Abstract
Variable temperature electrospray ionization (ESI) is coupled with mass spectrometry techniques in order to investigate structural transitions of monoclonal antibody immunoglobulin G (IgG) in a 100 mM ammonium acetate (pH 7.0) solution from 26 to 70 °C. At 26 °C, the mass spectrum for intact IgG shows six charge states +22 to +26. Upon increasing the solution temperature the fraction of low-charge states decreases and new, higher-charge state ions are observed. Upon analysis it appears that heating the solution aids in desolvation of the intact IgG precursor. Above, ~50 °C a cleavage event between the light and heavy chains is observed. An analysis of the kinetics for this processes at different temperatures yields transition state thermochemistry of ΔH‡ = 95 ± 10 kJ·mol−1, ΔS‡ = 8 ± 1 J·mol−1·K−1, and ΔG‡ = 92 ± 11 kJ·mol−1. The mechanism for light chain dissociation appears to involve disulfide bond scrambling that ultimately results in a non-native Cys199-Cys217 disulfide bond in the light chain product. Above ~70 °C, we are unable to produce a stable ESI signal. The loss of signal is ascribed to aggregation that is primarily associated with the remaining portion of the antibody after having lost the light chain.
Introduction
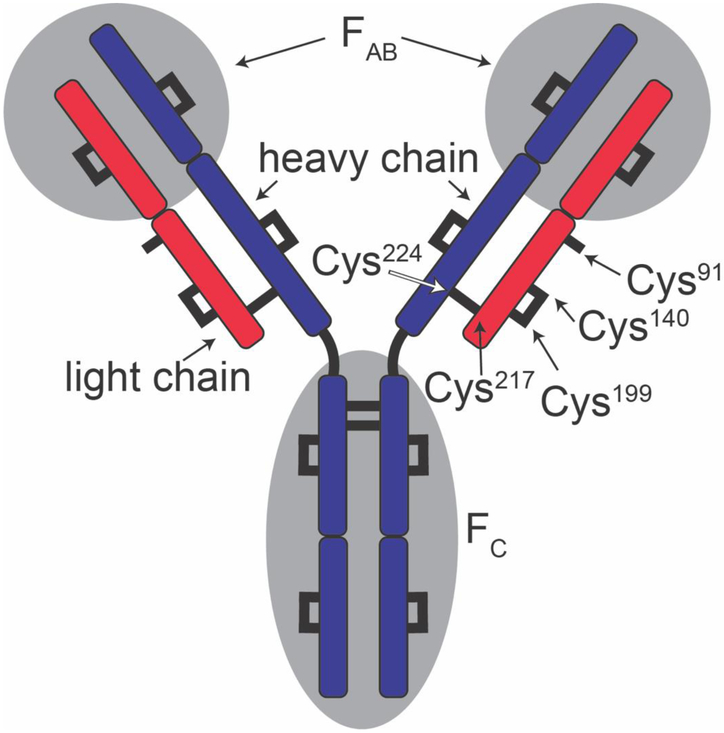
The immunoglobulin G antibody (IgG) is a ~147 kDa protein in the immune system that is involved in antigen recognition and binding.[1] This molecule is often visualized by the ‘Y’ shaped diagram shown in Scheme I. As shown, IgG is comprised of a dimer of heterodimers (the heavy and light chains). The heterodimers are linked by two disulfide bonds. The light and heavy chains of each heterodimer are bound by a single disulfide bond. Together these regions create a highly-specific antigen binding pocket called the FAB portion of the molecule that is critical for immune response.[2] In recent years, numerous monoclonal antibodies with therapeutic value have been introduced.[3-6] Because of this, an understanding of the structures and stabilities of these molecules is of fundamental importance.
Scheme 1:
IgG schematic showing light chain (red), heavy chain (blue), the antigen binding region (FAB) and the crystallizable region (FC). Cys or disulfide bonded cysteines are shown as black lines. Cys residues discussed in main text are labeled. Single free cysteine is shown on each light chain.
Although calorimetric studies of antibodies are routinely carried out in the development and testing of new therapeutic antibodies these methods provide information about only the stability of the ensemble average. That is, the structural change is observed as a two-state cooperative transition, and little is known about the nature of the configurations and mechanisms leading to denatured states.[7, 8] In the work presented below, we investigate the stability of IgG using a new, variable temperature electrospray ionization (vT-ESI) source coupled with mass-spectrometry (MS) measurements.[9, 10] At elevated temperatures (above 50 °C) the MS measurements reveal that the light chain of IgG dissociates, through a mechanism that involves scrambling of the disulfide bonds, resulting in the formation of a light-chain product that adopts non-native Cys199-Cys217 and Cys91-Cys140 disulfide bonds. From an Arrhenius analysis of the kinetics of dissociation at varying temperatures we derive transition state thermochemistry for dissociation process. This thermochemistry is discussed.
The present work builds on a number of new MS-based measurements that are being developed with the aim of understanding structures and structural transitions of biomolecules in solution as well as the gas phase. In the last decade ‘native ESI’ has enabled the study of large complexes.[11-16] Analyses of biomolecular conformations from solutions of varying composition and temperature now have an extensive history.[17-20] Differences in structures found under varying solution conditions can be investigated with a range of reaction chemistries and techniques, including: isotopic hydrogen-deuterium (H-D) exchange;[21-26] fast photochemical oxidation of proteins;[27-30] chemical cross-linking[31-33] and other residue-specific modifications;[34, 35] and ion mobility measurements.[36-46] Once ionized, an array of physical and chemical methods can be used to investigate biomolecular structure in vacuo. These include, low-energy and high-energy collisions with buffer gasses[47-52] and surfaces;[53-55] photodissociation techniques;[56-59] measurements of collision cross sections with many new ion mobility methods;[60-67] ion-molecule reactions, including proton-transfer[68-71] and H-D exchange reactions;[72-75] ion-ion reactions;[76-78] as well as electronic and vibrational spectroscopies.[79-82]
Experimental
Variable-temperature electrospray ionization.
The solution temperature of the ESI emitter, is controlled using a home-built variable temperature electrospray ionization emitter.[9] This interface holds a borosilicate glass ESI emitter which has been pulled to a narrow inner dia. of ~1 to 5 μm using a Flaming/Brown P-97 pipette tip puller (Sutter Instruments, Novato, CA, U.S.A.) The emitter interface is made of a thermally conductive ceramic block that is resistively-heated using a cartridge heater. An ESI voltage of 0.7 to 1.0 kV is applied to a platinum wire that is inserted into the back of the emitter, making electrical connection with the solution. The temperature is measured (to a precision of ± 0.5 °C) using a K-type thermocouple, that is inserted into the ceramic block.
Instrumentation.
Initial experiments were performed on Waters Synapt G2 and remaining kinetics experiments were preformed Waters Synapt G2s instrument. Both instruments were used in with the source interlocks overridden to accommodate the vT-ESI source.[9, 83] Source pressures and voltages that minimize ion activation were used for initial studies, summarized below.[84, 85] Backing pressures were increased to ~ 8 mbar using Speedivalve (Edwards, Burgess Hill, UK). Gas control were optimized to minimize ion activation and increase ion transmission; including flow rates in the source (20.9 mL/min), trap (10 mL/min), helium cell (180 mL/min) and IMS cell (90 mL/min). We additionally optimized and used instrument settings; including sampling cone voltage (62 V), extraction cone (1.6 V), source temperature (50 °C), cone gas (10 L/h), flow gas (0.6 bar), purge gas (100 L/hr), trap collision energy (12.6 collision energy). Finally, TriWave DC voltages were also optimized: entrance voltage (3.2 V), bias (45.3 V), trap DC (1.7 V) and exit voltage (1.3 V). We note that there are different source configurations between the Synapt G2 and the Synapt G2S that can contribute to charge state shifts, presumably due to collisional activation. This may be the origin of the charge state shift between the two sets of mass spectra presented. The analysis presented below uses only the time-of-flight mass analyzer. That is, the quadrupole is fixed to transmit all ions. The data were originally acquired as nested ion mobility mass spectra, however only the mass dimension was used for this analysis as there were no resolvable changes in drift time distributions for these species.
Analysis of the data.
Each dataset was collected by systematically increasing the temperature from 26 to 70 °C. Samples were allowed to incubate at each temperature for at least three minutes. After this time the mass spectra recorded at low solution temperatures (26 to 45 °C) do not appear to change for an extended period and it appears that we have reached an equilibrium (on this timescale). At higher temperatures the antibody dissociates. Kinetics experiments of this process were carried out by increasing the temperature to a set point, and then collecting a series of three minute acquisitions until spray was lost. Unidec[86] (Oxford, UK) was used for deconvolution of native charge state distributions. Data from both experiments were exported using TWIM extract (University of Michigan, Ann Arbor, MI) and processed using Origin2018 (OriginLab Corporation, Northampton, MA, U.S.A.). Kinetic data were fit using a first order reaction rate of formation. Measured rate constants, k, were plotted as a function of the temperature in an Arrhenius plot and fit linearly to obtain transition state chemistry.
Sample preparation.
Immunoglobulin G (IgG1, universal antibody standard, human, ≥ 90% purity) was purchased from Sigma Aldrich (St. Louis, MO, U.S.A.). IgG (1 mg) was resolubilized in 100 mM ammonium acetate solution (pH 7.0, 500 μL). Sample was buffer exchanged using a spin concentrator (molecular weight cut off = 30,000 Da, 100 to 500 μL × 3, Millipore Sigma, Burlington, MA, U.S.A.). IgG was brought to a final concentration of 80 μM in 100 mM ammonium acetate (Sigma Aldrich, St. Louis, MO, U.S.A).
Results and discussion
Changes in ESI charge state distribution of IgG precursor with temperature.
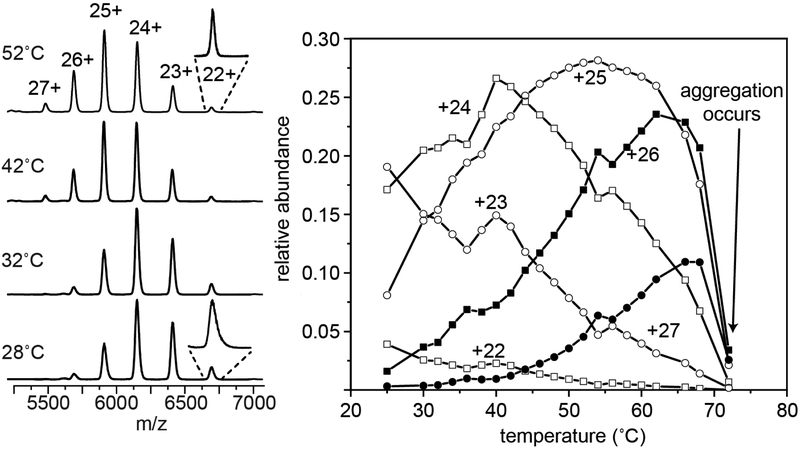
Figure 1 shows representative mass spectra for intact IgG recorded upon electrospraying a 100 mM ammonium acetate solution at 28, 32, 42 and 52 °C. At low temperatures (28 to 42 °C) a narrow distribution of IgG charge states from +22 to +26 and centered at +24 is observed. As the solution temperature is increased beyond this point, a new peak corresponding to the +27 species is observed and the +25 and +26 species increase in relative abundance. When the peaks in the mass spectrum are examined more carefully (see the inset in Figure 1) we find that at low temperatures peaks are broader and extend to higher masses. As the solution temperature is increased each charge state becomes noticeably shaper and the distribution of charge states changes. From ~28 to 42 °C the abundances of the lower-charge state +22 and +23 species decrease with increasing temperature; in this same temperature region the populations of +24 through +27 also increase. Above 42 °C, the +24 species decreases in abundance and the +25 species is favored. This ion reaches a maximum abundance at ~54 °C and decreases above this temperature. The +26 and +27 continue to increase until ~70 °C, where we no longer maintain a stable ion signal. At this temperature, the clear solution becomes turbid due to the formation of insoluble aggregates,[87, 88] likely originating from IgG unfolding. The slight shift in charge state observed here is similar to the that seen when tetrameric concanavalin A is heated, which was coupled to a structural change.[89] While we do not observe a change in the collision cross sections for IgG, there have been reports that IgG undergoes structural changes between 25 and 70 °C under acidic pH.[90, 91] In these cases, highly charged MS peaks emerged at elevated temperatures, indicating that the protein had unfolded. We also find that the peaks decrease in width at elevated temperatures, suggesting that IgG emerges from hot electrospray droplets with fewer species adducted.
Figure 1:
Mass spectrum showing the charge state shift observed for IgG charge states 22 to 27 at 28, 32, 42, and 52 °C (left). Inset shows +22 species at 28 and 52 °C. The relative abundance plotted as a function of temperature for charge states +22, +23, +24, +25, +26 and +27. Above ~70°C spray stability is lost.
High-temperature dissociation of the light chain.
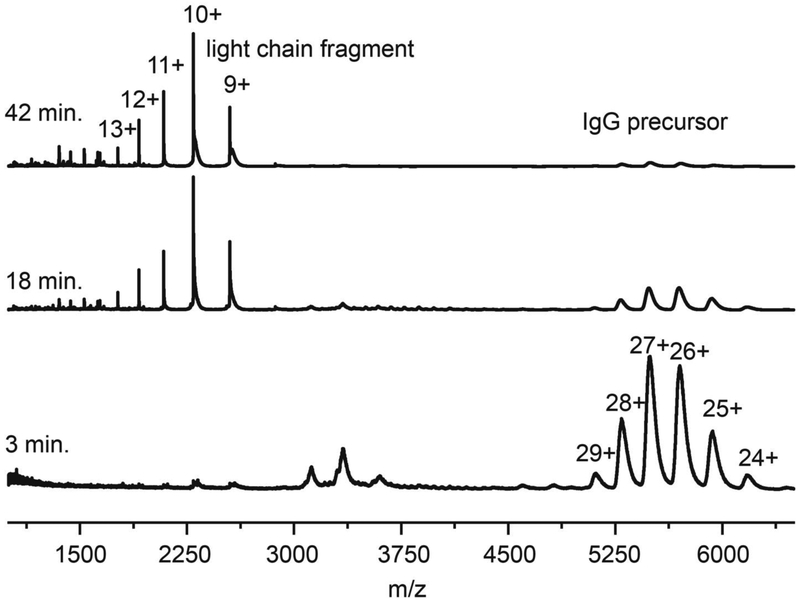
At low temperatures IgG remains stable for long times. We monitored the mass spectra of a solution at 45 °C for up to ~15 hours, and it shows no measureable change. At elevated temperatures IgG is known to dissociate by loss of the light chain.[92, 93] Figure 2 shows mass spectra acquired after incubating solutions at 65 °C for 3, 18 or 42 min. At relatively short incubation times (3 min.) the mass spectrum is dominated by peaks associated with the IgG precursor. At longer times, (18 min. as shown in Figure 2), the relative abundances of the IgG peaks decrease and a new, well-defined set of peaks m/z < 3000 are observed. These peaks increase in magnitude with increasing incubation time. The theoretical molecular weight of the light chain species is 22,942 Da. Using this value, we determine that the major peaks correspond to +9 through +13 charge states of the light chain. Once the charge states are assigned our experimental measurement yields m = 22,943 ± 1 Da, in close agreement with the theoretical value. It is interesting that we do not observe the complementary remaining IgG fragment, which should have m ~ 124 kDa, although aggregation of this species is known to occur rapidly.[88, 92]
Figure 2:
Mass spectrum showing formation of light chain charge states +9 through +13 after incubation at 65 °C for 3, 18, and 42 min.
Mechanism of light chain dissociation.
Before describing the kinetics experiments, we first present a possible mechanism for the covalent bond cleavage leading to the release of the light chain species. Dissociation of the light chain from the heavy chain must involve cleavage of the Cys217-Cys224 disulfide bond, between the two chains (Scheme 1). It is established that during the dissociation of the light chain, disulfide bonds can scramble.[92, 94, 95] The precursor IgG antibody used in our study has a single free Cys91 residue on each of the light chains. As we think about thermodynamic considerations we see that disulfide scrambling upon dissociation of the light chain could stabilize the products. That is, if only the native Cys224-Cys217 bond between the heavy and light chains were cleaved, three free, unbound Cys residues would be available (Cys224 on the heavy chain, and Cys217 and Cys91 on the light chain). Consider one scenario. After the Cys224-Cys217 bond is cleaved the freed light chain might refold and in doing so scramble its disulfide bonds in order to stabilize this product. For example, if the Cys199-Cys140 disulfide bond were also to cleave, we might form a non-native Cys199-Cys217 linkage. In this case the newly freed Cys140 residue could form a second disulfide bond with the Cys91 residue and the resulting covalent Cys140-Cys91 bond would further stabilize the light chain product. With this change, the only free Cys residue is located on the heavy chain (Cys224) and the light chain is no longer covalently linked to the heavy chain. Overall, this process is a disulfide mixing[96, 97] step with nearby Cys residues both making and breaking covalent bonds, a process that has been described previously for light-chain dissociation from IgG under mildly denaturing conditions.[92, 98-101] It is likely that at elevated temperatures a similar disulfide bond scrambling will occur.
With this idea in mind, we carried out studies to identify the location of the non-native disulfide bond. After incubation at 65 °C for 90 min. we alkylated the free Cys residues with iodoacetimide. The products of antibody dissociation were then proteolytically digested and the tryptic peptides that were formed were analyzed using a combination of chromatographic separations with MS detection (see supporting information). An analysis of the cross-linked peptides provides evidence for the non-native Cys199-Cys217 disulfide bond. Although we anticipate that the Cys91-Cys140 disulfide bond should also stabilize this fragment, we did not detect this cross linked peptide in our analysis. We note that it would be difficult for us to fragment this species with our experiment due to its size (60 amino acids); so, the dearth of experimental information does not rule it out entirely. Overall, this result supports the idea that the mechanism for light-chain dissociation involves disulfide bond scrambling.
Kinetics measurements at varying temperatures.
We next carried out a series of kinetics experiments at specified 57, 60, 62, and 65 °C using the vT-ESI source. Because we do not observe the IgG fragment that complements the light chain, we report kinetics based on only the increase in the light chain signal and the decrease of the intact IgG precursor (see supporting information for details). As mentioned above, it is interesting that we do not observe the complementary heavy-chain fragment. We suspect that this species aggregates soon after the dissociation process. At our longest times at each temperature, where the dissociation of the intact IgG precursor approaches completion, the ESI signal is lost, consistent with this idea, and the experiment is terminated.
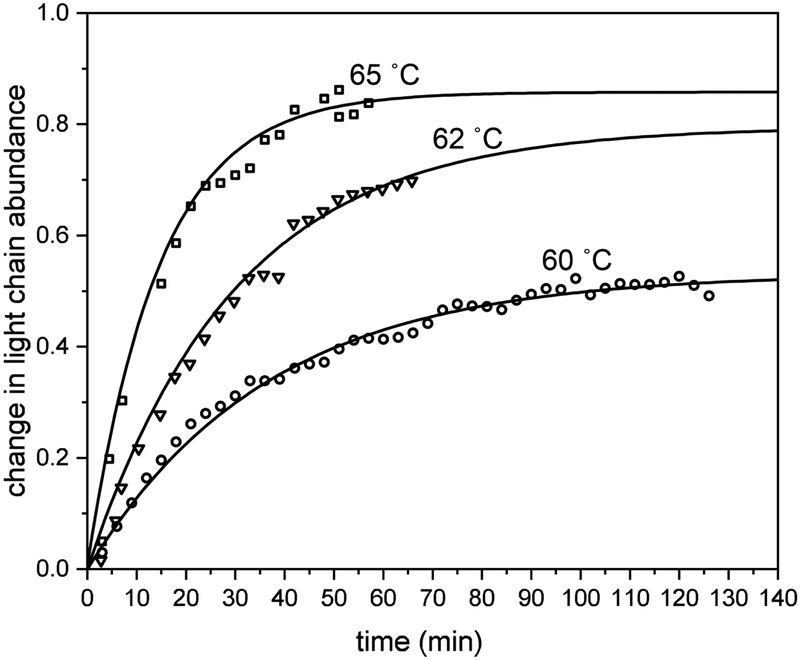
Examples of kinetics data recorded at several temperatures (60, 62, and 65 °C) are shown in Figure 3 for the increase in the light-chain abundance with increasing time (the 57 °C data are not included because the figure appears crowded; but, an example for these data can be found in the supporting information). Examination of the kinetics shows that this process follows a simple, first order reaction rate, and can be modeled with equation 1,
| (1) |
where Bt is the intensity of the light chain signal at time t, I0 is the final light chain signal upon reaction completion, and k is the rate constant.
Figure 3:
Light chain signal monitored over time at 60, 62, and 65 °C, as open squares, triangles, and circles, repectively. Change in light chain abundance with time is shown modeled with first order kinetics (black lines).
Transition state thermochemistry associated with formation of the light chain.
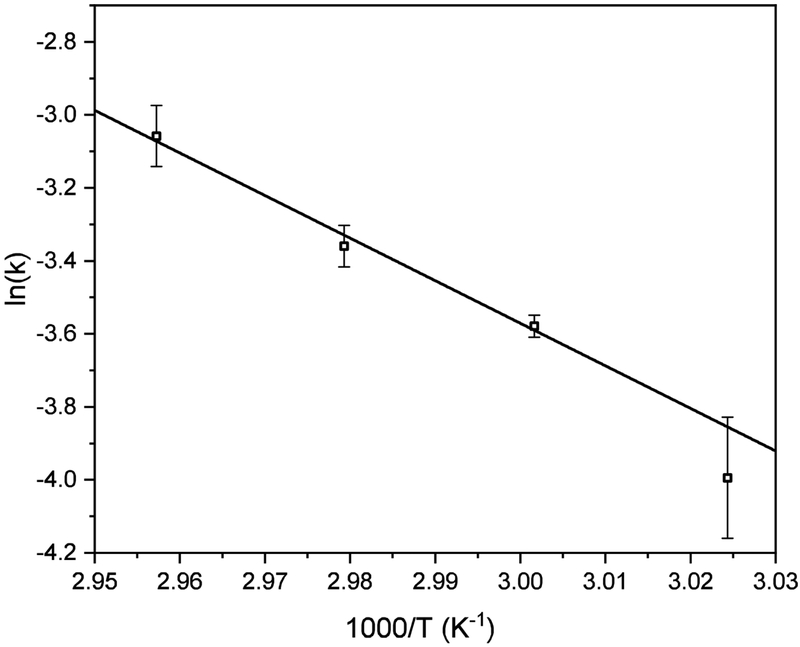
The rate constants obtained from the kinetics experiments shown above were used to generate the Arrhenius plot in Figure 4. This plot yields a pre-exponential factor, A = 4.33 ± 0.8 × 1013 s−1 and activation energy, Ea = 96 ± 28 kJ·mol−1. This value of A indicates that accessing the transition state is very efficient – occurring near the vibrational frequency that is expected for a simple bond cleavage of a small molecule. We suggest that this may indicate that the transition state involves a very localized motion associated with cleavage of the native Cys224-Cys217 bond in the intact precursor IgG. These values can be converted into transition state thermochemistry, yielding: ΔG‡ = 92 ± 11 kJ‡mol−1, ΔH‡ = 95 ± 10 kJ·mol−1, and ΔS‡ = 8 ± 1 J·mol·K−1. The large enthalpic barrier is consistent with the cleavage of a covalent disulfide bond. Furthermore, the relatively small entropy change at the transition state suggests little change in structure. Overall, this thermochemistry is consistent with a sequential process. First, the native Cys224-Cys217 bond in the IgG precursor is cleaved. Upon cleavage the light-chain fragment refolds such that at least one (and possibly two) new non-native disulfide bridges (the scrambled Cys217-Cys199 bond that was detected, and the Cys140-Cys91 bond that we anticipate could be formed, but was not directly detected) are formed, stabilizing the light chain product. The complementary heavy chain product of dissociation, having the single reduced free Cys224 rapidly aggregates, and is not detected in our experiments.
Figure 4:
Arrhenius plot showing triplicate reaction rate of formation versus inverse temperature. Error bars represent the standard deviation of the triplicate analysis.
Conclusions
Variable temperature ESI and mass spectrometry have been used to investigate thermal transitions in the IgG antibody. It is found that IgG dissociates through loss of the light chain, a process that involves disulfide bond scrambling. Kinetics studies at multiple temperatures were used to determine transition state thermochemistry of ΔH‡ = 95 ± 10 kJ·mol−1, ΔS‡ = 8 ± 1 J·mol−1K−1, and ΔG‡ = 92 ± 11 kJ·mol−1.
Supplementary Material
Acknowledgements
This work was supported in part from funds from the National Institutes of Health (5R01GM117207-04, and 5R01GM121751-02).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Vidarsson G, Dekkers G, Rispens T: IgG subclasses and allotypes: from structure to effector functions. Front. Immunol 5, 520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA: The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annu. Rev. Immunol 25, 21–50 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Weiner LM, Surana R, Wang S: Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat. Rev. Immuno 10, 317–327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellman I, Coukos G, Dranoff G: Cancer immunotherapy comes of age. Nature. 480, 480–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P, Allison JP: The future of immune checkpoint therapy. Science. 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Pardoll DM: The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer. 12, 1–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieberman RL, D’aquino JA, Ringe D, Petsko GA: Effects of pH and Iminosugar Pharmacological Chaperones on Lysosomal Glycosidase Structure and Stability. Biochemistry. 48, 4816–4827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez JMJ, Renisio JG, Prompers JJ, van Platerink CJ, Cambillau C, Darbon H, Frenken LGJ: Thermal Unfolding of a Llama Antibody Fragment: A Two-State Reversible Process. Biochemistry. 40, 74–83 (2001). [DOI] [PubMed] [Google Scholar]
- 9.El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, Russell DH, Clemmer DE: Melting Proteins: Evidence for Multiple Stable Structures upon Thermal Denaturation of Native Ubiquitin from Ion Mobility Spectrometry-Mass Spectrometry Measurements. J. Am. Chem. Soc 139, 6306–6309 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Abzalimov RR, Kaltashov IA: Direct monitoring of heat-stressed biopolymers with temperature-controlled electrospray ionization mass spectrometry. Anal. Chem 83, 2870–2876 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Chorev DS, Baker LA, Wu D, Beilsten-Edmands V, Rouse SL, Zeev-Ben-Mordehai T, Jiko C, Samsudin F, Gerle C, Khalid S, Stewart AG, Matthews SJ, Grünewald K, Robinson CV: Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science. 362, 829–834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liko I, Degiacomi MT, Mohammed S, Yoshikawa S, Schmidt C, Robinson CV: Dimer interface of bovine cytochrome c oxidase is influenced by local posttranslational modifications and lipid binding. Proc. Natl. Acad. Sci 113, 8230–8235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Loo RRO, Loo JA: Structural Characterization of a Thrombin-Aptamer Complex by High Resolution Native Top-Down Mass Spectrometry. J Am Soc Mass Spectrom. 28, 1815–1822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinkse MWH, Maier CS, Kim J-I, Oh B-H, Heck AJR: Macromolecular assembly of Helicobacter pylori urease investigated by mass spectrometry. J. Mass Spectrom 38, 315–320 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Heck AJR: Native mass spectrometry: a bridge between interactomics and structural biology. Nat Meth. 5, 927–933 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Susa AC, Xia Z, Tang HYH, Tainer JA, Williams ER: Charging of Proteins in Native Mass Spectrometry. J Am Soc Mass Spectrom. 28, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung EW, Henriques DA, Renzoni D, Morton CJ, Mulhern TD, Pitkeathly MC, Ladbury JE, Robinson CV: Probing the nature of interactions in SH2 binding interfaces - Evidence from electrospray ionization mass spectrometry. Protein Sci. 8, 1962–1970 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudgins RR, Woenckhaus J, Jarrold MF: High resolution ion mobility measurements for gas phase proteins: correlation between solution phase and gas phase conformations. Int. J. Mass. Spectrom 165, 497–507 (1997). [Google Scholar]
- 19.Winger BE, Hofstadler SA, Bruce JE, Udseth HR, Smith RD: High-Resolution Accurate Mass Measurements of Biomolecules Using a New Electrospray-Ionization Ion-Cyclotron Resonance Mass-Spectrometer. J Am Soc Mass Spectrom. 4, 566–577 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Li J, Taraszka JA, Counterman AE, Clemmer DE: Influence of solvent composition and capillary temperature on the conformations of electrosprayed ions: unfolding of compact ubiquitin conformers from pseudonative and denatured solutions. Int. J. Mass. Spectrom 185, 37–47 (1999). [Google Scholar]
- 21.Wales TE, Engen JR: Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spec Rev. 25, 158–170 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR: Characterization of IgG1 Conformation and Conformational Dynamics by Hydrogen/Deuterium Exchange Mass Spectrometry. Anal. Chem 81, 2644–2651 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houde D, Peng Y, Berkowitz SA, Engen JR: Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 9, 1716–1728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katta V, Chait BT: Hydrogen/deuterium exchange electrospray ionization mass spectrometry: a method for probing protein conformational changes in solution. J. Am. Chem. Soc 115, 6317–6321 (1993). [Google Scholar]
- 25.Konermann L, Pan J, Liu Y-H: Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR: Probing Protein Ligand Interactions by Automated Hydrogen/Deuterium Exchange Mass Spectrometry. Anal. Chem 78, 1005–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Li KS, Shi L, Gross ML: Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization. Acc. Chem. Res 51, 736–744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones LM, B Sperry J, A Carroll J, Gross ML: Fast Photochemical Oxidation of Proteins for Epitope Mapping. Anal. Chem 83, 7657–7661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gau BC, Sharp JS, Rempel DL, Gross ML: Fast Photochemical Oxidation of Protein Footprints Faster than Protein Unfolding. Anal. Chem 81, 6563–6571 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chea EE, Jones LM: Modifications generated by fast photochemical oxidation of proteins reflect the native conformations of proteins. Protein Science. 27, 1047–1056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl F-U, Ban N, Malmstroem L, Aebersold R: Structural Probing of a Protein Phosphatase 2A Network by Chemical Cross-Linking and Mass Spectrometry. Science. 337, 1348–1352 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R: Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 9, 1634–1649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauber MA, Reilly JP: Novel Amidinating Cross-Linker for Facilitating Analyses of Protein Structures and Interactions. Anal. Chem 82, 7736–7743 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Ji J, Chakraborty A, Geng M, Zhang X, Amini A, Bina M, Regnier F: Strategy for qualitative and quantitative analysis in proteomics based on signature peptides. J. Chromatogr. B Biomed. Sci. Appl 745, 197–210 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R: Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 17, 994–999 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE: Number of Solution States of Bradykinin from Ion Mobility and Mass Spectrometry Measurements. J. Am. Chem. Soc 133, 13810–13813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H, Pierson NA, Valentine SJ, Clemmer DE: Conformation Types of Ubiquitin [M+8H] 8+Ions from Water:Methanol Solutions: Evidence for the N and A States in Aqueous Solution. J. Phys. Chem. B 116, 3344–3352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi H, Clemmer DE: Evidence for Two New Solution States of Ubiquitin by IMS–MS Analysis. J. Phys. Chem. B 118, 3498–3506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L, Holliday AE, Shi H, Zhu F, Ewing MA, Russell DH, Clemmer DE: Characterizing intermediates along the transition from polyproline I to polyproline II using ion mobility spectrometry-mass spectrometry. J. Am. Chem. Soc 136, 12702–12711 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Shi H, Atlasevich N, Merenbloom SI, Clemmer DE: Solution Dependence of the Collisional Activation of Ubiquitin [M + 7H]7+ Ions. J Am Soc Mass Spectrom. 25, 2000–2008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi L, Holliday AE, Glover MS, Ewing MA, Russell DH, Clemmer DE: Ion Mobility-Mass Spectrometry Reveals the Energetics of Intermediates that Guide Polyproline Folding. J Am Soc Mass Spectrom. 27, 22–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemmer DE, Russell DH, Williams ER: Characterizing the Conformationome: Toward a Structural Understanding of the Proteome. Acc. Chem. Res 50, 556–560 (2017). [DOI] [PubMed] [Google Scholar]
- 43.El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, Russell DH, Clemmer DE: Melting Proteins: Evidence for Multiple Stable Structures upon Thermal Denaturation of Native Ubiquitin from Ion Mobility Spectrometry-Mass Spectrometry Measurements. J. Am. Chem. Soc 139, 6306–6309 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Fuller DR, Conant CR, El-Baba TJ, Brown CJ, Woodall DW, Russell DH, Clemmer DE: Conformationally Regulated Peptide Bond Cleavage in Bradykinin. J. Am. Chem. Soc 140, 9357–9360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein SL, Wyttenbach T, Baumketner A, Shea J-E, Bitan G, Teplow DB, Bowers MT: Amyloid β-Protein: Monomer Structure and Early Aggregation States of Aβ42 and Its Pro-19 Alloform. J. Am. Chem. Soc 127, 2075–2084 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT: Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nature Chemistry. 1, 326–331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian Y, Han L, Buckner AC, Ruotolo BT: Collision Induced Unfolding of Intact Antibodies: Rapid Characterization of Disulfide Bonding Patterns, Glycosylation, and Structures. Anal. Chem 87, 151015204733009–11515 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Tian Y, Ruotolo BT: Collision induced unfolding detects subtle differences in intact antibody glycoforms and associated fragments. Int. J. Mass. Spectrom 425, 1–9 (2018). [Google Scholar]
- 49.Dong S, Wagner ND, Russell DH: Collision-Induced Unfolding of Partially Metalated Metallothionein-2A: Tracking Unfolding Reactions of Gas-Phase Ions. Anal. Chem 90, 11856–11862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin SA, Biemann K: A comparison of keV atom bombardment mass spectra of peptides obtained with a two-sector mass spectrometer with those from a four-sector tandem mass spectrometer. Int J Mass Spectrom Ion Process. 78, 213–228 (1987). [Google Scholar]
- 51.Johnson RS, Martin SA, Biemann K: Collision-induced fragmentation of (M + H)+ ions of peptides. Side chain specific sequence ions. Int J Mass Spectrom Ion Process. 86, 137–154 (1988). [Google Scholar]
- 52.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth HR, Smith RD, Clemmer DE: An IMS–IMS Analogue of MS–MS. Anal. Chem 78, 4161–4174 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Zhou M, Jones CM, Wysocki VH: Dissecting the large noncovalent protein complex GroEL with surface-induced dissociation and ion mobility-mass spectrometry. Anal. Chem 85, 8262–8267 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Quintyn RS, Harvey SR, Wysocki VH: Illustration of SID-IM-SID (surface-induced dissociation-ion mobility-SID) mass spectrometry: homo and hetero model protein complexes. Analyst. 140, 7012–7019 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Dongré AR, Somogyi A, Wysocki VH: Surface-induced dissociation: an effective tool to probe structure, energetics and fragmentation mechanisms of protonated peptides. J. Mass Spectrom 31, 339–350 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW: Infrared Multiphoton Dissociation of Large Multiply Charged Ions for Biomolecule Sequencing. Anal. Chem 66, 2809–2815 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Shaffer CJ, Marek A, Pepin R, Slovakova K, Turecek F: Combining UV photodissociation with electron transfer for peptide structure analysis. J. Mass Spectrom 50, 470–475 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Wilson JJ, Brodbelt JS: MS/MS Simplification by 355 nm Ultraviolet Photodissociation of Chromophore-Derivatized Peptides in a Quadrupole Ion Trap. Anal. Chem 79, 7883–7892 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Valentine SJ, Reilly JP, Clemmer DE: Analyzing a mixture of disaccharides by IMS-VUVPD-MS. Int. J. Mass. Spectrom 309, 161–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ewing MA, Glover MS, Clemmer DE: Hybrid ion mobility and mass spectrometry as a separation tool. Journal of Chromatography A. 1439, 1–23 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Liu FC, Ridgeway ME, Park MA, Bleiholder C: Tandem trapped ion mobility spectrometry. Analyst. 143, 2249–2258 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Valentine SJ, Koeniger SL, Clemmer DE: A Split-Field Drift Tube for Separation and Efficient Fragmentation of Biomolecular Ions. Anal. Chem 75, 6202–6208 (2003). [DOI] [PubMed] [Google Scholar]
- 63.May JC, Russell DH: A Mass-Selective Variable-Temperature Drift Tube Ion Mobility-Mass Spectrometer for Temperature Dependent Ion Mobility Studies. J Am Soc Mass Spectrom. 22, 1134–1145 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Donohoe GC, Maleki H, Arndt JR, Khakinejad M, Yi J, McBride C, Nurkiewicz TR, Valentine SJ: A new ion mobility-linear ion trap instrument for complex mixture analysis. Anal. Chem 86, 8121–8128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez-Lima F, Kaplan DA, Suetering J, Park MA: Gas-phase separation using a trapped ion mobility spectrometer. Int J Ion Mobil Spectrom. 14, 93–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A Shvartsburg A, A Anderson G, D Smith R: Pushing the Frontier of High-Definition Ion Mobility Spectrometry Using FAIMS. Mass Spectrom (Tokyo). 2, S0011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyttenbach T, Bushnell JE, Bowers MT: Salt Bridge Structures in the Absence of Solvent? The Case for the Oligoglycines. J. Am. Chem. Soc 120, 5098–5103 (1998). [Google Scholar]
- 68.Ogorzalek Loo RR, Winger BE, Smith RD: Proton transfer reaction studies of multiply charged proteins in a high mass-to-charge ratio quadrupole mass spectrometer. J Am Soc Mass Spectrom. 5, 1064–1071 (1994). [DOI] [PubMed] [Google Scholar]
- 69.Valentine SJ, Counterman AE, Clemmer DE: Conformer-dependent proton-transfer reactions of ubiquitin ions. J Am Soc Mass Spectrom. 8, 954–961 (1997). [Google Scholar]
- 70.He F, Ramirez J, Lebrilla CB: Evidence for an Intermolecular Proton-Transfer Reaction Induced by Collision in Gas-Phase Noncovalently Bound Complexes. J. Am. Chem. Soc 121, 4726–4727 (1999). [Google Scholar]
- 71.Gross DS, Schnier PD, Rodriguez-Cruz SE, Fagerquist CK, Williams ER: Conformations and folding of lysozyme ions in vacuo. Proc. Natl. Acad. Sci 93, 3143–3148 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLafferty FW, Guan Z, Haupts U, Wood TD, Kelleher NL: Gaseous Conformational Structures of Cytochrome c. J. Am. Chem. Soc 120, 4732–4740 (1998). [Google Scholar]
- 73.Bohrer BC, Atlasevich N, Clemmer DE: Transitions between elongated conformations of ubiquitin [M+11H]11+ enhance hydrogen/deuterium exchange. J. Phys. Chem. B 115, 4509–4515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valentine SJ, Clemmer DE: H/D exchange levels of shape-resolved cytochrome c conformers in the gas phase. J. Am. Chem. Soc 119, 3558–3566 (1997). [Google Scholar]
- 75.Valentine SJ, Anderson JG, Ellington AD, Clemmer DE: Disulfide-intact and - reduced lysozyme in the gas phase: Conformations and pathways of folding and unfolding. J. Phys. Chem. B 101, 3891–3900 (1997). [Google Scholar]
- 76.Li H, Nguyen HH, Loo RRO, Campuzano IDG, Loo JA: An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nature Chemistry. 10, 139–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loo RRO, Loo JA: Salt Bridge Rearrangement (SaBRe) Explains the Dissociation Behavior of Noncovalent Complexes. J Am Soc Mass Spectrom. 27, 975–990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hogan Jason M, Pitteri Sharon J, Chrisman Paul A, A., McLuckey SA: Complementary Structural Information from a Tryptic N-Linked Glycopeptide via Electron Transfer Ion/Ion Reactions and Collision-Induced Dissociation. J. Proteome Res 4, 628–632 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rijs AM, Oomens J: IR Spectroscopic Techniques to Study Isolated Biomolecules. Top Curr Chem. 364, 1–42 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Forbes MW, Bush MF, Polfer NC, Oomens J, Dunbar RC, Williams ER, Jockusch RA: Infrared Spectroscopy of Arginine Cation Complexes: Direct Observation of Gas-Phase Zwitterions. J. Phys. Chem. A 111, 11759–11770 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Ben Faleh A, Warnke S, Rizzo TR: Combining ultrahigh-resolution ion-mobility spectrometry with cryogenic IR spectroscopy for the analysis of glycan mixtures. Anal. Chem 91, acs.analchem.9b00659–4882 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Banyasz A, Douki T, Improta R, Gustavsson T, Onidas D, Vayá I, Perron M, Markovitsi D: Electronic Excited States Responsible for Dimer Formation upon UV Absorption Directly by Thymine Strands: Joint Experimental and Theoretical Study. J. Am. Chem. Soc 134, 14834–14845 (2012). [DOI] [PubMed] [Google Scholar]
- 83.El-Baba TJ, Fuller DR, Woodall DW, Raab SA, Conant CR, Dilger JM, Toker Y, Williams ER, Russell DH, Clemmer DE: Melting proteins confined in nanodroplets with 10.6 μm light provides clues about early steps of denaturation. Chemical Communications. 54, 3270–3273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV: Ion mobility–mass spectrometry analysis of large protein complexes. Nat Protoc. 3, 1139–1152 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Sobott Frank, Hernández Helena, McCammon Margaret G, Tito Mark A, A., Robinson CV: A Tandem Mass Spectrometer for Improved Transmission and Analysis of Large Macromolecular Assemblies. Anal. Chem 74, 1402–1407 (2002). [DOI] [PubMed] [Google Scholar]
- 86.Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV: Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal. Chem 87, 4370–4376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL: Design of therapeutic proteins with enhanced stability. Proc. Natl. Acad. Sci 106, 11937–11942 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vermeer AWP, Norde W: The Thermal Stability of Immunoglobulin: Unfolding and Aggregation of a Multi-Domain Protein. Biophysical Journal. 78, 394–404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Baba TJ, Clemmer DE: Solution thermochemistry of concanavalin A tetramer conformers measured by variable-temperature ESI-IMS-MS. Int J Mass Spectrom. 443, 93–100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brader ML, Estey T, Bai S, Alston RW, Lucas KK, Lantz S, Landsman P, Maloney KM: Examination of Thermal Unfolding and Aggregation Profiles of a Series of Developable Therapeutic Monoclonal Antibodies. Mol. Pharmaceutics 12, 1005–1017 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Wang G, Bondarenko PV, Kaltashov IA: Multi-step conformational transitions in heat-treated protein therapeutics can be monitored in real time with temperature-controlled electrospray ionization mass spectrometry. Analyst. 143, 670–677 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Brody T: Multistep denaturation and hierarchy of disulfide bond cleavage of a monoclonal antibody. Analytical Biochemistry. 247, 247–256 (1997). [DOI] [PubMed] [Google Scholar]
- 93.Kamerzell TJ, Li M, Arora S, Ji JA, Wang YJ: The relative rate of immunoglobulin gamma 1 fragmentation. J Pharm Sci. 100, 1341–1349 (2011). [DOI] [PubMed] [Google Scholar]
- 94.Lakbub JC, Shipman JT, Desaire H: Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins. Anal Bioanal Chem. 410, 2467–2484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Lu Q, Wu SL, Karger BL, Hancock WS: Characterization and comparison of disulfide linkages and scrambling patterns in therapeutic monoclonal antibodies: using LC-MS with electron transfer dissociation. Anal. Chem 83, 3133–3140 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X, Kumar S, Singh SK: Disulfide scrambling in IgG2 monoclonal antibodies: insights from molecular dynamics simulations. Pharm. Res 28, 3128–3144 (2011). [DOI] [PubMed] [Google Scholar]
- 97.Salas-Solano O, Tomlinson B, Du S, Parker M, Strahan A, Ma S: Optimization and Validation of a Quantitative Capillary Electrophoresis Sodium Dodecyl Sulfate Method for Quality Control and Stability Monitoring of Monoclonal Antibodies. Anal. Chem 78, 6583–6594 (2006). [DOI] [PubMed] [Google Scholar]
- 98.Arosio P, Rima S, Morbidelli M: Aggregation mechanism of an IgG2 and two IgG1 monoclonal antibodies at low pH: from oligomers to larger aggregates. Pharm. Res 30, 641–654 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Roberts CJ: Therapeutic protein aggregation: mechanisms, design, and control. Trends in Biotechnology. 32, 372–380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vlasak J, Ionescu R: Fragmentation of monoclonal antibodies. mAbs. 3, 253–263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moore JM, Patapoff TW, Cromwell ME: Kinetics and thermodynamics of dimer formation and dissociation for a recombinant humanized monoclonal antibody to vascular endothelial growth factor. Biochemistry. 38, 13960–13967 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.