Abstract
RNA editing occurs in the organellar mRNAs of all land plants but the marchantioid liverworts, making liverworts a perfect group for studying the evolution of RNA editing. Here, we profiled the RNA editing of 42 exemplars spanning the ordinal phylogenetic diversity of liverworts, and screened for the nuclear-encoded pentatricopeptide repeat (PPR) proteins in the transcriptome assemblies of these taxa. We identified 7,428 RNA editing sites in 128 organellar genes from 31 non-marchantioid liverwort species, and characterized 25,059 PPR protein sequences. The abundance of organellar RNA editing sites varies greatly among liverwort lineages, genes, and codon positions, and shows strong positive correlations with the GC content of protein-coding genes, and the diversity of the PLS class of nuclear PPR proteins.
Keywords: GC content, liverworts, organellar genes, PLS PPR proteins, RNA editing, phylogeny
Introduction
RNA editing is a co- or posttranscriptional process that changes the content of the transcripts, resulting in sequences of mature RNA transcripts deviating from that of the genomic templates (Covello and Gray 1993; He et al. 2016). In plants, this process mainly converts cytidines (C) to uridines (U) in organellar transcripts, and occasionally U to C (Gray 2012; Ichinose and Sugita 2016). Plant organellar RNA editing plays important roles in gene expression and functioning, by restoring start/stop codons, removing internal stop codons, maintaining proper splicing of transcripts, or modifying amino acids (Sloan 2017). RNA editing occurs in organellar genomes of all land plant lineages except the marchantioid liverworts (Rüdinger et al. 2008). In general, angiosperms contain 200–500 RNA editing sites in their mitochondrial (mt) genomes, but only 30–50 editing sites in their plastid (pt) genomes (Oldenkott et al. 2014). Lycophytes possess the highest number of editing sites, with for example, the mt genome of Selaginella moellendorffii holding 2,139 C-to-U RNA editing sites in only 18 protein-coding genes (PCGs) (Hecht et al. 2011) and the pt genome of Selaginellauncinata holding 3,415 such sites in its 82 genes (Oldenkott et al. 2014). Mosses have the fewest RNA editing sites, with only 11 and 1 RNA editing sites in the mt (Rüdinger et al. 2009) and pt genes, respectively, of Physcomitrella patens (Takenaka et al. 2013). RNA editing sites have been characterized in many vascular plants (Richardson et al. 2013; Guo et al. 2015, 2017; Edera et al. 2018), but only in a few nonvascular plants, that is, the liverwort Marchantia polymorpha (Rüdinger et al. 2008), the hornworts Anthoceros formosae (Kugita et al. 2003) and Leiosporoceros dussii (Villarreal et al. 2018), and the moss Physcomitrella patens (Rüdinger et al. 2009). Thus, empirical data are still scarce to understand the phylogenetic distribution and evolution of RNA editing among nonvascular land plants.
The abundance of RNA editing in organellar genes may be determined by many factors. The RNA editing frequency is clearly associated with the function of the gene (Jobson and Qiu 2008; Edera et al. 2018), with genes encoding membrane-bound proteins or under strong functional selections holding more RNA editing sites (Mower and Palmer 2006; Edera et al. 2018). RNA editing frequency may also be positively linked to the genomic GC content, especially across second codon positions (Malek et al. 1996; Smith 2009; Hecht et al. 2011; Guo et al. 2017), and the diversity of nuclear-encoded pentatricopeptide repeat (PPR) proteins (Fujii and Small 2011). The PPR proteins, characterized by tandem arrays of a weakly conserved 35 amino-acid motif (Small and Peeters 2000), form one of the largest gene families in land plants (Cheng et al. 2016), with most angiosperms encoding 400–600 PPRs (Fujii and Small 2011). PPR proteins are divided into two major classes, the P and PLS classes, with the latter further subdivided into five groups, namely PLS, E1, E2, E+, and DYW, based on the characteristic of the carboxyterminus. The majority of PLS-class members may be dedicated to RNA editing processes (Fujii and Small 2011), especially the DYW type proteins, whose functional cytidine deaminase activities have recently been confirmed (Oldenkott et al. 2019). The diversity of DYW domains was reported to be well correlated to the frequency of RNA editing sites in plants (Salone et al. 2007; Rüdinger et al. 2008). Based on empirical and in silico predictive RNA editing data, the abundance of plastid RNA editing was proposed to be strongly and positively correlated (PCC = 0.9) to the diversity of PLS PPR proteins in 21 green plants (Fujii and Small 2011). As these correlationships were inferred with simple regression methods without accounting for phylogenetic relationships, which may yield trivial implications on biological relationships (Maddison and Fitzjohn 2015). Therefore, comparative methods incorporating the phylogenetic information (Martins and Hansen 1997) need to be implemented to further test such correlations.
With some 7,300 extant species (Söderström et al. 2016), liverworts compose a diverse lineage of land plants. Although their relationships to other bryophytes remains somewhat controversial (Morris et al. 2018), they undoubtedly arose early in the diversification of land plants, and hence might be critical to unravel the timing and subsequent evolution of various innovations such as RNA editing, a process possibly acquired by the common ancestor of land plants (Hiesel et al. 1994; Sabater et al. 2002). Here, we screened deep genomic and organellar transcripts enriched transcriptomic sequence data to critically analyze RNA editing in mt and pt genes of 42 liverworts from 13 of the 15 liverwort orders. We also characterized the diversity of six types of PPR proteins based on transcriptome assemblies. Our study aims to contrast RNA editing site abundance and variation 1) between mt and pt genomes, and 2) among different liverwort clades, and to test for a relationship of RNA editing site abundance 3) with the GC content, 4) and the diversity of PPR proteins.
Materials and Methods
Taxon Sampling and NGS Experiments
We sampled fresh collections of 42 liverworts (supplementary table S1, Supplementary Material online) representing all but 2 of the 15 orders of liverworts (Crandall-Stotler et al. 2009). Individual shoots and branches of each accession were separated under a dissecting microscope to avoid contamination from other organisms, cleaned with distilled water, and used for DNA and RNA isolations using the modified CTAB methods following Porebski et al. (1997) and Reid et al. (2006), respectively. Approximately 1 μg of genomic DNA and RNA were used to generate paired-end sequencing libraries with the insert fragment size of 300–500 and 200–300 bp, respectively. NGS sequencing was carried out on an Illumina HiSeq 2000 platform at Majorbio (Shanghai, China). The raw sequencing data in fastq format (∼10 and ∼6 G raw data for genomic and transcriptomic sequencing, respectively) were trimmed and filtered for adaptors, low quality reads, undersized inserts, and duplicate reads using Trimmomatic (Bolger et al. 2014).
Genome Assembly and Annotation
The filtered reads from each species were de novo assembled into contigs (≥1 kb) using the CLC Genomics Workbench v5.5 (CLC Bio, Aarhus, Denmark) using default parameters (word size = 20, bubble size = 50, minimum contig length = 1,000). All contigs were blasted to the Marchantia polymorpha pt and mt genomes (GenBank accession: NC_037507 and NC_037508), to retrieve the pt and mt contigs, respectively. These contigs were elongated and assembled into organellar genome scaffolds using overlapping methods (Dong et al. 2019). Ultimately, we assembled 29 and 40 complete mt and pt genomes, respectively. For the remaining 13 mt genomes and the 2 pt genomes, for which corresponding contigs could not be linked due to frequent occurrences of repeated sequences, we anchored the contigs by mapping them to the organellar genomes of Marchantia polymorpha and connected them with Ns to generate the draft genomes. The PCGs of the organellar genome were annotated following the steps described in Xue et al. (2010). The exact gene and exon/intron boundaries were further confirmed in Geneious v10.0.2 (www.geneious.com) by aligning orthologous genes with the available annotated liverwort organellar genomes (such as Pleurozia, GenBank accession no.: NC_013444) to those of the 42 liverworts.
RNA Editing Site Identification
The DNA-seq reads and RNA-seq reads were mapped to the corresponding draft organellar genome using Bowtie2 (Langmead and Salzberg 2012) and Tophat2 (Kim et al. 2013) separately as described by Edera et al. (2018). The resulting mapping files in bam format were sorted to generate vcf file using Samtools (Li et al. 2009) and Bcftools (Narasimhan et al. 2016), from which, the snp file were produced using a custom perl script (Dryad Digital Repository, accession 10.5061/dryad.nzs7h44ms). The RNA mapping files were processed using Redo (Wu et al. 2018) using default parameters. The Redo output of the RNA editing site annotation file was then manually filtered against the SNP sites by their positions on the genome sequence to remove potential genomic SNPs. The final annotation file of the RNA editing sites and the corresponding transcriptome mapping bam file were imported in Geneious v10.0.2 to manually check the bioinformatically identified editing sites and make necessary revisions to further reduce false positives and false negatives.
Transcriptome Assembly and PPR Protein Identification
All transcriptomes including those from the outgroups were de novo assembled using Trinity pipeline (Grabherr et al. 2011). The open reading frames were extracted and annotated from the transcriptome assembly using TransDecoder (http://sourceforge.net/projects/transdecoder/). The peptide file of each species was filtered for redundancy using Cd-hit (Li and Godzik 2006) with a similarity threshold of c = 0.9, and screened for PPR domains using the pipeline as described by Cheng et al. (2016).
Correlation Tests between the Number of RNA Editing Site and PPR Protein Diversity/GC Content
To assess the evolutionary correlationships between the abundance of RNA editing sites and the diversity of PPR proteins, as well as the GC content. We used the phylogenetic generalized least-squares (PGLS) regression, allowing λ to be fitted by maximum likelihoods (Mundry 2014). In all analyses, variables were log transformed to make them normally distributed. We calculated the variance–covariance matrix for the ML tree derived from 127 organellar genes using “corPagel” as implemented in the R package “ape” (Paradis et al. 2004), and then fit the regression by maximizing the restricted log-likelihood using “gls” as implemented in the R package “nlme” (Pinheiro et al. 2016).
Results and Discussions
RNA Editing Site Abundance in Liverworts
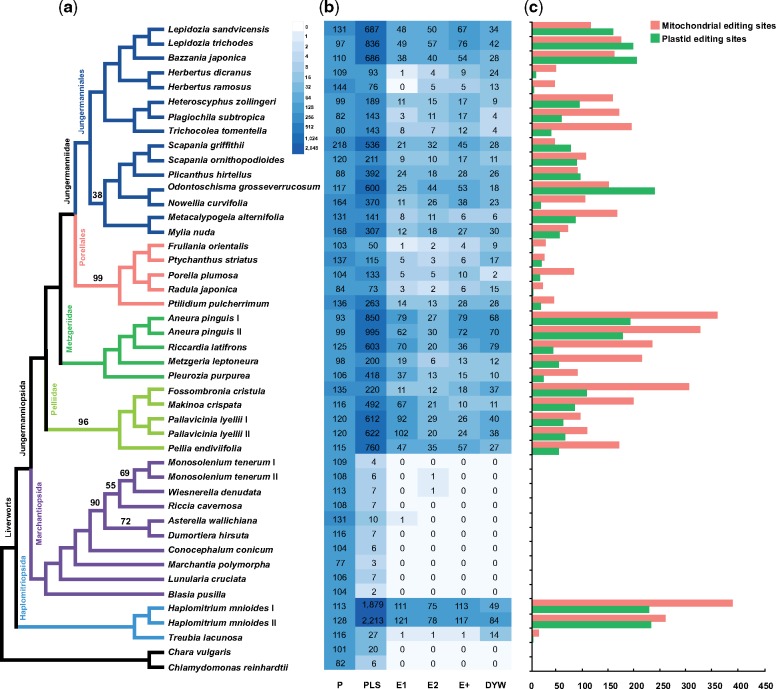
Our genomic and transcriptomic data yielded high read coverage for both mt and pt PCGs (supplementary fig. S1, Supplementary Material online). Altogether, we identified 7,428 C-to-U RNA editing sites from the 128 organellar PCGs of 33 liverworts, including 4,694 RNA editing sites for 42 mt PCGs (supplementary table S2, Supplementary Material online) and 2,734 RNA editing sites for 86 pt PCGs (supplementary table S3, Supplementary Material online). The average number of RNA editing sites per mt gene (3.39) is more than three times that per pt gene (0.96). No RNA editing was detected from either organellar genome of any complex thalloid (Marchantiopsida) species.
The number of organellar RNA editing sites varies significantly among liverwort lineages, and even among different accessions of the same species (fig. 1). The Haplomitriales (with Haplomitrium sampled here), widely recognized as emerging from the deepest splits in liverworts, has the highest editing level, whereas its sister group, the Treubiales, has the fewest editing sites among all liverworts with the exception of the complex thalloids (Marchantiopsida), which completely lack editing sites (fig. 1). The simple thalloids, that is, Pelliidae and Metzgeriidae, exhibit the second highest RNA editing frequency, followed by the Jungermanniales, except for two Herbertus species with a low editing level.
Fig. 1.
—(a) Phylogenetic tree of liverworts based on a concatenated nucleotide data set of 41 mt and 86 pt genes. Branches are maximally supported (i.e., 100% bootstrap frequencies) unless otherwise marked. (b) Heatmap of PPR protein diversity across liverworts. (c) Histogram of RNA editing site abundance in mt (pink) and pt (green) genomes for each liverwort accession.
The abundance of RNA editing also varies among genes and functional categories within liverworts (supplementary fig. S2, Supplementary Material online), with high editing level in the respiration complex and photosynthesis related protein genes, and low editing level in ribosomal protein genes, a pattern consistent with that observed in angiosperms (Edera et al. 2018). RNA editing sites show relatively higher editing efficiencies in the second and first codon positions compared with the third codon positions (supplementary fig. S3, Supplementary Material online), and occur primarily in the second codon positions of both mt and pt genes (supplementary fig. S4, Supplementary Material online), leading to amino acid changes that primarily increase the hydrophobicity of proteins (supplementary fig. S5, Supplementary Material online). Such modifications may be essential for protein folding, functioning, and interacting with other proteins (He et al. 2016). In Arabidopsis thaliana (Hammani et al. 2011), Physcomitrella patens (Ichinose et al. 2013), and maize (Liu et al. 2013; Yang et al. 2017), the disruption of RNA editing activity severely impacts plant growth at specific developmental stages. Testing the significance of RNA editing process on the fitness of liverworts may await the development of tools for genetically transforming liverworts, but the development of Marchantia as a model system (Bachtrog 2011; Shimamura 2016) contributes little to such understanding since it lacks organellar RNA editing ability. Hence, other liverwort model systems must be developed.
PPR Protein Diversity in Liverworts
We identified 25,059 PPR protein sequences (i.e., 165 P and 19,894 PLS class proteins) in the transcriptome assembly of the 43 liverworts (including the proteome of Marchantia polymorpha downloaded from phytozome.jgi.doe.gov; supplementary table S1, Supplementary Material online). The diversity of PPR proteins varies considerably among different liverwort lineages (fig. 1). Among the five groups of PLS class PPR proteins (PLS, E1, E2, E+, and DYW), the PLS group contributes most of the diversity (∼80%), and shows greatest variations in number (2–2,213) among liverworts. The other four groups of PLS PPR proteins (supplementary fig. S6, Supplementary Material online) are generally absent in charophycean algae and complex thalloid liverworts, with the exception of the occasional presence of E1 and E2 proteins in three complex thalloid liverworts. However, the E2 type proteins from the complex thalloid group (i.e., Wiesnerella denudata and Monosolenium tenerum II) form a structure with a tandem repeat of P type motifs plus a single E2 motif, and the E1 type protein from Asterella wallichiana forms a structure of L-S-E1. All these E-type PPR proteins from the complex thalloids lack the PLS triplet structure that characterizes the PLS PPR proteins. The absence of the functional E and DYW type PLS PPR proteins observed in complex thalloids might be associated with their exclusive absence of RNA editing sites. As Knoop and Rüdinger (2010) have suggested, plant E and E+ type PLS proteins are likely products of serial carboxyterminal domain deletions from the DYW type proteins, the presence of a few truncated E type PLS protein sequences in complex thalloids might imply they are from relics of functional genes, thus providing new evidence for a secondary loss of RNA editing machinery in this group.
Factors Correlated with the Number of RNA Editing Sites
Based on the in silico prediction of RNA editing sites in three mt PCGs and PCR amplifications of DYW type protein genes in over 100 bryophyte species, the diversity of the DYW type PPR protein genes appears to be positively correlated with the abundance of RNA editing sites (Rüdinger et al. 2012; Schallenberg-Rüdinger et al. 2013). Such a correlation of RNA editing site abundance and PPR protein diversity is here confirmed across all organellar genes within liverworts. In general, the number of RNA editing sites in mt and pt genes is strongly and positively correlated to the diversity of nuclear PLS class rather than P class PPR proteins (table 1). All the five types of PLS PPR proteins, but the DYW type, show significantly positive correlations with the RNA editing site abundance for both mt and pt genes. As there is an experimentally proved functional association between DYW type PPRs and RNA editing (Oldenkott et al. 2019), the lack of evidence for their correlationships is unclear and should be further tested with expanded samplings.
Table 1.
Phylogenetically Controlled Generalized Least Squares (PGLS) Regressions between RNA Editing Site Abundance and PPR Protein Diversity and GC Content
| Statistics | Lambda (λ) | Slope | P values |
|---|---|---|---|
| PPPR/MTE | 0.9963 | −0.1513 | 0.0622 |
| PPPR/PTE | 0.9996 | 0.0248 | 0.5409 |
| PLSPPR/MTE | 0.9318 | 0.7102 | 0.0000** |
| PLSPPR/PTE | 0.9720 | 0.6527 | 0.0000** |
| PLS/MTE | 0.9414 | 0.4922 | 0.0000** |
| PLS/PTE | 0.3769 | 0.9547 | 0.0000** |
| E1/MTE | 0.9696 | 0.5141 | 0.0000** |
| E1/PTE | 0.9912 | 0.7830 | 0.0000** |
| E2/MTE | 0.9705 | 0.4188 | 0.0014** |
| E2/PTE | 0.6575 | 1.0741 | 0.0000** |
| E+/MTE | 0.9674 | 0.5971 | 0.0001** |
| E+/PTE | 0.9973 | 0.7425 | 0.0000** |
| DYW/MTE | 0.9967 | −0.0933 | 0.3111 |
| DYW/PTE | 0.9993 | 0.0824 | 0.2383 |
| MGC/MTE | 0.9395 | 0.0126 | 0.0006** |
| MGC1/MTE1 | 0.9895 | 9.1152 | 0.0369* |
| MGC2/MTE2 | 0.9699 | 27.1628 | 0.0000** |
| MGC3/MTE3 | 0.9813 | 4.1102 | 0.0639 |
| PGC/PTE | 0.9715 | 0.0301 | 0.0003** |
| PGC1/PTE1 | 1.0000 | 0.0498 | 0.8204 |
| PGC2/PTE2 | 0.9835 | 14.2927 | 0.0000** |
| PGC3/PTE3 | 0.9795 | 3.8486 | 0.0006** |
Note.—Correlations were tested between each two of the following parameters: the diversity of the P and PLS class PPR proteins (PPPR/PLSPPR) and the diversity of the five types of PLS PPR proteins (PLS/E1/E2/E+/DYW) with the RNA editing site abundance of the mt and pt genes (MTE/PTE), respectively; GC content of all three and the first, second, and third codon positions for protein-coding genes of mt (MGC/MGC1/MGC2/MGC3) and pt (PGC/PGC1/PGC2/PGC3) with the number of RNA editing sites for all three and the first, second, and third codon positions of mitochondrial (MTE/MTE1/MTE2/MTE3) and plastid (PTE/PTE1/PTE2/PTE3) protein-coding genes, respectively.
P < 0.05, ** P < 0.01—significance levels.
The abundance of RNA editing site is considered to be positively correlated with the GC content of organellar PCGs based on a few discrete loci and/or several closely related taxa with simple regression analyses (Malek et al. 1996; Smith 2009; Hecht et al. 2011; Guo et al. 2017). Such correlationships are confirmed here in liverworts with the PGLS regressions for both mt and pt genes, and for most of their codon positions, but the third codon positions of mt genes and the first codon positions of the pt genes (table 1). The correlation between the GC ratio and the RNA editing abundance in organellar genes of liverworts may suggest that either the rise in GC content facilitated the evolution of RNA editing in land plants or vice versa.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors are grateful to the two anonymous reviewers for their constructive suggestions on the manuscripts. We thank David Glenny for collecting Treubia lacunosa from New Zealand, Qin Zuo for collecting Fossombronia cristula. We also thank Yang Peng for assisting with the lab experiments, Pu Liang, Fujuan Song, and Manhua Lin from the Shenzhen Volunteer Association for help cleaning the liverwort materials. This project is funded by the National Natural Science Foundation of China (NSFC-31470314 and NSFC-31600171).
Author Contributions
Y.L. conceived and designed the study. Y.L., S.D., C.Z., and B.G. collected the fresh liverwort samples. T.W. and S.D. drew the figures. W.M. and N.L. performed the PPR characterization analysis. S.D. carried out the lab experiments, analyses, and drafted the article. Y.L. and B.G. revised the article, and all the authors approved the article. The authors alone are responsible for the content and writing of the article.
Data deposition: The organellar transcriptomic sequence data have been deposited in the Sequence Read Archive database of NCBI under the study number of SRP170646. The newly generated organellar genomes have been deposited in the China National GeneBank DataBase (CNGBdb) under the accession numbers of N_000000002.1–N_000000085.1. This project has been deposited at the Dryad Digital Repository under the accession 10.5061/dryad.nzs7h44ms. All other relevant data are available from the authors.
Literature Cited
- Bachtrog D. 2011. Are all sex chromosomes created equal? Trends Genet. 27(9):350–357. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, et al. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85(4):532–547. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW.. 1993. On the evolution of RNA editing. Trends Genet. 9(8):265–268. [DOI] [PubMed] [Google Scholar]
- Crandall-Stotler B, Stotler RE, Long DG.. 2009. Phylogeny and classification of the Marchantiophyta. Edinb J Bot. 66(1):155–198. [Google Scholar]
- Dong S, et al. 2019. The mitochondrial genomes of Bazzania tridens and Riccardia planiflora further confirms conservative evolution of mitogenomes in liverworts. Bryologist 122(1):130–139. [Google Scholar]
- Edera AA, Gandini CL, Sanchez-Puerta MV.. 2018. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol Biol. 97(3):215–231. [DOI] [PubMed] [Google Scholar]
- Fujii S, Small I.. 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191(1):37–47. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2012. Evolutionary origin of RNA editing. Biochemistry 51(26):5235–5242. [DOI] [PubMed] [Google Scholar]
- Guo W, Grewe F, Mower JP.. 2015. Variable frequency of plastid RNA editing among ferns and repeated loss of uridine-to-cytidine editing from vascular plants. PLoS One 10(1):e0117075.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhu A, Fan W, Mower JP.. 2017. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 213(1):391–403. [DOI] [PubMed] [Google Scholar]
- Hammani K, et al. 2011. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J Biol Chem. 286(24):21361–21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Huang S, Xiao G, Zhang Y, Yu J.. 2016. Abundant RNA editing sites of chloroplast protein-coding genes in Ginkgo biloba and an evolutionary pattern analysis. BMC Plant Biol. 16(1):257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V.. 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 3:344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R, Combettes B, Brennicke A.. 1994. Evidence for RNA editing in mitochondria of all major groups of land plants except the Bryophyta. Proc Natl Acad Sci U S A. 91(2):629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M, Sugita C, Yagi Y, Nakamura T, Sugita M.. 2013. Two DYW subclass PPR proteins are involved in RNA editing of ccmFC and atp9 transcripts in the moss Physcomitrella patens: first complete set of PPR editing factors in plant mitochondria. Plant Cell Physiol. 54(11):1907–1916. [DOI] [PubMed] [Google Scholar]
- Ichinose M, Sugita M.. 2016. RNA editing and its molecular mechanism in plant organelles. Genes 8(1):5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson RW, Qiu YL.. 2008. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol Direct. 3(1):43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. 2013. Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Rüdinger M.. 2010. DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 584(20):4287–4291. [DOI] [PubMed] [Google Scholar]
- Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K.. 2003. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 31(9):2417–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map (sam) format and Samtools. Bioinformatics 25(16):2078–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC.. 2013. Empty Pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25(3):868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Fitzjohn RG.. 2015. The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst Biol. 64(1):127–136. [DOI] [PubMed] [Google Scholar]
- Malek O, Lättig K, Hiesel R, Brennicke A, Knoop V.. 1996. RNA editing in bryophytes and a molecular phylogeny of land plants. Embo J. 15(6):1403–1411. [PMC free article] [PubMed] [Google Scholar]
- Martins EP, Hansen TF.. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat. 149(4):646–667. [Google Scholar]
- Morris JL, et al. 2018. The timescale of early land plant evolution. Proc Natl Acad Sci U S A. 115(10):E2274–E2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Palmer JD.. 2006. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Genet Genomics. 276(3):285–293. [DOI] [PubMed] [Google Scholar]
- Mundry R. 2014. Statistical issues and assumptions of phylogenetic generalised least squares In: Garamszegi LZ, editor. Modern phylogenetic comparative methods and their application in evolutionary biology. New York: Springer; p. 131–153. [Google Scholar]
- Narasimhan V, et al. 2016. Bcftools/roh: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32(11):1749–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V.. 2014. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20(10):1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yang Y, Lesch E, Knoop V, Schallenberg-Rüdinger M.. 2019. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun Biol. 2(1):85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D. 2018. Nlme: Linear and nonlinear mixed effects models. R package version 3. 1–137. Retrieved from https://cran.r-proje ct.org/web/packa ges/nlme/nlme.pdf
- Porebski S, Bailey LG, Bernard RB.. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST.. 2006. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6(1):27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD.. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V.. 2009. RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genomics. 281(5):473–481. [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Polsakiewicz M, Knoop V.. 2008. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol Biol Evol. 25:1405–1414. [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Volkmar U, Lenz H, Grothmalonek M, Knoop V.. 2012. Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria. J Mol Evol. 7:37–51. [DOI] [PubMed] [Google Scholar]
- Sabater B, Martín M, Schmitz-Linneweber C, Maier RM.. 2002. Is clustering of plastid RNA editing sites a consequence of transitory loss of gene function? Implications for past environmental and evolutionary events in plants. Perspect Plant Ecol. 5(2):81–89. [Google Scholar]
- Salone V, et al. 2007. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581(22):4132–4138. [DOI] [PubMed] [Google Scholar]
- Schallenberg-Rüdinger M, Lenz H, Polsakiewicz M, Gott JM, Knoop V.. 2013. A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol. 10(9):1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura M. 2016. Marchantia polymorpha: taxonomy, phylogeny, and morphology of a model system. Plant Cell Physiol. 57(2):230–256. [DOI] [PubMed] [Google Scholar]
- Sloan DB. 2017. Nuclear and mitochondrial RNA editing systems have opposite effects on protein diversity. Biol Lett. 13(8):20170314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N.. 2000. The PPR motif – a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 25(2):46–47. [DOI] [PubMed] [Google Scholar]
- Smith DR. 2009. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 71(6):627–639. [DOI] [PubMed] [Google Scholar]
- Söderström L, et al. 2016. World checklist of hornworts and liverworts. Phytokeys 59:1–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A.. 2013. RNA editing in plants and its evolution. Annu Rev Genet. 47(1):335–352. [DOI] [PubMed] [Google Scholar]
- Villarreal et al. 2018. Genome-wide organellar analyses from the hornwort Leiosporoceros dussii show low frequency of RNA editing. PLoS One 13:e0200491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, et al. 2018. REDO: RNA editing detection in plant organelles based on variant calling results. J Comput Biol. 25(5):509–516. [DOI] [PubMed] [Google Scholar]
- Xue JY, Liu Y, Li L, Wang B, Qiu YL.. 2010. The complete mitochondrial genome sequence of the hornwort Phaeoceros laevis: retention of many ancient pseudogenes and conservative evolution of mitochondrial genomes in hornworts. Curr Genet. 56(1):53–61. [DOI] [PubMed] [Google Scholar]
- Yang YZ, et al. 2017. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 214(2):782–795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.