ABSTRACT
Peiai64S (PA64S) is a photo-thermo-sensitive genic male sterile line (PTGMS), with wide application in hybrid seed production in rice (Oryza sativa L.). Micro-RNAs are 21–24 nt, endogenously expressed small RNAs that have been characterized in various developmental stages of rice, but none have been studied with respect to the regulation of TGMS in rice. Here, we employed high-throughput sequencing to identify expression profiles of miRNAs in the anthers of PA64S at high (PA64S-H) and low temperature (PA64S-L). Two small RNA libraries from PA64S-H and PA64-L anthers were sequenced, and 263 known and 321 novel candidate miRNAs were identified. Based on the number of sequencing reads, a total of 133 known miRNAs were found to be differentially expressed between PA64S-H and PA64S-L. Target prediction showed that the target genes encode MYB and TCP transcription factors, and bHLH proteins. These target genes are related to pollen development and male sterility, suggesting that miRNA/targets may play roles in regulating TGMS in rice. Further, starch and sucrose metabolism pathways, sphingolipid metabolism, arginine and proline metabolism, and plant hormone signal transduction pathways were enriched by KEGG pathway annotation. These findings contribute to our understanding of the role of miRNAs during anther development and TGMS occurrence in rice.
KEYWORDS: Mirna, TGMS, Oryza sativa L
Introduction
Rice (O. sativa L.) is one of the most important global grain crops and provides a staple food for almost half of the world population.1 The development of hybrid rice is the main way to increase its yield potential, because hybrid rice varieties have a yield advantage of about 20% or higher than the improved inbred varieties.2 Hybrid rice technologies are mainly based on two male sterility systems, namely environmentally sensitive genic male sterile (EGMS) and cytoplasmic male sterile systems (CMS).3 Different from the three-line system (CMS), the two-line system is based on the discovery and application of thermosensitive genic male sterile (TGMS) lines, which serve as both the male sterile lines and maintainer lines under different environmental conditions.4 The advantages of the two-line system compared to the three-line system include the wider germplasm resources used for breeding parents, better grain quality, higher yields, and simpler breeding and hybrid seed production processes.5
PA64S is one of the most important indica rice genic male sterile (GMS) lines in the two-line system, which was first identified as the photoperiod-sensitive GMS line (PGMS) by Mingsong Shi in Hubei, China in 1973,6 where the development of its anthers are sensitive to length of photoperiod, with abortive pollen grains generated under long photoperiod condition, and with fertile pollen grains occurred under short photoperiod condition,during the meiosis period of anther development. PA64S was subsequently found to be thermal-sensitive in pollen development, and is also a TGMS line, where at high temperature, it is male-sterile and produces sterile pollen, but at low temperature it converts to male-fertile plants that produce fertile pollen.7 Compared to other TGMS lines, PA64S is a indica-type (O. sativa indica) TGMS line with an japonica-type NK58S-derived sterility gene,8 and has wide compatibility and good agronomic traits. With its lower critical point temperature of fertility alteration (~23°C), PA64S is a valuable male sterile germplasm source and has become the most widely used female parent for two-line hybrid rice breeding.8
Extreme temperatures are a key factor limiting global rice distribution. Super hybrid rice cultivars grown in tropical and subtropical climates are highly productive, but they are often harmed by low and/or high temperatures.9 Heat and cold stress seriously affect the normal growth of rice throughout its life span.10 During the rice life cycle, the reproductive phase is the most sensitive to temperature stress, and even small changes in temperature can cause significant losses to food crops during flowering.11 Cold and heat tolerance are complex traits regulated by multiple genes, and these traits are highly genotype dependent. As temperature stress and climatic changes become more prevalent, it is urgent to identify genes associated with temperature stress tolerance and to understand their regulatory mechanisms to develop crops with enhanced temperature stress tolerance through genetic manipulation.12
Recent studies have documented that MicroRNAs (miRNAs) are fundamental sequence-specific regulatory elements (~24 nucleotides) and play key roles in the regulation of growth and development of eukaryotic organisms.13,14 In plants, mature miRNAs are processed from stem-loop regions of long primary transcripts by a Dicer-like 1 (DCL1) enzyme, then incorporated into the RNA-induced silencing complexes (RISC), guiding RISC to target mRNA, where they silence the expression of specific miRNAs by directing either mRNAs cleavage or translational repression.14–18 The miRNAs are non-coding, single-stranded RNAs encoded by endogenous genes involved in post-transcriptional regulation of target gene expression in plants and animals, and most miRNA target genes are transcription factors.19–21 Both miRNA and transcription factors can regulate the expression of downstream genes. Most of the miRNAs down-regulate the expression of target genes, and transcription factors activate or inhibit genes to determine the fate of target genes.22,23 In short, miRNAs not only regulate transcription factor expression, but also interact with transcription factors, all of them forming a complex regulatory network that participates in plant response mechanisms under developmental and stress conditions.24 Accordingly, a number of miRNA functions have also been identified in various biological processes, including developmental regulation, metabolism, and response to biotic and abiotic stresses.25–27
As a class of negative regulators, miRNA biogenesis and activity is well characterized in sporophytic tissues,28 and play a broad regulatory role during male gametophyte development in plants.29 Studies have reported that miR390, miR166, miR159, miR530, miR167, miR164, miR168 and two new miRNAs are differentially expressed in photoperiod-sensitive male-sterile mutant 7B-1 and its wild type. The predicted targets of these miRNAs are potentially involved in anther development and regulation of male-sterility in tomatoes.30 Recent transcriptomics studies identified 24 conserved miRNAs that are differentially expressed in the pollen of a cytoplasmic male-sterile line of rice and its maintainer line. Targets of these miRNAs include the MYB family protein, PPR-containing proteins, and kinases, which are involved in carbohydrate metabolic pathways, signal transduction, and reproductive processes.31
There is increasing evidence showing that miRNAs play key roles in the regulation of anther development. In plants, miR159 generally regulates the expression of GAMYB or GAMYB-like genes, which are involved in the regulation of anther development and microsporogensis.32–34 In rice, the gamyb deletion mutant is aborted due to the inability of the tapetum to perform PCD (programmed cell death) and degradation.35,36 Overexpression of miR159 in Arabidopsis thaliana and cereal crops results in sterile anthers.37–39 Thus, the role of GAMYB in anthers is highly conserved and regulated by miR159. In addition, miR159 interacts with miR319 and miR167. The expression of the target genes GAMYB and TCP4 are inhibited by miR159 and miR319, respectively. They can regulate the expression of miR167a alone or in the form of a protein complex to regulate the activity of ARF6/8. Therefore, the miR159-miR167-miR319 network is very important for the development of reproductive organs, including the ripening of anthers.40,41
To our knowledge, there has been no report comparing miRNA expression profiles of TGMS during reproductive development under high and low temperatures in rice. To do this, we used high-throughput sequencing technology to investigate the differential miRNA expression and targets from rice anthers at the early uninucleate stage of PA64S at high (PA64S-H) and low temperature (PA64S-L). Differential expression patterns of miRNAs were analyzed between PA64S-H and PA64S-L. Targets were predicted, and their expression profiles were selectively validated. These results may shed light on the regulatory roles of miRNAs related to temperature during pollen development as well as the occurrence of TGMS in rice.
Materials and methods
The TGMS line, PA64S, was used in this study. Plants were grown in the field until the fertility sensitive period at Hunan Normal University (latitude 28°19ʹ’ N; longitude 112°95ʹ’ E) under standard growing conditions. At jointing-booting stage, rice (PA64S-H) were transferred to a growth chamber at 30°C for 2 weeks of high temperature treatment, whereas rice (PA64S-L) were placed in a cooling pool at 22°C for 2 weeks of low temperature treatment. In the early mononuclear stage anthers with a floret length of ~5 mm were manually collected from plants of both temperature treatments and stored at −80°C for RNA isolation.42 The resulting product was reverse transcribed and amplified by polymerase chain reaction (PCR) to produce a cDNA sequencing library. The cDNA libraries were sent to Kang Cheng Biotechnology Co (Shanghai China). Solexa sequencing was performed on the HiSeq 2,000 platform (BGI).
After the Illumina sequencing, the raw sequences were first obtained in fastq format. Subsequently, the raw sequence passes through a filtering process to produce high quality readings. Unique sequences represented the sum of small RNA sequences of different base compositions, each unique small RNA contains several small RNA tags of identical sequence. All unique sequences were aligned against the rice genome to analyze their expression in the genome. Unique sequences recognized as tRNA, rRNA, snRNA, and snoRNA by searching through GenBank from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) and Rfam (11.0) where RNA family databases were dislodged.
The sequence is paired with the miRNA sequence of the species in miRBase (http://mirbase.org/), to obtain known miRNAs already present in the sample, and the base distribution. To identify miRNAs with differential expression between PA64S-H and PA64S-L, the expression of miRNAs was first quantified in transcripts per million. P value and fold change were calculated to confirm the significance of expression difference. The missing criteria for differential expression of miRNA was that | fold – change | > 2 and P value < .05. The sequences of known miRNA、exist miRNA and novel miRNA were used to Patmatch software to predict the potential target genes. After obtaining the target gene of miRNA expressed in a single sample, we performed GO (Gene Ontology) function analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis on the part of the gene. In addition, we processed target gene enrichment analysis on the known miRNA、exist miRNA and novel miRNA.
Results
Sequencing and date analysis
To identify the differentially expressed miRNAs in an early stage of microspore development, two sRNA libraries were constructed and sequenced from PA64S-H and PA64S-L anthers at the early uninucleate stage, which produced about 14,030,132 and 16,844,884 raw reads, respectively. After removing sequences of low quality, adaptor contaminants, insert- null, RNAs smaller than 18 nucleotides, and polyA, 6,788,401 and 12,484,929 clear reads remained from PA64S-H and PA64S-L, respectively (Table 1). The clean reads were sequence-matched with the GenBank and Rfam databases, and the ncRNAs such as rRNA, tRNA, snRNA, and snoRNA, and the repeated sequences were filtered to obtain unannotated reads containing the miRNA (Table S1-S4).
Table 1.
Statistics of small RNA sequences from the PA64S-H and PA64S-L libraries.
| PA64S-H |
PA64S-L |
|||
|---|---|---|---|---|
| Type | Count | Percentage (%) | Count | Percentage (%) |
| Total-reads | 14030132 | 100% | 16844884 | 100% |
| High-quality | 12537263 | 89.3596% | 15112119 | 89.7134% |
| 3ʹadapter-null | 136949 | 1.0923% | 1052179 | 6.9625% |
| Insert-null | 104738 | 0.8354% | 10196 | 0.0675% |
| 5ʹadapter-contaminants | 127002 | 1.0130% | 24331 | 0.1610% |
| Smaller-than-18 nt | 5379902 | 42.9113% | 1540181 | 10.1917% |
| PolyA | 271 | 0.0022% | 303 | 0.0020% |
| Clean-reads | 6788401 | 54.1458% | 12484929 | 82.6153% |
The proportion of several non-coding miRNAs was low in PA64S-H and PA64S-L, among which rRNA accounted for the highest proportion and snRNA accounted for the least (Figure 1; Figure S1). The unannotated reads were aligned with the reference rice genome using miRDeep2 software to obtain positional information on the reference genome (Figure S2). In PA64S-H and PA64S-L, the number of miRNAs that could be aligned was 4,289,438 and 7,743,506, respectively, which accounted for about 60% of the total miRNAs (Table S5).The majority of sRNAs were 21–24 nt in both PA64S-H and PA64S-L libraries, which formed the typical size range for Dicer-derived products.43 Among these, sRNAs with a length of 24 nt were the most abundant (30.37% in PA64S-H and 39.97% in PA64S-L), followed by those of 21 nt (16.53% in PA64S-H and 15.60% in PA64S-L) (Figure 2). The size distribution of sRNAs from the TGMS was quite similar to that of other plant species, such as A. thaliana44 and Phaseolus vulgaris,45 in which the 24 nt class dominates the sRNA transcriptome. However, there are some exceptions, as in Brassica juncea, where the 21-nt class dominates the sRNA in total and unique reads.46
Figure 1.
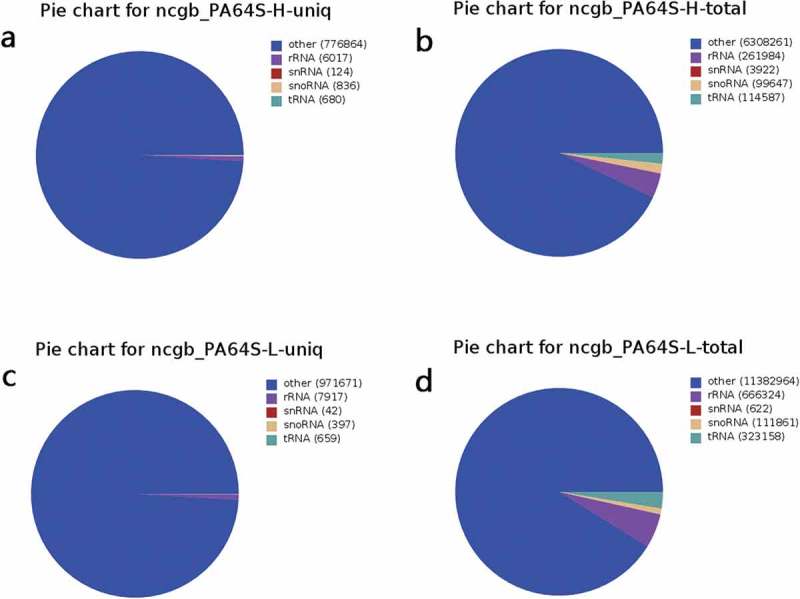
Summary of unique and total of small RNAs in non-coding RNAs in GenBank between PA64S-H and PA64S-L. a,b. Unique and total sRNAs in PA64S-H. c.d. Unique and total sRNAs in PA64S-L.
Figure 2.
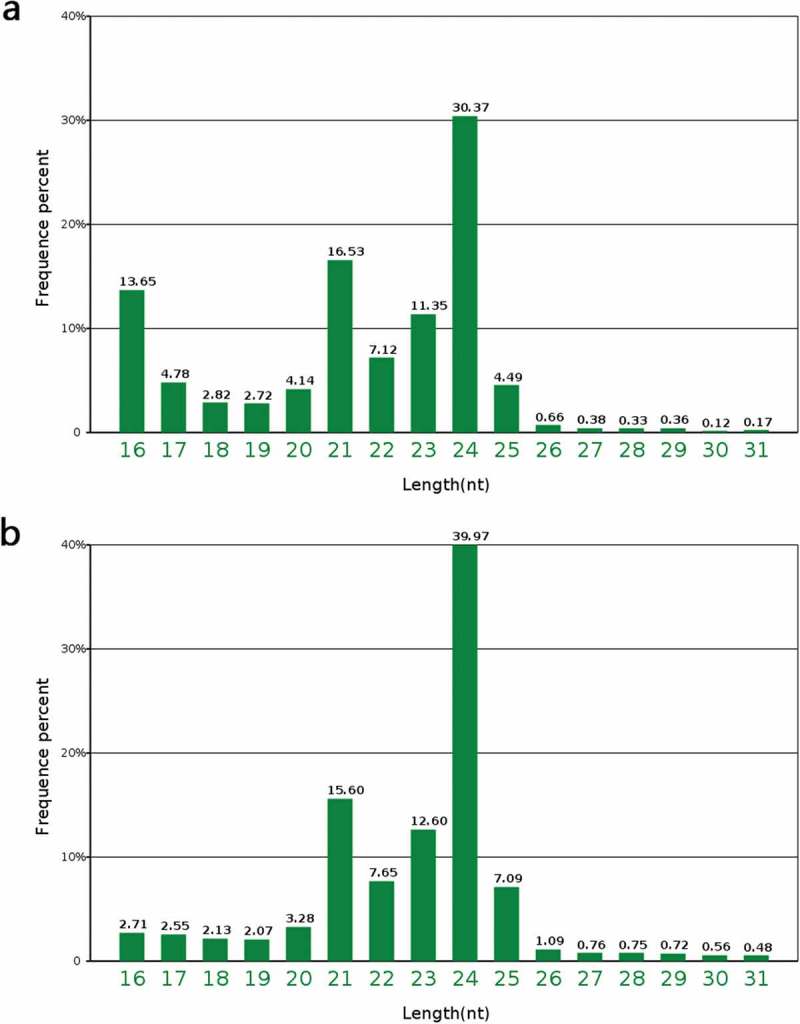
Length distribution of small RNAs in the different libraries. a. PA64-H. b. PA64-L.
Identification of known and new miRNAs
In the present study, a total of 263 known miRNAs (Table S6) were identified in the anthers of PA64S-H and PA64S-L at the early uninucleate stage. Among these known miRNAs, 151 were found in both sequence libraries, and 56 were specific to PA64S-H or PA64S-L (Figure 3(a)). The composition of miRNA families was slightly different in the two libraries, and there was a clear PA64S-H or PA64S-L-specific profile, for example, miR172 and miR158 was only expressed in PA64S-H, while miR169, miR4399, and miR9473 were only expressed in PA64S-L (Table S6). Further, sequencing frequencies of conserved miRNAs were also detected. Expression levels varied greatly between distinct miRNA families. Some of the miRNA family members, such as miR159, miR166, miR319, and miR168, were abundantly expressed in both libraries (Table S6), suggesting their conserved and essential roles for rice anther development. Compared to known reference data, these miRNAs are associated with pollen development. For example, miR159 regulates GAMYB or GAMYB-like expression, the gamyb deletion mutant is aborted due to the inability of the tapetum to perform PCD and degradation in rice;35,36 miR159 and miR319 regulate genital development by regulating the activity of ARF6/8, including sepals, petals and anthers.40,41 In contrast, other miRNA families, such as miR437, miR5509, miR5796, and miR171, were expressed at lower levels in both rice lines, suggesting the functional divergence among these miRNA families.
Figure 3.
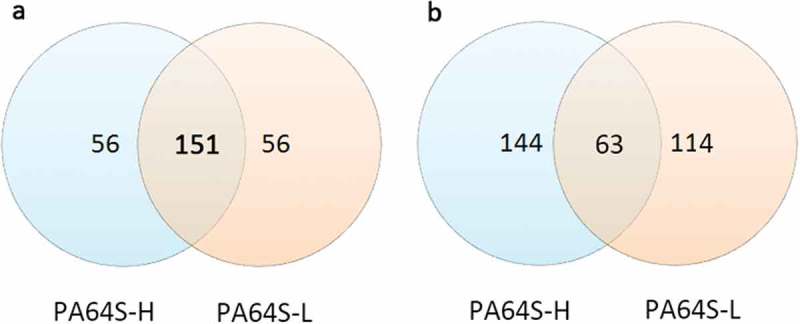
Venn diagram analysis of expressed genes of PA64S-H and PA64S-L. a. known miRNA. b. novel miRNA.
Generally, the first nucleotide of miRNAs is thought to be related with recruitment of the AGO protein. In Arabidopsis, AGO1 tends to recruit the miRNAs beginning with a uracil (U), while AGO2 and AGO5 show preferences for miRNAs initiating with adenosine (A) and cytosine (C), respectively.47 In this study, the first nucleotide bias of known miRNAs was “U”, followed by “A” (Figure 4). The first nucleotide bias of known miRNAs from TGMS was quite similar to that of the cytoplasmic male sterile line and its maintainer line.31 This indicated that the identified miRNAs in rice anthers are conserved, and could be recruited by different AGO proteins and enter different regulatory pathways. After excluding sRNA reads homologous to known miRNAs and other noncoding RNAs from GenBank and the Rfam databases, many unannotated sequences that could not match any of the above databases were analyzed using miRDeep (http://deepbase.sysu.edu.cn/miRDeep.php) to predict novel miRNA candidates. A total of 321 putative new miRNAs were identified in the two libraries (Table S7), where 144 were present only in PA64-H, 114 only in PA64-L, and 63 in both sequencing libraries (Figure 3(b)). These novel miRNAs formed typical stem-loop hairpin structures (Figure S5), and their corresponding new sequences were mainly derived from the same precursor gene when aligned with the rice genome. This suggests that the unidentified small fragments of miRNAs may be rice-specific miRNAs. The first nucleotide bias and nucleotide bias at each position in the mature sequence of novel miRNAs was further analyzed (Figure S4) and was similar to that of known miRNAs.
Figure 4.
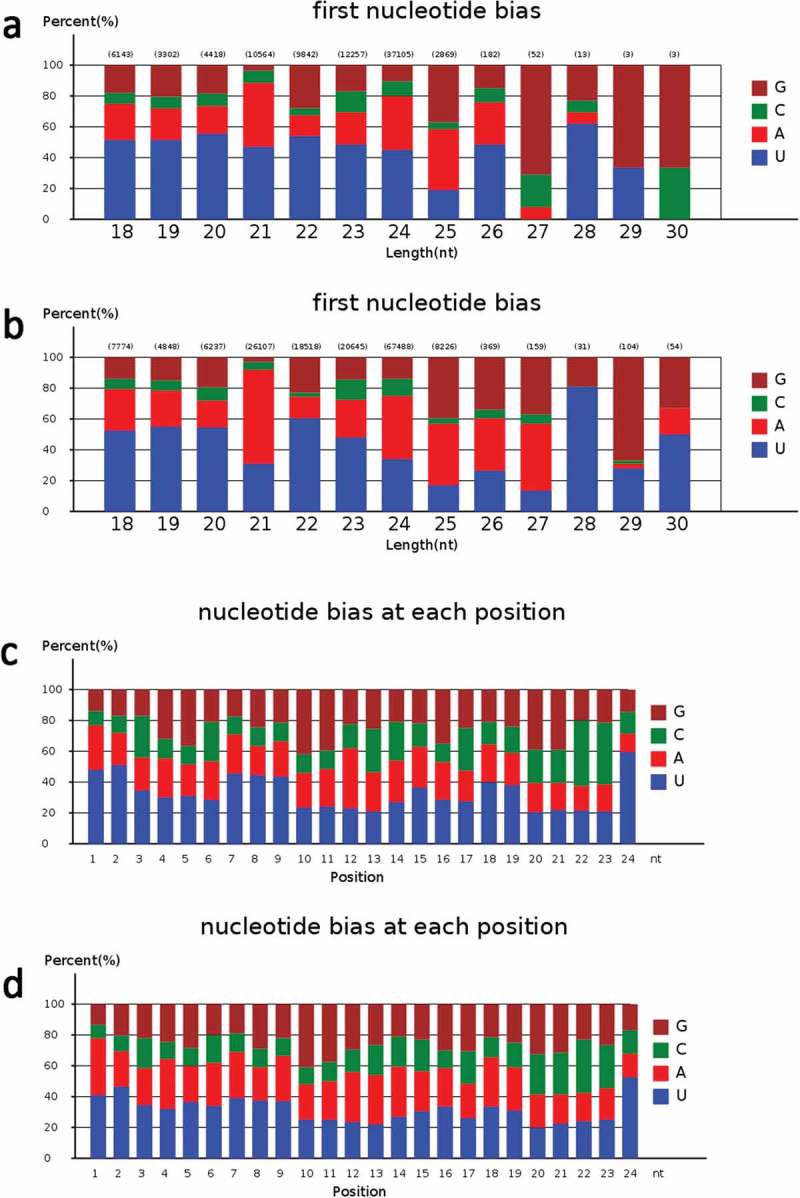
The analysis of miRNA first nucleotide bias and nucleotide bias at each position in known miRNAs.
a. The miRNA first nucleotide bias analysis of known miRNAs in PA64S-H.b. The miRNA first nucleotide bias analysis of known miRNAs in PA64S-L.c. The miRNA nucleotide bias at each position analysis of known miRNA in PA64S-H.d. The miRNA nucleotide bias at each position analysis of known miRNA in PA64S-L.
Differential expression of miRNAs during polloen development of PA64S-H and PA64S-L
To identify miRNAs related to anther development and regulation of male-sterility in PA64S under high and low temperature, a total of 263 miRNAs were used for differential expression analysis between
PA64S-H and PA64S-L (Table S6), and the normalized expression values of these miRNAs were used to draw the scatter plot (Figure 5(a)). The selection threshold were fold-change |(log2)| ≥2 and P-value < 0.05. Considering that extremely low abundances might be false results, the miRNAs with less than 15 norm reads were removed from expression. Among these differentially expressed, known miRNAs, 59 were up-regulated and 74 were down-regulated in PA64S-L compared to those in PA64S-H (Table S15), while other miRNAs were equally expressed (Figure 5(a)). Studies have shown that miR390 and miR164 are significantly differentially expressed in the photosensitive male sterile mutant 7B-1 and its wild type,30 and low temperature stress leads to the induction of miR171 and miR172 in Brachypodium serrata and Arabidopsis,48 respectively. To validate the differential expression data obtained from the high-throughput sequencing, we further tested the expression patterns of six selected, known miRNAs by quantitative real-time RT-PCR analysis. The expression levels of MIR531, MIR6300, and MIR9773 decreased, whereas MIR1118, MIR2275, and MIR9473 were up-regulated in PA64S-H compared with that of PA64S-L (Figure 5(b)). The qRT-PCR results were consistent with the high-throughput sequencing, implying that the high throughput sequencing results were reliable.
Figure 5.
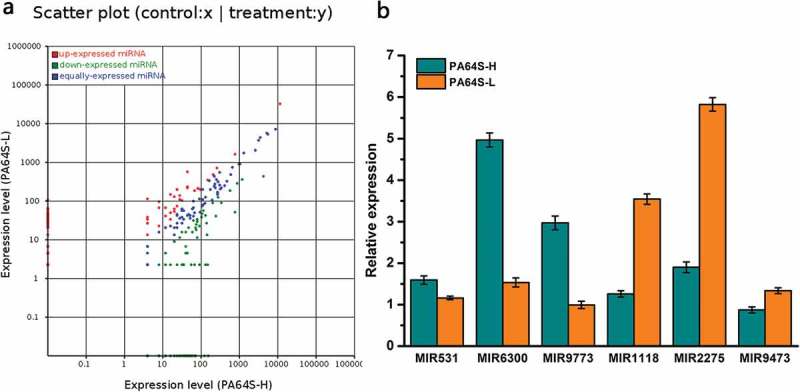
Comparison and validation of the differential expression of known miRNA in the different libraries. a. Scatter plot of expression rations for known in Pa64S-H and PA64S-L. Red points represent miRNAs with fold change>2, showing upregulated miRNAs in PA64S-L; blue points represent miRNAs with 1/2≤ fold change ≤2, indicating equally expressed miRNAs in both libraries; green points represent miRNAs with fold change<1/2, meaning downregulated miRNAs in PA64S-L. Fold change = normalized expression in PA64S-L/normalized expression in PA64S-H. b. Comparison of the miRNA expression levels between PA64S-H and PA64S-L by qPCR for MIR531, MIR6300, MIR9773, MIR1118, MIR2275, MIR9473. The experiments were performed with three biological replicates, and the error bars represent the standard error.
Target prediction and function analysis
Given that these miRNAs found during the reproductive stage may have functions associated with anther development, subsequent analyzes focused on their targets. A total of 5944 target genes were identified in 769 miRNAs (Table S10), including known miRNAs corresponding to 2226 target genes (Table S11), and novel miRNAs correspond to 233 target genes (Table S12). The number of target genes corresponding to novel miRNAs was significantly lower than that of conserved miRNAs. Most novel miRNAs did not predict any target gene, and some new miRNAs corresponded to the same target gene as the known miRNAs. For example, the target gene corresponding to novel-m0280-5p is ABH1 transcription factor, and the target gene corresponding to novel-m0098-5p is F-box domain containing protein (Table S12). This reflects, to some extent, that the functions of these novel miRNAs may be similar to those of conserved miRNAs, and regulate the development of rice flowers by regulating similar target genes.
Among the prediction targets, some miRNA targets related to anther development are worth mentioning. For example, growth-regulating factors (OSGRF1/2/3/6/7/9/10/12), kinases, MYB transcription factors, and TCP transcription factors have potential roles in anther development. API5, receptor-like kinase, and bHLH proteins are involved in tapetum development/degeneration31,36,41,49–52 (Table S10). In addition, 12 putative auxin response factors (ARFs) were predicted to be targeted by miRNAs (Table S10), which are quite similar to that mentioned in a previous study,29 and indicate that the miRNA-mediated silencing of the ARF gene has a potential role in pollen development. This result suggests that miRNAs can directly regulate gene expression at both transcription and translation level.
To gain more information on miRNA targets for TGMS at high and low temperature, the targets of differentially expressed miRNAs were further analyzed. Interestingly, many differentially expressed miRNA targets have been shown to be associated with cold or heat stress responses, as well as with anther development. MIR5809-y, which specifically expressed in PA64S-L, is predicted to target transcripts encoding a receptor-like protein kinase (Os01G0155500) (Table S15; Table S10). Mutations in receptor-like protein kinase have been reported to result in male sterility in Arabidopsis.53 The target gene bHLH (Os02g0116600), corresponding to MIR3440-y, is responsive to the regulation of cold stress in rice and associated with anther development10,54 (Table S15; Table S10). In addition, many heat shock proteins, which are the targets of MIR164-y, MIR419-x, and MIR8011-x, are responsible for protein folding, assembly, and translocation, and play pivotal roles in protecting plants against stress by stabilizing proteins and membranes55,56 (Table S15; Table S10). These significantly differentially expressed miRNAs are specifically expressed in PA64S-H (Table S15), and their target gene expression was downregulated under heat stress (Figure R2).57
GO terms and KEGG pathway enrichment analysis of targets
To illuminate the function of the differentially expressed miRNAs, gene ontology (GO) analysis and KEGG pathway annotation were used to enrich and classify the given target genes into specific informative groups.58 Here, the most frequent “biological process” terms were ‘‘metabolic process’’ and ‘‘cellular process’’ (Figure S3). “Cell”, “cell part”, and “organelle” were the top three “cellular components” (Figure S3), and the most abundant terms in ‘‘molecular function’’ were ‘‘binding’’, ‘‘catalytic activity’’, ‘‘transcription regulator activity’’, and ‘‘transcription activity’’ (Figure 6, Figure S3). Further, the targets of differential expression of known miRNAs were analyzed. The result showed that the targets were classified into 22 reference terms of “biological processes”, with the most frequent term being ‘‘metabolic process’’, followed by ‘‘cellular process’’, ‘‘response to stimulus’’, and “biological regulation” (Figure 6(a)). The results indicate a correlation between pollen development and low temperature. Comparatively, the results for ‘‘biological process’’ were classified into 18 reference terms in novel miRNAs, with the most common terms being ‘‘cellular process’’, ‘‘metabolic process’’, ‘‘response to stimulus’’, and ‘‘localization’’ (Figure 6(b)).
Figure 6.
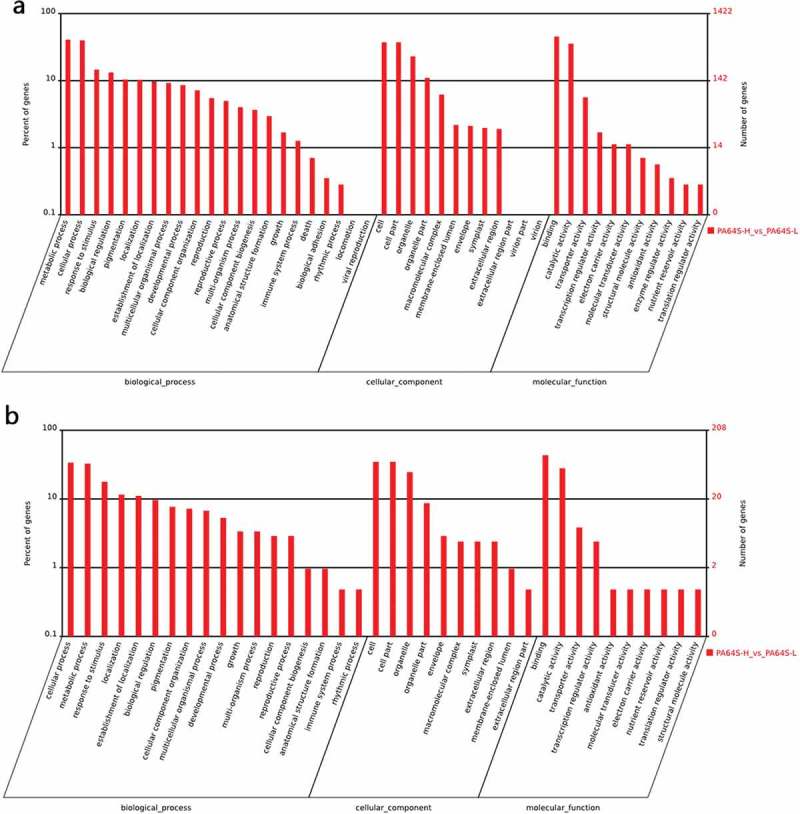
Gene ontology analysis for the targets of the differential expression of known and novel miRNAs based on the terms of cellular component, biological process and molecular function in PA64S-H and PA64S-L. a. known miRNAs. b. novel miRNAs.
To further illustrate our findings, the KEGG enrichment networks were also used for comparative analysis between differentially expressed target genes in PA64S-H and PA64S-L (Table S8, Table S9). As a result, most of the target genes corresponding to differentially expressed miRNAs were involved in metabolic pathways and biosynthesis of secondary metabolites (Table 2). Among them, several pathways are especially noteworthy. First, 13 target genes were enriched in the starch and sucrose metabolism pathway (ko00500) (Table 2, Table S8, Table S9). Starch and sucrose metabolism provide raw materials for all stages of plant growth, and the accumulation of soluble sugar is directly related to pollen fertility.59 Second, differentially expressed miRNA target genes were also heavily involved in sphingolipid metabolism (ko00600) (Table 2, Table S8, Table S9). Sphingolipids are key cellular membranes and signaling molecules involved in several cellular activities.60–62 Sphingolipid homeostasis is also reported to be involved in responses to ABA and cold temperatures,63,64 as well as regulation of PCD and pollen fertility.65,66 This indicates that miRNAs affect pollen fertility under low temperatures indirectly by silencing sphingolipid metabolism-related genes. Third, arginine and proline metabolism pathways were also annotated (ko00330) (Table S8). Retardation of glutamic and aspartic acid conversion to proline is considered to be a cause of pollen sterility in the male sterile line.67,68 In addition, the plant hormone signal transduction pathway was also enriched (ko04075) (Table S8), suggesting that miRNAs are involved in the regulation of plant growth and development and respond to environmental signals by affecting important transcription factors in the hormone signaling pathway.69
Table 2.
Some KEGG enrichment pathways analysis.
| KEGG Pathway | Target genes with pathway annotation (272) | All genes of the species with pathway annotation (5086) | P value | Q value | Pathway ID |
|---|---|---|---|---|---|
| Metabolic pathways | 106 (38.97%) | 1979 (38.91%) | 0.5155038 | 0.9952613 | ko01100 |
| Biosynthesis of secondary metabolites | 52 (19.12%) | 1050 (20.64%) | 0.7611412 | 0.9952613 | ko01110 |
| Pyruvate metabolism | 14 (5.15%) | 125 (2.46%) | 0.006478281 | 0.105095012 | ko00620 |
| Phenylpropanoid biosynthesis | 14 (5.15%) | 157 (3.09%) | 0.04023822 | 0.22131021 | ko00940 |
| Starch and sucrose metabolism | 13 (4.78%) | 196 (3.85%) | 0.2485438 | 0.781137657 | ko00500 |
| Sphingolipid metabolism | 7 (2.57%) | 32 (0.63%) | 0.001227664 | 0.036011477 | ko00600 |
| Plant hormone signal transduction | 6 (2.21%) | 254 (4.99%) | 0.9946631 | 0.9952613 | ko04075 |
| Endocytosis | 12 (4.41%) | 140 (2.75%) | 0.07010644 | 0.308468336 | ko04144 |
| Purine metabolism | 12 (4.41%) | 165 (3.24%) | 0.9946631 | 0.56361197 | ko00230 |
| Arginine and proline metabolism | 1 (0.37%) | 91 (1.79%) | 0.993576 | 0.9952613 | ko00330 |
Discussion
Peiai64S (O. sativa L) is a photoperiod temperature-sensitive genic male sterile line (PTGMS). Its fertility alternates between sterile and fertile at a certain day length and temperature, and has been widely used in hybrid seed production.6 The photosensitivity of PA64S is regulated by a long-chain non-coding RNA LDMAR. A SNP (G→C) mutation on LDMAR alters the secondary structure of LDMAR, reducing the transcriptional level of LDMAR under long-day conditions, which leads to premature PCD in pollen and results in photosensitive male sterility in rice.70 Zhou revealed the TGMS mechanism of RNase ZS1-mediated UbL40 mRNA processing in Annon S-1.71 So far, what confers the TGMS traits in PA64S is still unknown.
It has been reported that the “miRNA-target gene” regulatory pathway regulates the fertility of plants. For example, in Arabidopsis, miR167 regulates its target ARF transcription factor, miR159 regulates the target MYB transcription factor, and miR319 regulates the TCP transcription factor to participate in anther development and maturation.41,72,73 However, the relationship between miRNA and TGMS in rice has not been reported. In this study, we used high throughout sequencing technology to profile miRNAs expression and target gene function in PA64S anther under high and low temperatures. The result showed that a total of 263 known miRNAs was identified in the anthers of PA64S-H and PA64S-L at the early uninucleate stage (Table S6). As a result, many known miRNAs are abundantly expressed in two libraries (Table S6). For example, MiR159 and miR319 regulate the development of sepals, petals and anthers by modulating the activity of the target gene ARF6/8.40,41 The results suggested that abundantly expressed known miRNAs are conserved and play important roles in rice anther development. Among the 263 known miRNAs, 133 differentially expressed miRNAs were identified in PA64S-L compared with PA64S-H, of which 59 were up-regulated and 74 were down-regulated (Table S15). Interestingly, many male fertility-related miRNAs, including miR390 and miR164, were significantly, differentially expressed.30,74 Furthermore, miR172, which was only expressed in PA64S-H, is not only induced by low temperature stress, but also regulates male and female differentiation of floral organs48,75 (Table S15).
Targets of these differentially expressed miRNAs were also analyzed. Growth-regulating factors (GRFs), kinases, MYB transcription factors, and TCP transcription factors were characterized in this study (Table S10). OsGRF1 not only regulates leaf growth, but also controls flowering time.50 Overexpression of AtGRF1 and AtGRF2 in Arabidopsis also delayed flowering compared to the wild type.51 Kinases act as major regulators of various pathways. Mutations in receptor-like protein kinases have been reported to result in male sterility in Arabidopsis.53 MIR5809-y was specifically expressed in PA64S-H (Table S15) and is predicted to target a transcript encoding a receptor-like protein kinase (Os01G0155500) (Table S10). In Arabidopsis, the target MYB and TCP transcription factors corresponding to miR159 and miR319, respectively, are involved in the development and maturation of anthers.41,73 To further illuminate the functional role of the targets, the predicted targets were analyzed with GO and KEGG. Some metabolic and biosynthetic pathways that may be associated with anther development, include biosynthesis of secondary metabolites, the phenylpropanoid biosynthesis pathway, starch, sucrose, and sphingolipid metabolism. In addition, the plant hormone signal transduction pathway is also enriched, which is associated with anther development and stress responses.32,35,37,76 In addition, we used CRISPR-Cas9 technology to knock out target genes for some clearly differentially expressed miRNAs. Mutation phenotypic analysis showed that pollen fertility was transformed under high temperature and low temperature (data not show). These results suggest that the miRNA-target gene pathway may be involved in regulating pollen fertility at high or low temperatures.
Conclusions
We employed high-throughput sequencing to identify expression profiles of anther miRNAs from PA64S-H and PA64S-L, and found some clues that may be involved in pollen fertility under low temperature in PA64S. These findings may shed light on the possible mechanism of miRNAs regulating TGMS occurrence in rice during anther development. However, further research is needed to verify the miRNA/target gene function role by using the CRISPR-Cas9 technology to knock out for validation analysis. In addition, the functions of a large number of differentially expressed miRNAs/target genes are still unknown, which is an obstacle to clarify the fertility mechanism in PA64S.
Funding Statement
This work was supported by grants of NSFC [31570316; 31371244], Projects of Hunan Provincial Education Department [17C0974], and Hunan Provincial Innovation Foundation for Postgraduate [CX2017B230, CX2018B300].
Disclosure of potential conflict of interest
The authors declare no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Delseny M, Jm S, Cooke R, Sallaud C, Regad F, Lagoda P, Ghesquière A.. Rice genomics: present and future. Plant Physiol Biochem. 2001;39:1–11. [Google Scholar]
- 2.Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY.. Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot. 2007;100:959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan L. Purification and production of foundation seed of rice PGMS and TGMS lines. Hybrid Rice. 1994; 6:1-3. [Google Scholar]
- 5.Yang Q, Liang C, Zhuang W, Li J, Deng H, Deng Q, Wang B. Characterization and identification of the candidate gene of rice thermo-sensitive genic male sterile gene tms5 by mapping. Planta. 2007;225:321–330. doi: 10.1007/s00425-006-0353-6. [DOI] [PubMed] [Google Scholar]
- 6.Shi MS. The discovery and study of the photosensitive recessive male-sterile rice (Oryza sativa L. subsp. japonica). Sci Agric Sin. 1985;2:44–48. [Google Scholar]
- 7.Xu M. Response of fertility of Pei’ai 64 S to temperature and photoperiod in rice. Acta Agronomica Sin. 1999;25:772–776. [Google Scholar]
- 8.Zhou H, Liu Q, Li J, Jiang D, Zhou L, Wu P, Lu S, Li F, Zhu L, Liu Z, et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012;22:649–660. doi: 10.1038/cr.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Guo Z, Liu C, Xiao W, Wang R, Zhang L, Guan S, Zhang S, Cai L, Liu H, Huang X, et al. Comparative transcriptome profile analysis of anther development in reproductive stage of rice in cold region under cold stress. Plant Mol Biol Rep. 2019;37:129–145. doi: 10.1007/s11105-019-01137-6. [DOI] [Google Scholar]
- 11.Mittler R, Finka A, Goloubinoff P. How do plants feel the heat? Trends Biochem Sci. 2012;37:118–125. doi: 10.1016/j.tibs.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Yan S, Yang T, Zhang S, Chen YQ, Liu B. Small RNAs in regulating temperature stress response in plants. J Integr Plant Biol. 2017;59:11. doi: 10.1111/jipb.12571. [DOI] [PubMed] [Google Scholar]
- 13.Shukla LI, Chinnusamy V, Sunkar R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta. 2008;1779:743–748. doi: 10.1016/j.bbagrm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Voinnet O. Origin, biogenesis, and activity of plant MicroRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 15.Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJM. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X. A MicroRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong D-H, Nakano M, Cao S, Liu C, et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu B, Chen Z, Song X, Liu C, Cui X, Zhao X, Fang J, Xu W, Zhang H, Wang X, et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell. 2007;19:2705–2718. doi: 10.1105/tpc.107.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant MicroRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 20.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayushi K, Abira C, Mohan K, Asis D. Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci. 2015;6:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Istrail S, De-Leon SB-T, Davidson EH. The regulatory genome and the computer. Dev Biol. 2007;310:187–195. doi: 10.1016/j.ydbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh Y-F, Hou P-F, Yang T-Y, Chang W-C. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016;44:D1154–60. doi: 10.1093/nar/gkv1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samad AFA, Muhammad S, Nazaruddin N, FI A, MAM A, Zamri Z, Ismail I.. MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci. 2017;8:565. doi: 10.3389/fpls.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong D, Zhang L, Hang W, Liu Z, Zhang Z, Zheng Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot. 2009;103:29–38. doi: 10.1093/aob/mcn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campo S, Peris-Peris C, Sire C, Moreno AB, Donaire L, Zytnicki M, Notredame C, Llave C, San Segundo B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013;199:212–227. doi: 10.1111/nph.12292. [DOI] [PubMed] [Google Scholar]
- 27.Guo H, Xie Q, Fei J, Chua N. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root devel. Plant Cell. 2005;17:1376. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He H, Yang T, Wu W, Zheng B. Small RNAs in pollen. Sci China Life Sci. 2015;58:246–252. doi: 10.1007/s11427-015-4800-0. [DOI] [PubMed] [Google Scholar]
- 29.Peng H, Chun J, Ai T-B, Tong Y-A, Zhang R, Zhao -M-M, Chen F, Wang S-H. MicroRNA profiles and their control of male gametophyte development in rice. Plant Mol Biol. 2012;80:85–102. doi: 10.1007/s11103-012-9898-x. [DOI] [PubMed] [Google Scholar]
- 30.Omidvar V, Mohorianu I, Dalmay T, Fellner M. Identification of miRNAs with potential roles in regulation of anther development and male-sterility in 7B-1 male-sterile tomato mutant. BMC Genomics. 2015;16:878. doi: 10.1186/s12864-015-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan J, Zhang H, Zheng Y, Ding Y. Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta. 2015;241:109–123. doi: 10.1007/s00425-014-2167-2. [DOI] [PubMed] [Google Scholar]
- 32.Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA. The MicroRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010;154:757–771. doi: 10.1104/pp.110.160630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csukasi F, Donaire L, Casaal A, Martínez-Priego L, Botella MA, Medina-Escobar N, Llave C, Valpuesta V. Two strawberry miR159 family members display developmental-specific expression patterns in the fruit receptacle and cooperatively regulate Fa-GAMYB. New Phytol. 2012;195:47–57. doi: 10.1111/j.1469-8137.2012.04134.x. [DOI] [PubMed] [Google Scholar]
- 34.Li WF, Zhang S-G, Han S-Y. Regulation ofLaMYB33by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tissue Organ Culture. 2013;113:131–136. doi: 10.1007/s11240-012-0233-7. [DOI] [Google Scholar]
- 35.Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell. 2009;21:1453–1472. doi: 10.1105/tpc.108.062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko M. Loss-of-function mutations of the rice GAMYB gene impair ?-Amylase expression in aleurone and flower development. Plant Cell. 2004;16:33–44. doi: 10.1105/tpc.017327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyes JL, Chua N-H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2010;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 38.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulatedmicroRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 39.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of MicroRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 41.Rubio-Somoza I, Weigel D, Qu L-J. Coordination of flower maturation by a regulatory circuit of three MicroRNAs. PLoS Genet. 2013;9:e1003374. doi: 10.1371/journal.pgen.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng JH. LU Y, Liu XD, Guangzhou SC Emy. Pollen development and its stages in rice (Oryza sativa L.). Chin J Rice Sci. 2001;15:21–28. [Google Scholar]
- 43.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 44.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelaez P, Trejo MS, Iniguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, Sanchez F. Identification and characterization of microRNAs inPhaseolus vulgarisby high-throughput sequencing. BMC Genomics. 2012;13:83. doi: 10.1186/1471-2164-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Liu X, Xu B, Zhao N, Yang X, Zhang M. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines ofbrassica juncea. BMC Genomics. 2013;14:9. doi: 10.1186/1471-2164-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo AD, LIU L, TANG ZS, BAI XQ, CAO SY, CHU CC. Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. Bull Bot. 2005;47:745–752. [Google Scholar]
- 51.Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2010;36:94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 52.Pajoro A, Madrigal P, Mui?o JM, Matus J, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15,3(2014-03-03) 2014;15:R41. doi: 10.1186/gb-2014-15-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K. Receptor‐like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2010;50:751–766. doi: 10.1111/j.1365-313X.2007.03083.x. [DOI] [PubMed] [Google Scholar]
- 54.Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Nakashima K.. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Molgenetgenomics. 2010;284:173–183. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 56.Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/bf00039383. [DOI] [PubMed] [Google Scholar]
- 57.Xiang J, Xinbo C, Wei H, Yanci X, Mingli Y, Jieming W. Overexpressing heat-shock protein OsHSP50.2 improves drought tolerance in rice. Plant Cell Rep. 2018;37:1585–1595. doi: 10.1007/s00299-018-2331-4. [DOI] [PubMed] [Google Scholar]
- 58.Ye J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W7. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vriet C, Edwards A, Smith AM, Wang TL.. Sucrose and Starch Metabolism; 2014:97-115. doi:10.1007/978-3-662-44270-8_10.
- 60.Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Zuo J.. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Plant Signal Behav. 2009;17:1030–1040. [DOI] [PubMed] [Google Scholar]
- 61.Sperling P, Heinz E. Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. BBA - Mol Cell Biol Lipids. 2003;1632:1–15. doi: 10.1016/s1388-1981(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 62.Lynch DV, Dunn TM. An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 2010;161:677–702. doi: 10.1111/j.1469-8137.2004.00992.x. [DOI] [PubMed] [Google Scholar]
- 63.Coursol S, Fan L-M, Stunff HL, Spiegel S, Gilroy S, Assmann SM. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature. 2003;423:651–654. doi: 10.1038/nature01643. [DOI] [PubMed] [Google Scholar]
- 64.Liang G, Mishra G, Markham JE, Li M, Wang X. Connections between sphingosine kinase and phospholipase D in the abscisic acid signaling pathway in Arabidopsis. J Biol Chem. 2012;287:8286–8296. doi: 10.1074/jbc.M111.274274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chao DY, Gable K, Chen M, Baxter I, Dietrich CR, Cahoon EB, Guerinot ML, Lahner B, Lü S, Markham JE, et al. Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell. 2011;23:1061–1081. doi: 10.1105/tpc.110.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chutharat C, Ketsuwan C, Keasinee P, Sriprapai C, Numphet S, Kanidta S, Suksangpanomrung M, Michaelson LV, Napier JA, Muangprom A, et al. Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development. PLoS One. 2014;9:e106386. doi: 10.1371/journal.pone.0106386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang X, Fu H-F, Gong Z-H, Chai W-G. Involvement of a universal amino acid synthesis impediment in cytoplasmic male sterility in pepper. Sci Rep. 2016;6:23357. doi: 10.1038/srep23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Funck D, Winter G, Baumgarten L, Forlani G. Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 2012;12:191. doi: 10.1186/1471-2229-12-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Curaba J, Singh MB, Bhalla PL. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot. 2014;65:1425. doi: 10.1093/jxb/eru348. [DOI] [PubMed] [Google Scholar]
- 70.Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci U S A. 2012;109:2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Feng M.. RNase Z(S1) processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun. 2014;5:4884. doi: 10.1038/ncomms5972. [DOI] [PubMed] [Google Scholar]
- 72.Goetz M. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell. 2006;18:1873–1886. doi: 10.1105/tpc.105.037192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.CECCHETTI V. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell. 2008;20:1760–1774. doi: 10.1105/tpc.107.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Komatsuda T. Cleistogamous flowering in barley arises from the Suppression of MicroRNA-guided HvAP2 mRNA cleavage. Procnatlacadsciusa. 2010;107:490–495. doi: 10.1073/pnas.0909097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 76.Yu S, Galvao VC, Zhang Y-C, Horrer D, Zhang T-Q, Hao Y-H, Feng Y-Q, Wang S, Schmid M, Wang J-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


