ABSTRACT
Sugar acts as an important nutrient for plant development. Previously, we reported that Oryza sativa DNA BINDING WITH ONE FINGER 11 (OsDOF11) plays a crucial role in sucrose transport by binding promoters of sucrose transporter genes, OsSUT1, OsSWEET11, and OsSWEET14. Meanwhile, sucrose transport activity abnormal also involved susceptibility to infection of Xanthomonas oryzae pathovar oryzae (Xoo) by OsSWEET genes. Here, we provid an addendum, that OsDOF11 expression pattern in spikelet development stage, and transcript levels of OsSWEET12, OsSWEET13, OsSWEET15, and OsSWEET16 in the mutants of OsDOF11 at different developmental stages. This information further supplied a new insight that sucrose transport activity mediated susceptibility to Xoo infection.
KEYWORDS: Rice, sucrose transport, OsDOF11, OsSWEET, Xanthomonas oryzae pathovar oryzae
Sugar is the key element for plant development, and its transport activity is a crucial factor for crop biomass.This transport is initiated by different phloem loading processes in the source tissue and then move into sink tissues, especially in the reproductive organ.1 Sucrose is the main material for starch synthesis. Thus, sucrose transporter genes, SUT (Sucrose Transporter; also called SUC for Sucrose Carrier) and SWEET (Sugars Will Eventually be Exported Transporters)-type transporters, directly or indirectly mediate sink organ development processes.1-5 Previous reported data demonstrate that the OsSUTs genes, OsSUT1,OsSUT2, OsSUT3, OsSUT4, and OsSUT5, perform sucrose import by the membrane.6,7 OsSUT1 is preferentially expressed in leaf phloem cells along with the stamens of mature spikelet formation and OsSUT1 Tos17 insertional mutant is unable to produce homozygous progeny due to impaired pollen.3 OsSUT3 and OsSUT4 are highly expressed in developing pollen.3,8 But OsSUT2 expressed in the cell layers of seed coats.9 Meanwhile, OsSWEET genes, OsSWEET11 and OsSWEET14, function in sucrose export.10,11 OsSWEET11 is expressed in vascular cells at the reproductive stage and loss-of-function mutation shows stamen defects, reduced pollen viability, and smaller seeds.10 Mutation of OsSWEET14 also showed semi-dwarf phenotype, with a smaller grain.11 These reports suggest that the sucrose transporters function in reproductive organ development, possibly in consistent with vegetative stage in rice.
As the sucrose is transferred into the grain, it has been reported that sucrose reaches the caryopsis through phloem loading at the dorsal vascular bundle.12 Bai et al. (2017) reported that OsNF-YB1(Nuclear Factor Y B1) expressed in caryopses from 4 to 21 DAP, and mutants of OsNF-YB1 generated by CRSPER 9 and RNA interference (RNAi) showed chalky endosperm possibly due to transcript level reduction of OsSUT1, OsSUT3, OsSUT4, and OsMST4 (monosaccharide transporter 4).12 In our previous study, it has been reported that OsDOF11 is expressed in the phloem cells of leaves, mature spikelet, developmental seed, and root. T-DNA insertion mutants and RNAi transgenic plants displayed semi-dwarf phenotypes with smaller panicles and grains.13 We also reported that OsDOF11 involved in sucrose transport by OsSUTs and OsSWEETs, as shown in Figure 1a.13 In the main manuscript, the researchers showed the expression level of almost all the SUT and 2 SWEETs in different developmental stages in OsDOF11 T-DNA insertional mutant were reduced by binding were checked and validated in OsSUT1, OsSWEET11, and OsSWEET14, no all its promoter.13 osdof11 also showed smaller seeds as shown in Figure 1b, probably due to OsDOF11’s expression pattern during spikelet development stages as well as in the filling stage.
Figure 1.
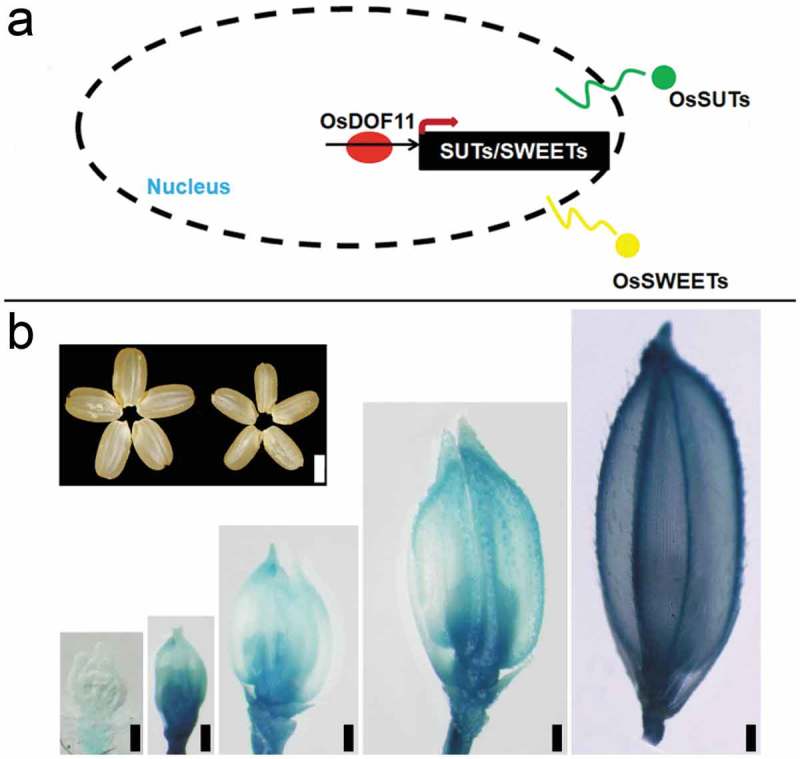
OsDOF11 functions in sucrose transport in rice. (a) OsDOF11 functions in sucrose transport by OsSUTs and OsSWEETs; (b) OsDOF11 functions during reproductive stage, the upper left picture showed seed phenotype, left: WT; right: osdof11-1. the bottom-right picture showed expression pattern. Bar: 2 mm, 200 um, 200 µm; 200 µm; 200 µm; 200 µm.
We found that OsDOF11 also affect susceptibility to infection of Xoo which might through OsSWEET11 and OsSWEET14 in leaf blade.13 Sugar acts as an important nutritent in bacterial proliferation. Published data showed that OsSWEET11, OsSWEET12, OsSWEET13, OsSWEET14, OsSWEET15 not only mediate sugar transport, but also involved in the response to the infection of Xoo.10,11,13-15 Mutants of OsSWEET11 and OsSWEET14 were less susceptible to infection. Also, the transcript levels of OsSWEET12, OsSWEET13, and OsSWEET15 in leaf were also increased to Xoo (PXO99-modified or PXO339 lines) by different effectors.15 Yuan et al. (2010) reported that OsSWEET11 is induced by PXO99.14 Xa25/OsWEET13 were induced by PXO399 but not PXO99 (Liu et al., 2011). And there is no native TAL effectors have been reported for finding OsSWEET12 and OsSWEET15, although TAL protein AAW76267 and Tal7b/Tal8b may be two putative candidates, respectively.15 Designed effectors of TALE-S1/TALE-S2/artTAL12-2 and artTAL15-1 were used for finding OsSWEET12 and OsSWEET15 functions.12,15As the infection of Xoo PXO99 in mutants of OsDOF11, relatively lower expression of all OsSWEET11, OsSWEET12, OsSWEET13, OsSWEET14, and OsSWEET15 were observed in osdof11-1and RNAi-9 lines than WT. However, other diseases affect rice panicle or grain development, such as Gibberellazeae and Fusarium graminearum.
Decreased expressions were observed for OsSUT1, OsSUT3, OsSUT5, OsSWEET11, and OsSWEET14 in the embryo of germination stage, leaf of vegetative stage, young panicle, and spikelet of reproductive stage. Here, we also found that the transcript level of OsSWEET13 was reduced (Figure 2). Also, OsSWEET15 transcript level was decreased (Figure 2), although there was no native effector. Recently, Yang et al. (2017) reported that ossweet11 single and ossweet11/ossweet15 double mutants showed defects in endosperm development and filling, which indicated that OsSWEET15 also contributes to the reproductive stage.16 Compared to the kit genes, OsSWEET12, OsSWEET13, OsSWEET15, and OsSWEET16, expression of OsSWEET15 was higher in 3 days after immersion (DAI), 1–2 cm inflorescence meristem (IF), and mature spikelet flower (MF)) than 35 days after germination (DAG) leaf, which means OsSWEET15 functions in sugar loading.
Figure 2.
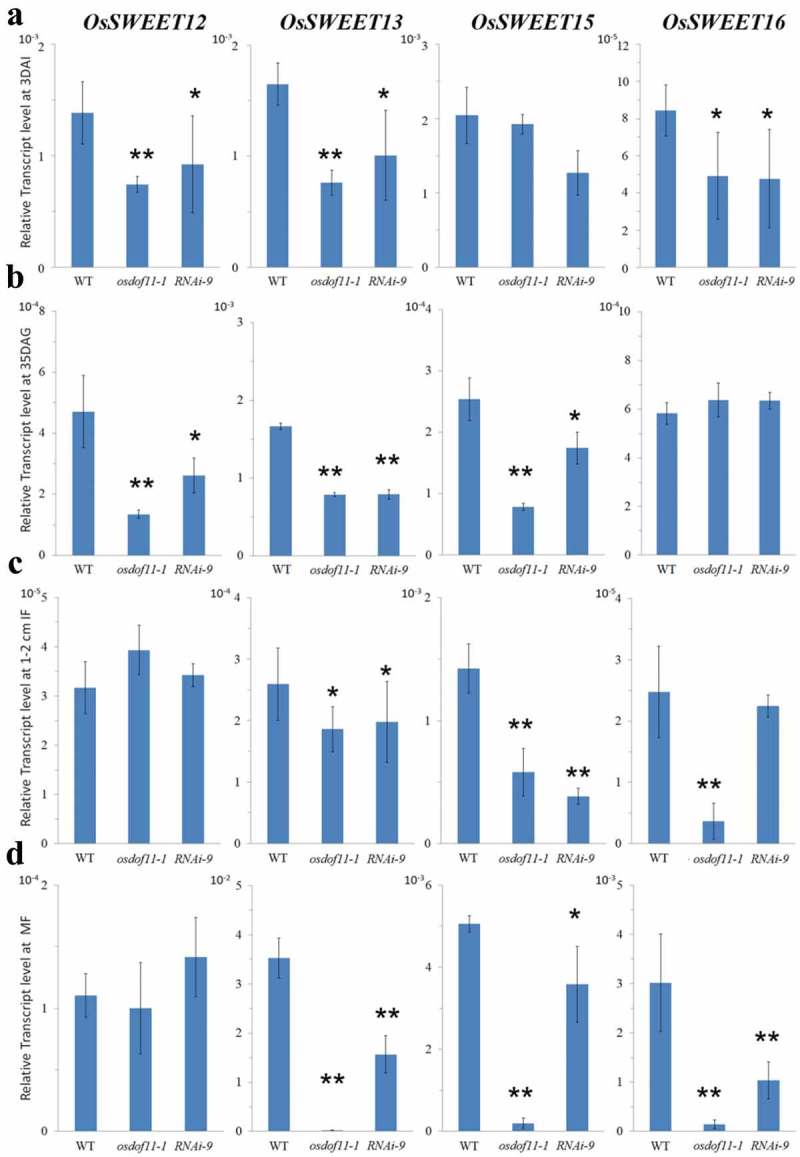
Relative transcript levels (RTL) of OsSWEETs in WT and OsDOF11 mutants (osdof11-1 and RNAi-9). (a) RTL at 3 DAI stage; (b) RTL at 35 DAG stage; (c) RTL at 1–2 mm IF stage; (d) RTL at MF stage.Error bars represent SE of at least three samples. *P < .05; **P < .01.
The present results reveal that sucrose transporter–related gene SWEET paralog can function as a sugar transporter involved in plant development and also supported bacterial (Xoo) to amplify and increase disease susceptibility. This inference gives a new insight into susceptibility to bacterial infection.
RT-PCR analyses
Total RNA was isolated from leaf blades of greenhouse-grown plants at 35 DAG as well as from 3 DAG seedlings and 1–2-cm-long young panicles. The complementary DNA were synthesized and quantitative real-time Reverse Transcription-Polymerase Chain Reaction was performed as previously described. The internal control was rice UBQ5 (LOC_Os01g22490). All experiments were conducted at least three times, with three or more samples taken at each point. To ensure primer specificity, we performed the experiments when the melting curve showed a single sharp peak. The PCR products were sequenced to verify the specificity of the reaction. All primers used for studying gene expression were listed in our previous report.13
Histochemical GUS Analysis and Subcellular Localization of OsDOF11
The β-glucuronidase assays were performed as previously described,13 using GUS-stained samples that were fixed in 3% (w/v) paraformaldehyde, 5% (v/v) acetic acid, and 63% (v/v) ethanol. Following ethanol dehydration, samples were embedded in Technovit Embedding Kits 7100 (Germany).
Funding Statement
This study was supported by a Project Funded by the Natural Science Foundation of Jiangsu Province (Grant BK20190889), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 19KJB180033), the Talent Support Program of Yangzhou University, and Yangzhou University Postdoctoral Science Foundation.
Disclosure statement
The authors declare that they have no competing interest.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- 1.Braun DM, Wang L, Ruan YL.. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot. 2014;65:1–4. doi: 10.1093/jxb/ert416. [DOI] [PubMed] [Google Scholar]
- 2.Rennie EA, Turgeon R.. A comprehensive picture of phloem loading strategies. Proc Natl Acad Sci USA. 2009;106:14162–14167. doi: 10.1073/pnas.0902279106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eom JS, Choi SB, Ward JM, Jeon JS. The mechanism of phloem loading in rice (Oryza sativa). Mol Cells. 2012;33:431–438. doi: 10.1007/s10059-012-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201:1150–1155. [DOI] [PubMed] [Google Scholar]
- 5.Julius BT, Leach KA, Tran TM, Mertz RA, Braun DM. Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol. 2017;58:1442–1460. doi: 10.1093/pcp/pcx090. [DOI] [PubMed] [Google Scholar]
- 6.Aoki N, Hirose T, Scofield N, Whitfeld PR, Furbank RT. The sucrose transporter gene family in rice. Plant Cell Physiol. 2003;44:223–232. doi: 10.1093/pcp/pcg030. [DOI] [PubMed] [Google Scholar]
- 7.Eom JS, Nguyen CD, Lee DW, Lee SK, Jeon JS. Genetic complementation analysis of rice sucrose transporter genes in Arabidopsis SUC2 mutant atsuc2. J Plant Biol. 2016;59:231–237. doi: 10.1007/s12374-016-0015-6. [DOI] [Google Scholar]
- 8.Chung P, Hsiao HH, Chen HJ, Chang CW, Wang SJ. Influence of temperature on the expression of the rice sucrose transporter 4 gene, OsSUT4, in germinating embryos and maturing pollen. Acta Physiol Plant. 2014;36:217–229. doi: 10.1007/s11738-013-1403-x. [DOI] [Google Scholar]
- 9.Eom JS, Cho JI, Reinders A, Lee SW, Yoo Y, Tuan PQ, Choi SB, Bang G, Park YI, Cho MH, et al. Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 2011;157:109–119. doi: 10.1104/pp.111.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 2010;22:3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai AN, Lu XD, Li DQ, Liu JX, Liu CM. NF‐YB1‐regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016;26:384–388. doi: 10.1038/cr.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Lee S, Yoo Y, Wei J, Lee S, Jeon J, An G. Transcription factor OsDOF11 modulates sugar transport by enhancing expression of sucrose transporter (SUT) and SWEET genes. Mol Plant. 2018;11(6):833–845. doi: 10.1016/j.molp.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Yuan M, Chu Z, Li X, Xu C, Wang S. Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 2009;50:947–955. doi: 10.1093/pcp/pcp046. [DOI] [PubMed] [Google Scholar]
- 15.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 16.Yang JI, Luo DP, Yang B, Frommer WB, Eom JS. SWEET11 and 15 as key players in seed filling in rice. New Phytol. 2017;218:604–615. doi: 10.1111/nph.15004. [DOI] [PubMed] [Google Scholar]


