differentiation. Curr Issues Mol Biol. 2013;15:7–18. [PubMed] [Google Scholar]
- 12.Zhou M, Li M, Yu X. Regulation of microRNA in osteoblast differentiation and its clinical significance. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2012;26:755–759. (In Chinese) [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Berenson JR. Myeloma bone disease. Best Pract Res Clin Haematol. 2005;18:653–672. doi: 10.1016/j.beha.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Diao Y, Ma Q, Cui F, Zhong Y. Human umbilical cord mesenchymal stem cells: Osteogenesis in vivo as seed cells for bone tissue engineering. J Biomed Mater Res A. 2009;91:123–131. doi: 10.1002/jbm.a.32186. [DOI] [PubMed] [Google Scholar]
- 16.Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- 17.Ell B, Kang Y. MicroRNAs as regulators of bone homeostasis and bone metastasis. Bonekey Rep. 2014;3:549. doi: 10.1038/bonekey.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Li H, Liu W, Yang L, Tan YF, Luo XH. Targeting miRNAs in osteoblast differentiation and bone formation. Expert Opin Ther Targets. 2010;14:1109–1120. doi: 10.1517/14728222.2010.512916. [DOI] [PubMed] [Google Scholar]
- 19.Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng YB, Li X, Li ZY, Zhao J, Yuan XB, Ren Y, Cui ZD, Liu YD, Yang XJ. microRNA-21 promotes osteogenic differentiation of mesenchymal stem cells by the PI3K/β-catenin pathway. J Orthop Res. 2015;33:957–964. doi: 10.1002/jor.22884. [DOI] [PubMed] [Google Scholar]
- 21.Arfat Y, Basra MAR, Shahzad M, Majeed K, Mahmood N, Munir H. miR-208a-3p Suppresses osteoblast differentiation and inhibits bone formation by targeting ACVR1. Mol Ther Nucleic Acids. 2018;11:323–336. doi: 10.1016/j.omtn.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Gao Y, Cai L, Li F, Lou Y, Xu N, Kang Y, Yang H. MicroRNA-221 is involved in the regulation of osteoporosis through regulates RUNX2 protein expression and osteoblast differentiation. Am J Transl Res. 2017;9:126–135. [PMC free article] [PubMed] [Google Scholar]
- 23.Derynck R, Zhang Y, Feng XH. Smads: Transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/S0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 24.Hackinger S, Trajanoska K, Styrkarsdottir U, Zengini E, Steinberg J, Ritchie GRS, Hatzikotoulas K, Gilly A, Evangelou E, Kemp JP, et al. Evaluation of shared genetic aetiology between osteoarthritis and bone mineral density identifies SMAD3 as a novel osteoarthritis risk locus. Hum Mol Genet. 2017;26:3850–3858. doi: 10.1093/hmg/ddx285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, Greenblatt MB, Yan G, Feng H, Sun J, Lotinun S, Brady N, Baron R, Glimcher LH, Zou W. SMURF2 regulates bone homeostasis by disrupting SMAD3 interaction with vitamin D receptor in osteoblasts. Nat Commun. 2017;8:14570. doi: 10.1038/ncomms14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Q, Tu ML, Li CJ, Zhang L, Jiang TJ, Liu T, Luo XH. GDF11 inhibits bone formation by activating Smad2/3 in bone marrow mesenchymal stem cells. Calcif Tissue Int. 2016;99:500–509. doi: 10.1007/s00223-016-0173-z. [DOI] [PubMed] [Google Scholar]
- 27.Borton AJ, Frederick JP, Datto MB, Wang XF, Weinstein RS. The loss of Smad3 results in a lower rate of bone formation and osteopenia through dysregulation of osteoblast differentiation and apoptosis. J Bone Miner Res. 2001;16:1754–1764. doi: 10.1359/jbmr.2001.16.10.1754. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo G, Xu B, Huang Y. Icariside II promotes the osteogenic differentiation of canine bone marrow mesenchymal stem cells via the PI3K/AKT/mTOR/S6K1 signaling pathways. Am J Transl Res. 2017;9:2077–2087. [PMC free article] [PubMed] [Google Scholar]
- 30.Tong Y, Feng W, Wu Y, Lv H, Jia Y, Jiang D. Mechano-growth factor accelerates the proliferation and osteogenic differentiation of rabbit mesenchymal stem cells through the PI3K/AKT pathway. BMC Biochem. 2015;16:1. doi: 10.1186/s12858-015-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu YX, Du J, Si MS, Mo JJ, Qiao SC, Lai HC. The roles of PI3K/Akt signaling pathway in regulating MC3T3-E1 preosteoblast proliferation and differentiation on SLA and SLActive titanium surfaces. J Biomed Mater Res A. 2013;101:748–754. doi: 10.1002/jbm.a.34377. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Chen X, Hu Y, Wu J, Cao Q, Chen S, Gao Y. All-trans retinoic acid modulates Wnt3A-induced osteogenic differentiation of mesenchymal stem cells via activating the PI3K/AKT/GSK3β signalling pathway. Mol Cell Endocrinol. 2016;422:243–253. doi: 10.1016/j.mce.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 33.You L, Gu W, Chen L, Pan L, Chen J, Peng Y. miR-378 overexpression attenuates high glucose-suppressed osteogenic differentiation through targeting CASP3 and activating PI3K/Akt signaling pathway. Int J Clin Exp Pathol. 2014;7:7249–7261. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.