Abstract
The role of long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1 (lncRNA HAND2-AS1) in tumor suppression has been reported in a number of cancer types, while its functionality in triple-negative breast cancer (TNBC) remains unclear. The present study aimed to investigate the involvement of lncRNA HAND2-AS1 in TNBC using different methodologies. HAND2-AS1 and RUNX2 expression was analyzed using reverse transcription-quantitative PCR and western blot analysis. Diagnostic analysis was performed using receiver operating characteristic curve. Overexpression experiments were performed to analyze the interaction between HAND2-AS1 and RUNX2 while the Cell Counting Kit-8 assay was performed to analyze cell proliferation. In patients with early-stage TNBC, the expression level of lncRNA HAND2-AS1 was downregulated, whilst runt-related transcription factor 2 (RUNX2) mRNA was upregulated in tumor tissues, compared with paired healthy tissues. Expression levels of lncRNA HAND2-AS1 and RUNX2 mRNA were inversely correlated in tumor tissues, but not in paired healthy tissues. Decreased plasma expression levels of lncRNA HAND2-AS1 were observed in TNBC patients compared with those in healthy controls, and the downregulation of lncRNA HAND2-AS1 distinguished patients with TNBC from healthy controls. lncRNA HAND2-AS1 overexpression inhibited RUNX2 expression, whilst RUNX2 overexpression did not significantly affect lncRNA HAND2-AS1 expression in the MDA-MB-231 and BT-20 cell lines. lncRNA HAND2-AS1 overexpression resulted in the inhibition of cell proliferation, while RUNX2 overexpression promoted the proliferation of TNBC cells. RUNX2 overexpression partially reversed the effects of lncRNA HAND2-AS1 overexpression on cancer cells. Therefore lncRNA HAND2-AS1 may inhibit the proliferation of cancer cells by inhibiting RUNX2 expression in TNBC.
Keywords: triple-negative breast cancer, runt-related transcription factor 2, long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1, proliferation
Introduction
Triple-negative breast cancer (TNBC) is one of the most frequently diagnosed malignancies among females (1–3), and is characterized by a lack of estrogen and progesterone receptors, and the absence of the human epidermal growth factor receptor (2). Chemotherapy is commonly used to treat TNBC (4), as due to its heterogeneity, the availability of other treatment options is limited (5). Although a number of molecular therapeutic targets have been identified, including poly ADP ribose polymerase, epidermal growth factor receptor, fibroblast growth factor receptor and the angiogenic pathway [some of which are currently being tested in clinical trials (3)], no reliable outcomes have been observed (6,7). Therefore identification of novel molecular targets is required.
Runt-related transcription factor 2 (RUNX2) is a transcription factor that participates in the regulation of cell proliferation by influencing the expression of a large set of downstream genes (8). A growing body of literature has demonstrated the critical role of RUNX2 in a number of human cancer types, including different types of breast cancer (9,10). RUNX2 expression is now considered a promising therapeutic target for cancer treatment (11).
Long non-coding RNAs (lncRNAs) are a group of non-protein coding RNAs involved in physiological and pathological processes (12,13). RUNX2 influences cancer biology, not only by affecting protein production, but also by its interaction with lncRNAs (12,13). lncRNA HAND2-AS1, a transcribed antisense (AS) lncRNA adjacent to heart and neural crest derivatives expressed 2 (HAND2), has been characterized as a tumor suppressor lncRNA in various types of malignancy (14–16). lncRNA HAND2-AS1 is involved in cancer biology through interactions with multiple signaling molecules, including microRNAs (miRNAs), hypoxia-inducible factor 1α (HIF1α) and neuromedin (14–16). However, its functions in TNBC are yet to be elucidated.
In the present study, the role of lncRNA HAND2-AS1 in TNBC was investigated, and revealed to be downregulated. Additionally, lncRNA HAND2-AS1 may inhibit the proliferation of cancer cells by reducing RUNX2 expression in TNBC, providing a potential therapeutic target for the disease.
Materials and methods
Cell lines and patient samples
Two human TNBC cell lines, MDA-MB-231 and BT-20, were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cutured with ATCC-formulated Eagle's minimum essential medium (cat. no. 30-2003) with 10% fetal bovine serum (cat. no. F2442-50ML, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) at 37°C in a 5% CO2 incubtaor.
The study included 63 female patients with TNBC and 43 healthy females. Participants were admitted to the International Peace Maternity and Child Health Hospital (Shanghai, China) between January 2016 and January 2018. The inclusion criteria were as follows: i) A diagnosis of TNBC through pathological examinations; ii) TNBC of American Joint Committee on Cancer stages (17) I and II at presentation; and iii) willingness to donate biopsies of tumor tissues and adjacent healthy tissues within 2 cm around the tumor site. Exclusion criteria: i) Patients suffering from multiple diseases; ii) treatment prior to admission; and iii) patients at advanced cancer stages. Biopsies of tumor and adjacent healthy tissues were confirmed by histopathological examination. Tissues were fixed in 4% formaldehyde overnight at 4°C. Subsequently, paraffin-embedded (8 µm) tissue sections were stained with hematoxylin and eosin at 37°C for 2 h and visualized using a light microscope (×40 magnification). Plasma samples derived from the blood of patients and healthy controls were also collected by centrifuging blood samples in EDTA tubes for 10 min at 1,200 × g. All samples were stored in liquid nitrogen before use. The 43 healthy females (control group) received a routine physical examination at the International Peace Maternity and Child Health Hospital during the same time period. The age range of the patient group was 30–69 years, with a mean age of 45.5±6.1 years, while that of the control group was 28–66 years, with a mean age of 43.9±5.7 years. The 2 groups had similar age distributions (revealed using Mann-Whitney U test). The present study was approved by the ethics committee of the International Peace Maternity and Child Health Hospital, and all patients and healthy volunteers provided written informed consent.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Following total RNA extraction using RNAzol RT® (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to the manufacturer's protocol, cDNA was obtained through RT using the QuantiTect RT kit (Qiagen GmbH, Hilden, Germany). The SuperScript III Platinum One-Step RT-qPCR kit (SYBR; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to prepare all reactions. Thermocycling conditions were as follows: 95°C for 48 sec, 95°C for 16 sec and 56.5°C for 28 sec for 40 cycles. The sequences of the primers used were: HAND2-AS1 forward, 5′-GGGTGTTTACGTAGACCAGAACC-3′ and reverse, 5′-CTTCCAAAAGCCTTCTGCCTTAG-3′; RUNX2 forward, 5′-GTTATGAAAAACCAAGTAGCCAGGTC-3′ and reverse, 5′-GTAATCTGACTCTGTCCTTGTGGAT-3′; and β-actin forward, 5′-GACCTCTATGCCAACACAGT3′ and reverse, 5′-AGTACTTGCGCTCAGGAGGA3′. Data were normalized using the 2−ΔΔCq method (18).
Vectors and transfection
The HAND2-AS1 and RUNX2 expression vectors were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were cultured overnight to achieve 80–90% confluence, and Lipofectamine 2000® reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect 10 nM of each vector into 5×105 cells. To verify the overexpression of HAND2-AS1 and RUNX2, untransfected cells and those transfected with empty vectors were used as controls. A HAND2-AS1 expression rate of >175% was confirmed prior to subsequent experimentation. The interval between transfection and following experimentation was 24 h.
Cell proliferation assay
Following transfection, cell proliferation was measured using Cell Counting Kit-8 (CCK-8) (Beyotime Institute of Biotechnology, Haimen, China). Briefly, cells were harvested and single cell suspensions (4×104 cells/ml) were prepared; 0.1 ml cell suspension was added to each well of a 96-well plate. Cells were cultured in a 5% CO2 incubator at 37°C, and 10 µl CCK-8 solution was added at 24, 48, 72 and 96-h time points. Cells were cultured for an additional 4 h, and the Fisherbrand™ accuSkan™ GO UV/Vis Microplate Spectrophotometer (Thermo Fisher Scientific, Inc.) was used to measure absorbance at 450 nm.
Western blotting
The ReadyPrep™ Protein Extraction kit (Total Protein; Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to extract total protein from cell lines and patient samples. Protein concentration was measured using bicinchoninic acid assay (Sigma-Aldrich; Merck KGaA). The lysates were denatured, and 20 µg protein per lane was separated using SDS-PAGE with a 12% gel. Following gel transfer to PVDF membranes, blocking was performed in 5% skimmed milk (PBS) at room temperature for 2 h. The membranes were subsequently incubated with primary rabbit anti-human antibodies against RUNX2 (1:1,200; cat. no. ab23981; Abcam, Cambridge, UK) and GAPDH (1:1,400; cat. no. ab9485; Abcam) overnight at 4°C. Membranes were subsequently washed twice using PBS at room temperature, 15 min each time, followed by further incubation with immunoglobulin G-horseradish peroxidase-conjugated secondary antibody (1:1,000; cat. no. MBS435036; MyBioSource, San Diego, CA, USA) at room temperature for 2 h. Signals were developed using enhanced chemiluminescence (Sigma-Aldrich, Merck KGaA), and ImageJ v1.6 software (National Institutes of Health, Bethesda, MD, USA) was used for data normalization.
Statistical analysis
GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA) was used to process all data. All experiments were performed in triplicate, and the data are expressed as the mean ± standard deviation. Comparisons between 2 groups were performed using the unpaired Student's t-test. Comparison of age between two groups was performed using Mann-Whitney U test while comparisons between tumor and adjacent-tumor tissues was performed using paired t-test. Comparisons between >2 groups were performed using one-way analysis of variance followed by Tukey's test. Diagnostic analysis was performed using receiver operating characteristic (ROC) curve analysis, and Pearson's correlation coefficient was used to determine correlations between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Altered expression of lncRNA HAND2-AS1 and RUNX2 is observed in the tumor tissues of patients with TNBC
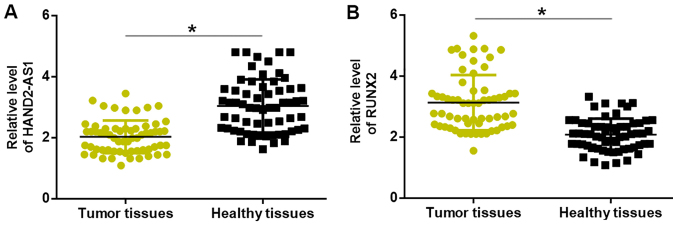
Compared with levels in adjacent healthy tissues, expression levels of lncRNA HAND2-AS1 were significantly downregulated (Fig. 1A), whilst those of RUNX2 mRNA were significantly upregulated (Fig. 1B), in the cancerous tissues of patients with TNBC.
Figure 1.
Altered expression of lncRNA HAND2-AS1 and RUNX2 in tumor tissues. Compared with that in adjacent healthy tissues, (A) significantly downregulated lncRNA HAND2-AS1 and (B) significantly upregulated RUNX2 mRNA expression was observed in triple-negative breast cancer tissues. *P<0.05. lncRNA HAND2-AS1, long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1; RUNX2, runt-related transcription factor 2.
Expression levels of lncRNA HAND2-AS1 and RUNX2 are correlated in tumor tissues, but not in paired healthy tissues
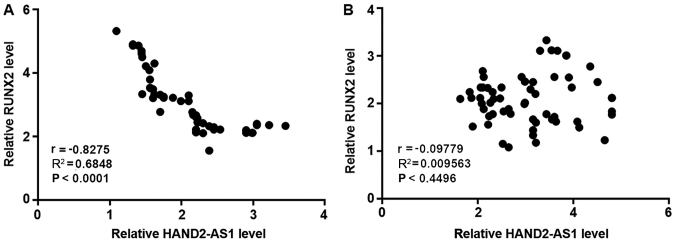
Pearson's correlation coefficient analysis revealed an inverse correlation between the expression levels of lncRNA HAND2-AS1 and RUNX2 in the tumor tissues of patients with TNBC (Fig. 2A). By contrast, expression levels of lncRNA HAND2-AS1 and RUNX2 were not significantly correlated in paired healthy tissues (Fig. 2B).
Figure 2.
Expression levels of lncRNA HAND2-AS1 and RUNX2 are correlated in tumor tissues, but not in paired healthy tissues. (A) Pearson's correlation coefficient analysis revealed an inverse correlation between the expression levels of lncRNA HAND2-AS1 and RUNX2 in tumor tissues, but not in (B) paired healthy tissues. lncRNA HAND2-AS1, long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1; RUNX2, runt-related transcription factor 2.
Downregulation of lncRNA HAND2-AS1 in the plasma distinguishes patients with TNBC from healthy controls
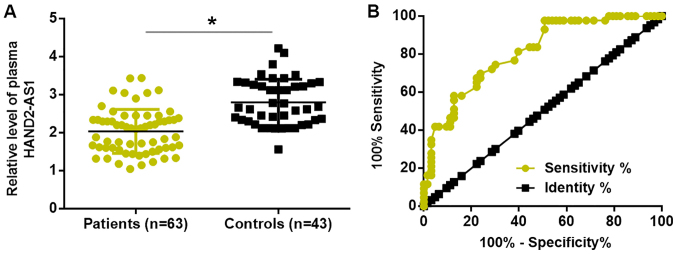
Compared with healthy controls, plasma levels of lncRNA HAND2-AS1 were significantly reduced in patients with TNBC (Fig. 3A). ROC curve analysis was performed to evaluate the diagnostic value of plasma lncRNA HAND2-AS1 for TNBC. As illustrated in Fig. 3B, downregulation of plasma lncRNA HAND2-AS1 distinguished patients with TNBC from healthy controls.
Figure 3.
Downregulation of lncRNA HAND2-AS1 in the plasma distinguishes TNBC patients from healthy controls. (A) Plasma levels of lncRNA HAND2-AS1 were significantly reduced in TNBC patients compared with those in healthy controls. (B) Downregulation of plasma lncRNA HAND2-AS1 distinguished TNBC patients from healthy controls. *P<0.05. lncRNA HAND2-AS1, long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1; TNBC, triple-negative breast cancer.
lncRNA HAND2-AS1 overexpression inhibits the expression of RUNX2
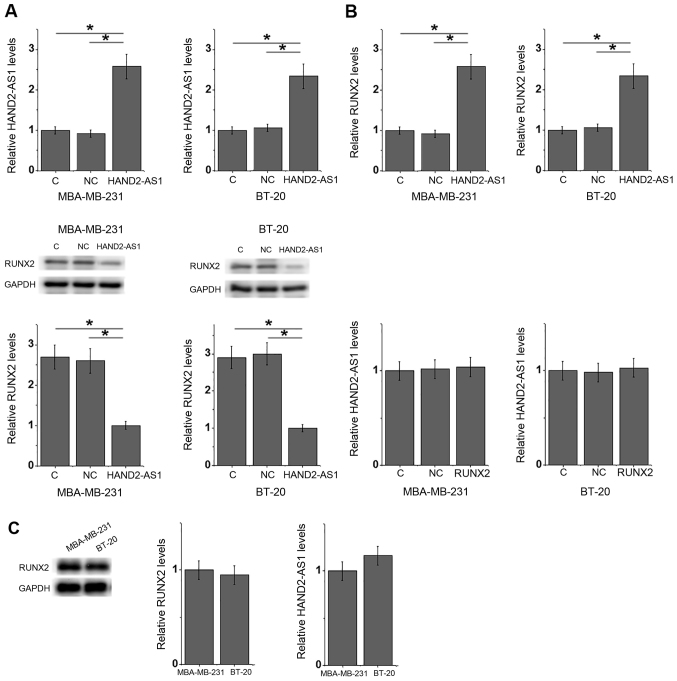
The expression level of lncRNA HAND2-AS1 was inversely correlated with that of RUNX2 in TNBC tumor tissues. Furthermore, compared with the negative control and control groups, lncRNA HAND2-AS1 transfection significantly downregulated RUNX2 expression in the MDA-MB-231 and BT-20 TNBC breast cancer cell lines (Fig. 4A). By contrast, RUNX2 transfection had no significant impact on lncRNA HAND2-AS1 expression in either cell line (Fig. 4B). No significant differences in the expression levels of HAND2-AS1 and RUNX2 were observed between MDA-MB-231 and BT-20 TNBC cells (Fig. 4C).
Figure 4.
lncRNA HAND2-AS1 overexpression inhibits RUNX2 expression. (A) lncRNA HAND2-AS1 transfection mediated a significant downregulation in RUNX2 expression in MDA-MB-231 and BT-20 TNBC breast cancer cells, whilst (B) RUNX2 transfection had no significant effects on lncRNA HAND2-AS1 expression. (C) No significant differences in the expression levels of HAND2-AS1 and RUNX2 were observed between the two cell lines. *P<0.05. lncRNA HAND2-AS1, long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1; RUNX2, runt-related transcription factor 2; C, control; NC, negative control.
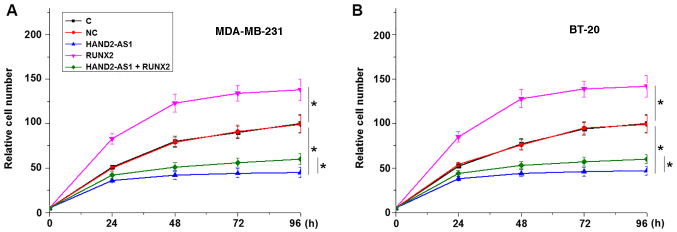
lncRNA HAND2-AS1 overexpression reduces cancer cell proliferation, potentially by inhibiting RUNX2 expression
The CCK-8 assay results revealed that lncRNA HAND2-AS1 transfection inhibited, while RUNX2 transfection promoted, the proliferation of MDA-MB-231 (Fig. 5A) and BT-20 TNBC cells (Fig. 5B) when compared with the negative control and control groups. In addition, RUNX2 overexpression partially reversed the effects of lncRNA HAND2-AS1 overexpression on cancer cells.
Figure 5.
lncRNA HAND2-AS1 overexpression reduces cancer cell proliferation by inhibiting RUNX2 expression. In (A) MDA-MB-231 and (B) BT-20 TNBC cells, lncRNA HAND2-AS1 transfection inhibited proliferation, while RUNX2 transfection promoted proliferation. *P<0.05. lncRNA HAND2-AS1, long non-coding RNA heart and neural crest derivatives expressed 2-antisense RNA 1; RUNX2, runt-related transcription factor 2; C, control; NC, negative control.
Discussion
Previous studies have characterized the functionality of lncRNA HAND2-AS1, a tumor suppressor gene in osteosarcoma (14), colorectal cancer (15) and endometrioid endometrial carcinoma (16). However, the involvement of lncRNA HAND2-AS1 in TNBC, a subtype of breast cancer, is unknown. The present study revealed that lncRNA HAND2-AS1 may also possess a tumor suppression role in TNBC by inhibiting cancer cell proliferation. The actions of lncRNA HAND2-AS1 in TNBC are potentially influenced by the inhibition of RUNX2 expression.
An early diagnosis of cancer is critical for the successful surgical resection of tumor tissues prior to metastasis, which is the principal cause of mortality among patients with breast cancer (19). The present study included TNBC patients at cancer stages I and II, which are considered to be the early stages of cancer. Differential gene expression in tumor and adjacent healthy tissues indicates the involvement of certain genes in cancer. As a tumor suppressor, the expression of lncRNA HAND2-AS1 has been reported to decrease in osteosarcoma (14), colorectal cancer (15) and endometrioid endometrial carcinoma (16). In the present study, significantly downregulated expression of lncRNA HAND2-AS1 was observed in tumor tissues compared with that in the adjacent healthy tissues of patients with TNBC. In addition, plasma expression levels of lncRNA HAND2-AS1 were also significantly lower in patients with TNBC compared with those in healthy females. The downregulation of lncRNA HAND2-AS1 was used to distinguish patients with TNBC from healthy controls; therefore, lncRNA HAND2-AS1 may act as tumor suppressor in TNBC, and the detection of circulating lncRNA HAND2-AS1 may provide guidance for the early diagnosis of the disease.
As an oncogene, RUNX2 is frequently upregulated during the development of human cancer (20,21). Consistent with previous studies, the present study also illustrated significantly upregulated expression of RUNX2 in tumor tissues compared with that in adjacent healthy tissues in patients with TNBC. Furthermore, an inverse correlation between the expression level of lncRNA HAND2-AS1 and RUNX2 in tumor tissues was observed. It was also revealed that lncRNA HAND2-AS1 may serve as an upstream regulator of RUNX2 in cancer cell proliferation; lncRNA HAND2-AS1 overexpression inhibited RUNX2 expression, although RUNX2 overexpression did not significantly affect HAND2-AS1 expression. Additionally, RUNX2 overexpression partially reversed the inhibitory effect of HAND2-AS1 overexpression in TNBC cells. Therefore, lncRNA HAND2-AS1 may serve as a potential therapeutic target for TNBC by inhibiting the expression of RUNX2.
It is worth noting that no correlations between lncRNA HAND2-AS1 and RUNX2 expression levels were observed in adjacent healthy tissues, indicating the indirect action of lncRNA HAND2-AS1 on RUNX2 expression. In addition, lncRNA HAND2-AS1 overexpression had no significant effect on cancer cell migration and invasion (data not shown), suggesting that lncRNA HAND2-AS1 may specifically participate in cell proliferation in TNBC.
lncRNA HAND2-AS1 is involved in different types of cancer through interactions with different signaling molecules, including miRNAs, HIF1α and neuromedin (14–16), indicating that lncRNA HAND2-AS1 may participate in different pathological processes between diseases. However, the present study failed to establish lncRNA HAND2-AS1 siRNA silencing in TNBC cells, which is a consideration for future studies.
In conclusion, lncRNA HAND2-AS1 may serve as a tumor suppressor gene in TNBC by inhibiting cancer cell proliferation through the downregulation of RUNX2.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MW and ZW designed the experiments. MW and LL performed the experiments and analyzed the data. ZW drafted the manuscript and all authors approved the manuscript for publication.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the International Peace Maternity and Child Health Hospital, and all patients and healthy volunteers provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zubeda S, Kaipa PR, Shaik NA, Mohiuddin MK, Vaidya S, Pavani B, Srinivasulu M, Latha MM, Hasan Q. Her-2/neu status: A neglected marker of prognostication and management of breast cancer patients in India. Asian Pac J Cancer Prev. 2013;14:2231–2235. doi: 10.7314/APJCP.2013.14.4.2231. [DOI] [PubMed] [Google Scholar]
- 2.Podo F, Buydens L, Degani H, Hilhorst R, Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J, Monleon D, et al. Triple-negative breast cancer: Present challenges and new perspectives. Mol Oncol. 2010;4:209–229. doi: 10.1016/j.molonc.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Wan YW, Allen GI, Pang K, Anderson ML, Liu Z. Molecular pathway identification using biological network-regularized logistic models. BMC Genomics. 2013;14(Suppl 8):S7. doi: 10.1186/1471-2164-14-S8-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, Pietenpol JA. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collignon J, Lousberg L, Schroeder H, Jerusalem G. Triple-negative breast cancer: Treatment challenges and solutions. Breast Cancer (Dove Med Press) 2016;8:93–107. doi: 10.2147/BCTT.S69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389:2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 7.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer-deciphering the heterogeneity. Clin Cancer Res. 2014;20:782–790. doi: 10.1158/1078-0432.CCR-13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucero CM, Vega OA, Osorio MM, Tapia JC, Antonelli M, Stein GS, van Wijnen AJ, Galindo MA. The cancer-related transcription factor Runx2 modulates cell proliferation in human osteosarcoma cell lines. J Cell Physiol. 2013;228:714–723. doi: 10.1002/jcp.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taipaleenmäki H, Browne G, Akech J, Zustin J, van Wijnen AJ, Stein JL, Hesse E, Stein GS, Lian JB. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75:1433–1444. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chai J, Guo D, Ma W, Han D, Dong W, Guo H, Zhang Y. A feedback loop consisting of RUNX2/lncRNA-PVT1/miR-455 is involved in the progression of colorectal cancer. Am J Cancer Res. 2018;8:538–550. [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Zhao J, He Y. Long non-coding RNA PVT1 functions as an oncogene in human colon cancer through miR-30d-5p/RUNX2 axis. J BUON. 2018;23:48–54. [PubMed] [Google Scholar]
- 14.Kang Y, Zhu X, Xu Y, Tang Q, Huang Z, Zhao Z, Lu J, Song G, Xu H, Deng C, Wang J. Energy stress-induced lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and inhibits osteosarcoma progression. Am J Cancer Res. 2018;8:526–537. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Lin J, Zhang H, Zhu F, Xie R. lncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem Biophys Res Commun. 2018;503:1848–1853. doi: 10.1016/j.bbrc.2018.07.125. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Wang CC, Lee WYW, Trovik J, Chung TKH, Kwong J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018;413:23–34. doi: 10.1016/j.canlet.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 17.Giuliano AE, Connolly JL, Edge SB, Mittendorf EA, Rugo HS, Solin LJ, Weaver DL, Winchester DJ, Hortobagyi GN. Breast cancer-major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:290–303. doi: 10.3322/caac.21393. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 20.Pratap J, Imbalzano KM, Underwood JM, Cohet N, Gokul K, Akech J, van Wijnen AJ, Stein JL, Imbalzano AN, Nickerson JA, et al. Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure: Implications for breast cancer progression. Cancer Res. 2009;69:6807–6814. doi: 10.1158/0008-5472.CAN-09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sancisi V, Manzotti G, Gugnoni M, Rossi T, Gandolfi G, Gobbi G, Torricelli F, Catellani F, Faria do Valle I, Remondini D, et al. RUNX2 expression in thyroid and breast cancer requires the cooperation of three non-redundant enhancers under the control of BRD4 and c-JUN. Nucleic Acids Res. 2017;45:11249–11267. doi: 10.1093/nar/gkx802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.