ABSTRACT
Plants growing in natural habitats have evolved a wide range of mechanisms to copy with environmental challenging, including biotic and abiotic stresses. Abiotic stresses-induced increases in Abscisic acid (ABA) levels in plants suffering from stresses, including drought, cold or heat stress. To explore the function of the core components in ABA signaling, we used the overexpression of RCARs transgenic plants to expose in heat or cold stress. In this study, overexpression of RCAR12 or RCAR13 (R12-OE or R13-OE) transgenic plants had higher germination and survival rate than the wild-type (WT) Arabidopsis, indicating that they are both positively responsive to the high temperature. And the heat shock genes HSP18.2 and HSP70 were significantly induced by RCAR12 or RCAR13. Further, the results inferred that the over-expression of RCAR12 or RCAR13 could tolerance the cold stress, through induction CBFs expressions, the cold-responsive genes when plants were challenged the cold tress. And when complementation of RCAR12 to the 1124 mutant (R12:1124), the results indicated that RCAR12 could recover the insensitivity of 1124 to heat and cold stresses. Hence, we propose that RCAR12 and RCAR13, the ABA receptors, may play the positive roles in regulating the extreme temperature, including cold and high temperature in Arabidopsis.
KEYWORDS: Abscisic acid (ABA) receptor, Heat stress, Heat shock protein, Cold stress, CBFs
1. Introduction
Plants as sessile organisms have evolved complex developmental and physiological strategies to adapt to the unfavorable and changing environments such as drought, high salinity, and temperature fluctuations.1,2 When extreme temperature emerges, plants should face balancing adaptation of heat and cold stresses. Heat stress causes cellular damage of plants, ultimately leading to severe growth retardation and possible death.3,4 Low temperatures can severely disrupt the metabolism and physiological homeostasis of plant cells, and also result in weakness of stem, root and leaf development.5 Under natural conditions, vegetable crops eventually face episodes of heat and cold stresses that cause a loss of yield and agricultural quality. Therefore, understanding extreme temperature stresses, and how they damage plants are of practical importance.
Previous studies on Arabidopsis have suggested that phytohormones are involved in stress adaptation, especially for abscisic acid (ABA), which is generally considered a stress hormone.6–8 ABA regulates many essential physiological and biochemical processes. Importantly, it mediates many stress responses, such as the inhibition of primary root growth, guard cell closure and stress gene expression.9–11 Numerous reports have showed that ABA plays an essential role of dehydration stress.12–14 However, it has also been suggested that the plant hormone ABA induces thermotolerance in maize,15 or regulates heat stress transcription factors (HSFs) and heat shock proteins (HSPs) in Festuca arundinacea Schreb.16 ABA-deficient and -insensitive mutants are sensitive to heat stress, whereas ABRE/ABF-OE plants showed enhanced thermotolerance.17–20 ABA as a signal molecule is not only important for adaption to high temperature, but also cold stress. The literature reports indicate that cold acclimation can be triggered by exposing plants to exogenous ABA.5,21 Meanwhile, cold stress is accompanied with increased levels of endogenous ABA in many plant, such as maize, rice, cucumber, and pepper.22–24
ABA core signal pathway includes three components: pyrabactin resistance (PYR)/pyrabactin resistance-like (PYL)/regulation component of ABA receptors (RCAR) (hereafter referred as RCARs), protein phosphatase 2C (PP2Cs) and sucrose non-fermenting (SNF1)-related protein kinase 2 (SnRK2).25,26 In the absence of ABA, SnRK2s physically interacts with PP2Cs, which inactivate SnRK2s to phosphorylate downstream transcription factors.27 In the presence of ABA, RCARs would bind with ABA and subsequently tend to inhibit the activity of PP2Cs phosphatase.7 This lead to the release of SnRK2s and then the kinases of SnRK2s could phosphorylate ABRE/ABF/ABI5 transcription factors or guard cell anion channel (SLAC1), all of which further activate ABA-induced gene expressions.28–30 ABA receptors constitute a 14-member family, all of which are able to activate ABA-responsive gene expression in protoplast transfection assays.28,31
According to the alignment of amino acid sequences and expression patterns, substantial functional differences and redundancy among them should be expected.32,33 For example, RCAR3 conferred greater ABA sensitivity to the PP2C regulation than RCAR1, whereas regulation of the protein phosphatase activity by RCAR1 and RCAR3 was more sensitive to ABA for ABI1 than for ABI2.34 CARK1 interacts and phosphorylates RCAR3 and RCAR11 to involve in the ABA signal pathway, respectively.35 Except for functions of regulation in the ABA signal pathway, ABA receptors also mediate abiotic stresses. The expression profile of ABA core signaling component genes shifts under heat and cold stress36 ABA receptor genes, including RCAR8 and RCAR9, were upregulated in UBP1b-ox plants.37 Constitutive overexpression of rice landrace Nagina 22 PYL3 showed improved cold stress tolerance in Arabidopsis.24 However, it is not known how ABA-related genes both regulate heat and cold stress.
Here, we report that overexpressing of ABA receptors, including RCAR12 and RCAR13 response to heat and cold stress, respectively. The results show that overexpression of RCAR12 or RCAR13 increases thermotolerance in Arabidopsis through accumulation of heat short proteins (HSP), HSP18.2 and HSP70. Interestingly, we also found that the gain-of-function of RCAR12 and RCAR13 plants could tolerance cold stress through the induction of C-repeat binding factor (CBF) genes.
2. Materials and methods
2.1. Plant materials and growth conditions
Arabidopsis ecotype Col-0 was used as the wild-type (WT) control in this study. The quadruple 1124 mutant from the lab of Prof. Rodriguez is genetic impairment of PYR1, PYL1 (RCAR12), PYL2 and PYL4.25,38 The transgenic plants (RCAR12 and RCAR13-OE) were from our lab. The transgenic plants were identified.39–41 For plate culture, seeds were surface sterilized by treatment with 75% ethanol for 5 min, followed by commercial bleach (20%) for 15 min, and finally, four washes with sterile-distilled water. Stratification of the seeds was conducted in the dark at 4°C for 3 d. Then, seeds were sown on MS medium containing 2% sucrose and 0.8% agar, pH 5.7 with KOH. All plants were incubated in the growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μE m−2 s−1.
2.2. RNA extraction and quantitative real-time PCR (RT-PCR) analysis
Total RNA was extracted from 10-d-old Arabidopsis seedlings treated with heat or cold stress using TRIzol RNA reagent (TaKaRa, China). The cDNA was synthesized using PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, China), following the manufacturer’s instruction. PR-PCR was carried out on a CFX96 TouchTM Real-Time PCR detection system (Bio-Rad, USA) with iQTM SYBR Green Supermix and gene-specific primers (Table S1). All reactions were performed in triplicate with the following cycling conditions: 95°C for 5 min; 45 cycles each at 95°C for 20 s and 52°C for 20 s, and 72°C for 20 s. Actin2/8 was used as an internal standard.
2.3. Phenotype analysis
Seven or 10-d seedling on Murashige and Skoog agar medium (MS) were heated in the water incubator at 42°C for 1 h. The seed heat tolerance assay, seeds after stratification were heated in the water incubator at 50°C for 2 h. The freezing tolerance assay, 7 or 10-d seedlings were put at −20°C for 70 min, followed by 4°C for 24 h, and then 23°C in the chamber for 2 d.
3. Results
ABA receptors RCARs are well known to mediate plants response to ABA-dependent drought signal pathway and function as a major part of the ABA signal pathway. In this study, we tested whether RCARs are involved in response to other abiotic stress, including heat stress and cold stress.
3.1. RCARs involve in the heat stress response
The transcript expressions of RCARs were detected in Arabidopsis before and after heat stress. The expression of RCAR12had a 6.5-fold increase in WT plants exposed to the high temperature at 38°C for 2 h and increased 12.5-fold at 42°C for 2 h (Figure S1). The expression of RCAR13 increased 17.9-fold at 52°C for 2 h, while it did not changed exposing to 38°C or 42°C. ABA receptors had a medium response to high temperature. Although ABA receptors are highly conserved in Arabidopsis,32 the response to heat stress are different.
To explore the roles of signal transduction pathway involved in heat-stressed pathway, we tested the heat stress tolerance of over-expressing RCARs transgenic plants. The survival rates of RCAR12-OE and RCAR13-OE were significantly higher (~40%) than that of the wild-type plants (~15%) after the seedlings were exposed to the high temperature at 42°C for 2 h (Figure 1). To detect the heat tolerance during seed germination, seeds were exposed to the high temperature at 50°C for 2 h. The analysis demonstrated that of RCAR12-OE and RCAR13-OE plants presented higher germination rates than WT (Figure 2). Therefore, ABA receptors RCAR12 and RCAR13 had positive roles in heat stress response of plants.
Figure 1.
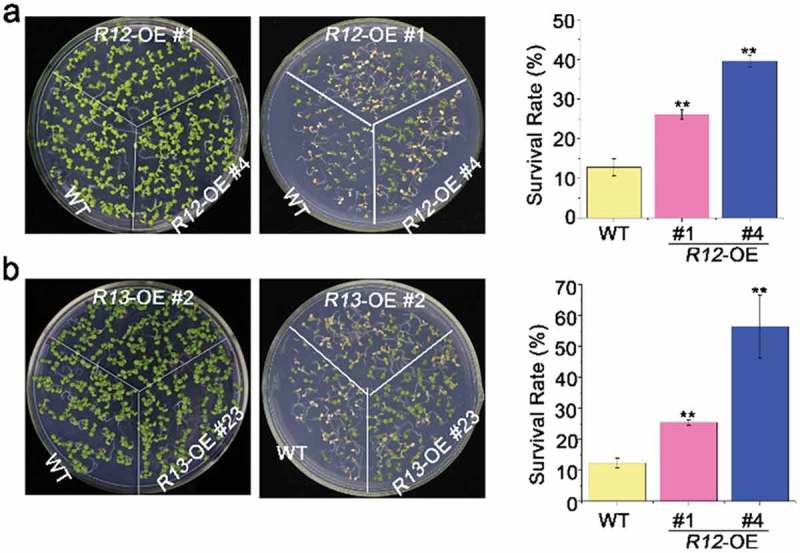
Heat tolerance of overexpressing of RCAR12 and RCAR13 transgenic plants in seedlings.
(a and b) 4-d-old seedlings in wild-type (WT), overexpression RCAR12 plants (a), overexpression RCAR13 plants on MS agar medium exposing to heat shock at 42°C for 2 h. Survival rates were cored at day 2 after heat stress treatment. Seedlings of plants on the control were 7-day-old plants. Data are means ± SD of the three independent biological experiments. ** Significant at P < .01 compared with the WT.
Figure 2.
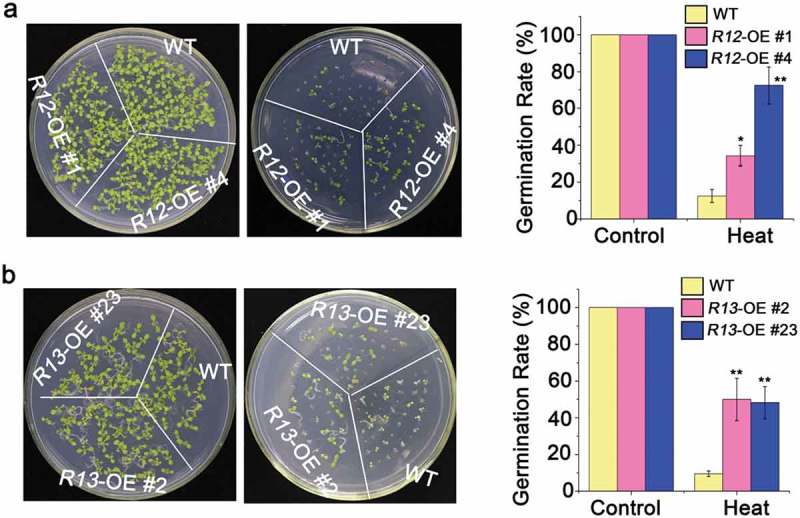
Heat tolerance of overexpressing of RCAR12 and RCAR13 transgenic plants.
(a and b) Seeds after stratification at 4°C for 3 days in the dark were sown on MS agar medium after 50°C for 2 h. The germination rates of plants (WT, RCAR12-OE (a) and RCAR13-OE (b)) were measured at day 10. Data are means ± SD of the three independent biological experiments. ** Significant at P < .01 compared with the WT, Student’s t-test.
The previous literature suggests a relationship between heat stress tolerance and heat shock proteins (HSPs) in plants.16,42 Thus, the transcript abundance of HSPs was detected in different RCARs transgenic plants by qRT-PCR. Our results revealed that the transcription expression levels of HSP18.2 and HSP70 in RCAR13-OE plants had 40 and 37-fold increase, comparing with the control under the normal condition after heat treatment (Figure 3a). RCAR12 also could induce the expressions of HSP18.2 and HSP70 (Figure 3a). In addition, the complementation of RCAR12 in the 1124 mutant showed higher germination rates than that of 1124 and WT (Figure 3b). Therefore, these results further demonstrated that ABA receptors RCAR12 and RCAR13 were involved in heat stress responses to plants by the induction of HSPs expressions.
Figure 3.
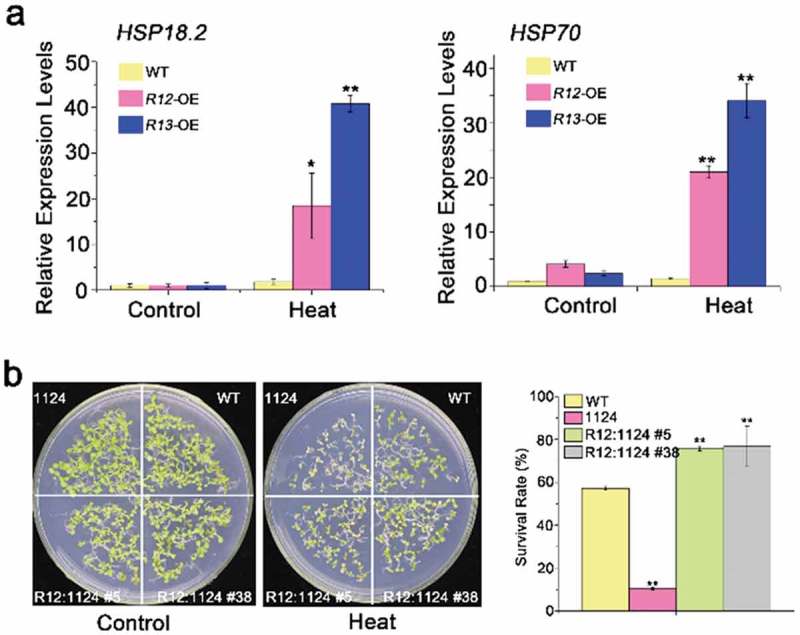
Heat-responsive genes expressions in overexpressing of RCAR12 and RCAR13 transgenic plants and RCAR12 recovers the heat-sensitive phenotype in 1124 mutant.
(a) Relative expression levels of heat-responsive genes HSP18.2 and HSP70 in wild-type and RCAR11, RCAR12 and RCAR13 overexpression transgenic plants. 10-d-old seedlings in MS agar medium were treated with or without heat stress at 42°C for 2 h. The transcriptional levels were determined by qRT-PCR analysis. Values are means ± SD (n = 3). ACTIN2/8 was used as an internal control. The experiments were repeated three times (*P < .05, **P < .01, Student’s t-test). (b) Seeds after stratification at 4°C for 3 days in the dark were sown on MS agar medium after 50°C for 2 h. The germination rates of plants were measured on the tenth day.
3.2. RCARs regulates CBFs during plant cold responses
Subsequently, we tested the ABA receptors response to the cold or freezing stress, including RCAR12 and RCAR13. After the chilling or freezing treatment, the expression levels of RCAR12 had about twofold increase, while RCAR13 was significantly increased in the WT plants after −20°C for 60 min comparing with that in normal conditions (Figure S2). However, phenotypic analysis showed that RCAR12/13-OE after the freezing treatment had a higher level of survival rate than WT (Figure 4a,b and Figure S3). C-repeat/DREB binding factor (CBFs) as key transcription factors are critical for cold acclimation in high plants.2,43 In this study, the qRT-PCR was employed to analyze the changes of expressions of CBFs in transgenic plants before and after cold treatment. The results indicated that CBF1/2/3 transcription levels in the overexpression RCAR12 or RCAR13 plants after cold stress had highly induced comparing with the control (Figure 4c). Furthermore, the transgenic plants with overexpressing RCAR12 in the 1124 mutant were generated and were cooled at −20°C for 70 min. The results showed that the R12:1124 transgenic plants had significantly higher survival rates than that of 1124 mutant and the wild type plants (Figure 5). Taken together, RCAR12 and RCAR13 mediated the cold stress response of plants in the manner of inducing CBFs expressions.
Figure 4.
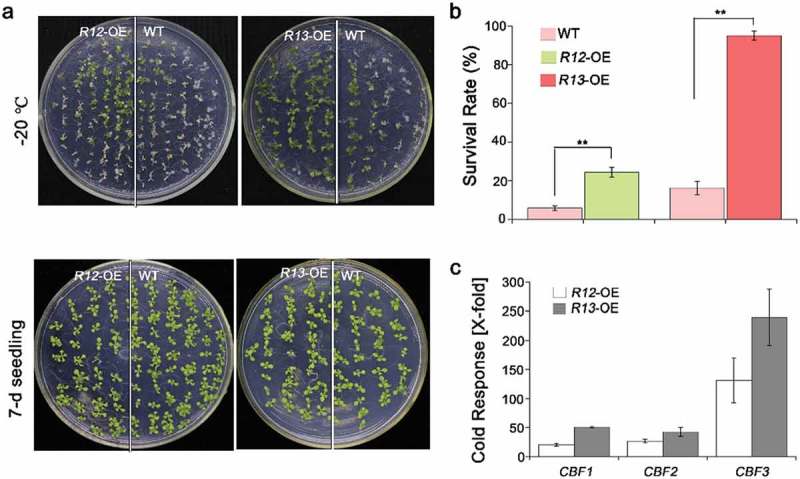
Freezing phenotypes of the plants.
(a) 7-d-old plants including WT, RCAR12-OE (R12-OE #1) and RCAR13-OE (R13-OE #4) under freezing stress for 70 min and then were recovered in the plant chamber. (b) Survival rates were cored at day 2 after recovery. Data are means ± SD of the three independent biological experiments. ** Significant at P< .01 compared with the WT. Independent biological experiments were performed. (c) Relative expression levels of heat-responsive genes ABF1, ABF2 and ABF3 in wild-type, RCAR12 and RCAR13 overexpression transgenic plants. 10-d-old seedlings in MS agar medium were treated with or without heat stress at 4°C for 12 h. The transcriptional levels were determined by qRT-PCR analysis. Values are means ± SD (n = 3). ACTIN2/8 was used as an internal control. The experiments were repeated three times (*P < .05, **P < .01, Student’s t-test).
Figure 5.
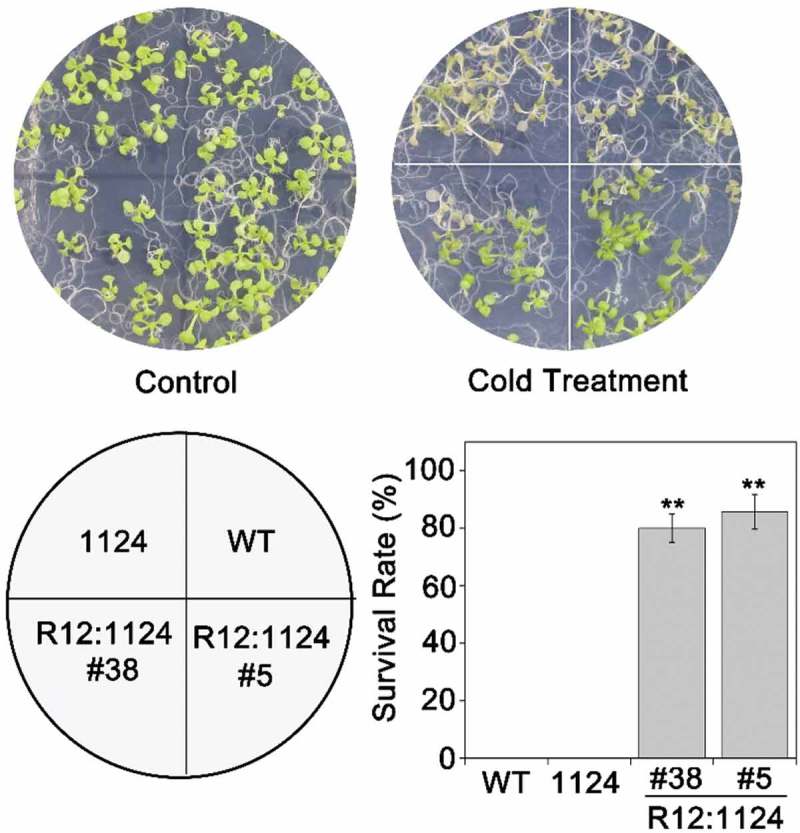
RCAR12 could recover the sensitivity of the 1124 mutant to cold stress.
7-d-old plants including WT, 1124 and RCAR12:1124 (R12:1124) under freezing stress for 70 min and then were recovered in the plant chamber and the survival rates were determined. Independent biological experiments were performed.** Significant at P < .01.
4. Discussion
The phytohormone abscisic acid (ABA) would accumulate under stressed conditions, including drought, high and low temperature.5 RCARs, as a key regulator, involve in the ABA signaling pathway and ABA-dependent drought-stress signal pathway.41,44 Here, we reported the functions of ABA receptors in extreme temperature.
Drought stress usually accompanies with high temperature. Therefore, we hypothesis that RCARs also play an essential role in plant response to high temperature. The seeds and seedlings of RCAR12/13-OE showed heat tolerance through induction of HSPs. These results indicate that RCAR12 and RCAR13 play a positive role in regulation of heat-stress signal pathway. This conclusion is consistent with that HSP6a, participates in ABA-mediated resistance, as a positive regulator in heat-stress response.20 In addition, ABA-responsive element binding protein 3 (AREB3), function as upstream trans-acting factors and regulate transcriptional activities of FaHSFA2c and the downstream FaHSPs leading to improve heat tolerance in Festuca arundinacea Schreb.16 However, it is interesting that RCAR1-OE could not bear high temperature. Although ABA receptors are conserved, their functions also have divergence. For example, the PP2C AHG1 is regulated by RCAR1, RCAR2, and RCAR3, but not other members.31 Therefore, it is reasonable to propose that the components in the ABA signal pathway also involve in heat-stress responses through regulation of heat shock proteins.
Another common environmental stress, a low temperature, severely inhibits plant growth and development. A recent report suggests that the phosphorylation of ICE1 by the kinase SnRK2.6 could depress the degradation of ICE1, ultimately positively mediates the plant cold-stress response.45 In plants, ABA accumulates under stress condition and ABA receptors bind to ABA, thus releasing the inhibition of SnRK2.6 by PP2Cs.25,26 Our results indicate that RCAR12-OE and RCAR13-OE have higher levels of survival rates than other plants, and induce the expressions of CBF1/2/3. Therefore, we postulate that RCAR12 and RCAR13 binding to ABA inhibit the activities PP2Cs, resulting that SnRK2.6 enhances ICE1 stability through phosphorylation.45 In polar, overexpression of PtPYR1 or PtPYL5 could increase the resistance to cold stress.1
5. Conclusion
In conclusion, RCAR12 and RCAR13 could positively regulate the plant responses to the extreme temperature through induction the expressions of downstream genes. In this study, we demonstrated that overexpression of RCAR12 or RCAR13 could enhance the plants in responses to the heat tolerance, through induction of the heat shock proteins, including HSP18.2 and HSP70. In addition, RCAR12 or RCAR13 mediated the cold-stress signal pathway through induction of CBFs in Arabidopsis (Figure 6). However, more information is needed to explore how ABA receptors have an effect on cold resistance.
Figure 6.
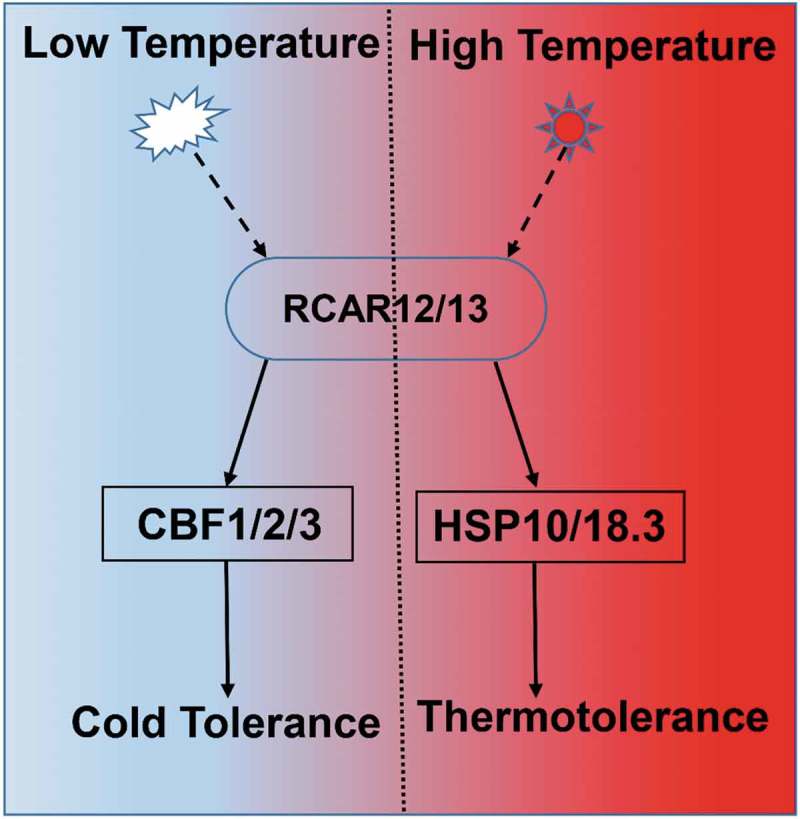
Model illustrating how RCARs response to heat and cold stresses in plants.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Yu J, Ge H, Wang X, Tang R, Wang Y, Zhao F, Lan W, Luan S, Yang L.. Overexpression of pyrabactin resistance-like abscisic acid receptors enhances drought, Osmotic, and Cold Tolerance in Transgenic Poplars. Front Plant Sci. 2017;8:1. doi: 10.3389/fpls.2017.01752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhtar M, Jaiswal A, Taj G, Jaiswal JP, Qureshi MI, Singh NK. DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J Genet. 2012;91:385–7. [DOI] [PubMed] [Google Scholar]
- 3.Huang YC, Niu CY, Yang CR, Jinn TL. The Heat Stress Factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016;172:1182–1199. doi: 10.1104/pp.16.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146:748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Yang S. ABA regulation of the cold stress response in plants. Netherlands: Springer, Dordrecht. 2014. doi: 10.1007/978-94-017-9424-4. [DOI] [Google Scholar]
- 6.Grossi M, Cattivelli L, Terzi V, Stanca AM. Modification of gene expression induced by ABA, in relation to drought and cold stress in barley shoots. Plant Physiol Biochem. 1992;30:97–103. [Google Scholar]
- 7.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 8.Hao Q, Yin P, Yan C, Yuan X, Li W, Zhang Z, Liu L, Wang J, Yan N. Functional Mechanism of the Abscisic Acid Agonist Pyrabactin. J Biol Chem. 2010;285:28946. doi: 10.1074/jbc.M110.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grill E, Himmelbach A. ABA signal transduction. Curr Opin Plant Biol. 1998;1:412–418. [DOI] [PubMed] [Google Scholar]
- 10.Riichiro Y, Taishi U, Tsuyoshi M, Seiji T, Fuminori T, Kazuo S. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 11.Tomo Y, Noriyuki N, Nobutaka K, Takashi K, Takuya I, Tadao A, Kazuo S, Takashi H. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K-I, et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci U S A. 2016;113:1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ming G, Li YJ, Chen SZ. Abscisic acid-induced thermotolerance in maize seedlings is mediated by calcium and associated with antioxidant systems. J Plant Physiol. 1998;153:488–496. doi: 10.1016/S0176-1617(98)80179-X. [DOI] [Google Scholar]
- 16.Wang X, Zhuang L, Shi Y, Huang B. Up-Regulation of HSFA2c and HSPs by ABA Contributing to Improved Heat Tolerance in Tall Fescue and Arabidopsis. Int J Mol Sci. 2017;18:1981. doi: 10.3390/ijms18091981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight MR. Protection against Heat Stress-Induced Oxidative Damage in Arabidopsis Involves Calcium, Abscisic Acid, Ethylene, and Salicylic Acid. Plant Physiol. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Kang JY, Cho DI, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2010;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 19.Larkindale J, Hall JD, Knight MR, Vierling E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki N, Bassil E, Hamilton JS, Inupakutika MA, Zandalinas SI, Tripathy D, Luo Y, Dion E, Fukui G, Kumazaki A, et al. ABA Is Required for Plant Acclimation to a Combination of Salt and Heat Stress. PLoS One. 2016;11:e0147625. doi: 10.1371/journal.pone.0147625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lång V, Palva ET. The expression of a rab -related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol. 1992;20:951–962. doi: 10.1007/BF00027165. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Shi H, Hu Z, Liu A, Amombo E, Chen L, Fu J. ABA Is Involved in Regulation of Cold Stress Response in Bermudagrass. Front Plant Sci. 2017;8:1613. doi: 10.3389/fpls.2017.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian LX, Li J. The effects of exogenous ABA applied to maize (Zea mays L.) roots on plant responses to chilling stress. Acta Physiologiae Plant. 2018;40:77. doi: 10.1007/s11738-018-2655-2. [DOI] [Google Scholar]
- 24.Lenka SK, Muthusamy SK, Chinnusamy V, Bansal KC. Ectopic Expression of Rice PYL3 Enhances Cold and Drought Tolerance in Arabidopsis thaliana. Mol Biotechnol. 2018;60:350–361. doi: 10.1007/s12033-018-0076-5. [DOI] [PubMed] [Google Scholar]
- 25.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow T-F-F, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 28.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu J-K. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita Y, Yoshida T, Yamaguchi‐Shinozaki K. Pivotal role of the AREB/ABF‐SnRK2 pathway in ABRE‐mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 30.Tan W, Zhang D, Zhou H, Zheng T, Yin Y, Lin H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018;14:e1007336. doi: 10.1371/journal.pgen.1007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischer SV, Wunschel C, Papacek M, Kleigrewe K, Hofmann T, Christmann A, Grill E. Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proc Natl Acad Sci USA 2017; 114: 10280-10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Zhang Z, Gao J, Wang P, Hu T, Wang Z, Hou Y-J, Wan Y, Liu W, Xie S, et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018;23:3340–51 e5. doi: 10.1016/j.celrep.2018.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2009;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Li X, Li D, Sun Y, Li Y, Luo Q, Liu Z, Wang J, Li X, Zhang H, et al. CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discovery. 2018;4:30. doi: 10.1038/s41421-018-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalal M, Inupakutika M. Transcriptional regulation of ABA core signaling component genes in sorghum (Sorghum bicolor L. Moench). Mol Breed. 2014;34:1517–1525. doi: 10.1007/s11032-014-0114-3. [DOI] [Google Scholar]
- 37.Nguyen CC, Nakaminami K, Matsui A, Watanabe S, Kanno Y, Seo M, Seki M. Overexpression of oligouridylate binding protein 1b results in ABA hypersensitivity. Plant Signal Behav. 2017;12:e1282591. doi: 10.1080/15592324.2017.1282591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li D, Zhang L, Li X, Kong X, Wang X, Li Y, Liu Z, Wang J, Li X, Yang, Y. AtRAE1 is involved in degradation of ABA receptor RCAR1 and negatively regulates ABA signaling in Arabidopsis. Plant Cell Environ. 2017,41:231-244. doi: 10.1111/pce.13086. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Zhang L, Li D, Liu Z, Wang J, Li X, Yang Y. The Arabidopsis F‐box E3 ligase RIFP1 plays a negative role in abscisic acid signalling by facilitating ABA receptor RCAR3 degradation. Plant Cell Environ. 2015;39:571–582. doi: 10.1111/pce.12639. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Li G, Li Y, Kong X, Zhang L, Wang J, Li X, Yang Y. ABA Receptor Subfamily III Enhances Abscisic Acid Sensitivity and Improves the Drought Tolerance ofArabidopsis. Int J Mol Sci. 2018;19:1938. doi: 10.3390/ijms19071938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat Shock Protein 101 Plays a Crucial Role in Thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batista R, Saibo N, Lourenço T, Oliveira MM. Microarray Analyses Reveal That Plant Mutagenesis May Induce More Transcriptomic Changes than Transgene Insertion. Proc Natl Acad Sci U S A. 2008;105:3640–3645. doi: 10.1073/pnas.0707881105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- 45.Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. OST1 Kinase Modulates Freezing Tolerance by Enhancing ICE1 Stability in Arabidopsis. Dev Cell. 2015;32:278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


