Abstract
Early life stress (ELS) is associated with an increased risk of depression and this association may be mediated by epigenetic mechanisms. A previous epigenome-wide DNA methylation (DNAm) study investigating human newborns and two animal models of ELS suggested that the epigenetic regulator MORC1 is differentially methylated following ELS. The ELS-induced DNAm alterations were long-lasting in the animal models. However, whether this finding is also transferable to humans experiencing ELS in childhood was not investigated. Further, MORC1 may provide a link between ELS and adult depression, as MORC1 DNAm and genetic variants were found to be associated with depressive symptoms in humans. In the present study, we investigated the validity of MORC1 DNAm as a biomarker of ELS in humans and its role in linking ELS to depression later in life by studying childhood maltreatment. We analyzed whole blood MORC1 DNAm in an adult cohort (N=151) that was characterized for both the presence of depressive symptoms and childhood maltreatment. Further, we investigated the association between MORC1 DNAm, depressive symptoms and childhood maltreatment in two additional cohorts (N=299, N=310). Overall, our data do not indicate an association of MORC1 DNAm with childhood maltreatment. An association of MORC1 DNAm with depressive symptoms was present in all cohorts, but was inconsistent in the specific CpG sites associated and the direction of effect (Tuebingen cohort: standardized β=0.16, unstandardized β=0.01, 95% CI [−0.0004, −0.0179], p=0.061, PReDICT cohort: standardized β=−0.12, unstandardized β=−0.01, 95% CI [−0.0258, −0.0003], p=0.045), Grady cohort: standardized β=0.16, unstandardized β=0.008, 95% CI [0.0019, 0.0143], p=0.01). Our study thus suggests that peripheral MORC1 DNAm cannot serve as biomarker of childhood maltreatment in adults, but does provide further indication for the association of MORC1 DNAm with depressive symptoms.
Keywords: Epigenetics, DNA methylation, childhood trauma, early life stress, depression
Introduction
DNA methylation (DNAm) is a chemical modification of cytosine bases in the DNA and a highly conserved epigenetic mark (Law and Jacobsen, 2010). It is present throughout the genome, but most frequently found and best studied in the context of CpG dinucleotides (Jones, 2012). DNAm affects gene transcription by changing chromatin structure and modifying the affinity to transcription factors and other DNA-binding proteins (Boyes and Bird, 1991; Eden and Cedar, 1994; Kass et al., 1993). DNAm in somatic tissues, including blood, is dynamic to some extent (Ziller et al., 2013). Factors influencing blood DNAm are cell type composition, genetic variants, age, sex, and also environmental exposures and lifestyle factors, such as smoking, nutrition and stress (Lam et al., 2012). Therefore, blood DNAm has been extensively studied as a predictor and biomarker for diseases that are characterized by a strong influence of environmental risk factors, such as stress-related psychiatric disorders (Cortessis et al., 2012; Klengel et al., 2014). For example, many studies report associations between blood DNAm and major depressive disorder (MDD) (Menke and Binder, 2014). Data from family and twin studies suggests that the heritability of MDD is approximately 40 %, while the remaining variance is attributed to environmental risk factors (Rice et al., 2002).
One important environmental risk factor for adult depression is early life stress (ELS) (Pietrek et al., 2013; Syed and Nemeroff, 2017), a broad term describing various forms of stress experienced early in life. Different types of ELS range from maternal stressors affecting the developing fetus including in utero exposure to harmful substances, childhood maltreatment or traumatic experiences early in life. The risk of depression later in life is particularly increased by the occurrence of childhood abuse, neglect, lack of caregiver, and parental loss (Brown et al., 1999; de Carvalho Tofoli et al., 2011). This effect is believed, in part, to be facilitated through long-lasting influences of ELS on the endogenous stress regulation system, the hypothalamus-pituitary-adrenal axis (de Carvalho Tofoli et al., 2011), and through immunological effects (Baumeister et al., 2016; Slavich and Irwin, 2014). It has been proposed that this translation of psychological stressors to long-lasting, biological changes is partly mediated by DNAm (Hornung and Heim, 2014).
In support of this theory, recent studies have identified a number of DNAm loci associated with ELS (Bustamante et al., 2016; Kang et al., 2013; Klengel et al., 2013; Nieratschker et al., 2014; Weder et al., 2014). These were found within genes that are known to be relevant to the pathogenesis of depression (i.e. SLC6A4 (Kang et al., 2013)) or the stress response (i.e. NR3C1 (Bustamante et al., 2016), FKBP5 (Klengel et al., 2013)), and also within genes that have no prior connection to stress-related psychiatric disorders (Labonte et al., 2012; Nieratschker et al., 2014; Suderman, 2014; Weder et al., 2014; Yang et al., 2013). One of these, MORC1, was identified in a cross-species epigenome-wide association study (EWAS), where its promoter was hypomethylated in rats, rhesus macaques, and human newborns that had experienced different forms of ELS (Nieratschker et al., 2014). The differential methylation was present in the brain and in peripheral tissue (blood cells). MORC1 is a highly conserved nuclear protein that is increasingly recognized as an epigenetic regulator (Li et al., 2013). In plants, the MORC1 homolog AtMORC1 plays an important role in heterochromatin condensation and gene silencing (Dong et al., 2018) and accumulating evidence suggest that it is implicated in gene silencing in humans and animals as well (Koch et al., 2017). Although human MORC1 was initially believed to be expressed exclusively in male testis, it is now known that MORC1 mRNA is present in nearly all tissues (Fishilevich et al., 2016). The protein may be linked to neuropsychiatric disorders, as has been suggested for other members of the MORC protein family (Boukas et al., 2018). Providing further support for this, MORC1 was found to be genetically associated with MDD in a European study cohort (N=1968) (Rietschel et al., 2010). Animal studies confirmed this association by showing that MORC1 knockout mice display a depression-like phenotype (Schmidt et al., 2016). In addition, epigenetic modification of MORC1 has been linked to depression by a study showing that MORC1 promoter methylation in buccal tissue was associated with depressive symptoms in healthy adults (N=60) (Mundorf et al., 2018). Even though additional evidence is needed, these data suggest that MORC1 may provide a link between ELS and depression later in life (Nieratschker et al., 2014). In support of this, assessment of DNAm across different time points in the animal models indicated that the established ELS responsive MORC1 DNAm pattern persisted into adulthood. However, the stability of the differential MORC1 DNAm in response to ELS was only investigated in animals and it has not yet been studied whether this pattern persists into adulthood in humans (Nieratschker et al., 2014). Therefore, we investigated MORC1 promoter DNAm in blood in a cohort of N=151 depressive patients and healthy controls and evaluated the association of DNAm with depressive symptoms, as assessed by the Beck Depression Inventory (BDI-II) and ELS, as assessed by the Childhood Trauma Questionnaire (CTQ). In addition, we acquired MORC1 DNAm data from a subset of the Grady Trauma Project Cohort (N=310) (Gillespie et al., 2009) and the PReDICT cohort (N=299) (Dunlop et al., 2012) to validate our findings.
Methods
In all steps of the analysis, samples were processed in balanced design in order to avoid batch effects.
MORC1 targeted bisulfite sequencing of human blood samples:
Study population:
N=151 participants were included in the study. Depressive patients were recruited at the Department of Psychiatry and Psychotherapy, University of Tuebingen and had received a diagnosis of major depression by trained psychiatrists and/or psychologists according to the fourth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) (American Psychiatric Association, 2013). Healthy controls were recruited via public announcements and screened to be free of any current and/or history of mental illness using the Mini-International Neuropsychiatric Interview (MINI) (Sheehan et al., 1998). Study participants were phenotypically characterized with the self-report questionnaires Symptom Checklist 90 - Revised (SCL90R) (Franke, 2002), Childhood Trauma Questionnaire (CTQ) (Bernstein et al., 2003) and Beck Depression Inventory II (BDI-II) (Beck, 1996). Further questionnaires assessed demographic information and information about smoking behavior. Complete blood counts were available from 56 % of the participants. All subjects were of Caucasian origin and gave written informed consent prior to participation in the study, which was approved by the ethics committee of the University of Tuebingen and was conducted in accordance with the Declaration of Helsinki. There was no overlap between the present sample and previous study cohorts where differential MORC1 DNAm had been analyzed (Mundorf et al., 2018; Nieratschker et al., 2014).
Sampling and DNA extraction:
Venous blood was obtained from all participants at study inclusion, collected in Ethylenediaminetetraacetic (EDTA) tubes and stored at −80 °C until further analysis. DNA was extracted using the QIAamp DNA Blood Maxi-Kit (Qiagen).
DNA methylation analysis:
We selected five different regions within the promoter of the MORC1 gene covering 56 CpG sites for methylation analysis (1.5kb upstream and 0.5kb downstream of the transcription start site, Chr3:108836490-108838489, hg19). The primer sequences, amplicons and genomic location of the analyzed CpG sites relative to the region analyzed by the previous study of Nieratschker and colleagues (Nieratschker et al., 2014) can be found in the supplementary material (Supplementary Data S4 and S5). Targeted bisulfite sequencing and processing were performed as previously described in Roeh and colleagues (Roeh et al., 2018). Briefly, sample DNAs as well as five methylated DNA standards (0, 25, 50 ,75, 100 % methylated) were bisulfite converted in triplicate using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions and then pooled. PCRs were performed for each sample (primer sequences in Supplementary Data S2) using the PyroMark PCR Kit (Qiagen), and successful amplification was verified via agarose gel electrophoresis. The five amplicons per sample were pooled equimolarly and purified with Agencourt AMPure XP beads (Beckman Coulter GmbH, Krefeld, Germany) to remove excess primers and genomic DNA. The Illumina TruSeq DNA PCR-Free HT Library Prep Kit (Illumina, San Diego, CA. Cat. No. 20015963) was used to prepare sequencing libraries (500 ng of bisulfite DNA per sample) according to the manufacturer’s protocol. Samples were multiplexed and indexed using Illumina TruSeq DNA CD Indexes (Illumina, San Diego, CA. Cat. No. 20015949). Quality assessment and quantification of the libraries were performed using an Agilent’s 4200 TapeStation (Agilent Technologies, Waldbronn, Germany) and the Kapa HIFI Library quantification kit - KK4824 (Kapa Biosystems Inc., Wilmington, MA. Cat. No. 07960140001). Libraries were then pooled equimolarly and treated with an adaptor blocking reagent before the denaturation step (Illumina Free Adapter Blocking Reagent. Cat. No. 20024144) to reduce index hopping rates. Sequencing was performed on an Illumina MiSeq using the Reagent Kit v3 – 2 x 300 bp paired-end reads (Illumina, San Diego,CA; Cat. No. MS-102-3003), with 30 % PhiX added. Read quality was verified using FastQC (Andrews, 2010), and adapter sequences were trimmed using cutadapt v.2.0 (Martin, 2012). Bismark v.0.20.0 was used to align reads to the reference sequence (Krueger and Andrews, 2011). Methylation levels were quantified using the R package methylKit (Akalin et al., 2012) with a minimum quality Phred score of 30. Two samples were excluded because they had less than 95 % bisulfite conversion efficiency (as assessed by C/T ratio in non-CpG context). A standard curve and regression line based on methylated standards was calculated for each site, and sites with R2 < 0.95 were excluded from the analysis (six sites). All sites and samples had a median coverage > 1000x.
Statistical analysis:
Statistical analysis was performed using R version 3.5.1. Multiple linear regression was used to predict DNAm at each CpG site, using CTQ and BDI scores as predictors and sex, age and nicotine consumption as covariates. When predicting DNAm from CTQ scores, BDI was treated as covariate and vice versa. Strength of the association was assessed by calculating partial correlation estimates using the R package RVAideMemoire (Hervé, 2019). For each CpG site, samples more than three times the interquartile range away from the 25th and 75th percentile were classified as outlier and excluded from the analysis. Correction for multiple testing for the number of CpG sites tested per dataset was performed using false discovery rate adjustment (FDR). To identify potential confounding effects of cellular blood composition, the correlations between blood cell counts and predictor variables (BDI and CTQ) were assessed in the 66 % of samples where information about blood cell composition was available, using the first principal component (PC) derived from lymphocyte, monocyte and neutrophil percentages. There was no significant correlation between blood composition and the predictors of interest. To compare the three datasets Tuebingen, PReDICT and Grady, a random-effects meta-analysis was performed on the standardized regression coefficient of interest using the R package meta (Schwarzer, 2007) and Empirical Bayes as τ2 estimator.
MORC1 methylation analysis in Grady and PReDICT cohort
Study population:
The GRADY cohort was recruited at the Grady Memorial Hospital in Atlanta, Georgia (Davis et al., 2008). All participants were from an urban population of low socioeconomic status and characterized by high prevalence and severity of trauma over the lifetime (Gillespie et al., 2009; Zannas et al., 2015). The PReDICT cohort consisted of treatment-naïve patients (18-65 years) with moderate-to-severe MDD, recruited at two sites in Atlanta associated with the Emory University School of Medicine (Dunlop et al., 2012). All participants met criteria for current major depressive disorder (MDD) as defined in the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) (American Psychiatric Association, 2000). All subjects provided written informed consent and ethical approval was given by the Institutional Review Board or Ethical Committee of each site participating in every study. Subjects were characterized for depressive symptoms by the BDI-I (PReDICT) or BDI-II (Grady) and for ELS by CTQ. Smoking scores in each cohort were calculated according to Elliott et al. (Elliott et al., 2014). From both cohorts, we selected all participants of which depression questionnaire, CTQ and DNAm was present. This resulted in 310 samples from Grady and 299 samples from PReDICT.
DNA methylation data:
Whole blood DNA was extracted using standard techniques. DNA methylation was measured using Illumina Infinium HumanMethylation450K BeadChips. Beta values were normalized using functional normalization (Aryee et al., 2014; Fortin et al., 2014). Batch effects were removed using ComBat (Johnson et al., 2007). Subsequently, all CpGs on sex chromosomes and CpGs with single nucleotide polymorphisms (SNPs) in the probe sequence were removed. Additionally, probes were removed if the detection p-value was > 0.01 in at least 25% of the samples, the probe contained SNPs in the single base pair (bp) extension or CpG position, the probe had missing beta values, or was a cross-reactive probe (Chen et al., 2013). We used the Houseman method to estimate cell type composition (Houseman et al., 2012). Methylation data were adjusted for cell type composition using probe-wise multiple linear regression normalization, wherein all principal components generated from the estimated cell type compositions were used as predictors. Modelling and regression were performed using the R package limma (Ritchie et al., 2015). Only CpG sites within the MORC1 promoter were selected for analysis (12 sites).
Genotype analysis:
In all cohorts, genome-wide SNP genotyping was performed using Illumina HumanOmniExpress BeadChips and subsequently exported via Illumina’s GenomeStudio. Quality control in all cohorts was independently performed in PLINK. Samples with low genotyping rate (< 98%) were removed. SNPs with high rate of missing data (> 2%), significant deviation from the Hardy-Weinberg equilibrium (HWE, p < 10−5), or a low minor allele frequency (MAF < 5%) were excluded from further analyses. Afterwards, we performed MDS-analysis on the qc-ed genotypes.
Statistical analysis:
Statistical analysis was performed equivalently to the analysis for the Tuebingen cohort. To assess for effects of genotype, we performed PCA on the genome-wide SNP data and included the first two axes of variation (PCs) as covariates. Additional covariates were sex, age and smoking behavior.
Results
I. Cohort statistics
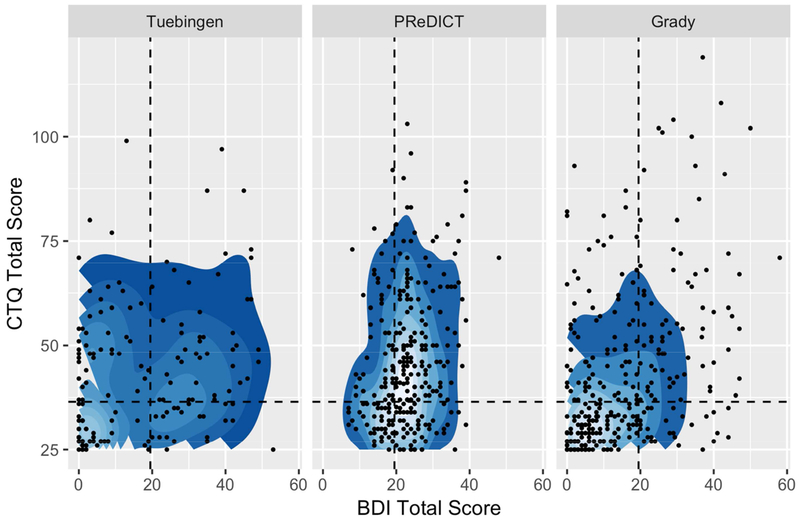
The three cohorts were similar with regards to CTQ total score and percentage of study participants with moderate to severe abuse/neglect in one or more subcategories. The average BDI total score was in the range of mild depression for the Tuebingen (full cohort: 17.5 ± 15.8; diagnosed depression: 27.7 ± 13.2, healthy controls: 4.3 ± 6.4) and Grady cohort (15.6 ± 12.6), and slightly higher (moderate depression) in the PReDICT cohort (22.7 ± 7.2). Further, there were differences in sex distribution and percentage of cases with diagnosed depression, with PReDICT containing only cases with diagnosed depression as compared to 56.3 % in the Tuebingen cohort (data not available in Grady). The Grady cohort contained the lowest amount of female participants (25.5 % vs. 57.5 % and 66.9 % in PReDICT and Tuebingen) (Table 1). Figure 1 shows that the distribution of study participants with regards to CTQ and BDI total score was most balanced in the Tuebingen cohort. The PReDICT cohort had a similar spread of CTQ scores, but a very narrow window of BDI values with low variance and a mean above the threshold for moderate to severe depression. Study participants in the Grady cohort were more balanced. In addition, this cohort contained more extreme cases (CTQ > 75 or BDI > 40).
Table 1:
Overview of cohort statistics in Tuebingen, PReDICT and Grady. Data given in percent with patient number in brackets or as mean ± standard deviation for average scores. For CTQ subscales, number of patients with moderate or severe abuse/neglect in this category were counted as cases (Bernstein et al., 2003).Data on neglect subscales was not available for the Grady cohort.
| Tuebingen (N=151) | PReDICT (N=299) | Grady (N=310) | ||
|---|---|---|---|---|
| Age | 32.7 ± 11.7 | 40.2 ± 11.8 | 42.0 ± 13.1 | |
| % Females | 66.9 % (101) | 57.5 % (172) | 25.5 % (79) | |
| % Diagnosed depression | 56.3 % (85) | 100 % (299) | n.a. | |
| Average BDI total score | 17.5 ± 15.8 | 22.7 ± 7.2 | 15.6 ± 12.6 | |
| Average CTQ total score | 43.4 ± 15.8 | 45.9 ± 15.8 | 43.0 ± 18.4 | |
| % | Emotional abuse | 34.4 % (52) | 46.2 % (138) | 39.7 % (123) |
| Cases | Physical abuse | 9.27 % (14) | 12.0 % (36) | 15.2 % (47) |
| Sexual abuse | 18.5 % (28) | 22.4 % (67) | 32.9 % (102) | |
| Emotional neglect | 35.1 % (53) | 32.3 % (97) | n.a. | |
| Physical neglect | 21.9 % (33) | 27.1 % (81) | n.a. | |
| One or more of the above | 58.3 % (88) | 65.6 % (196) | 51.0 % (158) | |
Fig. 1:
Overview of distribution of BDI and CTQ total scores in the three cohorts. Each datapoint represents one study participant. Blue shapes are 2D kernel density estimations performed using R package MASS (Venables W.N., 2002). Dotted lines are thresholds for moderate to severe abuse/neglect in at least one subcategory of CTQ (CTQ total > 36) and moderate to severe depressive symptoms (BDI total > 19)(Beck, 1996)).
II. No association of MORC1 DNAm with ELS
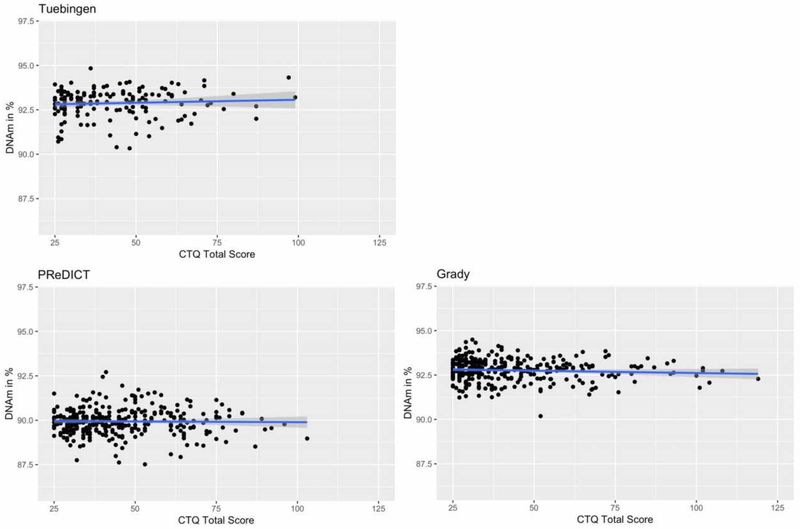
CTQ total score was not a significant predictor of DNAm at any CpG site tested within the MORC1 promoter, neither in the Tuebingen cohort, nor in PReDICT or Grady. The same was found for mean DNAm across all analyzed MORC1 CpG sites (12 sites in PReDICT and Grady, 50 in Tuebingen cohort) (Fig. 2). The interaction between lifetime diagnosis of depression and ELS on MORC1 DNAm could only be tested in the Tuebingen cohort and did not show any significant interaction (data not shown).
Fig. 2:
Mean MORC1 promoter DNAm in % plotted against the CTQ total score in Tuebingen, PReDICT and Grady cohort. In PReDICT and Grady, mean DNAm derives from 12 CpG sites within MORC1 promoter, whereas in the Tuebingen cohort, mean DNAm derives from 50 CpG sites. DNAm values are corrected for covariates.
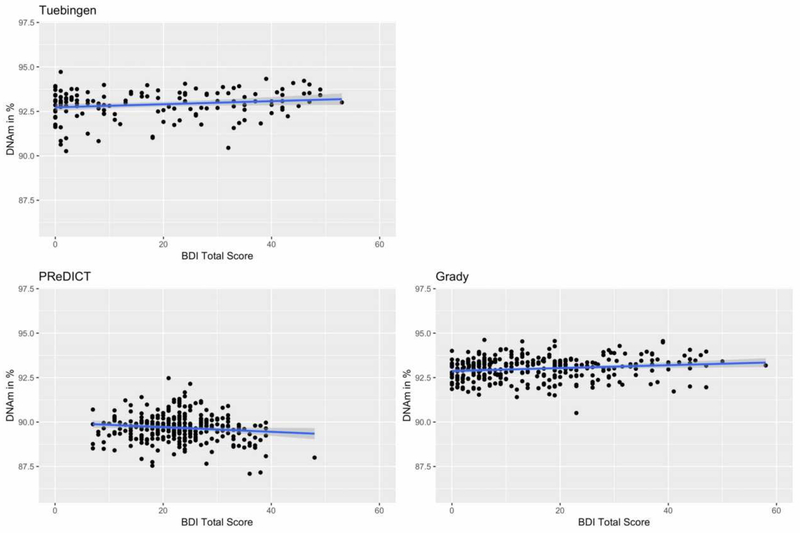
III. Association of MORC1 DNAm with depressive symptoms
Depression score was a nominally significant predictor of DNAm at one CpG site within the MORC1 promoter in the Tuebingen cohort and five sites within Grady (Table 2) (see Supplementary Data S4 and S5 for an overview of genomic location of these sites and those analyzed in previous studies (Mundorf et al., 2018; Nieratschker et al., 2014)). Two sites in the Grady cohort withstood FDR-correction. The mean DNAm across all analyzed sites was significantly associated with the depression score in PReDICT and Grady. However, this association was negative for PReDICT (standardized β=−0.12, unstandardized β=−0.01, 95% CI [−0.0258, −0.0003], p=0.045), and positive for Grady (standardized β=0.16, unstandardized β=0.008, 95% CI [0.0019, 0.0143], p=0.01). In the Tuebingen cohort, the mean DNAm was not significantly associated with depression score, though there was a trend towards a positive association (standardized β=0.16, unstandardized β=0.01, 95% CI [−0.0004, −0.0179], p=0.061) (Fig. 3). The absolute differences in DNAm between the group of individuals without depressive symptoms and those with severe depressive symptoms were small (< 2 %) (Supplementary Data S6). A random-effects meta-analysis for the effect of depressive symptoms on mean DNAm in the three datasets was not significant (standardized β estimate = 0.06, p=0.58).
Table 2:
Overview of partial correlation estimates between the depression score and the DNAm at single CpG sites, using PC1+2 generated from genotype data (for Grady and PReDICT only), sex, age, smoking behavior and CTQ total score as covariates. Results shown for Tuebingen (N=151), PReDICT (N=299) and Grady (N=310) datasets. Only sites that were nominally significant in at least one of the three datasets are listed. Total number of analyzed sites was 50 for Tuebingen and 12 for Grady and PReDICT cohort. Unstandardized betas for depression score are shown in brackets and in italic letters. Both estimates are rounded to two decimal places, abs<x denotes that the absolute value is smaller than x. Statistical significance of depression score as predictor of DNAm was assessed using multiple regression analysis. Nominally significant associations are marked in bold and designated with * =p-value<=0.05, **=p-value<=0.01, ***=p-value<=0.001. Sites are described by genomic position (hg19). Results that withstand FDR-correction are underlined. Mean DNAm was calculated from all analyzed sites (50, 12, 12).
| Tuebingen | PReDICT | Grady | |
|---|---|---|---|
| Ch3:108.836.796 | 0.17 * (0.03) | NA | NA |
| Chr3:108.836.878 | 0.06 (0.01) | −0.1 (−0.01) | 0.17 * (0.01) |
| Chr3:108.837.069 | 0.01 (abs<0.01) | −0.05 (−0.01) | 0.1 * (0.01) |
| Chr3:108.837.086 | abs<0.01 (abs< 0.01) | abs<0.01 (−0.01) | 0.16 ** (0.02) |
| Chr3:108.837.088 | 0.03 (abs<0.01) | −0.07 (−0.02) | 0.19 *** (0.02) |
| Chr3:108.837.620 | NA | abs<0.01(abs<0.01) | 0.13 * (0.01) |
| Mean of all analyzed sites | 0.15 (0.01) | −0.06 * (−0.01) | 0.15 * (0.01) |
Fig. 3:
Mean MORC1 promoter DNAm in % plotted against the depression score in Tuebingen, PReDICT and Grady cohort. In PReDICT and Grady, mean DNAm derives from 12 CpG sites within the MORC1 promoter, whereas in the Tuebingen cohort, mean DNAm derives from 50 CpG sites. DNAm values are corrected for covariates.
The detailed results of all analyses are in the supplementary materials (Supplementary Data S1). Age, sex, smoking and genotype (if available) were included as covariates in all models. Of the identified hits, only one CpG site (Chr.108836796 ) was also significantly associated with one of the covariates (age) and this effect was only present in the Tuebingen cohort (standardized β=−0.21, unstandardized β=−0.02, 95% CI [−0.0343, −0.0182], p=0.048). Further, age was a significant predictor of mean DNAm in the Grady cohort (standardized β=--0.15, unstandardized β=−0.01, 95% CI [−0.0135, −0.0022], p=0.048).
Discussion
Our study is the first to investigate the effect of childhood trauma on blood MORC1 DNAm in adults and its potential link to depression. Although we did not find evidence for an association of MORC1 DNAm with childhood trauma, our data indicates that MORC1 DNAm status is associated with current depressive symptoms. It was previously reported that MORC1 was differentially methylated in cord blood cells of newborns whose mothers had experienced stress during pregnancy, in the T cells of rhesus macaques subjected to maternal separation, and in prefrontal cortex tissue of adult rats whose mothers had experienced chronic restraint stress during pregnancy (Nieratschker et al., 2014). These combined findings provided reason to hypothesize that MORC1 might be a general, system-wide indicator of ELS that is valid for different types of stressors in multiple species and persists into adulthood. However, our results do not support this hypothesis among human adults that experienced ELS. Important differences between the current and previous studies are the method of DNAm analysis, the type and timing of stressor in the human cohort (prenatal vs. postnatal), and the type of tissue investigated. While previous results had been generated using methylated DNA immunoprecipitation (MeDIP) (Mohn et al., 2009), the results presented here were generated using single CpG site resolution methods, such as high-accuracy targeted bisulfite sequencing (Roeh et al., 2018) and 450k Illumina Bead Chip analysis (Bibikova et al., 2011). Despite their strong positive overall correlation (Clark et al., 2012), results obtained from immunoprecipitation vs. bisulfite conversion based methods are not necessarily comparable (Jeong et al., 2016). Most importantly, these methods also differ in specificity. While signals in bisulfite sequencing and 450k Illumina array derive from 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC), MeDIP is selective for 5-mC. Therefore, it is possible that the differential MORC1 DNAm signal was masked by 5-hmC, an epigenetic mark whose significance is increasingly acknowledged (Branco et al., 2011).
It is also plausible, however, that the previously observed association of MORC1 DNAm (Nieratschker et al., 2014) with ELS is not transferable to childhood trauma in humans. We assessed childhood trauma using the CTQ, a self-report questionnaire that is well-established and frequently used for this purpose (Martins et al., 2011). Nevertheless, the type of stress induced by abuse or neglect in childhood is different from prenatal stress and maternal separation, which were investigated in the previous study. In addition, there is a long time interval (several years) between stress exposure and MORC1 DNAm assessment in our study. It is therefore possible that other environmental factors, such as nutrition, lifestyle factors, stressful events later in life, as well as pharmacotherapy or psychotherapy have overridden potential ELS-responsive DNAm patterns within the MORC1 promoter. These confounding factors were not present in the previous study, where animals were kept in a controlled environment after ELS exposure (Nieratschker et al., 2014). Lastly, while the previous results were obtained in hematopoietic stem cells and T cells, here, we analyzed DNAm in whole blood. Hematopoietic stem cells are present through the entire lifespan and may, therefore, carry methylation patterns that have been established early in life. Similarly, T cells are likely to encode epigenetic ELS marks, as their lifespan is among the longest of all blood cells and they inherit patterns from hematopoietic progenitor cells. Further, T cells may be of high relevance to ELS since it is believed that ELS exposure induces long-lasting immune alterations which contribute to vulnerability to stress-related psychiatric disorders later in life (Fagundes et al., 2013). Such immune alterations can be encoded by DNAm patterns in T cells (Morales-Nebreda et al., 2019), as has already been shown for rhesus macaques that experienced maternal deprivation (Provencal et al., 2012), humans with low early life socioeconomic status (Borghol et al., 2012) and PTSD patients (Smith et al., 2011). In whole blood, MORC1 ELS signature in T cells might be masked by methylation patterns of neutrophils, monocytes and other short-lived cell types that are unlikely to encode long-term responses to ELS. We validated our findings in two additional whole blood cohorts, PReDICT and Grady, and found that all three datasets did not show an association of ELS and MORC1 methylation. Most importantly, if MORC1 links ELS and vulnerability to depression later in life, we would expect to see an interaction between the two in predicting MORC1 DNAm. This could only be tested in the Tuebingen cohort, where no such effect was evident. In conclusion, our data suggest that there is no association between childhood trauma and MORC1 DNAm in whole blood of adults. However, further studies are needed in order to elucidate whether ELS responsive MORC1 DNAm patterns are present in specific immune cell types, such as T cells.
In contrast to the ELS data, we found that the average methylation calculated over all analyzed CpG sites was significantly associated with depressive symptoms in the Grady and PReDICT cohort. Further, we identified single CpG sites associated with depression score in the PReDICT and Tuebingen cohorts. These results are consistent with previous findings indicating a role of MORC1 in depression (Mundorf et al., 2018; Nieratschker et al., 2014; Schmidt et al., 2016). Previously, positive correlations of MORC1 DNAm and depressive symptoms were reported for buccal cells (Mundorf et al., 2018) and our results indicate the same for blood. It is not known how MORC1 DNAm in these peripheral tissues is related to DNAm in the brain. However, both tissues contain immune cells and are therefore relevant for the study of depression independently due to the important role of stress and inflammation in the pathogenesis of depression (Slavich and Irwin, 2014; Strawbridge et al., 2017). The mechanisms underlying the association of MORC1 and depression are not yet understood, but possibly involve the action of MORC1 as epigenetic regulator of neuropsychiatric genes. In support of this hypothesis, it was shown that MORC1 knockout mice displayed altered expression of hippocampal brain-derived neurotrophic factor (BDNF), a protein that is widely implicated in psychiatric disorders (Autry and Monteggia, 2012). Further, MORC1 itself may be directly relevant for neuropsychiatric disease, as has been suggested for other members of the MORC protein family (Boukas et al., 2018). Lastly, MORC1 plays an important role in plant immunity and may also be involved in the human immune response (Koch et al., 2017). At the same time, disturbance of the immune system is often reported in depressive patients (Strawbridge et al., 2017) and may be part of the pathogenesis of depression (Slavich and Irwin, 2014). Therefore, it is also plausible, that MORC1 is linked to depression through its role in the immune system.
However, a meta-analysis of the three studies did not reveal a significant effect of depression score on DNAm due to the differences in sites associated with depressive symptoms in different cohorts and the inconsistent direction of the effect. Positive associations, i.e. higher DNAm in individuals with higher depression scores, were found in the Tuebingen and Grady cohort. These results are consistent with a previous study that reported a positive association of BDI scores with buccal MORC1 DNAm in a cohort of N=60 healthy adults (Mundorf et al., 2018). Since higher promoter methylation is generally associated with less gene transcription (Chodavarapu et al., 2010), these results suggest that individuals with depressive symptoms express lower levels of MORC1 protein. This is consistent with animal experiments showing that MORC1 knockout leads to a depressive-like phenotype in mice (Schmidt et al., 2016), though it remains to be proven that the observed effects indeed affect MORC1 gene expression. In contrast, DNAm was negatively associated with depressive symptoms in PReDICT. As PReDICT contains almost no participants without symptoms of clinical depression (BDI-I ≥ 10 for 97 % of the subjects), this could indicate that the association of depressive symptoms with MORC1 DNAm is not linear. A nonlinear, adaptive relationship between environmental exposures and DNAm has been suggested previously (Vaiserman, 2010) and this pattern was supported by a study reporting an inverted U-shape association of DNAm with exposure to organic pollutants (Na et al., 2014). In addition, PReDICT is the only treatment-naive cohort and information on medication intake in Grady and Tuebingen was not available. It is therefore possible, that intake of antidepressant medication in the Grady and Tuebingen cohort have affected MORC1 DNAm (Menke and Binder, 2014). Lastly, the least evidence for an association of depressive symptoms with MORC1 DNAm was found in the Tuebingen cohort, where only one site was significantly associated with depressive symptoms; however this result did not withstand correction for multiple testing. There was a trend towards an association of depressive symptoms with the mean DNAm across all 50 analyzed sites, but the effect was not significant (p=0.06). This may be because of lack of statistical power, as this cohort is the smallest one in size. Further, cell type composition was available only for a subset of the cohort and therefore not considered in the analysis. Even though we did not find evidence for an association of cell type composition with BDI score, it is still possible that the results in the full cohort are confounded by differences in cell type composition. In addition, genotype data were not available in this cohort. However, genotype was not a significant predictor of MORC1 DNAm in Grady and PReDICT and removing it as covariate did not change the results obtained in these cohorts (data not shown). In line with this, we found that the database of methylation quantitative trait loci (mQTLdb) (Gaunt et al., 2016) lists only small effects of SNPs on DNAm within the MORC1 promoter (difference in median DNAm between homozygote groups <1 %). Therefore, we assume that the lack of genotype data in the Tuebingen cohort did not confound the analysis. Altogether, strongest evidence for an association of MORC1 DNAm with depressive symptoms was found in the Grady cohort, where multiple sites were significantly associated with depressive symptoms and two sites withstood correction for multiple testing. While, the absolute differences in DNAm between study participants with low and high levels of depressive symptoms were small (< 2 %), such effect sizes are very common in observational studies of human cohorts and therefore not unexpected. Therefore, the overall results in the three independent cohorts indicate that there might be an association of MORC1 with depression and the inconsistent findings may be attributed to differences in the cohorts. In conclusion, there is previous evidence for an association of MORC1 DNAm and depression and our study supports these findings, but further validation in additional datasets is necessary.
Supplementary Material
Acknowledgements
We thank the patients, staff, and faculty of the Grady Trauma Project and the PReDICT study for sharing their data. We also thank the study participants of the Tuebingen cohort and the medical staff of the University Hospital Tuebingen for their support. We thank all funding agencies for their financial support of this study.
Conflict of Interest Statement
Financial disclosures for CEN:
Research/Grants: National Institutes of Health (NIH), Stanley Medical Research Institute
Consulting (last three years): Xhale, Takeda, Taisho Pharmaceutical Inc., Bracket (Clintara), Fortress Biotech, Sunovion Pharmaceuticals Inc., Sumitomo Dainippon Pharma, Janssen Research & Development LLC, Magstim, Inc., Navitor Pharmaceuticals, Inc., TC MSO, Inc., Intra-Cellular Therapies, Inc.
Stockholder: Xhale, Celgene, Seattle Genetics, Abbvie, OPKO Health, Inc., Antares, BI Gen Holdings, Inc., Corcept Therapeutics Pharmaceuticals Company, TC MSO, Inc., Trends in Pharma Development, LLC
Scientific Advisory Boards: American Foundation for Suicide Prevention (AFSP), Brain and Behavior Research Foundation (BBRF), Xhale, Anxiety Disorders Association of America (ADAA), Skyland Trail, Bracket (Clintara), Laureate Institute for Brain Research (LIBR), Inc.
Board of Directors: AFSP, Gratitude America, ADAA, Xhale Smart, Inc.
Income sources or equity of $10,000 or more: American Psychiatric Publishing, Xhale, Bracket (Clintara), CME Outfitters, Takeda, Intra-Cellular Therapies, Inc., Magstim, EMA Wellness
Patents:Method and devices for transdermal delivery of lithium (US 6,375,990B1) Method of assessing antidepressant drug therapy via transport inhibition of monoamine neurotransmitters by ex vivo assay (US 7,148,027B2)
Compounds, Compositions, Methods of Synthesis, and Methods of Treatment (CRF Receptor Binding Ligand) (US 8,551, 996 B2)
Financial disclosers for WEC:
WEC is a board member of Hugarheill ehf, an Icelandic company dedicated to the prevention of depression, and he receives book royalties from John Wiley & Sons. His research is also supported by the Mary and John Brock Foundation and the Fuqua family foundations. He is a consultant to the George West Mental Health Foundation and is a member of the Scientific Advisory Boards of the ADAA and AIM for Mental Health.
Financial disclosers for BD:
BD has served as a consultant to Aptinyx and Myriad Neuroscience. He receives research support from Takeda
All other authors declare that they have nothing to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Availability of data and material
The Tuebingen dataset used in the current study is available from the corresponding author on request. The raw methylation data and all related phenotypes for the Grady Trauma Project cohort have been deposited into NCBI GEO (GSE72680).
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE, 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13(10), R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association, 2000. Diagnostic and Statistical Manual of Mental Disorders (4th edition, Text revision), Washington, DC. [Google Scholar]
- American Psychiatric Association, 2013. Diagnostic and Statistical Manual of Mental Disorders (5th edition). Washington, DC. [Google Scholar]
- Andrews S, 2010. FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10), 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, Monteggia LM, 2012. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol Rev 64(2), 238–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister D, Akhtar R, Ciufolini S, Pariante CM, Mondelli V, 2016. Childhood trauma and adulthood inflammation: a meta-analysis of peripheral C-reactive protein, interleukin-6 and tumour necrosis factor-alpha. Mol Psychiatry 21(5), 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AT, Steer RA, Brown GK, 1996. Manual for the Beck Depression Inventory-II. Psychological Corporation, San Antonio, Texas. [Google Scholar]
- Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T, Stokes J, Handelsman L, Medrano M, Desmond D, Zule W, 2003. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl 27(2), 169–190. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth P, Gunderson KL, Fan JB, Shen R, 2011. High density DNA methylation array with single CpG site resolution. Genomics 98(4), 288–295. [DOI] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, Hertzman C, Power C, Szyf M, 2012. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol 41(1), 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukas L, Havrilla JM, Quinlan AR, Bjornsson HT, Hansen KD, 2018. Co-expression patterns define epigenetic regulators associated with neurological dysfunction. bioRxiv, 219097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J, Bird A, 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64(6), 1123–1134. [DOI] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W, 2011. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 13(1), 7–13. [DOI] [PubMed] [Google Scholar]
- Brown J, Cohen P, Johnson JG, Smailes EM, 1999. Childhood abuse and neglect: specificity of effects on adolescent and young adult depression and suicidality. J Am Acad Child Adolesc Psychiatry 38(12), 1490–1496. [DOI] [PubMed] [Google Scholar]
- Bustamante AC, Aiello AE, Galea S, Ratanatharathorn A, Noronha C, Wildman DE, Uddin M, 2016. Glucocorticoid receptor DNA methylation, childhood maltreatment and major depression. J Affect Disord 206, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R, 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8(2), 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M, 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466(7304), 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C, Palta P, Joyce CJ, Scott C, Grundberg E, Deloukas P, Palotie A, Coffey AJ, 2012. A comparison of the whole genome approach of MeDIP-seq to the targeted approach of the Infinium HumanMethylation450 BeadChip((R)) for methylome profiling. PLoS One 7(11), e50233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW, 2012. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet 131(10), 1565–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RG, Ressler KJ, Schwartz AC, Stephens KJ, Bradley RG, 2008. Treatment barriers for low-income, urban African Americans with undiagnosed posttraumatic stress disorder. J Trauma Stress 21(2), 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho Tofoli SM, Von Werne Baes C, Martins CMS, Juruena M, 2011. Early life stress, HPA axis, and depression. Psychology & Neuroscience 4(2), 229–234. [Google Scholar]
- Dong W, Vannozzi A, Chen F, Hu Y, Chen Z, Zhang L, 2018. MORC Domain Definition and Evolutionary Analysis of the MORC Gene Family in Green Plants. Genome Biol Evol 10(7), 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop BW, Binder EB, Cubells JF, Goodman MM, Kelley ME, Kinkead B, Kutner M, Nemeroff CB, Newport DJ, Owens MJ, Pace TW, Ritchie JC, Rivera VA, Westen D, Craighead WE, Mayberg HS, 2012. Predictors of remission in depression to individual and combined treatments (PReDICT): study protocol for a randomized controlled trial. Trials 13, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Cedar H, 1994. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev 4(2), 255–259. [DOI] [PubMed] [Google Scholar]
- Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, Davey Smith G, Hughes AD, Chaturvedi N, Relton CL, 2014. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics 6(1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes CP, Glaser R, Kiecolt-Glaser JK, 2013. Stressful early life experiences and immune dysregulation across the lifespan. Brain Behav Immun 27(1), 8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S, Zimmerman S, Kohn A, Iny Stein T, Olender T, Kolker E, Safran M, Lancet D, 2016. Genic insights from integrated human proteomics in GeneCards. Database (Oxford) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD, 2014. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 15(12), 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke GH, 2002. Symptom-Checkliste von L.R. Derogatis - Deutsche Version (SCL-90-R). Beltz Test, Göttingen. [Google Scholar]
- Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, Zheng J, Duggirala A, McArdle WL, Ho K, Ring SM, Evans DM, Davey Smith G, Relton CL, 2016. Systematic identification of genetic influences on methylation across the human life course. Genome Biol 17, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ, 2009. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry 31(6), 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé M, 2019. RVAideMemoire: Testing and Plotting Procedures for Biostatistics, R package version 0.9-73. https://CRAN.R-project.org/package=RVAideMemoire. [Google Scholar]
- Hornung OP, Heim CM, 2014. Gene–Environment Interactions and Intermediate Phenotypes: Early Trauma and Depression. Frontiers in Endocrinology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson H, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. Bmc Bioinformatics 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HM, Lee S, Chae H, Kim R, Kwon MJ, Oh E, Choi YL, Kim S, Shin YK, 2016. Efficiency of methylated DNA immunoprecipitation bisulphite sequencing for whole-genome DNA methylation analysis. Epigenomics 8(8), 1061–1077. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A, 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1), 118–127. [DOI] [PubMed] [Google Scholar]
- Jones PA, 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7), 484–492. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, Shin IS, Shin MG, Yoon JS, 2013. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol Biol Psychiatry 44, 23–28. [DOI] [PubMed] [Google Scholar]
- Kass SU, Goddard JP, Adams RL, 1993. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol 13(12), 7372–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB, 2013. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 16(1), 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D, 2014. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology 80, 115–132. [DOI] [PubMed] [Google Scholar]
- Koch A, Kang HG, Steinbrenner J, Dempsey DA, Klessig DF, Kogel KH, 2017. MORC Proteins: Novel Players in Plant and Animal Health. Front Plant Sci 8, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR, 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27(11), 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, Mechawar N, Szyf M, Meaney MJ, Turecki G, 2012. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry 69(7), 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, Kobor MS, 2012. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A 109 Suppl 2, 17253–17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE, 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3), 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DQ, Nair SS, Kumar R, 2013. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics 8(7), 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2012. Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinformatics in Action 17(1), 10–12. [Google Scholar]
- Martins CMS, Tofoli a.M.d.C., Baes CVW, Juruena M, 2011. Analysis of the occurrence of early life stress in adult psychiatric patients: a systematic review. J Psychology & Neuroscience 4, 219–227. [Google Scholar]
- Menke A, Binder EB, 2014. Epigenetic alterations in depression and antidepressant treatment. Dialogues Clin Neurosci 16(3), 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Schubeler D, Roloff TC, 2009. Methylated DNA immunoprecipitation (MeDIP). Methods Mol Biol 507, 55–64. [DOI] [PubMed] [Google Scholar]
- Morales-Nebreda L, McLafferty FS, Singer BD, 2019. DNA methylation as a transcriptional regulator of the immune system. Transl Res 204, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundorf A, Schmitz J, Gunturkun O, Freund N, Ocklenburg S, 2018. Methylation of MORC1: A possible biomarker for depression? J Psychiatr Res 103, 208–211. [DOI] [PubMed] [Google Scholar]
- Na YK, Hong HS, Lee DH, Lee WK, Kim DS, 2014. Effect of body mass index on global DNA methylation in healthy Korean women. Mol Cells 37(6), 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieratschker V, Massart R, Gilles M, Luoni A, Suderman MJ, Krumm B, Meier S, Witt SH, Nothen MM, Suomi SJ, Peus V, Scharnholz B, Dukal H, Hohmeyer C, Wolf A, Cirulli F, Gass P, Sutterlin MW, Filsinger B, Laucht M, Riva MA, Rietschel M, Deuschle M, Szyf M, 2014. MORC1 exhibits cross-species differential methylation in association with early life stress as well as genome-wide association with MDD. Transl Psychiatry 4, e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrek C, Elbert T, Weierstall R, Muller O, Rockstroh B, 2013. Childhood adversities in relation to psychiatric disorders. Psychiatry Res 206(1), 103–110. [DOI] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, Bennett J, Pierre PJ, Friedman DP, Cote SM, Hallett M, Tremblay RE, Suomi SJ, Szyf M, 2012. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci 32(44), 15626–15642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice F, Harold G, Thapar A, 2002. The genetic aetiology of childhood depression: a review. J Child Psychol Psychiatry 43(1), 65–79. [DOI] [PubMed] [Google Scholar]
- Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R, Steffens M, Mier D, Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Herms S, Wichmann HE, Schreiber S, Jockel KH, Strohmaier J, Roeske D, Haenisch B, Gross M, Hoefels S, Lucae S, Binder EB, Wienker TF, Schulze TG, Schmal C, Zimmer A, Juraeva D, Brors B, Bettecken T, Meyer-Lindenberg A, Muller-Myhsok B, Maier W, Nothen MM, Cichon S, 2010. Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry 68(6), 578–585. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7), e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeh S, Wiechmann T, Sauer S, Kodel M, Binder EB, Provencal N, 2018. HAM-TBS: high-accuracy methylation measurements via targeted bisulfite sequencing. Epigenetics Chromatin 11(1), 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Brandwein C, Luoni A, Sandrini P, Calzoni T, Deuschle M, Cirulli F, Riva MA, Gass P, 2016. Morc1 knockout evokes a depression-like phenotype in mice. Behav Brain Res 296, 7–14. [DOI] [PubMed] [Google Scholar]
- Schwarzer G, 2007. meta: An R package for meta analysis. R News 7(3), 40–45. [Google Scholar]
- Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, Hergueta T, Baker R, Dunbar GC, 1998. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 59 Suppl 20, 22–33;quiz 34-57. [PubMed] [Google Scholar]
- Slavich GM, Irwin MR, 2014. From stress to inflammation and major depressive disorder: a social signal transduction theory of depression. Psychol Bull 140(3), 774–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, Tang Y, Gillespie CF, Cubells JF, Ressler KJ, 2011. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet 156B(6), 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawbridge R, Young AH, Cleare AJ, 2017. Biomarkers for depression: recent insights, current challenges and future prospects. Neuropsychiatr Dis Treat 13, 1245–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman MB,N; Pappas JJ; Pereira SMP; Pembrey M; Hertzman C; Power C; Szyf M, 2014. Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Medical Genomics 7(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed SA, Nemeroff CB, 2017. Early Life Stress, Mood, and Anxiety Disorders Chronic Stress (Thousand Oaks: ) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiserman AM, 2010. Hormesis, adaptive epigenetic reorganization, and implications for human health and longevity. Dose Response 8(1), 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, R. BD, 2002. Modern Applied Statistics with S, Fourth ed. Springer, New York. [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, Lipschitz D, Douglas-Palumberi H, Ge M, Perepletchikova F, O’Loughlin K, Hudziak JJ, Gelernter J, Kaufman J, 2014. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J Am Acad Child Adolesc Psychiatry 53(4), 417–424 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F, Gelernter J, Kaufman J, 2013. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med 44(2), 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Roh S, Ressler KJ, Nemeroff CB, Smith AK, Bradley B, Heim C, Menke A, Lange JF, Bruckl T, Ising M, Wray NR, Erhardt A, Binder EB, Mehta D, 2015. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol 16, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, Gnirke A, Meissner A, 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500(7463), 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.