Abstract
The polystomes (Monogenea, Polystomatidae) radiated across semi-aquatic tetrapods including all three amphibian orders, freshwater turtles and the hippopotamus. Prior to this study, phylogenetic analyses revealed that the most diverse and widespread genus, Polystoma, was not monophyletic; a lineage comprising four undescribed species from the bladder of Zhangixalus spp. (Rhacophoridae) in Asia occupied a deep phylogenetic position. Regarding vicariance biogeography and molecular dating, the origin of this lineage is correlated with the breakup of Gondwanaland in the Mesozoic period. Based on a Bayesian analysis of four concatenated genes (18S, 28S, COI and 12S) and morphological evidence, one new genus, Indopolystoma n. gen., and three new species, sampled in Japan and China, are described here: Indopolystoma viridi n. sp. from Z. viridis of Japan, Indopolystoma elongatum n. sp. from Z. arboreus of Japan, and Indopolystoma parvum n. sp. from Z. omeimontis of China. Indopolystoma is unique amongst polystome genera infecting anurans by possessing a small haptor relative to the body size, posteriormost marginal hooklet C1 much bigger than hooklets C2–C8 with conspicuous broad blade and guard and a pair of hamuli lacking a deep notch. Eight species of Asian Polystoma, all from rhacophorids, are transferred as Indopolystoma carvirostris (Fan, Li & He, 2008) n. comb., I. hakgalense (Crusz & Ching, 1975) n. comb., I. indicum (Diengdoh & Tandon, 1991) n. comb., I. leucomystax (Zhang & Long, 1987) n. comb., I. mutus (Meng, Song & Ding, 2010) n. comb., I. pingbianensis (Fan, Wang & Li, 2004) n. comb., I. rhacophori (Yamaguti, 1936) n. comb., and I. zuoi (Shen, Wang & Fan, 2013) n. comb.
Keywords: Indopolystoma, Polystoma, Asia, Neobatrachia, Rhacophoridae
Abstract
Les polystomes (Monogenea, Polystomatidae) se sont diversifiés sur des tétrapodes semi-aquatiques, notamment les trois ordres d’amphibiens, les tortues d’eau douce et l’hippopotame. Avant cette étude, des analyses phylogénétiques avaient révélé que le genre le plus diversifié et le plus répandu, Polystoma, n’était pas monophylétique ; une lignée comprenant quatre espèces non décrites de la vessie de Zhangixalus spp. (Rhacophoridae) en Asie occupait une position phylogénétique profonde. En ce qui concerne la biogéographie de vicariance et la datation moléculaire, l’origine de cette lignée est corrélée à l’éclatement du Gondwana au Mésozoïque. D’après une analyse bayésienne de quatre gènes concaténés (18S, 28S, COI et 12S) et des preuves morphologiques, un nouveau genre, Indopolystoma n. gen. et trois nouvelles espèces échantillonnées au Japon et en Chine sont décrites ici : Indopolystoma viridi n. sp. de Z. viridis du Japon, Indopolystoma elongatum n. sp. de Z. arboreus du Japon et Indopolystoma parvum n. sp. de Z. omeimontis de Chine. Indopolystoma est unique parmi les genres de polystomes infectant les anoures. Il possède un hapteur petit par rapport à la taille du corps, un crocheton le plus postérieur C1 beaucoup plus gros que les crochetons C2 à C8, avec lame et garde bien visibles, ainsi qu’une paire d’hamuli dépourvus d’encoche profonde. Huit espèces de polystomes asiatiques, toutes issues de Rhacophoridae, sont transférées comme Indopolystoma carvirostris (Fan, Li & He, 2008) n. comb., I. hakgalense (Crusz et Ching, 1975) n. comb., I. indicum (Diengdoh & Tandon, 1991) n. comb., I. leucomystax (Zhang & Long, 1987) n. comb., I. mutus (Meng, Song & Ding, 2010) n. comb., I. pingbianensis (Fan, Wang et Li, 2004) n. comb., I. rhacophori (Yamaguti, 1936) n. comb. et I. zuoi (Shen, Wang et Fan, 2013) n. comb.
Introduction
In contrast to the digeneans that can be found in all groups of vertebrates, monogeneans are mostly parasites of marine and freshwater fishes [52]. With the exception of a few monogeneans that were assigned to the Gyrodactylidae Cobbold, 1864, Iagotrematidae Mañé-Garzón & Gil 1962, and Lagarocotylidae Kritsky, Hoberg & Aubry, 1993, only a single family, the Polystomatidae Gamble, 1896, radiated across semi-aquatic tetrapods including all three amphibian orders (anurans, salamanders and caecilians), freshwater turtles and the common hippopotamus. The Polystomatidae, in modern classification, belong to the order Polyopisthocotylea Odhner, 1912. Nowadays, polystomatids are globally in excess of 180 described species in 26 genera, most of which are endo-parasitic in the bladder of amphibian adults (18 genera) and in the pharyngeal cavity, bladder or conjunctival sacs of freshwater turtles (five genera). Whereas fish usually harbor a high diversity of monogeneans on their gills [28, 41], no more than two species of polystomes have thus far been recorded per species of anuran host [7, 15]. Finally, a high degree of host-specificity was assumed for polystomatids of especially anuran hosts (see [49] for a review on the diversity of polystomatids).
The Polystomatidae thus provided the opportunity to trace host-parasite co-evolution over an exceptionally long period of time, namely from the ecological transition from marine to terrestrial life at about 425 million years ago (Mya) [50]. Whereas flatworm groups often display diverse body plans, monogeneans and in particular polystomatids show limited interspecies variation [45]. Although hardly any information is known about ancestral forms, the molecular phylogenies published in Bentz et al. [4, 5], Verneau et al. [50, 51], Badets et al. [2] and Héritier et al. [22] gave an invaluable timescale to date evolutionary events and to infer origins of major monophyletic groups within the family. The extant anuran polystomatids in Asia are less than 20 species that belong to five genera, Diplorchis Ozaki, 1931, Eupolystoma Kaw, 1950, Neoriojatrema Imkongwapang & Tandon, 2010, Polystoma Zeder, 1800 and Sundapolystoma Lim & Du Preez, 2001. Of these, the ubiquitous Polystoma, which is the most speciose-polystome genus known from anurans of the suborder Neobatrachia Reig, 1958, encompasses 14 parasite species (six from China, four from Japan and one each from Sri Lanka, India, Iran and Turkey). Earlier studies based on the phylogeny and historical biogeography of polystomes infecting species of the Neobatrachia [2] revealed that Polystoma was not a monophyletic taxon and that the deep-branched lineage including Polystoma species sampled from rhacophorids of India, Japan and China was strongly correlated with the breakup of Gondwanaland in the Mesozoic period. Badets et al. [2] suggested from cophylogenetic and vicariance analyses supplemented by molecular dating that this lineage probably arose on the Indian subcontinent about 177 Ma when western and eastern Gondwanan components were fully separated, and later colonized southeast Asia following host dispersal after India collided with Asia close to 86 Mya.
India is one of the largest landmass countries in Asia and also well-recognized as a rich biogeographic area in terms of species diversity and endemic species, with its boundaries falling in Himalaya, Western Ghats, Indo-Burma and Sundaland biodiversity “hot spots” [34, 37]. For instance, India has a striking anuran diversity with 395 known species [21] of which 286 (73%) are endemic [1]. As shown by many authors, the geological history of India played a crucial role in shaping the current diversity, endemicity, and distribution patterns of amphibian lineages. Before joining Laurasia, India was part of Gondwanaland and gradually became detached from other landmasses during its northward journey across the Tethys Sea [29]. It broke off from Africa about 130 Mya [29] and subsequently from Madagascar about 88 Mya [42]. Its collision with southern Asia occurred during the Paleocene or Early Eocene at 66–56 Mya [3] and gave rise to biotic exchange [6].
According to plate tectonics, rifting and drifting of continents following the breakup of Gondwana provided ample time for animal differentiation. Therefore, the long period of isolation of the Polystoma lineage in the Indian subcontinent [2] should have been sufficient to restrict specific morphological marks for this higher taxon. In the present study, we focused on several specimens of the three undescribed Asian Polystoma species reported in Verneau et al. [51], Badets et al. [2] and Héritier et al. [22] to provide formal descriptions of this new taxon and species. Based on genetic and morphological characters, we bring some evidence that this lineage, which includes polystomes of Asian rhacophorids, is a new genus within the Polystomatidae, and we also reassign eight polystomes previously described as Polystoma to this genus.
Materials and methods
Polystome sampling and morphology
Polystomes were recovered from the bladder of three Asian rhacophorids belonging to Zhangixalus Jiang et al. 2019 [21, 27], namely Z. viridis (Hallowell) and Z. arboreus (Okada & Kawano) that were both collected in Japan by Hideo Hasegawa on 8 February 1986, and 27 June 2003, respectively, and Z. omeimontis (Stejneger) that was collected in China by Annemarie Ohler on 11 May 2004. A single parasite specimen from each host species was fixed in alcohol for molecular analyses and processed in Badets et al. [2] and Héritier et al. [22]. Whereas some of the material collected in Japan was stained and mounted in Canada balsam, all specimens collected in China were preserved in alcohol. We therefore stained all of them but one with acetocarmine and mounted them permanently in Canada balsam. Specimens were examined using a Nikon NiE compound microscope (Nikon, Netherlands) fitted with a Nikon DS-Ri1 digital camera and drawn using Adobe Illustrator software. Measurements were taken, in micrometers, using a Nikon NIS elements D software program and expressed as the mean, followed by the range in parentheses.
Sequence collection
In order to establish the phylogenetic relationships of the polystomes assumed to belong to a new genus, namely polystomes recovered from Zhangixalus frogs, we selected the four Asian Polystoma species reported in Badets et al. [2] and Héritier et al. [22] as well as one to three species of the main genera infecting neobatrachian frogs, namely Diplorchis ranae Ozaki, 1931, Eupolystoma alluaudi (de Beauchamp, 1913), Kankana manampoka Raharivololoniaina et al. 2011, Madapolystoma sp., Parapolystoma bulliense (Johnston, 1912), Polystoma cuvieri Vaucher, 1990, P. gallieni Price, 1939 and P. naevius Caballero & Cerecero, 1941, for which sequences were available in GenBank. Two other species were also selected for outgroup comparisons according to Héritier et al. [22], namely Pseudodiplorchis americanus (Rodgers & Kuntz, 1940) and Pseudopolystoma dendriticum (Ozaki, 1948). All these species with their respective accession numbers for two nuclear (18S and 28S) and two mitochondrial (12S and COI) genes are reported in Table 1. Prior to running phylogenetic analyses, we noticed that COI sequences reported in Héritier et al. [22] for polystomes infecting Rhacophorus maximus Günther (known today as Zhangixalus smaragdinus (Blyth)) (JF699303) and Z. viridis (KR856171) were almost identical, differing by only two substitutions, while pairwise comparisons of 12S, 18S and 28S sequences showed higher molecular divergences. This suggested inversion of DNA samples during the amplification process. Using primers L-CO1p and H-Cox1R and following the PCR procedure described in Héritier et al. [22], we therefore re-amplified the COI fragment from both polystome DNA samples recovered by these authors and selected the new sequences for phylogenetic and genetic analyses.
Table 1.
Species of polystomes investigated with their 18S, 28S, 12S and COI GenBank Accession numbers. Indopolystoma spp. were considered earlier as Polystoma spp. in Verneau et al. [51], Badets et al. [2] and Héritier et al. [22].
| Polystome species | Host species | Family | Country | 18S | 28S | 12S | COI |
|---|---|---|---|---|---|---|---|
| Diplorchis ranae | Glandirana rugosa | Ranidae | Japan | AM157184 | AM157198 | KR856070 | JF699304 |
| Eupolystoma alluaudi | Bufo sp. | Bufonidae | Togo | AM051066 | AM157199 | KR856072 | FR667558 |
| Kankana manampoka | Platypelis pollicaris | Microhylidae | Madagascar | HM854292 | HM854293 | KR856074 | JF699307 |
| Madapolystoma sp. | Blommersia wittei | Mantellidae | Madagascar | FM897290 | FM897273 | KR856075 | JF699308 |
| Indopolystoma elongatum n. sp. | Zhangixalus arboreus | Rhacophoridae | Japan | AM157190 | AM157213 | KR856094 | KR856170 |
| Indopolystoma parvum n. sp. | Z. omeimontis | Rhacophoridae | China | AM157189 | AM157212 | KR856093 | KR856169 |
| Indopolystoma viridi n. sp. | Z. viridis | Rhacophoridae | Japan | AM157191 | AM157214 | KR856095 | MN564839 |
| Indopolystoma* sp. | Z. smaragdinus | Rhacophoridae | India | AM157193 | AM157216 | KR856085 | MN564838 |
| Parapolystoma bulliense | Litoria gracilenta | Hylidae | Australia | AM157186 | AM157202 | KR856079 | KR856166 |
| Polystoma cuvieri | Physalaemus cuvieri | Leptodactylidae | Paraguay | AM051068 | AM157203 | KR856080 | AM913862 |
| Polystoma gallieni | Hyla meridionalis | Hylidae | France | AM051070 | AM157205 | KR856084 | JF699305 |
| Polystoma naevius | Smilisca baudinii | Hylidae | Costa Rica | AM157187 | AM157209 | KR856089 | AM913864 |
| Pseudodiplorchis americanus | Scaphiopus couchii | Scaphiopodidae | USA | AM051079 | AM157219 | KR856097 | KR856173 |
| Pseudopolystoma dendriticum | Onychodactylus japonicus | Hynobiidae | Japan | FM992700 | FM992707 | KR856122 | KR856180 |
Sequence analyses
18S and 28S sequences were aligned according to the procedure described in Badets et al. [2] and Héritier et al. [22] who took into account the rRNA secondary structure (stems and loops) of both genes. Partial COI and 12S gene sequences were aligned independently using Clustal W under default parameters [44] implemented in MEGA7 software [30]. Because it was too difficult to assess homologous characters within a highly variable region in the 12S, that specific region was deleted prior to running phylogenetic analyses. Using ModelTest implemented in PAUP* version 4.0b9 [43], evolutionary models were estimated independently for the four partitions from the Akaike Information Criterion [38]. All partitions with their own evolutionary model (18S: nst = 6; rates = invgamma; ngammacat = 4; 28S: nst = 6; rates = invgamma; ngammacat = 4; COI: nst = 2; rates = invgamma; ngammacat = 4; 12S: nst = 6; rates = gamma; ngammacat = 4) were subsequently concatenated and a Bayesian analysis was conducted using MrBayes 3.04b [24], with four chains running for one million generations and sampled every 100 cycles. Convergence was assessed with the program Tracer v1.7.1 (http://beast.community/tracer) [39]. A consensus tree was then reconstructed after removing the first 1000 trees (10%) as the burn-in phase. Finally, COI and 28S genetic divergences (p-distances) as well as total differences were also computed for species delimitations following thresholds designed in Du Preez et al. [17]. When all positions containing missing data and/or gaps were eliminated, there were a total of 342 and 1300 positions in the final COI and 28S datasets, respectively.
Results
Phylogenetic relationships and genetic differentiation within polystomes
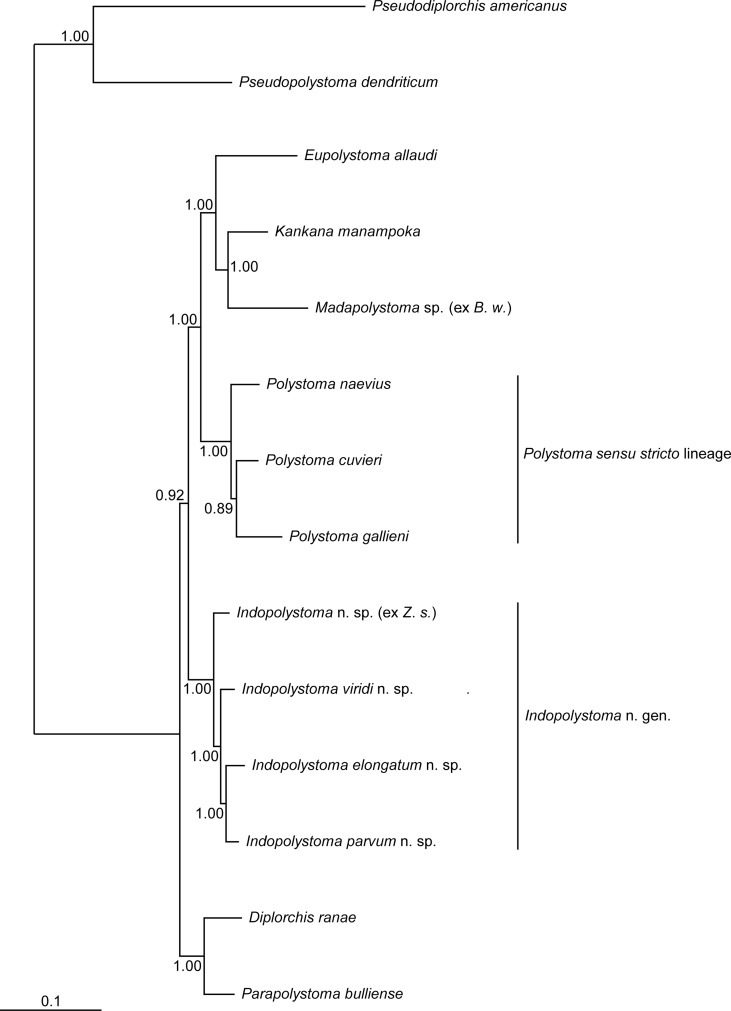
The Bayesian tree (Fig. 1) shows phylogenetic relationships within polystomes of the Neobatrachia. As previously illustrated by Badets et al. [2], Polystoma appears paraphyletic, the Polystoma sensu stricto lineage being more closely related to a clade grouping Eupolystoma, Kankana Raharivololoniaina et al. 2011 and Madapolystoma Du Preez et al. 2010 than it is to the other Polystoma lineage called here for more convenience Indopolystoma n. gen. Regarding the genetic differentiation between that clade and the Polystoma sensu stricto lineage, which is about 18.2% in the COI (Table 2) and 3.7% in the 28S (Table 3), we can indeed consider it is a new genus according to its morphological characteristics (see below).
Figure 1.
Bayesian tree for neobatrachian polystomes inferred from the analysis of four concatenated genes, namely 18S, 28S, COI and 12S. Numbers on nodes indicate Bayesian Posterior Probabilities. Indopolystoma spp. were regarded earlier as Polystoma spp. in Badets et al. [2] and Héritier et al. [22]. B. w. refers to Blommersia wittei and Z. s. to Zhangixalus smaragdinus. See also Table 1 for other host species. Scale bar represents 0.1 substitution/site.
Table 2.
Matrix of COI p-distances (lower left) and total differences (upper right) inferred from pairwise comparisons in MEGA7. B. w. refers to B. wittei and Z. s. to Z. smaragdinus. See also Table 1 for other host species.
| [1] | [2] | [3] | [4] | [5] | [6] | [7] | [8] | [9] | [10] | [11] | [12] | [13] | [14] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polystoma cuvieri [1] | 0 | 50 | 49 | 65 | 72 | 64 | 69 | 78 | 70 | 72 | 64 | 67 | 75 | 69 |
| Polystoma gallieni [2] | 0.146 | 0 | 55 | 64 | 66 | 61 | 64 | 78 | 67 | 73 | 67 | 71 | 80 | 69 |
| Polystoma naevius [3] | 0.143 | 0.161 | 0 | 51 | 55 | 54 | 64 | 70 | 69 | 67 | 60 | 66 | 77 | 70 |
| Indopolystoma sp. (ex Z. s.) [4] | 0.19 | 0.187 | 0.149 | 0 | 35 | 43 | 47 | 79 | 64 | 71 | 70 | 56 | 74 | 74 |
| Indopolystoma viridi n. sp. [5] | 0.211 | 0.193 | 0.161 | 0.102 | 0 | 36 | 50 | 75 | 67 | 75 | 69 | 64 | 70 | 71 |
| Indopolystoma parvum n. sp. [6] | 0.187 | 0.178 | 0.158 | 0.126 | 0.105 | 0 | 49 | 80 | 68 | 80 | 63 | 69 | 79 | 75 |
| Indopolystoma elongatum n. sp. [7] | 0.202 | 0.187 | 0.187 | 0.137 | 0.146 | 0.143 | 0 | 78 | 68 | 75 | 75 | 67 | 74 | 73 |
| Eupolystoma alluaudi [8] | 0.228 | 0.228 | 0.205 | 0.231 | 0.219 | 0.234 | 0.228 | 0 | 71 | 77 | 76 | 78 | 89 | 78 |
| Kankana manampoka [9] | 0.205 | 0.196 | 0.202 | 0.187 | 0.196 | 0.199 | 0.199 | 0.208 | 0 | 56 | 65 | 66 | 77 | 77 |
| Madapolystoma sp. (ex B. w.) [10] | 0.211 | 0.213 | 0.196 | 0.208 | 0.219 | 0.234 | 0.219 | 0.225 | 0.164 | 0 | 78 | 77 | 71 | 87 |
| Diplorchis ranae [11] | 0.187 | 0.196 | 0.175 | 0.205 | 0.202 | 0.184 | 0.219 | 0.222 | 0.19 | 0.228 | 0 | 65 | 72 | 76 |
| Parapolystoma bulliense [12] | 0.196 | 0.208 | 0.193 | 0.164 | 0.187 | 0.202 | 0.196 | 0.228 | 0.193 | 0.225 | 0.19 | 0 | 71 | 78 |
| Pseudodiplorchis americanus [13] | 0.219 | 0.234 | 0.225 | 0.216 | 0.205 | 0.231 | 0.216 | 0.26 | 0.225 | 0.208 | 0.211 | 0.208 | 0 | 82 |
| Pseudopolystoma dendriticum [14] | 0.202 | 0.202 | 0.205 | 0.216 | 0.208 | 0.219 | 0.213 | 0.228 | 0.225 | 0.254 | 0.222 | 0.228 | 0.24 | 0 |
Table 3.
Matrix of 28S p-distances (lower left) and total differences (upper right) inferred from pairwise comparisons in MEGA7. B. w. refers to B. wittei and Z. s. to Z. smaragdinus. See also Table 1 for other host species.
| [1] | [2] | [3] | [4] | [5] | [6] | [7] | [8] | [9] | [10] | [11] | [12] | [13] | [14] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polystoma cuvieri [1] | 0 | 29 | 17 | 42 | 41 | 42 | 41 | 58 | 55 | 79 | 59 | 58 | 232 | 206 |
| Polystoma gallieni [2] | 0.022 | 0 | 38 | 57 | 54 | 55 | 54 | 79 | 73 | 94 | 69 | 69 | 231 | 206 |
| Polystoma naevius [3] | 0.013 | 0.029 | 0 | 49 | 48 | 49 | 48 | 64 | 60 | 80 | 69 | 65 | 233 | 205 |
| Indopolystoma sp. (ex Z. s.) [4] | 0.032 | 0.044 | 0.038 | 0 | 5 | 6 | 5 | 60 | 56 | 82 | 49 | 43 | 233 | 195 |
| Indopolystoma viridi n. sp. [5] | 0.032 | 0.042 | 0.037 | 0.004 | 0 | 3 | 2 | 59 | 57 | 81 | 50 | 44 | 221 | 196 |
| Indopolystoma parvum. n. sp. [6] | 0.032 | 0.042 | 0.038 | 0.005 | 0.002 | 0 | 3 | 60 | 58 | 82 | 51 | 45 | 223 | 196 |
| Indopolystoma elongatum n. sp. [7] | 0.032 | 0.042 | 0.037 | 0.004 | 0.002 | 0.002 | 0 | 59 | 57 | 81 | 50 | 44 | 223 | 196 |
| Eupolystoma alluaudi [8] | 0.045 | 0.061 | 0.049 | 0.046 | 0.045 | 0.046 | 0.045 | 0 | 50 | 79 | 75 | 70 | 241 | 214 |
| Kankana manampoka [9] | 0.042 | 0.056 | 0.046 | 0.043 | 0.044 | 0.045 | 0.044 | 0.038 | 0 | 66 | 69 | 59 | 239 | 211 |
| Madapolystoma sp. (ex B. w.) [10] | 0.061 | 0.072 | 0.062 | 0.063 | 0.062 | 0.063 | 0.062 | 0.061 | 0.051 | 0 | 89 | 89 | 231 | 205 |
| Diplorchis ranae [11] | 0.045 | 0.053 | 0.053 | 0.038 | 0.038 | 0.039 | 0.038 | 0.058 | 0.053 | 0.068 | 0 | 20 | 227 | 197 |
| Parapolystoma bulliense [12] | 0.045 | 0.053 | 0.05 | 0.033 | 0.034 | 0.035 | 0.034 | 0.054 | 0.045 | 0.068 | 0.015 | 0 | 228 | 193 |
| Pseudodiplorchis americanus [13] | 0.178 | 0.178 | 0.179 | 0.172 | 0.17 | 0.172 | 0.172 | 0.185 | 0.184 | 0.178 | 0.175 | 0.175 | 0 | 176 |
| Pseudopolystoma dendriticum [14] | 0.158 | 0.158 | 0.158 | 0.15 | 0.151 | 0.151 | 0.151 | 0.165 | 0.162 | 0.158 | 0.152 | 0.148 | 0.135 | 0 |
Taking into account that uncorrected p-distances estimates within Asian polystomes are well beyond 10% in the COI (Table 2) and 0.2% in the 28S (Table 3), we can assume that there are four separate species according to the 1.2% and 0.07% genetic divergences that were considered as the species-level threshold within polystomes of amphibians from COI and 28S sequences, respectively [17]. Furthermore, though several substitutions were found between Asian polystomes, at least one unique change (autapomorphy) was observed in each of the four undescribed species, regardless of the gene of interest, COI or 28S. These results reinforced our hypothesis of four distinct polystome species.
Finally, a Bayesian tree inferred from the analysis of a dataset comprising 14 full-length 18S sequences (Table 1), which was supplemented by unpublished partial 18S sequences obtained from Polystoma carvirostris Fan, Li & He, 2008 (EU734835), P. zuoi Shen, Wang & Fan, 2013 (KF850147) and two other undescribed Polystoma spp. infecting Rana chaochiaoensis Liu (U734834) and Hyla annectans (Jerdon) (EU979386) of China, showed that the last two species were more closely related to species of Polystoma than they were to species of Indopolystoma (results not shown).
Morphological analyses with the description of a new genus and three new species within polystomes
Indopolystoma n. gen.
urn:lsid:zoobank.org:act:35517B78-23E5-4976-8D47-776F62A5D82F
Generic diagnosis
Body large and oblong. Intestical caeca bifurcate, diverticulated, confluent posteriorly with posterior diverticulum barely entering haptor. Intestinal anastomoses usually absent but at most a single anastomosis may be present. Vas deferens extends antero-medially, opens into seminal vesicle that opens into genital bulb, armed with 8–9 genital spines. Ovary comma shaped and prominent, sinistral, in anterior 20% of body. Oviduct arises from posterior region of ovary, connected by genito-intestinal canal to sinistral caecum, receives common vitelline duct, ascends giving rise to short tubular uterus that often holds a single egg but as many as 40. Vitellaria distributed throughout body proper except in region around ovary and reproductive ducts, extending marginally into haptor; left and right vitelline ducts join to form common vitelline reservoir near ovary, with duct to oviduct. Two prominent vaginae, antero-lateral to ovary; left and right vaginal ducts connected to respective vitelline ducts. Egg operculate, oval and lacking a filament. Haptor short relative to body size (haptor/total body length ratio < 0.15 for most species) with three pairs of suckers, one pair of hamuli and 16 marginal hooklets. Hamuli curved, unbranched in base (handle and guard not well separated) and with short recurved hook. Prominent big posteriormost marginal hooklet C1 (see numbering, [36]) with prominent broad blade and guard, in contrast with smaller hooklets C2–C8. Indopolystoma spp. are parasites of the bladder of rhacophorid frogs from Asia.
Etymology: The prefix indo refers to India, which was assumed to be the center of origin for this new genus [2].
Gender: neuter.
Type-species: Indopolystoma viridi n. sp.
Other species: Indopolystoma elongatum n. sp., Indopolystoma parvum n. sp., Indopolystoma carvirostris (Fan, Li & He, 2008) n. comb., Indopolystoma hakgalense (Crusz & Ching, 1975) n. comb., Indopolystoma indicum (Diengdoh & Tandon, 1991) n. comb., Indopolystoma leucomystax (Zhang & Long, 1987) n. comb., Indopolystoma mutus (Meng, Song & Ding, 2010) n. comb., Indopolystoma pingbianensis (Fan, Wang & Li, 2004) n. comb., Indopolystoma rhacophori (Yamaguti, 1936) n. comb., Indopolystoma zuoi (Shen, Wang & Fan, 2013) n. comb., and Indopolystoma sp.
Differential diagnosis
Within Asia, Indopolystoma can be distinguished from other polystomatid genera infecting anurans by a combination of characteristics. Unlike Diplorchis, Eupolystoma and Sundapolystoma that all have an extensive uterus, it has a short uterus like Polystoma and Neoriojatrema. Unlike Eupolystoma and Neoriojatrema that lack hamuli, it has a single pair of hamuli like Polystoma, Diplorchis and Sundapolystoma. The haptor/total body length ratio is, for all species but one, less than 0.15 while it is usually far greater for all other anuran polystomes, namely Polystoma (0.19–0.27), Diplorchis (0.15–0.29), Eupolystoma (0.15–0.34), Neoriojatrema (0.34) and Sundapolystoma (0.28). Whereas Eupolystoma, Neoriojatrema and Sundapolystoma all have marginal hooklets of equal length, posteriormost marginal hooklet C1 in Indopolystoma, Polystoma and Diplorchis is bigger than the remainder. However, if the posteriormost marginal hooklet C1 is the same shape as hooklets C2–C8 in Polystoma and Diplorchis, it is far more developed with prominent broad blade and guard in Indopolystoma (Table 4, Figs. 2–8).
Table 4.
Body measurements of Indopolystoma spp.
| Parasite species | Indopolystoma viridi n. sp. | Indopolystoma elongatum n. sp. | Indopolystoma parvum n. sp. | Indopolystoma carvirostris n. comb. | Indopolystoma hakgalense n. comb. | Indopolystoma indicum n. comb. | Indopolystoma leucomystax n. comb. | Indopolystoma mutus n. comb. | Indopolystoma pingbianensis n. comb. | Indopolystoma rhacophori n. comb. | Indopolystoma zuoi n. comb. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Host species | Zhangixalus viridis | Zhangixalus arboreus | Zhangixalus omeimontis | Kurixalus bisacculus or Kurixalus verrucosus | Taruga eques | Rhacophorus nigropalmatus | Polypedates leucomystax | Polypedates mutus | Zhangixalus dugritei | Zhangixalus arboreus | Feihyla palpebralis |
| Site | Bladder | Bladder | Bladder | Bladder | Bladder | Bladder | Bladder | Bladder | Bladder | Bladder | Bladder |
| Locality | Japan | Japan | China | China | Sri Lanka | India | China | China | China | Japan | China |
| Number of mature specimens | 15 | 3 | 2 | 10 | 12 | 8 | 5 | 2 | 6–7 | – | 3 |
| Body length | 8550 (5532–11,907) | 14,791 (12,847–14,878) | 4714 (4536–4891) | 5070 (4030–7790) | (3870–9230) | 12,556 (6732–16,614) | 7568 (6665–8514) | 7338 (6875–7800) | 9428 (6000–12,000) | (4500–6000) | 2716 (1624–3533) |
| Greatest width | 2112 (1534–2859) | 3182 (3170–3270) | 1916 (1701–2130) | 1130 (810–1400) | (890–1790) | 4164 (2222–5641) | 2732 (2542–3311) | 3063 (2550–3575) | 1990 (1640–2390) | (1000–1900) | 1280 (1202–1904) |
| Width at vagina | 1481 (1175–1800) | 1967 (1902–2031) | 1402 (1279–1525) | 830 | – | 2057 (1197–2622) | – | – | 1572 | – | – |
| Haptor length | 773 (526–1354) | 916 (734–1098) | 667 (584–749) | 680 (500–750) | (635–794) | 1298 (977–1710) | 824 (753–989) | 913 (875–950) | 869 (424–1253) | (800–1100) | 701 (632–924) |
| Haptor width | 1184 (588–1592) | 1661 (1512–1810) | 1258 (1022–1494) | 890 (640–1500) | (1016–1509) | 2104 (1584–2466) | 1693 (1355–2040) | 1650 (1425–1875) | 784 (415–1374) | (1100–1600) | 1063 (831–1317) |
| False oral sucker width | 394 (266–465) | 551 (528–573) | 395 (358–431) | – | (189–402) | 466 (236–574) | 392 (194–424) | 388 (275–500) | – | – | – |
| Pharynx length | 210 (124–268) | 237 (227–247) | 191 (187–194) | – | – | 291 (164–395) | 252 (219–286) | 245 (210–280) | 253 (218–328) | (188–240) | 164 |
| Pharynx width | 209 (164–244) | 259 (256–268) | 190 (174–205) | – | (135–197) | 249 (164–349) | 216 (194–246) | 235 (200–270) | 229 (189–252) | (188–220) | 151 |
| Ovary length | 775 (577–925) | 1072 (1062–1108) | 545 (483–606) | 497 | (492–874) | 1049 (504–1368) | 608 (451–683) | 588 (550–625) | 711.5 (453–927) | (420–630) | 369 (329–400) |
| Ovary width | 401 (286–536) | 520 (514–563) | 269 (231–306) | 390 | (207–349) | 499 (288–630) | 259 (191–294) | 400 (375–425) | – | (210–340) | – |
| Length of eggs | 241 (191–268) | 240 (224–256) | 221 (219–222) | – | – | 205 (190–214) | – | – | 307 | (238–300) | – |
| Width of eggs | 125 (74–165) | 118 (106–130) | 103 (93–113) | – | – | 132 (125–140) | – | – | 165 | (140–163) | – |
| Number of intra-uterine eggs | 1 | 1 | 1 | 0 | – | up to 40 | 0 | 0 | 1–8 | Usually 1 but sometimes more than 10 | 0 |
| Genital bulb width | 105 (67–124) | 139 (129–211) | 83 (75–90) | – | – | – | (69–76) | – | – | – | – |
| Number of genital spines | 8–9 | 8 | 8 | 8 | – | 8 | 8 | 9 | – | 8 | 8 |
| Length of genital spine | 40 (27–49) | 41 (38–44) | 17 (16–18) | – | – | 39 (38–40) | – | 39 (38–40) | – | 42 | – |
| Haptoral sucker width | 333 (242–423) | 420 (380–459) | 335 (281–389) | 259 (212–339) | (250–320) | 353 (264–465) | 289 (207–356) | 310 (270–350) | 434 (349–491) | (320–400) | 251 (209–311) |
| Hamulus handle length | 313 (276–373) | 407 (303–419) | 326 (311–340) | 285 (257–326) | (380–440) | 320 (178–414) | 332 (260–410) | 340 (330–350) | 408 (332–441) | (350–420) | 240 (185–307) |
| Hamulus guard length | – | – | – | 249 (208–306) | – | – | – | 290 (280–300) | 382 (340–461) | – | 205 (173–265) |
| Hamulus hook length | 66 (48–74) | 78.5 (72–85) | 52 (39–64) | – | – | – | – | 40 (38–43) | – | – | – |
| Marginal hooklet length | – | – | – | (16–18) only on suckers | (20.5–36.9) C1–C2 | – | – | – | – | (24–42) | – |
| Marginal hooklet C1 length | 40 (31–44) | 36 | 32 | – | – | – | – | – | – | (38–42) | – |
| Marginal hooklet C2–C8 length | 21.5 (16–31) | 23 (18–32) | (19–20) | – | – | – | – | – | – | (24–27) | – |
| Haptoral length /Body length | 0.09 (0.05–0.17) | 0.06 (0.05–0.07) | 0.14 (0.12–0.165) | 0.13 | 0.11 | 0.10 | 0.11 | 0.12 | 0.09 | 0.18 | 0.26 |
| Number of anastomoses | 0 | >1 | 1 | >1 | >1 | 1–2 | >1 | >1 | 0–1 | >1 | 3–4 |
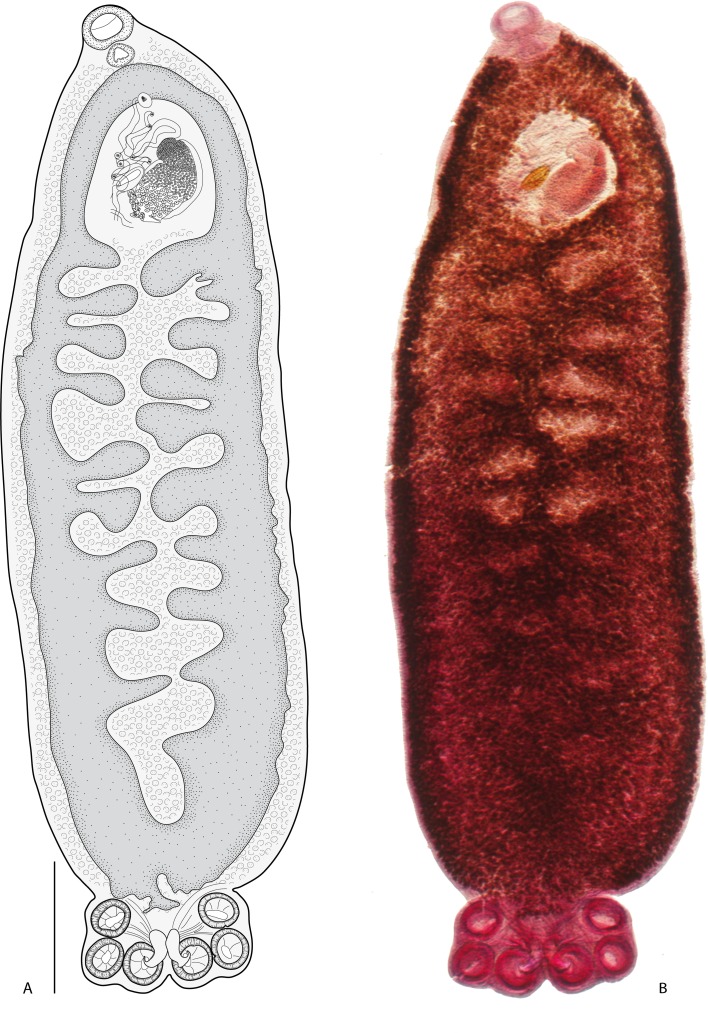
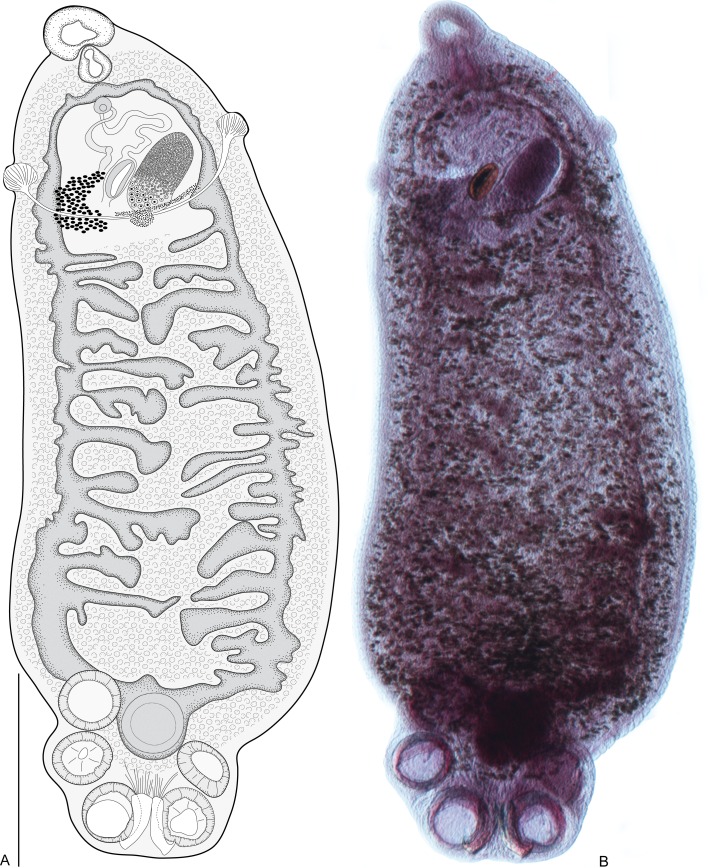
Figure 2.
Indopolystoma viridi n. gen. n. sp. from Zhangixalus viridis. Dorsal view of holotype. (A) A drawing; (B) A photograph. Scale bar: 1 mm.
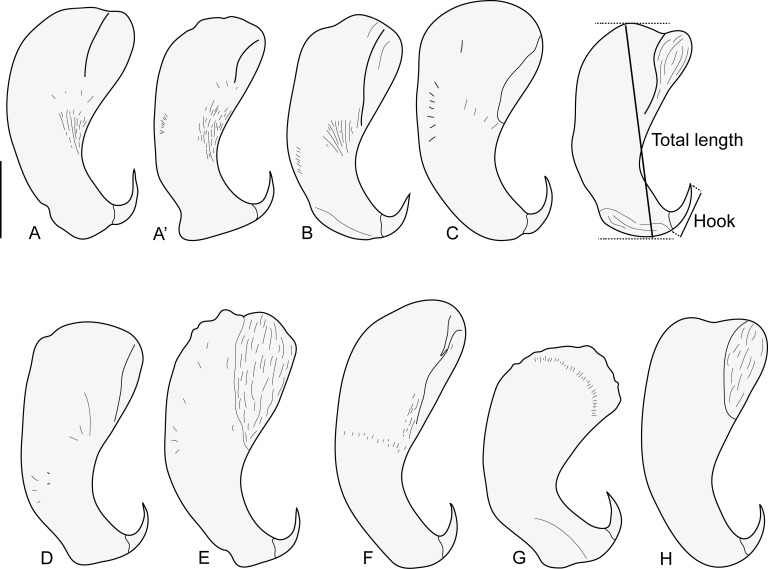
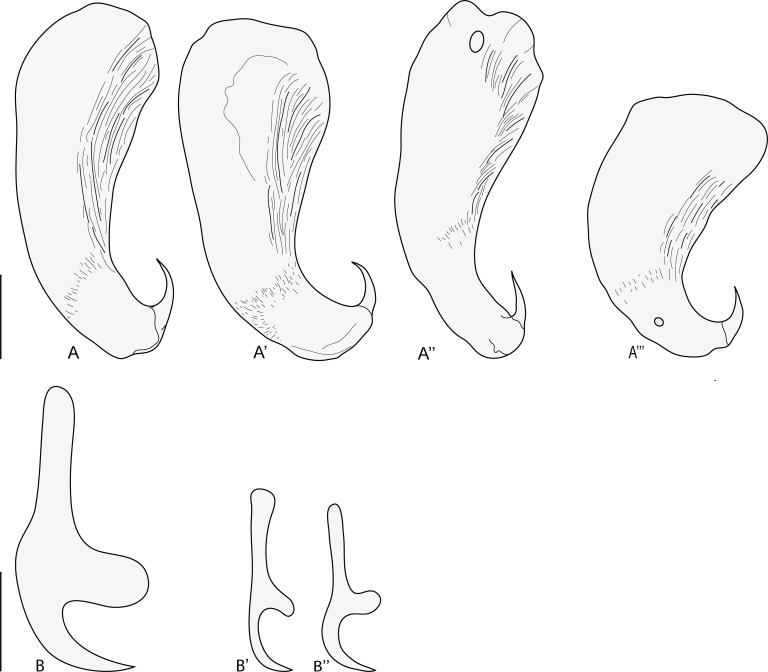
Figure 8.
Indopolystoma parvum n. gen. n. sp. from Zhangixalus omeimontis. Hamuli and marginal hooklets from holotype and paratype. (A–A’) Holotype; (A’’–A’’’) Paratype; (B) Marginal hooklet C1; (B’) Marginal hooklet C2; (B’’) Marginal hooklet C6 or C7. Scale bar: 100 μm (A–A’’’), 20 μm (B–B’’).
Indopolystoma viridi n. gen. n. sp. (Figs. 2, 3 and 4; Table 4)
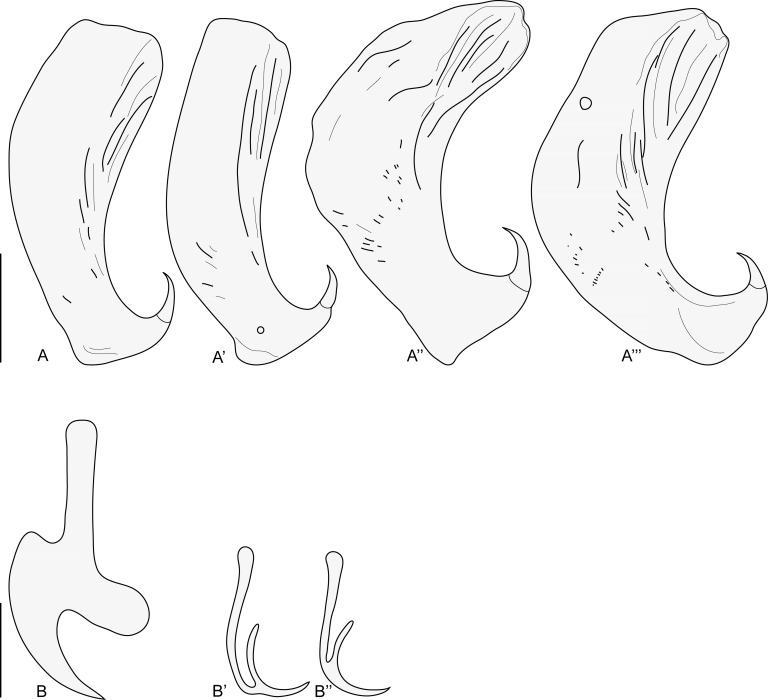
Figure 3.
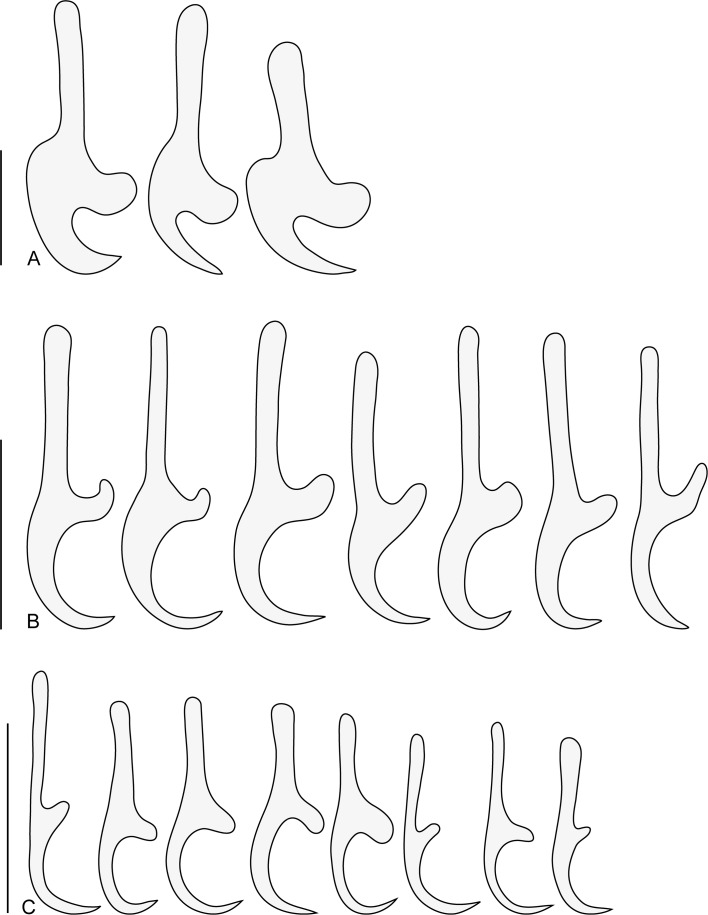
Indopolystoma viridi n. gen. n. sp. from Zhangixalus viridis. Hamuli from holotype and paratypes. (A–A’) Holotype; (B–H) Paratypes. Scale bar: 100 μm.
Figure 4.
Indopolystoma viridi n. gen. n. sp. from Zhangixalus viridis. Marginal hooklets from paratypes. (A) Marginal hooklet C1; (B) Marginal hooklet C8; (C) Marginal hooklets C2–C7. Scale bar: 20 μm.
urn:lsid:zoobank.org:act:E314A3B7-A5CE-48BA-9A21-B100556A34B9
Synonym: Polystoma sp. of Verneau et al. [51], Badets et al. [2] and Héritier et al. [22].
Type-host: Rhacophorus viridis (Hallowell). Now Zhangixalus viridis (Hallowell) [21, 27].
Site: Bladder.
Type-locality: Tokunoshima Island, Kagoshima prefecture, Japan.
Collector: Professor Hideo Hasegawa, Department of Biology, Oita Medical University, Hasama, Oita 879-5593, Japan.
Type-specimens: Holotype (MNHN HEL1173) and 10 paratypes (MNHN HEL1174–HEL1183) deposited in the Parasite Collection, Muséum national d’Histoire naturelle, Paris, France and four paratypes (NMB P512–P515) in the Parasitic Worm Collection, National Museum, Aliwal Street, Bloemfontein, South Africa.
Etymology: The name viridi refers to the species name of its host.
Description
Description based on 15 unflattened specimens stained in carmine and mounted in Canada balsam. Body elongate, total length 8550 (5532–11,907), including haptor; greatest width 2112 (1534–2859); width at vagina 1481 (1175–1800). Tegument smooth. Haptor sub-rectangular, 773 (526–1354) long, 1184 (588–1592) wide. Haptor/total body length ratio 0.09 (0.05–0.17). Suckers 333 (242–423) in diameter. Hamuli 313 (276–373) long; with hook 66 (48–74) long (Fig. 3A–H). Marginal hooklet C1 40 (31–44) long; C2–C8 21.5 (16–31) long (Fig. 4A–C). Mouth ventral, sub-terminal and surrounded by false oral sucker; false oral sucker 394 (266–465) wide. Pharynx pyriform, 210 (124–268) long, 209 (164–244) wide. Oesophagus not visible. Lateral intestinal caeca with medial diverticula branched lacking prehaptoral and haptoral anastomoses. Testis not visible hidden by digestive tract and vitellaria. Seminal vesicle prominent and packed with sperm. Genital bulb slightly sclerotized, medio-ventral, 105 (67–124) in diameter, with eight to nine sclerotized genital spines; genital spines 40 (27–49) long. Ovary prominent, sinistral and packed with oocytes; ovary 775 (577–925) long, 401 (286–536) wide. Ootype well developed. Genito-intestinal canal present on the same side of body as ovary, joining intestinal caecum posterior to ovary. Uterus confined to area anterior to ovary holding one egg; egg 241 (191–268) long, 125 (74–165) wide. No intrauterine development of eggs observed (Fig. 2).
Differential diagnosis
Indopolystoma viridi is similar to I. elongatum and I. parvum in terms of body shape, haptor/total body length ratio and shape of haptoral sclerites. However, it differs from the same two species by the general morphology of intestinal caeca and its body size (8550 μm vs. 14,791 μm for I. elongatum and 4714 μm for I. parvum). It differs from all other species of Indopolystoma in having intestinal diverticula without anastomoses.
Indopolystoma elongatum n. gen. n. sp. (Figs. 5 and 6; Table 4)
Figure 5.
Indopolystoma elongatum n. gen. n. sp. from Zhangixalus arboreus. Ventral view of holotype. (A) A drawing; (B) A photograph. Scale bar: 1 mm.
Figure 6.
Indopolystoma elongatum n. gen. n. sp. from Zhangixalus arboreus. Hamuli and marginal hooklets from holotype and paratypes. (A–A’) Holotype; (A’’–A’’’) Paratypes; (B) Marginal hooklet C1; (B’) Marginal hooklet C3; (B’’) Marginal hooklet C8. Scale bar: 100 μm (A–A’’’), 20 μm (B–B’’).
urn:lsid:zoobank.org:act:EDFE29A4-9B26-4A25-A92A-BB3CF106D4EC
Synonym: Polystoma sp. of Badets et al. [2] and Héritier et al. [22].
Type-host: Rhacophorus arboreus (Okada & Kawano). Now Zhangixalus arboreus (Okada and Kawano) [21, 27].
Site: Bladder.
Type-locality: Upstream of Kunigami-gun, city of Nago, Okinawa prefecture, Japan.
Other localities: Sado Island, Niigata prefecture, Japan.
Collector: Professor Hideo Hasegawa, Department of Biology, Oita Medical University, Hasama, Oita 879-5593, Japan.
Type-specimens: Holotype (MNHN HEL1184) and 2 paratypes (MNHN HEL1185–HEL1186) deposited in the Parasite Collection, National Museum of Natural History, Paris, France.
Etymology: The species name elongatum refers to its general body shape, which is elongate.
Description
Description based on three flattened specimens stained in carmine and mounted in Canada balsam. Body elongate, tapered anteriorly, total length 14,791 (12,847–14,878), including haptor; greatest width 3182 (3170–3270); width at vagina 1967 (1902–2031). Tegument smooth. Haptor sub-rectangular, 916 (734–1098) long, 1661 (1512–1810) wide. Haptor/total body length ratio 0.06 (0.05–0.07). Suckers 420 (380–459) in diameter. Hamuli 407 (303–419) long; with hook 78.5 (72–85) long (Fig. 6A–A’’’). Marginal hooklet C1 36 long; C2–C8 23 (18–32) long (Fig. 6B–B’’). Mouth ventral, sub-terminal and surrounded by false oral sucker; false oral sucker 551 (528–573) wide. Pharynx pyriform, 237 (227–247) long, 259 (256–268) wide. Oesophagus not visible. Intestine bifurcate with medial diverticula highly branched giving rise to prehaptoral anastomoses (up to two). Testis not visible hidden by the digestive tract and vitellaria. Seminal vesicle prominent and packed with sperm. Genital bulb slightly sclerotized, medio-ventral, 139 (129–211) in diameter, with eight sclerotized genital spines; genital spines 41 (38–44) long. Ovary prominent, submedian and packed with oocytes; ovary 1072 (1062–1108) long, 520 (514–563) wide. Ootype well developed. Genito-intestinal canal present on same side of body as ovary, joining intestinal caecum posterior to ovary. Uterus confined to area anterior to ovary holding one egg; egg 240 (224–256) long, 118 (106–130) wide. No intrauterine development of eggs observed (Fig. 5).
Differential diagnosis
Indopolystoma elongatum is well characterized by its body size and shape. This species is much bigger and more elongated (body length 14,791 μm) than any other species of Indopolystoma, though there is an overlap of size values with I. indicum. Indopolystoma elongatum can be easily distinguished from the later by the number of intrauterine eggs. None of the specimens of I. elongatum have more than a single egg in utero while I. indicum has as many as 40.
Remarks: Zhangixalus arboreus hosts two polystomes, namely I. elongatum and I. rhacophori (see below), which is uncommon within anuran polystomes. However, Z. arboreus and Z. schlegelii occur sympatrically in Japan [1]. The possibility of a misidentification can thus not be excluded especially since molecular evidence on host identity is currently not available. We consider for now that both I. elongatum and I. rhacophori are separate species primarily on the basis of body length and haptor/total body length ratio (0.06 for I. elongatum vs. 0.18 for I. rhacophori).
Indopolystoma parvum n. gen. n. sp. (Figs. 7 and 8; Table 4)
Figure 7.
Indopolystoma parvum n. gen. n. sp. from Zhangixalus omeimontis. Dorsal view of holotype. (A) A drawing; (B) A photograph. Scale bar: 1 mm.
urn:lsid:zoobank.org:act:6C7F74C6-BFEE-4277-903B-2A531FD09C63
Synonym: Polystoma sp. of Verneau et al. [51], Badets et al. [2] and Héritier et al. [22].
Type-host: Rhacophorus omeimontis (Stejneger). Now Zhangixalus omeimontis (Stejneger) [21, 27].
Site: Bladder.
Type-locality: Wawu Shan, Hongya Xian, Sichuan province, China.
Collector: Professor Anne Marie Ohler, Muséum National d’Histoire naturelle (MNHN), Paris, France.
Type-specimens: Holotype (MNHN HEL1187) and paratype (MNHN HEL1188) deposited in the Parasite Collection, National Museum of Natural History, Paris, France.
Etymology: The species name parvum refers to its small body size.
Description
Description based on two unflattened specimens stained in carmine and mounted in Canada balsam. Body elongate, total length 4714 (4536–4891), including haptor; greatest width 1916 (1701–2130); width at vagina 1402 (1279–1525). Haptor circular in outline, 667 (584–749) long, 1258 (1022–1494) wide. Haptor/total body length ratio 0.14 (0.12–0.165). Suckers 335 (281–389) in diameter. Hamuli 326 (311–340) long; with hook 52 (39–64) long (Fig. 8A–A’’’). Marginal hooklet C1 32 long; C2–C8 (19–20) long (Fig. 8B–B’’). Mouth ventral, sub-terminal and surrounded by false oral sucker; false oral sucker 395 (358–431) wide. Pharynx pyriform, 191 (187–194) long, 190 (174–205) wide. Oesophagus not visible. Intestine bifurcate with medial diverticula highly branched lacking prehaptoral anastomoses. Caeca confluent posteriorly, extending into haptor forming haptoral anastomosis. Testis lying on body midline anterior to ovary. Seminal vesicle prominent and packed with sperm. Genital bulb slightly sclerotized, medio-ventral, 83 (75–90) in diameter, with eight sclerotized genital spines; genital spines 17 (16–18) long. Ovary prominent, sinistral and packed with oocytes; ovary 545 (483–606) long, 269 (231–306) wide. Ootype well developed. Genito-intestinal canal present on same side of body as ovary, joining intestinal caecum posterior to ovary. Uterus confined to dextral and anterior to ovary holding one egg; egg 221 (219–222) long, 103 (93–113) wide. No intrauterine development of eggs observed (Fig. 7).
Differential diagnosis
Indopolystoma parvum can be easily distinguished from I. viridi and I. elongatum by its body size, haptor shape and general morphology of intestine. This species is much smaller than I. elongatum (4714 μm vs. 14,791 μm) while it is only half the size of I. viridi (4714 μm vs. 8550 μm). It shows haptor sub-spherical (vs. sub-rectangular) and intestinal caeca with haptoral anastomosis. It differs from all other congeners, apart from I. pingbianensis, in lacking medial anastomoses. Indopolystoma parvum is smaller than I. pingbianensis (4714 μm vs. 9428 μm).
Indopolystoma carvirostris (Fan, Li & He, 2008) n. comb. (Table 4)
Synonym: Polystoma carvirostris Fan, Li & He, 2008 [19].
Type-host: Polypedates cavirostris Günther. Now Kurixalus bisacculus (Taylor) (Previously Rhacophorus bisacculus Taylor) or Kurixalus verrucosus (Boulenger) (Previously Rhacophorus verrucosus Boulenger) [21, 26].
Site: Bladder.
Type-locality: Pingbian county (22°56′ N, 103°42′ E), Yunnan province, China.
Remarks: Although the authors of the original description did not draw the marginal hooklets [20], the general morphology of this species, including haptor/total body length ratio (0.13) and hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma. Furthermore, a phylogeny based on partial 18S sequences only (unpublished results) showed that this species fell within the clade of Indopolystoma species.
Indopolystoma carvirostris was originally recorded in China from P. cavirostris. However, P. cavirostris only occurs in Sri Lanka [21]. According to Inger et al. [26], Chinese records of P. cavirostris likely apply to R. bisacculus or R. verrucosus.
Indopolystoma hakgalense (Crusz & Ching, 1975) n. comb. (Table 4)
Synonym: Polystoma hakgalense Crusz & Ching, 1975 [12].
Type-host: Rhacophorus cruciger eques Kirtisinghe. Now Taruga eques (Günther) [21].
Site: Bladder.
Type-locality: Hakgala Strict Natural Reserve, Sri Lanka.
Remarks: Although the authors of the original description did not draw the marginal hooklets [12], the general morphology of this species, including haptor/total body length ratio (0.11) and hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma.
Indopolystoma indicum (Diengdoh & Tandon, 1991) n. comb. (Table 4)
Synonym: Polystoma indicum Diengdoh & Tandon, 1991 [13].
Type-host: Rhacophorus nigropalmatus Boulenger [21].
Site: Bladder
Type-locality: Cherrapunji (25°18′ N: 91°46′ E), East Khasi Hills District, Meghalaya state, India.
Remarks: Although the authors of the original description did not draw the marginal hooklets [13], the general morphology of this species, including haptor/total body length ratio (0.10) and hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma.
Indopolystoma leucomystax (Zhang & Long, 1987) n. comb. (Table 4)
Synonym: Polystoma leucomystax Zhang & Long, 1987 [54].
Type-host: Polypedates leucomystax (Gravenhorst) [21].
Site: Bladder.
Type-locality: Hangzhou, Zhejiang province, China.
Remarks: Although the authors of the original description did not draw the marginal hooklets [54], the general morphology of this species, including haptor/total body length ratio (0.11) and hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma.
Indopolystoma mutus (Meng, Song & Ding, 2010) n. comb. (Table 4)
Synonym: Polystoma mutus Meng, Song & Ding, 2010 [33].
Type-host: Rhacophorus mutus Smith. Now Polypedates mutus (Smith) [21].
Site: Bladder.
Type-locality: Jianfengling, Hainan Island, China.
Remarks: Although the authors of the original description did not draw the marginal hooklets [33], the general morphology of this species, including haptor/total body length ratio (0.12) and hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma.
Indopolystoma pingbianensis (Fan, Wang & Li, 2004) n. comb. (Table 4)
Synonym: Polystoma pingbianensis Fan, Wang & Li, 2004 [20].
Type-host: Polypedates dugritei David. Now Zhangixalus dugritei (David) [21, 27].
Site: Bladder.
Type-locality: Yunnan province, China.
Remarks: Although the authors of the original description did not draw the marginal hooklets [20], the general morphology of this species, including haptor/total body length ratio (0.09) and hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma.
Indopolystoma rhacophori (Yamaguti, 1936) n. comb. (Table 4)
Synonym: Polystoma rhacophori Yamaguti, 1936 [53].
Type-host: Rhacophorus schlegelii var. arborea Okada. Now Zhangixalus arboreus (Okada and Kawano) [21].
Site: Bladder.
Type-locality: Kurama, near Kyoto, Japan.
Remarks: Although the author of the original description did not draw the marginal hooklets [53], the general morphology of this species, including hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma. The haptor/total body length ratio of about 0.18 is bigger than that of any other Indopolystoma spp. with the exception of I. zuoi (Shen, Wang & Fan, 2013) n. comb. As discussed earlier for I. elongatum, which infests the same host, we consider for now that both I. elongatum and I. rhacophori are two separate species primarily on the basis of body length and haptor/ total body length ratio.
Indopolystoma zuoi (Shen, Wang & Fan, 2013) n. comb. (Table 4)
Synonym: Polystoma zuoi Shen, Wang & Fan, 2013 [40].
Type-host: Philautus palpebralis Smith. Now Feihyla palpebralis (Smith) [21].
Site: Bladder.
Type-locality: Pingbian county, Yunnan province (22°57.295′ N, 103°41.959′ E).
Remarks: Although the authors of the original description did not draw the marginal hooklets [40], the general morphology of this species, including hamuli shape (unbranched), is consistent with the diagnosis of Indopolystoma. The haptor/total body length ratio of about 0.26 is so much bigger than that of any other Indopolystoma spp. Nevertheless, a phylogeny based on partial 18S sequences only (unpublished results) showed that this species fell within the clade of Indopolystoma species.
Indopolystoma n. gen. sp.
Synonym: Polystoma indicum of Verneau et al. [51], Badets et al. [2] and Héritier et al. [22].
Type-host: Rhacophorus maximus Günther. Now Zhangixalus smaragdinus (Blyth) [21, 27].
Site: Bladder.
Type-locality: India.
Remarks: Indopolystoma sp. was tentatively assigned to P. indicum from Z. smaragdinus by Verneau et al. [51], Badets et al. [2] and Héritier et al. [22]. However, because we did not have any information on morphological characteristics of this species, which is nested in a clade with I. viridi, I. elongatum and I. parvum (Fig. 1; see also [2, 22, 51]), we must for now consider it as an undescribed species of Indopolystoma.
Discussion
In this paper, one genus and three new species are being described, and eight previously described species of Polystoma as well as an undescribed species from Z. smaragdinus are being transferred to the new genus. Whereas species of Polystoma in Asia infect mostly ranids and hylids, species of Indopolystoma are only reported from species assigned to rhacophorid genera, such as Feihyla, Kurixalus, Polypedates, Rhacophorus, Taruga and Zhangixalus. These results clearly illustrate that rhacophorids are frequent hosts for Indopolystoma in Asia in which polystome speciation and diversification would have occurred during the long isolation of India from Madagascar and Africa. The Rhacophoridae is currently represented by 422 valid species arranged in 20 genera [1, 21]. As such, they account for roughly 6% of the living anurans of the world. These neobatrachians occur almost exclusively in India as well as in southeast Asia, with only one genus, Chiromantis, having species known from Africa [1, 21]. Therefore, we may expect a greater diversity of polystomes within Indopolystoma which should be restricted to Asia, where rhacophorids have undergone spectacular radiation “out of India” [32].
The interspecific morphological variation of polystomes is generally limited [45]. Herein, the haptor along with sclerotized structures (or sclerites) makes Indopolystoma a unique genus amongst all polystome genera infecting anuran hosts. Despite their morphological plasticity, the haptoral sclerites which are the “hallmark of monogeneans” [9] remain a significant character for morphological identification. Within amphibian polystomes, the haptoral sclerites are typically represented by 16 marginal hooklets and one pair of hamuli, although a few exceptions are known [14, 25, 31]. These characters have been largely investigated because of their usefulness in polystome delimitation [8, 10, 11, 14, 16, 25, 35, 36, 46–48]. The species of Indopolystoma are characterized by a posteriormost marginal hooklet C1, with prominent broad blade and guard, much larger than those of hooklets C2–C8, unlike that of Polystoma and Diplorchis spp. where the hooklets are all morphologically similar, although posteriormost marginal hooklet C1 is also larger than hooklets C2–C8. On the other hand, all marginal hooklets are equal in length and similar in shape within species of Eupolystoma, Neoriojatrema and Sundapolystoma. Whereas the presence of hamuli within Indopolystoma allows the differentiation of that genus from Eupolystoma and Neoriojatrema in which hamuli are lacking, their particular structure with a handle not separated from the guard, i.e. they lack a deep notch in base, is not unique as it is similar with some species of Polystoma. Finally, the haptor/total body length ratio is also of value for delimitating Indopolystoma. For all species of Indopolystoma, with the exception of I. rhacophori and I. zuoi, this value is less than 0.15, while it is greater for most other anuran polystomes. Chiromantis rufescens (Günther) is currently the only rhacophorid frog in Africa known to host a polystome, namely Polystoma chiromantis Dupouy & Knoepffler, 1978. Although marginal hooklets were not described in the original description [18], this parasite shares the elongated body and small haptor of Indopolystoma. According to Imasuen (unpublished thesis), marginal hooklet C1 of P. chiromantis has the typical shape as seen in Polystoma species. Therefore, in the absence of molecular evidence, we herein consider this species as belonging to Polystoma, which could have arisen from host-switching in Africa.
In conclusion, even though three main characters, i.e. the shape of the posteriormost marginal hooklet C1, the haptor/total body length ratio, and host species belonging to Rhacophoridae, constitute key characters for the morphological delimitation of Indopolystoma, it is important that genotyping of several polystome worms be conducted prior to the description process, as recommended by Héritier et al. [23].
Acknowledgments
We are indebted to Annemarie Ohler and Hideo Hasegawa for providing parasite specimens to OV.
Cite this article as: Chaabane A, Verneau O & Du Preez L. 2019. Indopolystoma n. gen. (Monogenea, Polystomatidae) with the description of three new species and reassignment of eight known Polystoma species from Asian frogs (Anura, Rhacophoridae). Parasite 26, 67.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.AmphibiaWeb. 2019. https://amphibiaweb.org. Berkeley, CA, USA: University of California; Accessed 31 May 2019. [Google Scholar]
- 2.Badets M, Whittington I, Lalubin F, Allienne J-F, Maspimby J-L, Bentz S, Du Preez LH, Barton D, Hasegawa H, Tandon V, Imkongwapang R, Ohler A, Combes C, Verneau O. 2011. Correlating early evolution of parasitic platyhelminths to Gondwana breakup. Systematic Biology, 60, 762–781. [DOI] [PubMed] [Google Scholar]
- 3.Beck RA, Burbank DW, Sercombe WJ, Riley GW, Barndt JK, Berry JR, Afzal J, Khan AM, Jurgen H, Metje J, Cheema A, Shafique NA, Lawrence RD, Khan MA. 1995. Stratigraphic evidence for an early collision between northwest India and Asia. Nature, 373, 55–58. [Google Scholar]
- 4.Bentz S, Leroy S, Du Preez L, Mariaux J, Vaucher C, Verneau O. 2001. Origin and evolution of African Polystoma (Monogenea, Polystomatidae) assessed from molecular methods. International Journal for Parasitology, 31, 697–705. [DOI] [PubMed] [Google Scholar]
- 5.Bentz S, Sinnappah-Kang ND, Lim LHS, Lebedev B, Combes C, Verneau O. 2006. Historical biogeography of amphibian parasites, genus Polystoma (Monogenea: Polystomatidae). Journal of Biogeography, 33, 742–749. [Google Scholar]
- 6.Bossuyt F, Milinkovitch MC. 2001. Amphibians as indicators of early tertiary “Out-of-India” dispersal of vertebrates. Science, 292, 93–95. [DOI] [PubMed] [Google Scholar]
- 7.Bourgat R, Murith D. 1980. Polystoma lamottei n. sp. et P. aeschlimanni n. sp. deux polystomes (Monogènes) de la même espèce d’amphibien: Ptychadena pumilio (Boulenger, 1920). Zeitschrift für Parasitenkunde, 62, 293–301. [Google Scholar]
- 8.Caballero YCE, Cerecero C. 1941. Una nueva especie de Polystoma (Trematoda: Polystomatidae) parasito de la vejiga urinaria de Hyla baudinii (Dum. y. Bibr.). Anales del Instituto de Biologia, Universidad de Mexico, 12, 615–621. [Google Scholar]
- 9.Chisholm LA, Whittington ID. 1998. Morphology and development of the haptors among the Monocotylidae (Monogenea). Hydrobiologia, 383, 251–261. [Google Scholar]
- 10.Combes C, Laurent RF. 1978. Deux nouveaux Polystomatidae (Monogenea) de République Argentine. Acta Zoologica Lilloana, 33(1), 85–91. [Google Scholar]
- 11.Combes C, Laurent RF. 1979. Les monogènes Polystomatidae de République Argentine: description de deux nouvelles espèces et essai de synthèse. Revista Iberica de Parasitologia, 79, 545–557. [Google Scholar]
- 12.Crusz H, Ching CC. 1975. Parasites of the relict fauna of Ceylon. VI. More new helminths from amphibians and reptiles, a new host-record and redescription of Acanthocephalus serendibensis Crusz and Mills 1970. Annales de Parasitologie Humaine et Comparée, 50(5), 531–558. [PubMed] [Google Scholar]
- 13.Diengdoh CR, Tandon V. 1991. A new species of Polystoma (Monogenea) parasitic in rhacophorid amphibians in Meghalaya, India. Helminthologia, 28, 173–178. [Google Scholar]
- 14.Du Preez LH. 2015. Eupolystoma namibiensis n. sp. (Monogenea: Polystomatidae) parasitic in Poyntonophrynus hoeschi (Ahl, 1934) of Namibia. African Zoology, 50(2), 141–145. [Google Scholar]
- 15.Du Preez LH, Kok DJ. 1992. Synoptic occurrence of new species of Polystoma and Metapolystoma (Monogenea: Polystomatidae) in Ptychadena porosissima in South Africa. Systematic Parasitology, 22, 141–150. [Google Scholar]
- 16.Du Preez LH, Martiz MF. 2006. Demonstrating morphometric protocols using polystome marginal hooklet measurements. Systematic Parasitology, 63, 1–15. [DOI] [PubMed] [Google Scholar]
- 17.Du Preez LH, Verneau O, Gross TS. 2007. Polystoma floridana n. sp. (Monogenea: Polystomatidae) a parasite in the green tree frog, Hyla cinerea (Schneider), of North America. Zootaxa, 1663, 33–45. [Google Scholar]
- 18.Dupouy J, Knoepffler LP. 1978. Polystoma chiromantis n. sp. (Monogenea) chez Chiromantis rufescens (Gunther, 1868) du Cameroun, du Gabon et de Centrafrique. Bulletin du Muséum National d’Histoire Naturelle, Paris, 3(514), 217–220. [Google Scholar]
- 19.Fan LX, Wang ZL, Li J. 2004. A new species of Polystoma (Polystomatidae, Monogenea) parasitic in Polypedates dugritei. Acta Zootaxonomica Sinica, 29(3), 451–454. [Google Scholar]
- 20.Fan LX, Li JH, He ZY. 2008. A new species of the genus Polystoma parasitic in the host Rhacophorus carvirostris Guenther. Acta Zootaxonomica Sinica, 33(2), 340–343. [Google Scholar]
- 21.Frost DR. 2019. Amphibian species of the world: an online reference. Version 6.0 (Date of access). New York, USA: American Museum of Natural History; Electronic Database accessible at http://research.amnh.org/herpetology/amphibia/index.html. [Google Scholar]
- 22.Héritier L, Badets M, Du Preez LH, Aisien MS, Lixian F, Combes C, Verneau O. 2015. Evolutionary processes involved in the diversification of chelonian and mammal polystomatid parasites (Platyhelminthes, Monogenea, Polystomatidae) revealed by palaeoecology of their hosts. Molecular Phylogenetics and Evolution, 92, 1–10. [DOI] [PubMed] [Google Scholar]
- 23.Héritier L, Verneau O, Smith KG, Coetzer C, Du Preez LH. 2018. Demonstrating the value and importance of combining DNA barcodes and discriminant morphological characters for polystome taxonomy (Platyhelminthes, Monogenea). Parasitology International, 67(1), 38–46. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 25.Imkongwapang R, Tandon V. 2010. Neoriojatrema mokokchungensis n. g., n. sp. (Monogenea: Polystomatidae: Polystomatinae) from the urinary bladder of a megophryid frog, Xenophrys glandulosa, in Nagaland, north-eastern India. Journal of Helminthology, 84(4), 375–380. [DOI] [PubMed] [Google Scholar]
- 26.Inger RF, Orlov N, Darevsky I. 1999. Frogs of Vietnam: a report on new collections. Fieldiana Zoology. New Series, 92, 1–46. [Google Scholar]
- 27.Jiang D, Jiang K, Ren J, Wu J, Li J. 2019. Resurrection of the genus Leptomantis, with description of a new genus to the family Rhacophoridae (Amphibia: Anura). Asian Herpetological Research, 10, 1–12. [Google Scholar]
- 28.Justine J-L. 2007. Parasite biodiversity in a coral reef fish: twelve species of monogeneans on the gills of the grouper Epinephelus maculatus (Perciformes: Serranidae) off New Caledonia, with a description of eight new species of Pseudorhabdosynochus (Monogenea: Diplectanidae). Systematic Parasitology, 66(2), 81. [DOI] [PubMed] [Google Scholar]
- 29.Krause DW, Rogers RR, Forster CA, Hartman JH, Buckley GA, Sampson SD. 1999. The late Cretaceous vertebrate fauna of Madagascar: implications for Gondwanan paleobiogeography. GSA Today, 9(8), 1–7. [Google Scholar]
- 30.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamothe Argumedo R. 1985. Monogeneos de Los Anfibios de Mexico VII. Hallazgo de Pseudodiplorchis Americanus (Rodgers Y Kuntz, 1940) Yamaguti, 1963 en Baja California sur, Mexico. Anales del Instituto de Biología Universidad Nacional Autónoma de México Serie Zoología, 56, 291–300. [Google Scholar]
- 32.Li JT, Li Y, Klaus S, Rao DQ, Hillis DM, Zhang YP. 2013. Diversification of rhacophorid frogs provides evidence for accelerated faunal exchange between India and Eurasia during the Oligocene. Proceedings of the National Academy of Sciences of the United States of America, 110(9), 3441–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng YY, Song L, Ding XJ. 2010. A new species of the genus Polystoma (Monogenea, Polystomatidae) parasitic in Polypedates mutus from Hainan Island, China. Acta Zootaxonomica Sinica, 35(4), 842–845. [Google Scholar]
- 34.Mittermeier RA, Turner WR, Larsen FW, Brooks TM, Gascon C. 2011. Global biodiversity conservation: the critical role of hotspots, in Biodiversity hotspots, Zachos F, Habel J, Editors. Springer, Berlin: Heidelberg. [Google Scholar]
- 35.Murith D. 1979. Identité des larves de polystomes (Monogenea) parasites du têtard de Dicroglossus occipitalis (Günther) en Côte-d’Ivoire. Zeitschrift für Parasitenkunde, 59(2), 187–194. [DOI] [PubMed] [Google Scholar]
- 36.Murith D. 1981. Contribution à l’étude de la systématique des polystomes (Monogènes, Polystomatidae) parasites d’amphibiens anoures de basse Côte d’Ivoire. Revue Suisse de Zoologie, 88, 475–533. [Google Scholar]
- 37.Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature, 403, 853–858. [DOI] [PubMed] [Google Scholar]
- 38.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 39.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen JF, Wang ZL, Fan LX. 2013. A new species of the genus Polystoma (Monogenea: Polystomatidae) parasitic in tree frog Philautus palpebralis Smith, 1924. Acta Zootaxonomica Sinica, 38(4), 765–772. [Google Scholar]
- 41.Simkova A, Desdevises Y, Gelnar M, Morand S. 2000. Co-existence of nine gill ectoparasites (Dactylogyrus: Monogenea) parasitising the roach (Rutilus rutilus L.): history and present ecology. International Journal for Parasitology, 30, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 42.Storey M, Mahoney JJ, Saunders AD, Duncan RA, Kelley SP, Coffin MF. 1995. Timing of hotspot-related volcanism and the break-up of Madagascar and India. Science, 267, 852–855. [DOI] [PubMed] [Google Scholar]
- 43.Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0b9. Sinauer Associates: Sunderland (MA). [Google Scholar]
- 44.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tinsley RC. 1973. Observations on Polystomatidae (Monogenoidea) from east Africa with a description of Polystoma makereri n. sp. Zeitschrift für Parasitenkunde, 42, 251–263. [DOI] [PubMed] [Google Scholar]
- 46.Vaucher C. 1983. Helminthes parasites du Paraguay V: Polystoma lopezromani Combes et Laurent, 1979 et utilisation d’une technique inédite d’observation des hamuli chez les Polystomatidae (Monogenea). Bulletin de la Société Neuchâteloise des Sciences Naturelles, 106, 101–107. [Google Scholar]
- 47.Vaucher C. 1987. Polystomes d’Équateur, avec description de deux nouvelles espèces. Bulletin de la Societe Neuchâteloise des Sciences Naturelles, 110, 45–56. [Google Scholar]
- 48.Vaucher C. 1990. Polystoma cuvieri n. sp. (Monogenea: Polystomatidae), a parasite of the urinary bladder of the leptodactylid frog Physalaemus cuvieri in Paraguay. Journal of Parasitology, 76(4), 501–504. [PubMed] [Google Scholar]
- 49.Verneau O. 2004. Origine et Évolution des monogènes Polystomatidae, parasites d’amphibiens et de chéloniens d’eau douce. Perpignan: Habilitation à Diriger des Recherches, Université de Perpignan. [Google Scholar]
- 50.Verneau O, Bentz S, Sinnappah ND, Du Preez L, Whittington I, Combes C. 2002. A view of early vertebrate evolution inferred from the phylogeny of polystome parasites (Monogenea: Polystomatidae). Proceedings of the Royal Society of London Series B, Biological Sciences, 269, 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verneau O, Du Preez LH, Laurent V, Raharivololoniaina L, Glaw F, Vences M. 2009. The double odyssey of Madagascan polystome flatworms leads to new insights on the origins of their amphibian hosts. Proceedings of the Royal Society Series B, Biological Sciences, 276(1662), 1575–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whittington ID, Kearn GC. 2011. Hatching strategies in monogenean (Platyhelminth) parasites that facilitate host infection. Integrative and Comparative Biology, 51(1), 91–99. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguti S. 1936. Studies on the helminth fauna of Japan. Part 14. Amphibian trematodes. Japanese. Journal of Zoology, 6(4), 551–576. [Google Scholar]
- 54.Zhang SY, Long S. 1987. Three new monogenetic trematodes of the family Polystomatidae. Acta Zootaxonomica Sinica, 12(3), 230–237. [Google Scholar]