Abstract
Alzheimer’s disease (AD) is a pervasive neurodegenerative disorder, the molecular and cellular complexity of which remains poorly understood. Here, we profiled and analysed 80,660 single-nucleus transcriptomes from prefrontal cortex of 48 individuals with varying degrees of AD pathology. We identified transcriptionally-distinct subpopulations across six major brain cell-types, including those associated with pathology and characterized by regulators of myelination, inflammation, and neuron survival. The strongest AD-associated changes appeared early in pathological progression and were highly cell-type-specific, whereas genes upregulated in late-stage were common across cell types and primarily involved in global stress response. Surprisingly, we found an overrepresentation of female cells in AD-associated subpopulations, and substantially different transcriptional responses between sexes in multiple cell types, including oligodendrocytes. Overall, myelination-related processes were recurrently perturbed in multiple cell types, suggesting a key role in AD pathophysiology. Our single-cell transcriptomic resource provides a first blueprint for interrogating the molecular underpinnings and cellular basis of AD.
Alzheimer’s disease (AD) is a slowly-progressing neurodegenerative disorder, starting with mild memory loss and culminating in severe impairment of broad executive and cognitive functions1–3. AD pathophysiology involves neuron-glia interactions, supported by transcriptomic and epigenomic analyses that reveal downregulation of neuronal functions and upregulation of innate immune responses in AD brains4–14. However, bulk-tissue level resolution likely masks the complexity of alterations across cells and within cell groups, especially for lowly-represented cell types4. Potential changes in cell composition during neurodegeneration also confound the distinction between composition and activity changes in a given cell type. Finally, the complex interplay between protective and damaging molecular processes, within and across cell types, further contributes to the difficulty in interpreting tissue-resolution disease signatures.
Single-cell RNA sequencing (scRNA-seq) provides an alternative to study the cellular heterogeneity of the brain15–17, by profiling tens of thousands of individual cells15,18,19. With the goal of characterizing the complex cellular changes in AD brain pathology, here we provide the first single-cell view of AD pathology, profiling 80,660 droplet-based single-nucleus cortical transcriptomes across 48 individuals with varying degrees of AD pathology, and across both sexes. The resulting resource paints a unique cellular-level view of transcriptional alterations associated with AD pathology, and reveals cell type-specific and shared gene expression perturbations, disease-associated cellular subpopulations, and sex-biased transcriptional responses.
Single-nucleus RNA-seq profiling of prefrontal cortex in human subjects with AD pathology
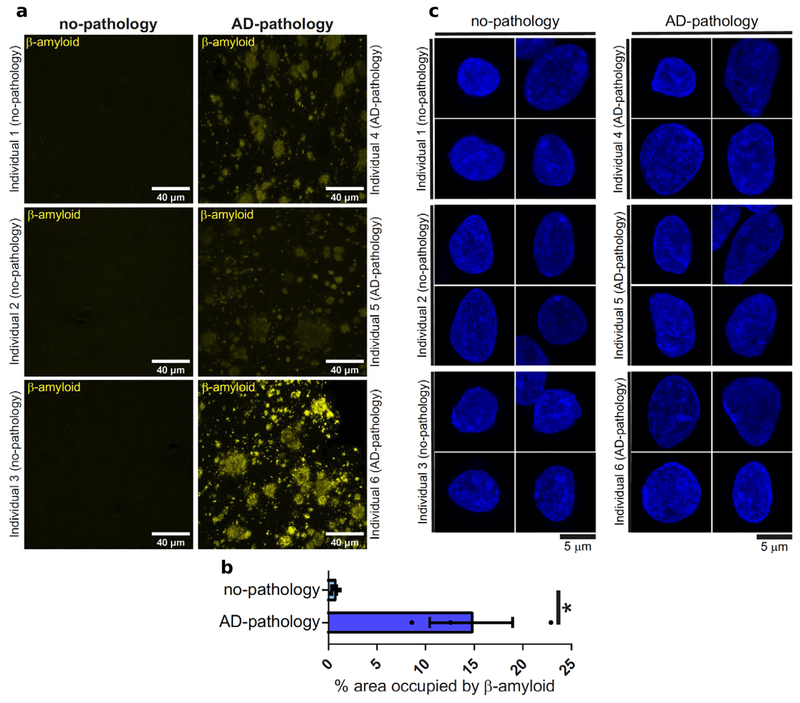
Postmortem human brain samples came from 48 participants in the Religious Order Study (ROSMAP), a longitudinal cohort of aging and dementia that includes clinical data, detailed post-mortem pathological evaluations, and omics tissue profiling20. We selected 24 individuals with elevated β-amyloid (Aβ) and other pathological hallmarks of AD (AD-pathology), and 24 individuals with no or very low Aβ burden or other pathologies (no-pathology). We profiled tissue from prefrontal cortex (Brodmann area 10) from each individual, given its major role in AD affected traits, including cognition. Immunohistochemistry for β-amyloid confirmed the pathological status of the samples (Extended Data Fig. 1a,b). Bright field and high-resolution confocal microscopy did not show any apparent physical damage to nuclei isolated from AD-pathology relative to no-pathology samples (Extended Data Fig. 1c). We report a total of 80,660 droplet-based single-nucleus RNA-seq (snRNA-seq) profiles (Extended Data Fig. 2a), which will be publicly available on the ROSMAP data compendium (see Data availability).
Cellular diversity of the aged human prefrontal cortex
To classify the major cell types in the aged cortex, we pre-clustered all cells jointly across the 48 individuals (Extended Data Fig. 2b) to produce 20 transcriptionally distinct pre-clusters with highly consistent expression patterns across individuals (Extended Data Fig. 2c, d). We identified and annotated the major cell-types of the human brain by interrogating the expression patterns of known gene markers18,21, excitatory neurons (Ex; marked by NRGN), inhibitory neurons (In; GAD1), astrocytes (Ast; AQP4), oligodendrocytes (Oli; MBP), microglia (Mic; CSF1R, CD74), oligodendrocyte progenitor cells (Opc; VCAN), endothelial cells (End; FLT1), and pericytes (Per; ITI) (Extended Data Fig. 3a,b). The cell types, markers, and cell type proportions matched previous single nucleus Drop-seq data from adult human cortex18, indicating that our results are robust to the inclusion of pathologically-affected brains (Extended Data Fig. 3c–e). We next collapsed the pre-clusters into 8 broad cell-type clusters using annotations supported by both direct marker expression and significant overlap with previously curated single-cell populations. We used these cell-type categories to characterize the specificity of AD-pathology gene expression perturbations, to quantify gene-trait associations, and to assess qualitative differences in cell-type-specific pathological responses between sexes.
Systematic differential expression analysis reveals common and cell-type-specific alterations of gene expression to Alzheimer’s pathology
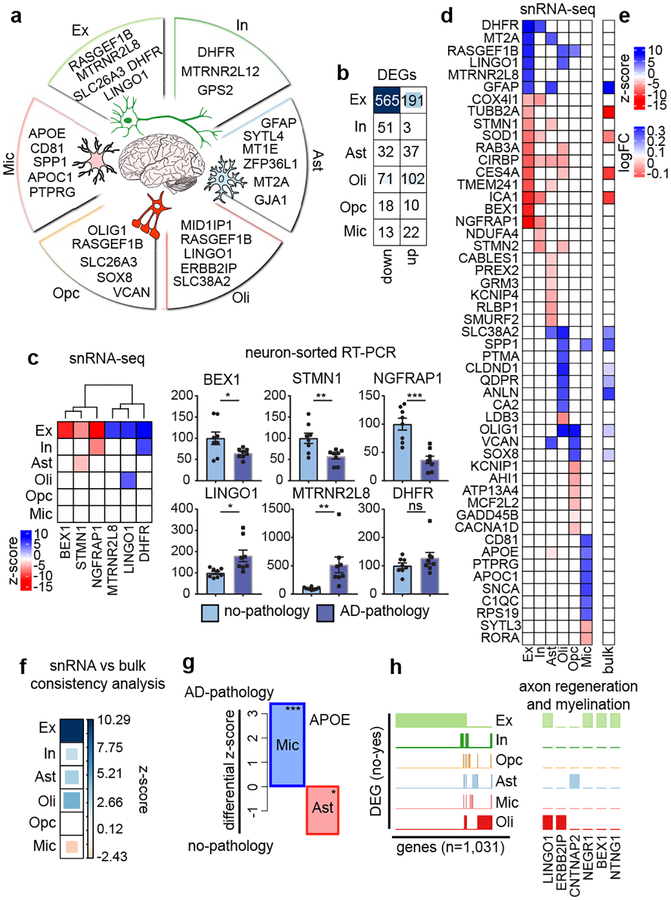
We compared gene expression levels for cells isolated from AD-pathology versus no-pathology individuals by cell type (methods). We identified 1,031 unique differentially-expressed genes (DEGs) that implicate all major cell types (Fig. 1a, b, Extended Data Table S2). Neurons showed a strong signature of repression, with 75% of DEGs in Ex and 95% in In showing downregulation, while most DEGs in Oli, Ast, and Mic showed upregulation (53%–63%). The number of significant DEGs for non-neuronal populations were substantially smaller, likely reflecting reduced power in lower-abundance cell types. These contrasting observations on the number and dominant directionality of DEGs reveal a heterogeneous response to AD pathology between cell types, a recurrent theme that we observed throughout the study. Importantly, DEGs were robustly detected at different levels of expression (Extended data Fig. 4a). Quantitative reverse transcription PCR (RT-qPCR) in NeuN-positive nuclei isolated by fluorescence-activated cell sorting corroborated significant differential expression for five of the six genes tested, including up- and down-regulated genes (Fig. 1c).
Fig. 1 |. Cell-type specific gene expression changes in AD pathology.
a, Genes most upregulated: excitatory (Ex) and inhibitory (In) neurons, astrocytes (Ast), oligodendrocytes (Oli), oligodendrocyte precursor cells (OPC), and microglia (Mic) (Font size, -log10 P-value). b, Differentially-expressed gene (DEG) counts (2-sided Wilcoxon-rank-sum test, FDR<0.01, logFC>0.25, Poisson mixed-model FDR<0.05). c, RT-qPCR validation. snRNA-seq differential scores for Ex and In DEGs (z-score, Poisson mixed-model) (left) and qPCR validation (right), FACS-sorted NeuN-positive nuclei (no-pathology n=8, pathology n=8 individuals) (mean ± s.e.m.; ***, P < 0.001; **, P < 0.01, *; P < 0.05; ns; P > 0.05; Student’s two-tailed t-test). d, Top snRNA-seq DEGs differential scores (z-score, Poisson mixed-model) and corresponding values in bulk RNA-seq (e) (ROSMAP cohort, n=484, P-Value <0.01). f, Global snRNA-seq and bulk RNA-seq consistency. (Dis)Agreement is estimated by deviation from random expectation (z-score) of single-cell DEG average rank scores in the ranked list of bulk differential analysis. g, APOE differential expression in Mic (n=955 cells, 24 pathology individuals; n=965 cells, 24 no-pathology individuals) and Ast (n=1830 cells, 24 pathology individuals; n=1562 cells, 24 no-pathology individuals) (z-score Poisson mixed-model, ***, P-Value = 0.001, *, P-Value = 0.05, two-sided, standard normal). h, Binary plot indicating with bars whether a gene (column) is a DEG in a cell-type (rows) or not (left, n=1,031 DEGs). Six genes associated with myelination and/or axon regeneration (right).
Analogous analysis of DEGs using high-quality bulk RNA-seq data from the ROSMaP cohort (n=484) validated DEGs identified in snR-NA-seq data (Fig. 1d, e). Rank permutation test (methods) revealed that bulk data is dominated by expression changes observed at the single-cell level in Ex and Oli (Fig. 1f). Consequently, changes in other cell-types are not well captured, with Mic particularly underrepresented. Bulk RNA-seq data also cannot capture DEGs with opposite directionality in different cell types. With snRNA-seq, we found that APOE was strongly upregulated in Mic but downregulated in Ast (FDR-corrected P < 0.01, 2-sided Wilcoxon-rank-sum test) (Fig. 1g), consistent with AD model studies in human induced pluripotent stem cells22 and mouse23–25.
The vast majority of DEGs (95%) were perturbed only in neurons or only in one glial cell type, indicating strong cell-type specificity of the perturbations (Fig. 1h). However, we found that top DEGs are involved in related processes, even across cell types. One such example is myelination, axonal outgrowth and regeneration. Top DEGs included LINGO1, which was upregulated in Ex and Oli and is a negative regulator of neuronal survival, axonal integrity, oligodendrocyte differentiation and myelination26,27, ERBB2IP, required for remyelination of axons28, CNTNAP2, a cell-adhesion molecule that mediates axon-myelin interactions29, NEGR1, a cell-adhesion molecule mediating myelin-stimulated axonal outgrowth30, BEX1, a gene involved in regeneration of axons after injury31, and NTNG1, a factor promoting neurite outgrowth of axons and dendrites (Fig. 1h)32. Consistent with observed downregulation of NTNG1 in Ex, RNA in situ hybridization revealed significantly fewer Ex, marked by expression of SLC17A7, with detectable NTNG1 expression in brain sections from AD-pathology individuals, compared to no-pathology individuals (Fig Extended data Fig. 4b).
Overall, these results indicate that all major cell types are affected at the transcriptional level by AD pathology and that single cell-level resolution is critical as changes in gene expression, including directionality, can be conditional on cell type.
Cell-type-specific changes during pathological progression
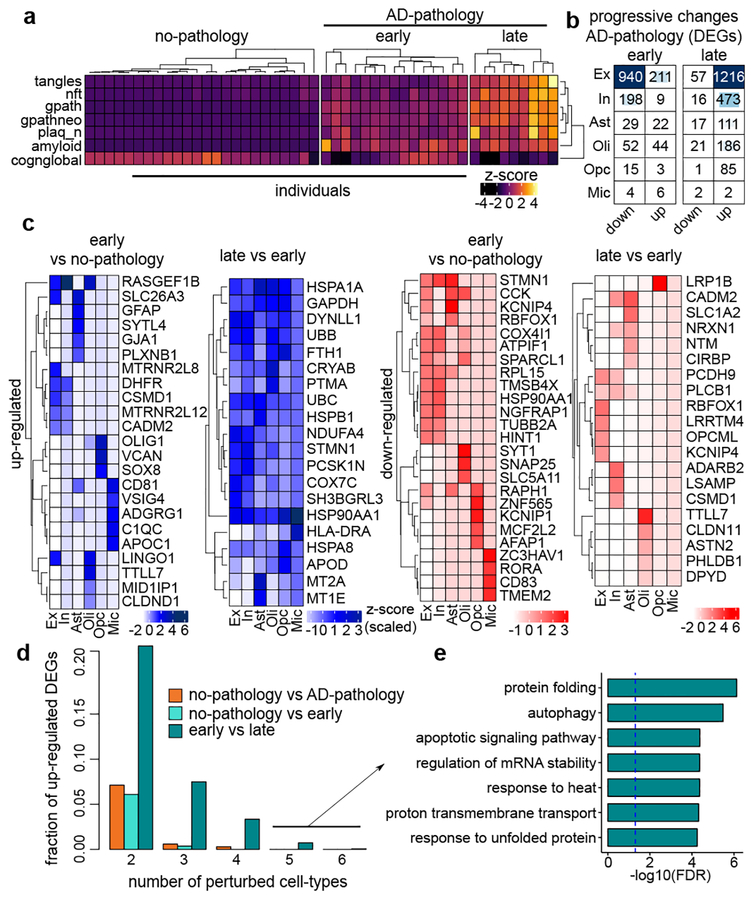
We next examined if there are qualitative changes in gene expression perturbations during early versus late stage AD pathology. To define pathology groups, we clustered individuals based on 9 clinico-pathological traits (Extended Data Table S3). AD-pathology individuals segregated in two subgroups, corresponding with AD pathological progression: 1) early-pathology1 - amyloid burden, but modest neurofibrillary tangles and cognitive impairment; and 2) late-pathology33 - higher amyloid, and also elevated neurofibrillary tangles, global pathology, and cognitive impairment (Fig. 2a). We then quantified gene expression changes in pairwise comparisons between these groups.
Fig. 2 |. Gene expression changes in AD pathology progression.
a, Phenotypic clustering of 48 individuals (columns) using clinico-pathological variables (rows) measuring neuronal neurofibrillary tangle density (tangles), neurofibrillary tangle burden (nft), global AD pathology burden (gpath), neuritic plaque burden (plaq_n), overall amyloid level (amyloid), and global cognitive function (cogn_global_lv). b, DEG counts (2-sided Wilcoxon-rank-sum test, FDR<0.01, |log-Fold-Change|>0.25, Poisson mixed-model FDR<0.05). c, Most-significantly-altered genes (rows) for each cell type (columns) and comparison, based on p-value rank (FDR<0.01, 2-sided Wilcoxon-rank-sum test, z-scores Poisson mixed-model, column-scaled). d, Fraction of total up-regulated genes (y-axis) as a function of the total number of cell-types in which the up-regulation occurs e, GO terms associated with genes upregulated in late-pathology common to ≥5 cell types (n=11 genes, hypergeometric test, FDR correction).
Comparing early versus no-pathology groups revealed large-scale transcriptional changes that occur before individuals develop severe pathological features (Fig. 2b, c). Both up- and down-DEGs were highly cell-type-specific with nearly all genes (96%) perturbed in either neurons (Ex and In) or in a single glial cell type. These changes were similar to the changes found between no-pathology and AD-pathology (Extended Data Fig. 5a), suggesting that major transcriptional changes appear early in pathological progression. Comparing late- versus early-pathology revealed common upregulated genes shared across cell types (Fig. 2d), contrasting with the earlier cell-type-specificity. Shared upregulated genes are involved in proper protein folding (e.g. HSP90AA1, HSPA1A), including molecular chaperones (e.g. HSPB1, CRYAB), and are associated with autophagy, apoptosis, and generalized stress response (Fig. 2c, e). These processes are collectively involved in the proteostasis network -- i.e., the molecular machinery that operates to maintain protein integrity34–36. In contrast to upregulated genes, downregulated genes mostly showed cell-type-specific changes.
Cell-type-specific associations of gene expression with AD-related neuropathological traits
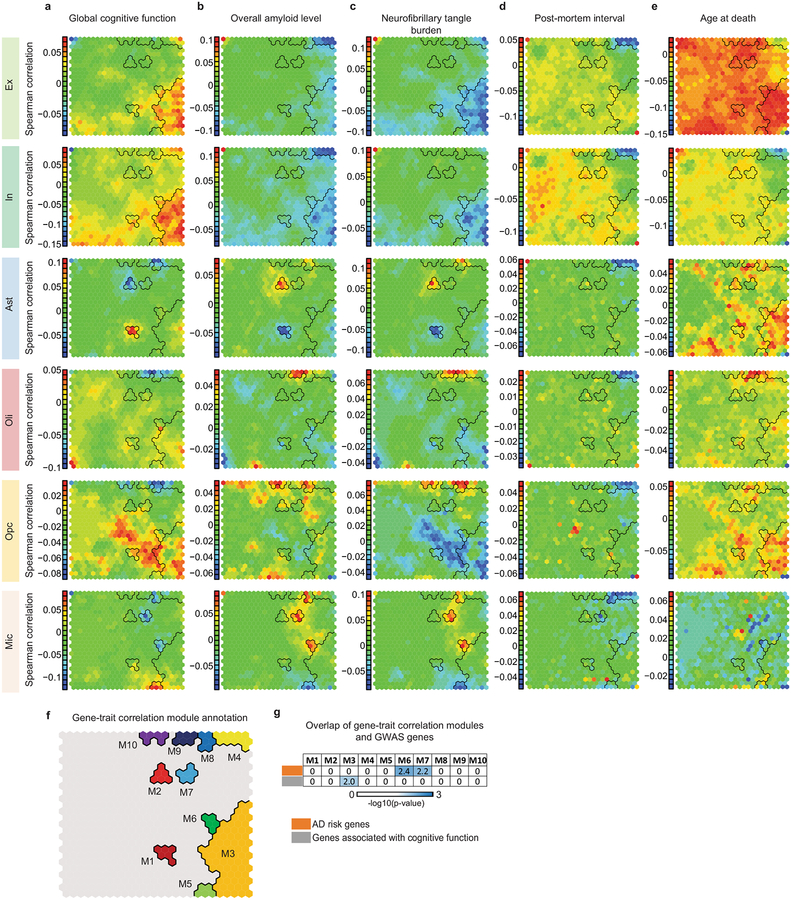
Given the complexity and heterogeneity of the AD phenotype, we next aimed to quantify the association between gene expression in specific cell types and variability of pathological traits. We used the major pathological quantitative traits in ROS: Aβ level, neurofibrillary tangle burden, neuritic plaque count, tangle density, global AD pathology, and global cognition. For each individual, we first computed the correlation between each genes expression profile for a given cell type and pathological trait. We then analyzed the resultant gene-trait correlation patterns using the self-organizing map (SOM) approach37 to discover gene sets with similar expression patterns that most strongly correlated with each phenotype (methods). Genes with similar phenotypic correlations are grouped in the same SOM grid unit, with similar units clustered nearby (Extended Data Fig. 6a–e). We observed that Ex, In, Ast, Mic, and Oli, each showed distinct SOM units, indicating that different gene groups respond to AD pathology in each cell type. Importantly, gene sets correlated with post-mortem interval and age-at-death were highly distinct from those correlated with AD pathological signatures, indicating they are orthogonal to each other.
To identify gene groups displaying similar trait correlations, we used an image segmentation method to identify and manually curate 10 SOM territories most strongly associated with AD phenotypes across cell types (Extended Data Fig. 6f). We found that gene groups defining these territories (gene-trait correlation modules M1–M10), are involved in common functional pathways relevant to the pathophysiology of AD (Extended Data Table S4). For example, M7 showed positive pathological correlation in Mic and was enriched in immune/inflammatory pathways and Aβ clearance. M9 was positively correlated with pathology in Oli and enriched in oligodendrocyte differentiation and myelination pathways, possibly reflecting an oligodendrocytic response to myelin loss.
To link genetic risk with coordinated gene activity, we quantified the overrepresentation of GWAS genes38 and identified modules implicating genetic risk factors for AD and for general cognitive function. M6 and M7 overlapped with AD risk factors (including APOE, TREM2, MEF2C, PICALM, and the MHC class II genes HLA-DRB1 and HLA-DRB5), whose expression in Mic positively correlates with measures of AD pathology. In contrast, M3 overlapped with genes that are associated with cognition39 and whose expression negatively correlates with pathology in neurons (Extended Data Fig. 6g, Extended Data Table S4). These observations provide a link between genetic risk factors and Mic and neuronal transcriptional response to pathology, which may partially explain some of the risk conferred by genetic variants.
Identification of AD-associated cellular subpopulations
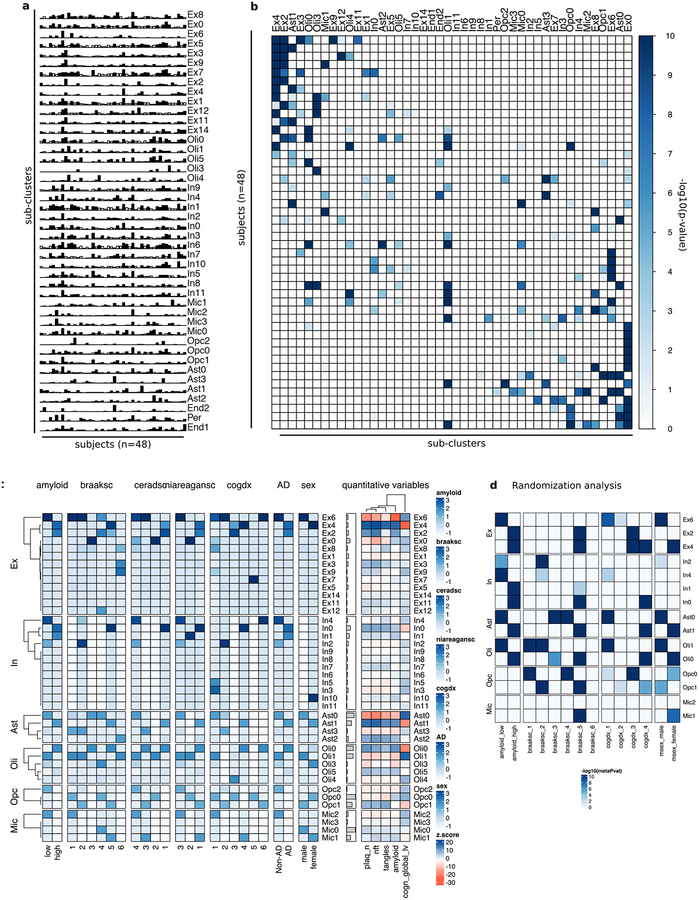
To dissect cell-type heterogeneity, we next sub-clustered each major cell type40, resulting in 13 Ex, 12 In, 4 Ast, 5 Oli, 3 Opc, and 4 Mic subclusters. The identified subpopulations were not exclusively enriched with cells from any single individual (Extended Data Fig. 7a, b). We examined the cellular composition of each subpopulation in relation to the pathological features of source brains (methods). We observed an overrepresentation of cellular subtypes in pathology for most major cell types. For example, cellular subpopulations Ex4, In0, Ast1, Oli0 were associated with cells isolated from subjects with AD pathology traits - i.e., high amyloid, Braak stage (V), CERAD score, NIA-Reagan score, and cognitive decline. By contrast, subpopulations Ex6, In2, Ast0, and Oli1 were associated with cells from subjects with no-pathology (FDR<0.01, hypergeometric test; Fig. 3a–d, Extended Data Fig. 7c, d). These observations were robust to randomization analysis, choosing 3 female and 3 male subjects at random from each pathological category for 100 trials (FDR< 0.01, Extended Data Fig. 7d, Extended Data Table S5). Consistent differences were also reflected in the enrichment of quantitative pathological features (Fig. 3f, FDR<0.01, permutation test). Thus, both categorical assignments and unbiased direct measurements of clinico-pathological variables support a strong association between the uncovered cell-type subpopulations and AD pathological status that is robust to individual selection.
Fig. 3 |. Cellular subpopulation trait associations.
a–d, Overrepresentation (hypergeometric test) within each sub-cluster of cells isolated from individuals with varying (a) amyloid levels, (b) Braak stages, (c) cognitive status at time of death (cogdx, 1=no impairment; 4=impairment), and (d) sex. Only significant associations after FDR correction over all variables and subpopulations are displayed. e, Fraction of cells in each sub-cluster. f, Quantitative clinico-pathological feature enrichment across sub-clusters for: global AD pathology burden (gpath); neurofibrillary tangle burden (nft); neuritic plaque count (plaq_n); overall amyloid levels (amyloid); global cognitive function (global_cogn). Z-score estimated using resampling. Square size proportional to numeric entry. g, Immunohistochemistry with anti-OLIG2 (red) and anti-CRYAB (green) antibodies in white matter of Brodmann area 10 of a no-pathology and an AD-pathology individual (scale bar: 10μm). The experiment was performed once. h, Similar to (g) for anti-OLIG2 (red) and anti-QDPR (green). The experiment was performed once.
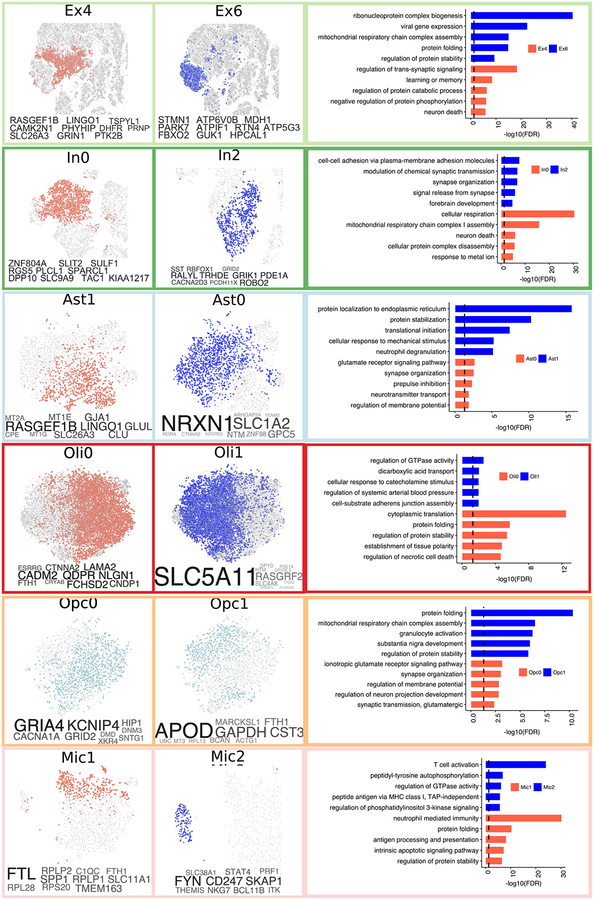
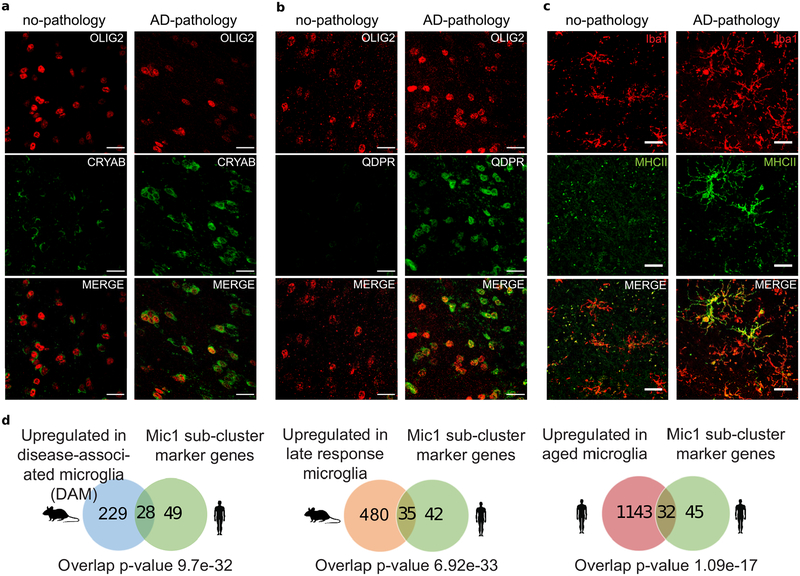
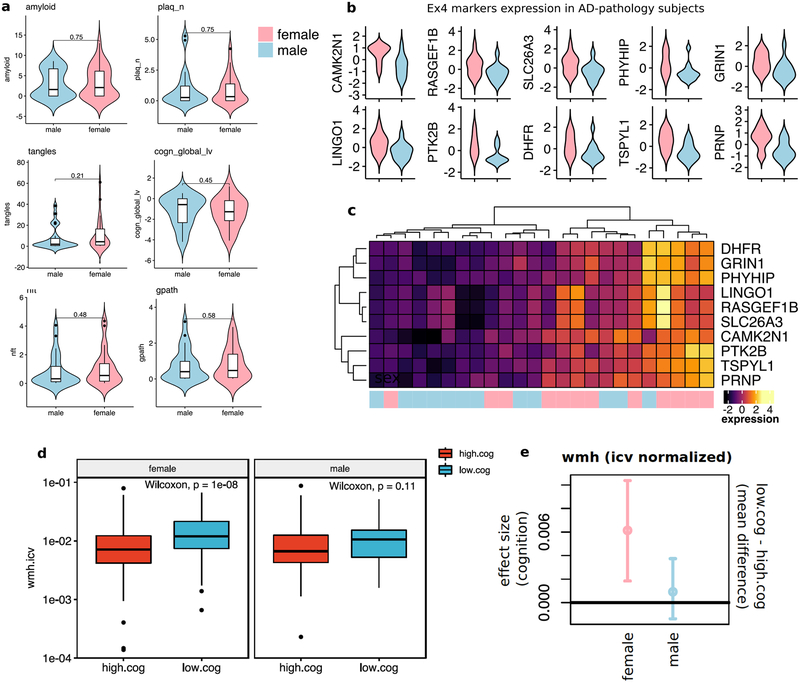
To gain further insight into the molecular processes discriminating these subpopulations, we identified marker genes for each cellular subpopulation (Extended Data Fig. 8 and Table S6, FDR<0.01, 2-sided Wilcoxon-rank-sum test). AD pathology-associated Ex4 neurons were marked by LINGO1, RASGEF1B, and SLC26A3; suggesting that subpopulations preferentially observed in neuropathology may underlie the differences in gene expression observed at the cell-type level. AD pathology-associated Oli0 cells were marked by CADM2, QDPR, NLGN1, and CRYAB -- the latter an anti-apoptotic and neuroprotective chaperone, whose dysfunction could exacerbate inflammation and demyelination41. Immunohistochemistry confirmed that subsets of oligodendrocyte lineage cells (Oli and Opc) express high levels of CRYAB or QDPR in the white matter of AD-pathology individuals (Fig. 3g, h, Extended Data Fig. 9a, b). The AD pathology-associated astrocyte subpopulation Ast1 showed preferential expression of GLUL and of the AD risk gene CLU, which has been shown to be up-regulated in reactive Ast in response to neurodegeneration42. AD pathology-associated In0, Opc1, and Mic1 were respectively marked by genes playing roles in protein folding and stability, neuronal and necrotic death, and T-cell activation and immunity, suggesting cell-type-specific responses to a global cellular stress (Extended Data Fig. 8). Thus, in addition to previously reported roles of neurons and Mic in AD pathophysiology, the disease-associated signatures of Oli and Ast reveal additional glial transcriptional responses to pathology.
Previous studies have profiled microglia from mouse models of AD23,25. We tested whether the expression signatures associated with AD pathology uncovered here in the human brain overlap with the reported states in mouse. We found that AD-associated Mic1 subpopulation marker genes significantly overlap (p ≤ 0.01; one-sided Fisher’s exact test) with mouse disease-associated25 and late-response microglia23 marker genes, including MHC-II genes CD74 and HLA-DRB1 (Extended Data Fig. 9d, Extended Data Table S7). Immunohistochemistry confirmed the presence of a Mic subpopulation expressing high levels of MHC class II in samples from AD-pathology individuals (Extended Data Fig. 9c). The Mic subpopulation identified here in human revealed AD-associated genes not seen in the animal models, including the complement component C1QB and the pattern recognition receptor CD14. Next, we tested to what extent Mic1 markers also overlap with non-AD human aged microglia43. Although we found a significant overlap (p ≤ 0.01; Fisher’s exact test, Extended Data Fig. 9d), many Mic1 marker genes, including APOE, were specific to AD-pathology and not identified in aged microglia. Our observations suggest that the Mic1 subpopulation represents a distinct microglia state that shares features with, but is also distinct from, previously reported microglia cell states in mouse models. However, more extensive single-cell references of glial cells in the human brain will be required to contextualize pathological vs normal heterogeneity.
Sex-specific differential response to AD pathology
We identified robust differences in the association of AD pathology between cells from female versus male individuals (Fig. 3d), with AD-associated cell subpopulations enriched with female cells (Ex4, Ast1, Oli0, and Mic1) and no-pathology subpopulations enriched with male cells (Ex6, Ast0, and Oli1). This overrepresentation was not due to the disproportional cell contribution of particular individuals (Extended Data Fig. 7a, b), nor to more severe AD pathology in female individuals (Extended Data Fig. 10a). Differences between sexes were also reflected in the marker genes of AD-associated subpopulations, which showed expression patterns that partially segregate female and male AD individuals, with higher expression in females (Extended Data Fig. 10b, c). We thus hypothesized that differences might stem from a sex-specific differential transcriptional response to AD pathology.
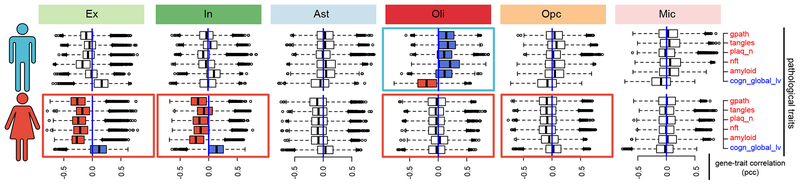
To discern whether female and male individuals present global differential responses to AD pathology, and whether such variability preferentially involves specific cell-types, we recomputed individual-level gene-trait correlations, splitting the dataset by sex (methods). The resulting data enables direct comparison of correlation distributions by cell-type and pathological feature (Fig. 4). Remarkably, we found contrasting, qualitatively distinct responses between sexes involving multiple cell-types (Extended Data Supplementary Table S8). The most extreme differences were observed in neuronal and oligodendrocyte cells. In males, increased pathology correlated with a global transcriptional activation in Oli, as reflected in positive correlation shifts (median correlation, nft=0.203, tangles=0.165, amyloid=0.108, plaques=0.101) (Fig. 4, top). In sharp contrast, shifts in Oli were not observed in females, where instead correlations with pathological traits and with cognition remained centered around zero (median correlation, nft= −0.04, tangles= −0.042, amyloid=0.028, plaques=–0.031) (Fig. 4). Extreme responses were also observed in neuronal cells, in particular in response to amyloid. In females, increased pathology correlated with a global down-regulation (median negative correlation) of gene activity in both Ex (median = −0.272, amyloid) and In (median = −0.225, amyloid). This response is much stronger than that observed in males, where Ex show a qualitatively similar but much less pronounced response (Ex = −0.01, amyloid). In, on the other hand, do not show a clear overall shift in response to pathology or a correlation with cognition in males, except for a slight increase in response to amyloid (median=0.1). We also observed differences in other cell-types, albeit more subtly. In particular, Opc in females present a down-regulation shift in response to pathology not observed in males, as reflected in a shift in directionality of correlation for all pathological variables. Bootstrap estimated median sex-difference estimations supporting these observations are provided in (Extended Data Supplementary Table S8).
Fig. 4 |. Sex-specific differential response to AD pathology.
Individual-level transcriptome-wide gene-trait correlation analysis. Boxplots show the distribution of correlation values (PCC, Pearson correlation coefficient) computed between gene expression profiles (n= 17,926 genes) averaged for cells of each type across each individual and the corresponding pathological measurements across individuals (female n=24, male n=24 individuals). Boxplots are centered around the median, with interquartile ranges defining the box. Pathological traits are represented in red font and cognition in blue. Rectangles around plots highlight the most contrasting differences observed between male and female transcriptional responses, with blue indicating dominant positive correlation and red negative correlation.
To examine a biological role of possible sex bias in relation to white matter pathology, we compared total volume of white matter hyperintensities from MRI data from 505 individuals in the ROSMAP cohort. We found a significant association between the volume of white matter lesions in female subjects and lower cognition (cogn_global_lv), that is not observed in male subjects (Extended Data Fig. 10d, e). These observations are consistent with a scenario of reduced transcriptional response, particularly in Oli, in females with AD pathology. Although these analyses support an underlying sex bias in molecular processes linking white matter changes in AD pathophysiology, further single-cell level analysis of larger sample size and additional experimental follow-up studies are needed to fully understand the relationship between transcriptional and pathological differences between sexes.
Discussion
We report 80,660 single-cell transcriptomes across 48 ROS cohort men and women with a range of AD-associated pathology. Whereas previous studies on AD pathophysiology have focused primarily on neurons and Mic, and considering that bulk RNA is dominated by neuron and Oli expression signal, here we provide pathology-responsive transcriptional signatures across 6 major cell types - Ex, In, Ast, Oli, Opc, Mic, - as well as 40 transcriptionally distinct subpopulations of cells, some of which are preferentially overrepresented in AD pathology and differentially between sexes.
Although most genes presented distinct cell-type-specific perturbations, many top DEGs were involved in related processes across cell types. Myelination related genes were recurrently perturbed not only in Oli and Opc, but in most major cell types, possibly indicating a major regulatory response to maintain proper myelin integrity. Although white matter pathologies have been documented in AD44, single-cell resolution enabled us to identify regulators of myelination, such as LINGO1, which are perturbed across neuronal and glial cells, as well as other myelin-related genes perturbed only in neurons (PRNP, CNTNAP2, ERBB2IP, NEGR1, and BEX1), or only in glial cells (CRYAB). Consideration for cell-type-specific regulatory complexity may be needed in the design of therapeutic interventions, and genes with a more homogeneous response, such as LINGO1, might have more potential for intervention26.
Whereas perturbations in gene expression were largely cell-type-specific at an early stage of pathology, genes up-regulated in late-stage pathology tended to be common across cell types and were associated with a global stress response. Previous studies at the bulk tissue level have implicated the downregulation of aggregation-prone proteins as a mechanism to mitigate compromised protein homeostasis in AD45. The global cell-agnostic upregulation of proteostasis pathways observed herein may similarly constitute an intrinsic adaptive response in an attempt to balance impaired protein homeostasis in late-stage pathology. Alternatively, it might reflect a disruption of the base level functioning of the proteostasis network, a factor known to contribute to disease progression34–36.
We found that transcriptional alterations seem to stem from cell state changes, with certain cell-type subpopulations more readily captured in AD pathology. Observed alterations are consistent with a scenario where existing subtypes of the normal brain are preferentially responsive to pathology, overexpressing a set of responsive genes in addition to constitutive markers of cell subtypes. However, changes in the relative abundance of subtypes as a consequence of differential vulnerability to pathology is an alternative explanation that we currently cannot rule out. Intriguingly, we observed that cells isolated from female individuals were overrepresented in many of the AD pathology-associated cell subpopulations, with contrasting and qualitatively distinct responses between sexes, particularly in neurons and oligodendrocytes. Our data highlight myelination-related processes in AD pathogenesis, and provides support for a sexual dimorphism in AD that manifests at the transcriptional level, even for individuals matched for age and pathology. The transcriptional sex differences may be interpreted as a greater transcriptional disease burden in female individuals. Alternatively, our findings may be viewed as indicating greater resilience in the female individuals in our study, that is, despite strong alterations at the transcriptional level, females match with males in the degree of pathophysiological and cognitive decline.
Overall, our single-cell resolution analysis highlights the complexity of glial-neuronal interactions in response to AD pathology. Additional future applications of our data include the need to distinguish between neuroprotection versus pathogenicity and the responsive versus driving nature of the transcriptional alterations observed. The mechanistic basis of the observed changes during the course of AD progression remains unknown.
METHODS
Selection of individuals from the Religious Order Study (ROS) cohort.
We selected a total of 48 individuals from the Religious Order Study (ROS) cohort Religious Order Study (ROSMAP), a longitudinal cohort of aging and dementia in elderly nuns, priests, and brothers. The cohort includes rich clinical data collected annually, detailed post-mortem pathological evaluations, and extensive genetic, epigenomic, transcriptomic, proteomic, and metabolomic bulk tissue profiling20. For analyses in this paper, we took into account a total of 12 clinical, cognitive, and pathological hallmarks of AD collected through ROS (Extended Data Table S3; ROS clinico-pathological variables in Extended Data Text S1). We selected 24 control individuals with no or very little pathology (no-pathology) and 24 age-matched individuals with a spectrum of mild to severe Aβ and other pathologies (AD-pathology) (Extended Data Table S1). The no-pathology group includes some individuals with clinical cognitive impairment as we selected study participants solely based on pathology and let all other variables freely associate. Details of the clinical and pathologic data collection methods have been previously reported46–50. Individuals were balanced between sexes (12 each), and matched for age (median: 86.7 AD-pathology, 87.1 no-pathology) and years of education (median: 19.5 AD-pathology, 18 no-pathology). Informed consent was obtained from each subject and the Religious Orders Study and Rush Memory and Aging Project were approved by an IRB of Rush University Medical Center.
Isolation of nuclei from frozen postmortem brain tissue.
The protocol for the isolation of nuclei from frozen postmortem brain tissue was adapted from Swiech et al. (2014)51. All procedures were carried out on ice or at 4 °C. Briefly, postmortem brain tissue was homogenized in 2 ml Homogenization Buffer (320 mM Sucrose, 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris-HCl pH 7.8, 0.1 mM EDTA pH 8.0, 0.1% IGEPAL CA-630, 1 mM β-mercaptoethanol, 0.4 U/microliter Recombinant RNase Inhibitor (Clontech) using a Wheaton Dounce Tissue Grinder (10 strokes with the loose pestle). 3 ml of Homogenization Buffer was added (final volume 5 ml) and the homogenized tissue was incubated on ice for 5 minutes. Then the homogenized tissue was filtered through a through 40 mm cell strainer, mixed with an equal volume of Working Solution (83% OptiPrep Density Gradient Medium (Sigma-Aldrich), 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris HCl pH 7.8, 0.1 mM EDTA pH 8.0, 1 mM β-mercaptoethanol) and loaded on top of an OptiPrep density gradient (10 ml 29% OptiPrep solution (29% OptiPrep Density Gradient Medium, 134 mM Sucrose, 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris-HCl pH 7.8, 0.1 mM EDTA pH 8.0, 1 mM β-mercaptoethanol, 0.04% IGEPAL CA-630, 0.17 U/microliter Recombinant RNase Inhibitor) on top of 5 ml 35% OptiPrep solution (35% OptiPrep™ Density Gradient Medium, 96 mM Sucrose, 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris-HCl pH 7.8, 0.1 mM EDTA pH 8.0, 1 mM β-mercaptoethanol, 0.03% IGEPAL CA-630, 0.12 U/microliter Recombinant RNase Inhibitor)). The nuclei were separated by ultracentrifugation using an SW32 rotor (20 minutes, 9000 rpm, 4 °C). 3 ml of nuclei were collected from the 29%/35% interphase and washed with 30 ml of PBS containing 0.04% BSA. The nuclei were centrifuged at 300 g for 3 minutes (4 °C) and washed with 20 ml of PBS containing 0.04% BSA. Then the nuclei were centrifuged at 300 g for 3 minutes (4 °C) and re-suspended in 500 microliter PBS containing 0.04% BSA. The nuclei were counted and diluted to a concentration of 1000 nuclei per microliter in PBS containing 0.04% BSA.
Droplet-based single-nucleus RNA-Seq.
For droplet-based single-nucleus RNA sequencing (snRNA-seq), libraries were prepared using the Chromium Single Cell 3’ Reagent Kits v2 according to the manufacturer’s protocol (10 × Genomics). The generated scRNA-seq libraries were sequenced using NextSeq 500/550 High Output v2 kits (150 cycles).
Analysis of droplet-based single-nucleus RNA-sequencing data.
Gene counts were obtained by aligning reads to the hg38 genome (GRCh38.p5 (NCBI:GCA_000001405.20) using CellRanger software (2.0.0 version) (10× Genomics). To account for unspliced nuclear transcripts, reads mapping to pre-mRNA were counted. After quantification of pre-mRNA using the CellRanger count pipeline on each of the 48 individual libraries, the CellRanger aggr pipeline was employed to aggregate all libraries to equalize the read depth between libraries before data merging (with the default parameter) to generate a gene count matrix. Then, a UMI cutoff of200 was used to select single cells for further analysis. From our pilot sample analysis, we realized that the default cell detection method employed by 10× Genomics (assuming UMI values - a reflection of the RNA content - varies by roughly an order of magnitude among cells) failed to detect a large fraction of the microglia population. Therefore, to determine a more appropriate UMI cutoff value, we plotted a histogram showing cell density as a function of UMI values. Based on this analysis we determined 200 UMIs as the lower cutoff for cell filtering. This resulted in an initial dataset that was then further examined to exclude low quality libraries (see Quality control for cell inclusion).
External Data sources.
Markers of human cortical layers were obtained from He et al.21. Marker genes of cell-type clusters were obtained from Lake et al.18. Disease-associated microglia cell state signatures where obtained from Mathys et al.23 and Keren-Shaul et al.25.
Quality control for cell inclusion.
The initial dataset contained 80,660 cells with a median value of 1,496 counts. As initial reference, the entire dataset was projected to the 2D space using t-distributed stochastic neighbor embedding (t-SNE) on the top 10 principal components. The tSNE coordinates were used to visualize potential biases in apparent cell similarity due to differential cell quality. For each cell, the following quality measures were quantified: (i) the number of genes for which at least one read was mapped, which is indicative of library complexity, (ii) the total number of counts, (iii) the percentage of counts mapping to the top 50 genes, and (iv) the percentage of reads mapped to mitochondrial genes, which may be used to approximate the relative amount of endogenous RNA, and it is commonly used as a measure of cell quality. Cells with a high ratio of Mitochondrial (MT) relative to endogenous RNAs had low starting amounts of RNA, which might indicate that source cells were dead or stressed, resulting RNA degradation. Outlier cells in these quality metrics were found to cluster together in the tSNE 2D space. Based on these observations and subsequent scatter plot analyses, cells with less than 200 detected genes, and cells with abnormally high ratio of counts mapping to MT genes, relative to the total number of detected genes, were removed. Specifically, given a highly skewed empirical distribution of the MT ratio values (i.e., having an elbow shape clearly separating high and low scores) outlier cells were classified in two groups using the k-means clustering algorithm (k=2) on the MT ratio, and subsequently removed. Only counts associated with protein-coding genes were considered; mitochondrially encoded genes, and genes detected in less than 2 cells were excluded. After applying these filtering steps, the dataset included 17,926 genes profiled in 75,060 nuclei.
Cell clustering.
All 75,060 cells were combined into a single dataset. Normalization and clustering were done with the scanpy package52. Briefly, counts for all nuclei were scaled by the total library size multiplied by 10,000, and transformed to log space. 3,188 highly variable genes were identified based on dispersion and mean, the technical influence of the total number of counts was regressed out, and the values rescaled. These preprocessing steps were performed by sequentially using the functions normalize_per_cell, filter_genes_dispersion, log1p.regress_out, and scale in scanpy. PCA was performed over the variable genes, and tSNE was run over the top 10 PCs using the MulticoreTSNE package by Ulanov (2017). The top 50 PCs were used to build a k-nearest-neighbors cell-cell graph with k=30 neighbors, subsequently spectral decomposition over the graph was performed with 15 components, and the Louvain graph clustering algorithm was applied to identify cell clusters. These analyses were performed using the functions pca, neighbors, and louvain in scanpy. We confirmed that a number of PCs greater that 30 captures 100% of the variance of the data. The initial pre-clustering analysis resulted in 20 pre-clusters with a median number of 2,990 cells, ranging from 413 to 15,900 cells, after excluding 2 pre-clusters of 360 and 791 cells that reflected low-quality cells (these showed mixed cell-type markers, extreme complexity with many more genes expressed than other cells, either too many or too few reads, and in one case cells isolated almost exclusively from one individual). For each pre-cluster, differentially expressed genes were detected using the variance adjusted t-test as implemented in the function rank_genes_groups in scanpy. The top 500 ranking genes were extracted for each cluster, and used to test for overlap with markers as previously reported18. The same clustering protocol was used for both pre- and sub-clustering analyses. During sub-clustering, additional potentially spurious clusters representing low-quality or doublet cells were detected on the basis of extreme separation from the rest of sub-clusters from the same cell-type. From these, those having distinctly high number of total counts and mixed expression of markers from different cell-types were tagged as potential doublets and not considered for downstream analyses, resulting in a total of 70634 cells.
Cell type annotation and sub-clustering.
For each pre-cluster, we assigned a cell-type label using statistical enrichment for sets of marker genes18,21, and manual evaluation of gene expression for small sets of known marker genes. Enrichment was statistically assessed using the hypergeometric distribution (Fisher’s exact test) and FDR correction over all gene sets and pre-clusters. Broad cell-type clusters were defined by grouping together all pre-clusters corresponding to the same cell type. Sub-clustering analysis was performed independently over each broad cell-type cluster.
Sub-cluster trait associations.
Cell sub-clusters were annotated for phenotypic traits and AD pathology status by assessing the overrepresentation within the sub-cluster of cells isolated from individuals annotated with the corresponding trait. For categorical phenotypic variables, enrichment was evaluated using the hypergeometric distribution (Fisher’s exact test) and FDR correction over all gene sets and sub-clusters. Enrichment or depletion of quantitative pathological variables was assessed individually by contrasting the average observed value across the cells of a given sub-cluster with a corresponding null distribution estimated by randomly resampling sub-cluster label assignments and computing an average score 10,000 times. The deviation of the observed value from the random expected distribution was quantified using a z-score statistic. Statistical overrepresentation analysis for cells isolated from each individual across the sub-clusters (Fisher’s exact test) corroborated that individual sub-clusters are not exclusively composed of cells from one or a few subjects (Extended Data Fig. 7b). Robustness of sub-cluster trait associations was assessed using a randomization study computing enrichment analysis over cells from 3 female and 3 male subjects randomly chosen from each pathological category (AD-pathology, no-pathology) for 100 trials. Aggregate p-value calculations were conducted by computing a meta-pvalue using the R package metap (Extended Data Fig. 7d).
Phenotypic analysis.
Postmortem scores on AD-related pathologic indices for amyloid, Braak staging, CERAD score, cognitive diagnosis, global cognitive function, global AD pathology burden, neurofibrillary tangle burden, NIA-Reagan score and diffuse and neuritic plaque burden, and neuronal neurofibrillary tangle density, were used to quantitatively describe the pathological phenotype of each individual (Extended Data Text S1). The quantitative values tangles, ntf, gpath (aggregate of neuritic plaque, diffuse plaque, and neurofibrillary tangle scores), plaq_n, amyloid, and cogn_global_Iv were selected as the primary markers for quantifying AD progression. To identify AD pathological groups, distance-based kmeans clustering was applied to the quantitative pathological features using k=3. The number of clusters was selected by testing multiple increasing values of k, and taking the value at which the within-cluster sum of square distance drops.
Marker identification.
For sub-clusters a set of markers (specifically overexpressed) genes was defined by differential expression analysis of the cells grouped in each sub-cluster against the remaining cells within the corresponding broad cell-type cluster. This analysis was applied to all cell types independently. Significantly over-expressed genes were defined based on the Wilcoxon-rank-sum test with a FDR corrected p-value <=0.01 and a logFC of 0.5. Only genes detected in at least 25% of the cell within the given subcluster were considered.
Gene Ontology (GO) enrichment analyses were performed using Metascape53–55 and the p-value ranked gene lists as input. For all cases we used the set of 17,926 protein-coding genes included in the QC data as background.
Gene differential expression analysis.
snRNAseq-based differential expression analysis was assessed using two tests. First, a cell-level analysis was performed using the Wilcoxon-rank-sum test and FDR multiple testing correction. Second, a Poisson mixed model accounting for the individual of origin for nuclei and for unwanted sources of variability was performed using the R packages lme4 and RUV-seq, respectively. Consistency of DEGs detected using the cell-level analysis model with those obtained with the Poisson mixed model was assessed by comparing DEGs directionality and rank for in the two models. Consistency in directionality for all cell-types was measured by counting what fraction of the top 1000 DEGs (ranked by FDR scores) detected in cell-level analysis show consistent direction in the mixed model. High consistency was found, with a median fraction of 0.99 (Extended Data Table S2). Global consistency between the two models was assessed statistically using a resampling test. We tested whether the differential p-value and z-score ranks corresponding to genes detected as up-regulated or down-regulated in the cell-level analysis was significantly higher or lower than expected by chance when computed using the mixed model. Expected scores were estimated by randomly sampling same-sized gene sets (n=1000, replicates). Both significance rank and direction deviate significantly from expectation, with directionality consistent for up- or down-regulated genes. Results of consistency tests are included in (Extended Data Table S2). Results from both cell-level and mixed-model differential tests are reported in Extended Data Table S2. For analyses involving DEG counts, only genes that are significantly supported by both models using the criteria FDR-corrected P < 0.01 in two sided Wilcoxon-rank-sum test, absolute Log-fold > 0.25, and FDR-corrected P < 0.05 in Poisson mixed model are considered. Per and End populations were excluded from differential analyses due to their small cell counts.
Bulk RNA-seq differential analyses was performed by fitting a linear model using the R package limma54, while accounting for the covariates age, RNA integrity number, post-mortem interval, and plate batches. The pathological definition of AD groups as provided by ROSMAP was used. This classification defines AD or Non-AD groups based exclusively on the overall pathological burden, without considering clinical diagnosis. We used the labels low- AD-pathology groups for consistency.
Consistency of gene expression perturbations in the different cell-types observed in snRNA data with those detected in tissue-level bulk was assessed using a resampling approach. To test whether the genes identified as DEGs in single-cell data are also detected as high-ranking in the differential analysis in bulk, a z-score statistic was computed to quantify the deviation of the observed differential (p-value) rank scores obtained in the bulk analysis for the genes detected as DEGs in single-cells, relative to those observed in 1000 randomly chosen gene sets (Extended Data Table S2). This analysis was performed for each cell-type independently.
Correlation analysis of gene expression and AD-related neuropathological traits using self-organizing maps (SOM).
First, for each major cell-type, a gene-wise correlation coefficient (Spearman’s rank correlation coefficient) was computed using gene expression and AD-related neuropathological trait values across all the annotated cells as variables. The AD-related neuropathological traits included in this analysis were cogn_global_lv (global cognitive function - last valid score), age_death (age at death), educ (years of education), msex (self-reported sex), parksc_lv (global parkinsonian summary score - last valid score), gpath (global AD pathology burden), gpath_3neocort (global measure of neocortical pathology), pmi (post-mortem interval), amyloid (Overall amyloid level), plaq_d (diffuse plaque burden), plaq_n (neuritic plaque burden), nft (neurofibrillary tangle burden), and tangles (tangle density). Only significant correlations after Bonferroni correction at P<0.01 were considered. The resulting correlation matrices for each major cell-type were concatenated and analyzed using a computational algorithm (self-organizing map, SOM)37. All SOMs were created using the Kohonen R package56. To identify the territories of the SOM most strongly correlated with AD-related neuropathological traits, we used an image segmentation method and further manual curation to identify territories (gene-trait correlation modules) based on all the individual cell-type-specific SOM plots for each neuropathological trait. Enrichment analysis for Gene Ontology (GO) terms among the genes of a gene-trait correlation module was performed using Metascape53. The robustness of gene-trait associations to single-cell heterogeneity and noise was confirmed examining individual-level correlations for the genes in gene-trait module. Individual-level correlations were computed by first averaging for each individual normalized gene expression profiles across cells of the same cell. This resulted in cell-type-specific averaged gene expression profiles across the 48 individuals. Average profiles were subsequently mean-centered and scaled to finally compute gene-wise correlation coefficients versus corresponding pathological values. Individual-level gen-trait correlations were computed independently for all 48 individuals, for only 24 male individuals, and for only 24 female individuals. The robustness of gene-trait associations to potential confounding variables was corroborated by confirming the cell-type specific recovery of identified gene-trait modules using partial correlation. Briefly, the partial Pearson’s correlation coefficient between average gene expression and each pathological trait, after correcting for the effect of PMI, age, gender, and education level of each individual; was computed by first orthogonalizing the normalized expression with respect to the normalized covariates and then computing the correlation in the residual subspace.
Immunohistochemistry.
Fixed human brain tissue (prefrontal cortex, BA10) was sectioned at 50 μm using a vibratome (Leica). To retrieve the antigens, sections were incubated at 95°C in IHC Antigen Retrieval Solution (ThermoFisher Scientific; catalog number 00-4955-58) containing 0.05% Tween-20 for 45 minutes and then placed in PBS for 20 minutes at room temperature. After washing with PBS (three times 10 minutes), the brain sections were incubated in quenching solution (50mM ammonium acetate, 100mM CuSO4) at room temperature overnight. After washing with ddH20 (one wash for 15 minutes) and PBS (three times 15 minutes), the sections were permeabilized in PBS containing 0.3% Triton X-100 for 10 minutes and blocked in PBS containing 0.3% Triton X-100 and 5% normal donkey serum at room temperature for 1 h. The sections were incubated overnight (anti-β-Amyloid (D54D2) antibody) or for 48 hours (anti-Iba1 and anti-Human HLA-DP, DQ, DR antibodies) at 4°C in primary antibody in PBS with 0.3% Triton X-100 and 5% normal donkey serum. Primary antibodies were anti-Iba1 (1:500; Synaptic Systems; Cat. No. 234 004, Polyclonal Guinea pig antiserum), anti-Human HLA-DP, DQ, DR antigen, clone CR3/43 (1:100, Agilent, M077501–2), and anti-β-Amyloid (D54D2) (1:500, Cell Signaling Technology, #8243). The sections were washed with PBS containing 0.1% Triton X-100 at room temperature (four times 15 minutes), and then incubated with secondary antibodies (dilution 1:2000) overnight at 4°C. Primary antibodies were visualized with Alexa-Fluor 488, Alexa-Fluor 594, and Alexa-Fluor 647 antibodies (Molecular Probes), and cell nuclei visualized with Hoechst 33342 (Sigma-Aldrich; 94403). The sections were washed with PBS containing 0.1% Triton X-100 at room temperature (four times 15 minutes) and then mounted on Fisherbrand Superfrost Plus Microscope Slides in ProLong Gold Antifade Mountant. Images were acquired using a confocal microscope (LSM 880; Zeiss) with a 20× or 63× objective.
For the Tyramide Signal Amplification (TSA) labelling, sections were incubated at 95°C in IHC Antigen Retrieval Solution (ThermoFisher Scientific; catalog number 00-4955-58) containing 0.05% Tween-20 for 45 minutes and then placed in PBS for 20 minutes at room temperature. After washing with PBS (three times 10 minutes), the brain sections were incubated in quenching solution (50mM ammonium acetate, 100mM CuSO4) at room temperature overnight. After washing with ddH20 (one wash for 15 minutes) and PBS (three times 15 minutes), endogenous peroxidases were quenched with 0.3% H2O2 for 20 minutes at RT followed by washing with PBS (three times 10 minutes). Then the sections were blocked in TNB Blocking Buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, 0.5% TSA Blocking Reagent (PerkinElmer, catalog number FP1020)) for 30 minutes at room temperature. Primary antibody incubation was performed for 48 hours at 4°C in TNB Blocking Buffer. The sections were washed with PBS containing 0.3% Triton X-100 at room temperature (four times 15 minutes) and then incubated with HRP-labeled secondary antibody (1:1500) in TNB Blocking Buffer for 30 minutes at room temperature. Then the sections were washed with PBS containing 0.3% Triton X-100 at room temperature (four times 15 minutes) and the fluorophore reaction was performed using the TSA Plus Cyanine 5 and Fluorescein System (PerkinElmer, catalog number NEL754001KT) for 3 minutes at room temperature. Then the sections were washed with PBS containing 0.3% Triton X at room temperature (four times 15 minutes) and incubated at 95°C in IHC Antigen Retrieval Solution (ThermoFisher Scientific; catalog number 00-4955-58) containing 0.05% Tween-20 for 45 minutes. After washing with PBS containing 0.3% Triton X-100 at room temperature (four times 15 minutes) the sections were either mounted or blocked in TNB Blocking Buffer for another round of TSA labeling. The following primary antibodies were used: anti-OLIG2 (1:1000, Atlas, HPA003254, rabbit polyclonal), anti-CRYAB (1:200; LSBio; LS-B3696, rabbit polyclonal)), and anti-QDPR (1:2500, Atlas Antibodies, HPA065649, rabbit polyclonal). The following secondary antibody was used: anti-Rabbit IgG (goat), HRP-labeled (PerkinElmer, NEF812001EA). Images were acquired using a confocal microscope (LSM 880; Zeiss) with a 20× or 63× objective.
RNA in situ hybridization.
Frozen brain tissue was embedded in Tissue-Tek® O.C.T. Compound (VWR, catalog number: 25608–930), cryosectioned to 16 μm thickness, and placed onto Fisherbrand™ Superfrost™ Plus Microscope Slides (ThermoFisher Scientific, catalog number 12-550-15). The RNAscope® 2.5 HD Duplex Detection Kit (Chromogenic) was used according to the manufacturer’s instructions, with minor modifications to tissue preparation. Briefly, sections frozen at −80°C were washed with 1× PBS, baked at 60°C for 15 min and fixed in 10% neutral buffered formalin (NBF) for 90 min at room temperature. Following fixation, the tissue was dehydrated using 50%, 70% and 100% ethanol and then baked at 60°C for 30min to avoid detachment. Tissues were then treated with H2O2 for 10min, followed by Protease IV for 30min. Brain sections were hybridized with two mRNA probes: SLC17A7 (RNAscope® Probe - Hs-SLC17A7-C2, ACD, catalog number 415611-C2) and NTNG1 (RNAscope® Probe - Hs-NTNG1, ACD, catalog number 446101). Probes in channel 1 (NTNG1) were labeled with HRP enzyme and visualized with a green substrate and probes in channel 2 (SLC17A7) were labeled with AP enzyme and a red substrate. Hematoxylin was used to mark cell nuclei in blue. Slides were then imaged using a Leica Dmi8 microscope to take 40× bright field images. For the quantification a blinded researcher manually counted the cells labeled with red substrate (SLC17A7) and determined how many of these cells are co-labeled with green substrate (NTNG1). The quantification is based on the analysis of BA10 tissue sections from four low amyloid individuals and four high amyloid individuals, with five to six images per individual.
Isolation of neuronal nuclei, cDNA synthesis and RT-qPCR analysis.
Postmortem brain tissue was homogenized in 2 ml NF1 Buffer (0.5% Triton X-100, 0.1M Sucrose, 5mM MgCl2, 1mM EDTA, 10mM Tris-HCl, pH 8.0, 1 mM β-mercaptoethanol, 0.4 U/microliter Recombinant RNase Inhibitor (Clontech)) using a Wheaton Dounce Tissue Grinder (15 strokes with the loose pestle). After homogenization, an additional 8 ml of NF1 Buffer was added and the samples were inverted 10 times to mix. The homogenate was passed through a 40μm cell strainer (VWR, catalog number 21008–949) and centrifuged at 300 g for 3 minutes at 4°C. Then the supernatant was removed and the nuclei were incubated with Alexa Fluor®488 conjugated anti-NeuN antibody (MilliporeSigma, catalog number MAB377X) in PBS containing 1% BSA for 15 minutes at 4°C on a shaker. The nuclei were washed by adding 35 ml PBS containing 1% BSA, spun down at 300 g for 3 minutes at 4°C, re-suspended in 0.5 ml PBS containing 0.04% BSA and 0.4 U/microliter Recombinant RNase Inhibitor (Clontech), and stained with NucBlue Live ReadyProbes™ Reagent (ThermoFisher Scientific, catalog number R37605). NeuN-positive nuclei were then directly sorted into RNA lysis buffer (Qiagen, 74134) using Fluorescence-activated cell sorting. Total RNA was extracted using the RNeasy Plus Mini kit (Qiagen, 74134) according to manufacturer’s instruction and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, 170–8891). For gene expression analysis cDNA was quantitatively amplified on a thermal cycler (BioRad) using SsoFast EvaGreen Supermix (BioRad, 1725202) and gene-specific primers (DHFR forward, CTGGACTATGTTCCGCCCAC; DHFR reverse, CTGGACTATGTTCCGCCCAC; STMN1 forward, ATACACTGCC TGTCGCTTGT; STMN1 reverse, CTTTTGACCGAGGGCTGAGA; MTRNR2L8 forward, ATTGACCTGCCCGTGAAGAG; MTRNR2L8 reverse, AGGGCCTG TGGACTTGTTAAG; LINGO1 forward, ACCGCATCAAAACGCTCAAC; LINGO1 reverse, CTAGCGGGATGAGCTTCAGG; NGFRAP1 forward, ATGGGCCATACCCAATAGGC; NGFRAP1 reverse, AGAGAGCTCC CCCATAAGGA; BEX1 forward, CGTCACTCGTGTCTCGCTAC; BEX1 reverse, CCATTACTCCTGGGCCTATCC; RPL13 forward, CCTTTCCGCT CGGCTGTTTT; RPL13 reverse, GGCCTTACGTCTGCGGATCT). The comparative Ct method was used to examine differences in gene expression and values were normalized to expression levels of RPL13.
Statistical analysis of white matter data from ROSMAP cohort studies.
Measures of white matter hyperintensities (WMH) were obtained through in-vivo brain MRI data collected through the ROSMAP cohort studies. Participants undergo MRI in the RADC cohort studies and substudies biennially on 3T scanners including the T1-weighted 3D Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) and T2-weighted 2D Fluid-Attenuated Inversion Recovery (FLAIR) sequences. Raw MPRAGE images were processed to generate total volumes including gray matter, white matter, CSF and intracranial volumes using SPM57. White matter lesions appearing hyperintense in T2-weighted images were segmented based on FLAIR and MPRAGE data using BIANCA58. A mask of white matter hyperintensities was generated and the total volume of hyperintensities calculated. Cognitive scores were taken from the recorded variable cogn_global_lv (last visit) (Extended Data Text S1), which measures averaged z-scored values from a battery of 19 cognitive tests, yielding a global cognitive function summary. A negative z-score simply means that someone has an overall score that is lower than the average of the entire cohort at baseline. High- and low-cog groups were defined based on weather a subject has an overall score lower (low-cog, z-scoe<0) or larger (high-cog, z-scoe>0) than the average. WMH values were compared between low- and high-cog groups within male and female subjects using the using the Wilcoxon-rank-sum test (Extended Data Fig. 10d). Statistical estimation of significant difference in WMH between low-cog and high-cog groups in females, and between low-cog and high-cog groups in males was assessed by bootstrap point and 95% confidence interval estimation of the effect size (mean difference) between groups, correcting for sample size (resampling n=40 observations per group 1000 times) (Extended Data Fig. 10e).
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The single-nucleus RNA-Sequencing data is available at Synapse (https://www.synapse.org/#!Synapse:syn18485175). The DOI for this dataset is: 10.7303/syn18485175. The DOI for the ROSMAP metadata is: 10.7303/syn3157322.
Extended Data
Extended Data Fig. 1 |. Pathological status verification physical integrity of isolated neuronal nuclei.
a, Immunohistochemistry with anti-amyloidbeta (D54D2, yellow) antibody in the grey matter of Brodmann area 10 of no-pathology and pathology individuals. b, Quantification of the amyloid-beta immunostaining shown in (a). Error bars show the standard error of the mean, asterisks denote statistical significance as assessed by Student’s two-tailed t-test (*, P-Value < 0.05, exact P value 0.030). c, High resolution confocal microscopy images of neuronal nuclei isolated from no-pathology and pathology individuals and stained with Hoechst. The experiment was performed once.
Extended Data Fig. 2 |. Single-nucleus RNA-seq profiling and cell type characterization.
a, Study cohort and sample preparation. b, Clustering analysis workflow. c, 2-dimensional t-SNE projection of all annotated cells (n=75,060 from 24 pathology and 24 no-pathology individuals). d, Correlation matrix (Pearson correlation coefficient) of the average expression profiles by cell-type for each individual.
Extended Data Fig. 3 |. Consistency of cells of the same type across individuals.
a, Known cell-type marker gene expression for each cell-type. b, Expression of known cell-type marker genes in each cluster (left) and fraction of cells in each cluster expressing each marker gene (right). Vertical blue line represents a scale bar referencing 0.5. c, over-representation analysis (hypergeometric test) within each of the pre-cluster marker sets (rows) of genes identified as markers in Lake et al. 2018 (n=1729 total genes, columns, left) and genes identified as markers of cortical layers in He et al. 2017 (n=3400 total genes, columns, right). d, fraction of cells of each type isolated across all (n=48, left), no-pathology (n=24, center), and AD-pathology (n=24, right) individuals. e, fraction of cells of each type isolated from each individual (n=48, columns).
Extended Data Fig. 4 |. Expression values and validation of top DEGs.
a, mean expression values of genes across the nuclei isolated from each individual. Each point represents one individual. DEGs were classified into low, mid, or highly expressed based on their median expression level across the cells of the corresponding cell-type. Groups were defined based on k-means clustering (k=3). Top 3 genes for each group and cell-types are shown. For Opc and Mic only 1 and 2 genes were classified within the high expression group. Box Plots are centered around the median, with interquartile ranges defining the box. b, RNA in-situ hybridization (RNAscope) with probes detecting excitatory neuron marker SLC17A7 (red) and NTNG1 (blue) in the grey matter of Brodmann area 10 (BA10) of a no-pathology and a pathology individual (left). The tissue was counterstained with hematoxylin. Right, quantification of RNA in situ hybridization on BA10 tissue sections (mean ± s.e.m.; *, P-Value < 0.05, Student’s two-tailed t-test, exact P value = 0.047; n=4 no-pathology and n=4 pathology individuals; n= 5, 6 images per individual).
Extended Data Fig. 5 |. Overlap of genes altered in AD pathology and progression.
Quantification of the overlap (jaccard coefficient) between pairs of gene sets identified as differentially expressed (DE) in each of the major cell-types when comparing cells isolated from AD pathology individuals with cells isolated from no-pathology, and combinations of early and late pathology.
Extended Data Fig. 6 |. Cell-type-specific and phenotype-specific gene-trait correlation analysis.
a–e, Self-organizing map (SOM) generated from transcriptome-wide gene expression correlation of each gene with neuropathological signatures of AD (a: cogn_global_lv; b: amyloid; c: nft d, pmi; e, age_death). Genes with similar correlation patterns are mapped to the same SOM unit and similar units group close together. SOM grid layout is common and built jointly across all phenotypes and all cell types (including Fig. 4). Color: Average Spearman’s rank correlation for genes in each unit. f, Selected SOM territories (M1–M10). g, Overlap (one-sided Fisher’s exact test) between gene-trait correlation module genes (M3 n = 1472, M6 n = 70, M7 n = 80 genes) and AD GWAS risk genes (top) (n = 28 genes), as well as genes associated with general cognitive function (bottom) (n = 709 genes). The p-values have been adjusted for multiple hypothesis testing, -log10(Bonferroni corrected p-values) are shown.
Extended Data Fig. 7 |. Sub-cluster cell over-representation analyses.
a, Cell composition of each identified subclusters (rows) across individuals (columns). Bars represent the fraction of cells corresponding to each individual. Bar color indicates whether the corresponding value exceeds (black) or not (white) the average value measured across all the entries in the row. b, Over-representation analysis (hypergeometric test) within each pre-cluster (columns) of cells isolated from each individual (rows). c, Over-representation analysis within each sub-cluster of cells isolated from individuals with different values of discrete clinico-pathological variables (overall amyloid level [amyloid], Braak stage [braaksc], CERAD score [ceradsc], NIA-Reagan score [niareagansc], clinical consensus diagnosis of cognitive status at the time of death [cogdx], sex). The scale bars on the right indicate the significance of the over-representation (hypergeometric test, −log10(p-value), z-scaled, FDR multiple testing correction). For quantitative variables enrichment was computed based on an estimated z-score quantifying the deviation from random expected values using resampling (see methods). The quantitative variables considered are neuritic plaque count (plaq_n), neurofibrillary tangle burden (nft), tangle density (tangles), overall amyloid level (amyloid), and global cognitive function (cogn_global_lv, last valid score). For a detailed description of clinico-pathological variables see Extended Data Text S1. d, Over-representation analysis (hypergeometric test) similar to (a) but computed across only cells isolated from randomly chosen female and male individuals for pathology and no-pathology groups (see methods). Scores represent aggregated p-values (meta-p values, meanp method, metap R package) computed across 100 random realizations. Only scores passing a FDR<0.01 (correction across traits × subpopulations) are plotted.
Extended Data Fig. 8 |. Cell-type subpopulations.
Cells from sub-clusters enriched (red) or depleted (blue) with cells for individuals with AD pathology and cognitive decline shown using t-SNE for major cell types (Ast1 n=1134, Ast0 n=1728, Oli0 n=8310, Oli1 n=8032, Ex4 n=3198, Ex6 n=2757, In0 n=2368, In2 n=984, Opc0 n= 1,589, Opc1 n= 976, Mic1 n=509, Mic2 n=169 cells), corresponding marker genes (font proportional to enrichment level) (left), and enriched gene ontology terms (right). GO enrichment based on FDR-corrected cumulative hypergeometric P-values, with P-value ranked gene marker lists (FDR<0.01, LFC>0.5, two-sided Wilcoxon-rank-sum test) used as input (Ex4=783, Ex6=2438, In0=1702, In2=350, Ast1=574, Ast0=73, Oli0=227, Oli1= 73, Opc0 n=19, Opc1 n=536, Mic1 n=487, Mic2 n=646).
Extended Data Fig. 9 |. Immunohistochemistry of subpopulation markers in oligodendrocyte lineage cells and microglia.
a, oligodendrocyte lineage cell subpopulation marked by alpha B-crystallin (CRYAB). Immunohistochemistry with anti-OLIG2 (red) and anti-CRYAB (green) antibodies in the white matter of Brodmann area 10 of a no-pathology and a pathology individual (scale bar: 20μm). A selected area of these images is shown in Fig. 3g. The experiment was performed once. b, oligodendrocyte lineage cell subpopulation marked by quinoid dihydropteridine reductase (QDPR). Immunohistochemistry with anti-OLIG2 (red) and anti-QDPR (green) antibodies in the white matter of Brodmann area 10 of a no-pathology and a pathology individual (scale bar: 20μm). A selected area of these images is shown in Fig. 3h. The experiment was performed once. c, Immunohistochemistry with anti-Iba1 (red) and anti-MHC class II (green) antibodies in the white matter of Brodmann area 10 of a no-pathology and a pathology individual (scale bar: 20μm). The experiment was performed once. d, Overlap (one-sided Fisher’s exact test) between Mic1 marker genes and genes upregulated in mouse disease-associated microglia (left), in mouse late response microglia (middle), and in aged human microglia (right).
Extended Data Fig. 10 |. Sex comparisons in pathology, expression and white matter.
a, Quantitative clinico-pathological measurement comparison between male and female individuals (female n=24, male n=24, two-sided Wilcoxon-rank-sum test). Violin plots are centered around the median with interquartile ranges with the shape representing individual distribution. The quantitative clinico-pathological variables considered are overall amyloid level (amyloid), neuritic plaque burden (plaq_n), neurofibrillary tangle burden (nft), tangle density (tangles), global cognitive function (cogn_global_lv, last valid score) and global AD pathology burden (gpath). Violin plots are centered around the median with interquartile ranges with the shape representing individual distribution. b, Violin plots showing aggregate expression levels (z-scaled) across Ex neurons in female (red) vs. male (blue) individuals (n= 12 each) of top 10 marker genes of AD-associated Ex4 subpopulation of excitatory neurons. Violin plots are centered around the median with interquartile ranges with the shape representing individual distribution. c, Hierarchical clustering of pathology-affected individuals (columns) based on average expression level (color) of top 10 marker genes (rows) of AD-enriched Ex4 subpopulation of excitatory neurons for female vs. male individuals. d–e Statistical comparison of in-vivo brain MRI imaging from ROSMAP cohort. d., Intracranial volume-normalized white matter hyperintensities (wmh.icv) measures for female (n=399) and male (n=106) subjects and high- (female n=252, male n=63) and low-cog (female n= 147, male n=43) groups. Groups were defined based on whether subjects have an overall cognition score lower (low-cog, z-scoe<0) or larger (high-cog, z-scoe>0) than the average. Mean rank difference values between cog groups were compared using the two-sided Wilcoxon rank sum test. Boxplots are centered around the median, with interquartile ranges defining the box. e., Statistical estimation of significant difference in WMH between low-cog and high-cog groups in females, and between low-cog and high-cog groups in males, assessed by bootstrap point and 95% confidence interval estimation of the effect size (mean difference) between groups. Bootstrap resampling was performed by resampling n=40 observations per group 1000 times. Horizontal line highlights zero difference. The positive effect size point and confidence interval estimates do not overlap the zero line in the female group, which indicates statistical evidence of an increment of white matter hyperintensities (wmh.icv) in the low.cog group relative to high.cog in females but not in males.
Supplementary Material
Acknowledgements
We thank study participants and staff of the Rush Alzheimer’s Disease Center. We are grateful to Thorvald F. Andreassen for technical assistance. We thank Scarlett J. Barker for discussions and comments. This work was supported in part by the Cure Alzheimer’s Fund (CAF), the JBP Foundation and by NIH grants RF1AG054321, RF1AG062377, RF1AG054012, U01NS110453, R01AG062335, and R01AG058002 to L.-H.T. D.A.B. is supported by grants P30AG10161, R01AG15819, R01AG17917, U01AG46152, R01AG57473. H.M. was supported by an Early Postdoc Mobility fellowship from the Swiss National Science Foundation (P2BSP3_151885). M.K. was supported by NIH grants: U01NS110453, R01AG062335, R01AG058002, R01MH109978, R01HG008155, RF1AG054012, RF1AG062377, and U01MH119509.
Footnotes
Reviewer information Nature thanks Hongjun Song and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's Disclaimer: This is a PDF file of a peer-reviewed paper that has been accepted for publication. Although unedited, the content has been subjected to preliminary formatting. Nature is providing this early version of the typeset paper as a service to our customers. The text and figures will undergo copyediting and a proof review before the paper is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers apply.
Competing interests The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41586-019-1195-2.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-019-1195-2.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Masters CL et al. Alzheimer’s disease. Nat Rev Dis Primers 1, 15056 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hardy J & Selkoe DJ The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Braak H & Braak E Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 4.De Strooper B & Karran E The Cellular Phase of Alzheimer’s Disease. Cell 164, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Canter RG, Penney J & Tsai L-H The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature 539, 187–196 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Heneka MT et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop NA, Lu T & Yankner BA Neural mechanisms of ageing and cognitive decline. Nature 464, 529–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pimenova AA, Raj T & Goate AM Untangling Genetic Risk for Alzheimer’s Disease. Biological Psychiatry 83, 300–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher DW, Bennett DA & Dong H Sexual dimorphism in predisposition to Alzheimer’s disease. Neurobiol. Aging (2018). 10.1016/j.neurobiolaging.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nativio R et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci 21, 497–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossers K et al. Concerted changes in transcripts in the prefrontal cortex precede neuropathology in Alzheimer’s disease. Brain 133, 3699–3723 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Zhang B et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JA, Woltjer RL, Goodenbour JM, Horvath S & Geschwind DH Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 5, 48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gjoneska E et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habib N et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake BB et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong S et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Lake BB et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett DA et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers. Dis 64, S161–S189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci 20, 886–895 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Lin Y-T et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 98, 1294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathys H et al. Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell Rep. 21, 366–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krasemann S et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keren-Shaul H et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Enright F & Andrews J Lingo-1: a novel target in therapy for Alzheimer’s disease? Neural Regeneration Res. 11, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi S et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat. Neurosci 8, 745–751 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Liang C et al. Erbin is required for myelination in regenerated axons after injury. J. Neurosci 32, 15169–15180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott R et al. Loss of Cntnap2 Causes Axonal Excitability Deficits, Developmental Delay in Cortical Myelination, and Abnormal Stereotyped Motor Behavior. Cereb. Cortex (2017). 10.1093/cercor/bhx341. [DOI] [PubMed] [Google Scholar]
- 30.Poplawski GHD et al. Adult rat myelin enhances axonal outgrowth from neural stem cells. Sci. Transl. Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khazaei MR et al. Bex1 is involved in the regeneration of axons after injury. J. Neurochem 115, 910–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seiradake E et al. Structural basis for cell surface patterning through NetrinG-NGL interactions. EMBO J. 30, 4479–4488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holtzman DM, Morris JC & Goate AM Alzheimer’s Disease: The Challenge of the Second Century. Sci. Transl. Med 3, 77sr1–77sr1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade WA et al. Early endosome localization and activity of RasGEF1b, a toll-like receptor-inducible Ras guanine-nucleotide exchange factor. Genes Immun. 11, 447–457 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Balch WE, Morimoto RI, Dillin A & Kelly JW Adapting proteostasis for disease intervention. Science 319, 916–919 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Labbadia J & Morimoto RI The Biology of Proteostasis in Aging and Disease. Annu. Rev. Biochem 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohonen T The self-organizing map. Proc. IEEE 78, 1464–1480 (1990). [Google Scholar]
- 38.Karch CM & Goate AM Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies G et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun 9, 2098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blondel VD, Guillaume J-L, Lambiotte R & Lefebvre E Fast unfolding of communities in large networks. J. Stat. Mech: Theory Exp 2008, P10008 (2008). [Google Scholar]
- 41.Ousman SS et al. Protective and therapeutic role for aB-crystallin in autoimmune demyelination. Nature 448, 474–479 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Shin Y-J et al. Clusterin enhances proliferation of primary astrocytes through extracellular signal-regulated kinase activation. Neuroreport 17, 1871–1875 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Olah M et al. A transcriptomic atlas of aged human microglia. Nat. Commun 9, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caso F et al. White Matter Degeneration in Atypical Alzheimer Disease. Radiology 277, 162–172 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Ciryam P et al. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proceedings of the National Academy of Sciences 113, 4753–4758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett DA et al. Natural history of mild cognitive impairment in older persons. Neurology 59, 198–205 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Bennett DA et al. Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer’s disease. Neurology 60, 246–252 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Bennett DA, Schneider JA, Wilson RS, Bienias JL & Arnold SE Neurofibrillary tangles mediate the association of amyloid load with clinical Alzheimer disease and level of cognitive function. Arch. Neurol 61, 378–384 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Bennett DA et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66, 1837–1844 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Bennett DA et al. Decision Rules Guiding the Clinical Diagnosis of Alzheimer’s Disease in Two Community-Based Cohort Studies Compared to Standard Practice in a Clinic-Based Cohort Study. Neuroepidemiology 27, 169–176 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Swiech L et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol 33, 102–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolde R & Vilo J GOsummaries: an R Package for Visual Functional Annotation of Experimental Data. F1000Res. 4, 574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wehrens R & Buydens LMC Self- and Super-organizing Maps inR: ThekohonenPackage. J. Stat. Softw 21, (2007). [Google Scholar]
- 57.Friston KJ et al. Spatial registration and normalization of images. Hum. Brain Mapp. 3, 165–189 (1995). [Google Scholar]
- 58.Griffanti L et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage 141, 191–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-nucleus RNA-Sequencing data is available at Synapse (https://www.synapse.org/#!Synapse:syn18485175). The DOI for this dataset is: 10.7303/syn18485175. The DOI for the ROSMAP metadata is: 10.7303/syn3157322.