ABSTRACT
Salt stress adversely affects plant growth and development. Multiple adaptive mechanisms have been used for plant salt tolerance. We previously reported that membrane trafficking-related protein patellin1 (PATL1) negatively regulates plant salt tolerance. Here, we characterized that Arabidopsis PATL1 negatively modulates nitric oxide (NO) accumulation upon salt exposure. Our work revealed a functional link between salt response and NO signaling.
KEYWORDS: Plant salt tolerance, PATL1, NO accumulation
As sessile organisms, plants encounter various environmental stresses during their life cycle. Soil salinization is one of the major adverse factor that limits crop growth and production.1 Plants have evolved multiple signaling networks for effective salt tolerance regulation, including ionic homeostasis maintenance, osmotic adjustment as well as other mechanisms.1 It is believed that elucidation of the mechanisms by which plants cope with salt stress will definitely promote the generation of salt-tolerant crops, thus providing a possibility to make full use of saline soil widespread globally.
Nitric oxide (NO) can be induced by numerous environmental stimuli in plants and functions as a versatile messenger in multiple signaling networks.2,3 In plants, the production of NO occurs mainly through two different enzymatic pathway.4 Subsequently, NO reacts with GSH to form GSNO, a major bioactive NO species, which is then reversibly metabolized by a highly conserved GSNOR (S-nitrosoglutathione reductase), making the NO level being dynamically modulated.5,6 The principal bioactivity of NO is executed through S-nitrosylation by covalently adding a NO group on the reactive cysteine (Cys) thiol of a substrate protein, resulting in S-nitrosothiols which might alter the functions of target proteins by diverse mechanisms.7,8 Interestingly, Arabidopsis GSNOR1 itself also undergoes NO-mediated S-nitrosylation at Cys10, causing local conformational changes and inducing selective autophagy of GSNOR1 through directly interacting with ATG8.9 S-nitrosylation is a redox-based post-translational modification (PTM) on proteins, and it also plays a role in reactive oxygen species (ROS) production in plants. This reciprocal regulation of ROS and NO are critical for early signaling in both abiotic and biotic stress responses.7,8,10,11 As aforementioned, GSNOR is the master regulator of NO germination in plants, and mutations in the single copied GSNOR1/HOT5/PAR2 gene in Arabidopsis genome cause elevated NO concentration and enhanced S-nitrosylation level, leading to severe growth defects and altered responses environmental stresses.12–14 Specifically, GSNOR1 loss-of-function mutant gsonr1-3 exhibits a salt-tolerant phenotype with low Na+/K+ ratio under salt stress condition, and salt-induced Ca2+ and calcium-binding protein CaM4 have been reported to function in GSNOR-mediated salt tolerance regulation.15 In fact, salt stress can effectively induce endogenous NO germination, which then results in salt resistance by increasing K+/Na+ ratio in two ecotypes of reed.16 Exogenous application of GSNO has been found to effectively enhance tonoplast H+-ATPase and H+-PPase activity, resulting in elevated Na+/H+ exchange activity across vacuole membrane and lower the Na+ toxicity in cytoplasma.17 In Arabidopsis, NOA1-dependent NO production and heme oxygenase1 expression are also associated with plant salt response.18,19 These studies together support a conclusion that NO and its dynamic regulation play a crucial role in plant salt stress signaling networks.
Arabidopsis patellin (PATL) protein family members are putative membrane trafficking-related proteins.20,21 These proteins are characterized by two conserved domains universally observed in membrane trafficking-related proteins: a Sec14 lipid-binding domain and a Golgi dynamics domain (GOLD) followed the N-terminal variable domain.20,21 Arabidopsis PATLs are expressed in distinct but partly overlapping patterns, which might represent the redundant and diversified functions of PATLs in Arabidopsis.21 Recently, we have characterized PATL1 as a novel negative regulator of plant salt tolerance.22 There exists a direct interaction between PATL1 and SOS1, a major component of Salt Overly Sensitive (SOS) pathway. PATL1 is implicated in ion homeostasis regulation in plants possibly through SOS1, and it also functions in cellular redox homeostasis during salt stress.22 PATL1 also forms a complex with calmodulin4 (CaM4) and they two together paly a regulatory role in plant freezing response.23 Interestingly, CaM4 is reported to participate in plant salt tolerance regulation through activating GSNOR and promoting NO accumulation upon salt exposure.15 Thus there might exist a possible unidentified mechanism that PATL1 regulates plant salt tolerance through the action of GSNOR and NO.
In the present study, we demonstrate that Arabidopsis PATL1 negatively modulates plant salt tolerance through attenuating NO accumulation. Consistent to our previous study,22 data showed that the patl1-1 mutant (SALK_103668) with elevated PATL1 transcription exhibits salt-sensitive phenotype (Figure 1(a)). To further determine the underlying mechanisms for the negative effect of PATL1 on plant salt tolerance, we thus examined intracellular NO accumulation in Col-0 and patl1 mutants at the seedling stage using the method of 4-amino-5-methylamino-2ʹ,7ʹ-difluorofluorescein diacetate (DAF-FM DA) staining. Data revealed that the NO levels were remarkably increased in both tested plants in the presence of 125 mM NaCl (Figure 1(b)). Interestingly, NO accumulated to a significantly lower level in patl1-1 seedlings than that in Col-0 (Figure 1(b)), which is consistent to the salt-sensitive phenotype of patl1 plants.
Figure 1.
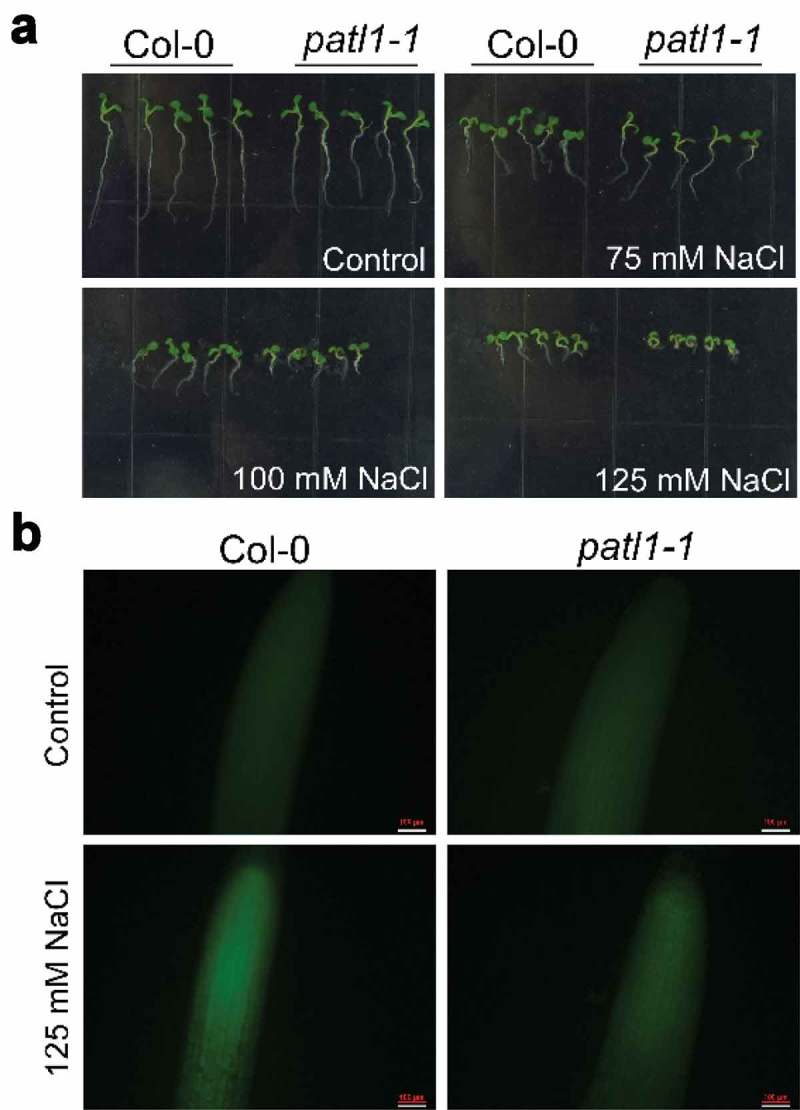
Patellin1 negatively regulates plant salt tolerance by attenuating NO accumulation in Arabidopsis.
(a) Salt sensitivity analysis of 7-d-old seedlings of Col-0 and patl1-1 grown on MS medium under control condition or supplemented with different concentrations of NaCl (75, 100 and 125 mM). Photographs were taken when phenotypic differences were observed. (b) NO accumulation in the roots of 7-d-old seedlings. Col-0 and patll-1 plants were incubated in 1/2 liquid MS medium (pH = 5.8) with 10 μM DAF-FM DA for 20 min. After incubation, the roots were washed three times for 15 min each in 1/2 liquid MS medium prior to observation using a fluorescence microscope. Bar = 100 μm.
Considering that GSNOR1 is implicated in NO homeostasis and there exists a possible link between PATL1 and GSNOR1, we propose that PATL1 might interact GSNOR directly and significantly enhances its activity, leading to decreased accumulation of NO under salt stress condition. Future assays to determine whether PATL1 and GSNOR1 together participate in plant salt tolerance are required. Since our data have shown that PATL1 plays a negative role in NO production upon salt exposure (Figure 1) and GSNOR1 serves as a negative regulator of plant salt tolerance,15 we therefore hypothesized that PATL1 might play a positive regulatory role on GSNOR activity, and that PATL1 attenuates NO accumulation through the action of GSNOR activity upon salt exposure. NO is significantly induced upon salt exposure, indicating a regulatory role of NO in plant salt tolerance. Previous studies have reported that NO alleviates salt toxicity in reeds and maize through the activation of H+-ATPase localized in the PM and vacuole membrane, promoting Na+ efflux to the apoplast and/or vacuolar Na+ compartmentation.17,24 In Arabidopsis, NO is associated with salt resistance via attenuation of the salt-induced Na+/K+ ratio increase.15 However, there exists no direct evidence concerning whether specific Na+ transporters are involved in NO-mediated salt tolerance network. Considering that PATL1 is linked to PM Na+/H+ antiporter SOS1 activity regulation as well as NO signaling, it is possible that NO might play a regulatory role in SOS1-mediated Na+ efflux.
Funding Statement
This work was supported by the NSFC [31600201, 31870241].
Acknowledgments
X.S. conducted the experiments. Y.Z. contributed to manuscript preparation. H.Z. wrote the manuscript. H.Z. and H.L. supervised the project. All the authors contributed to discussion. This work was supported by the National Natural Science Foundation of China (31870241 and 31600201 to H.Z.) and the Innovation Spark Fund of Sichuan University (Grant 2019SCUH0011 to H.Z.).
References
- 1.Yang Y, Guo Y.. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018;217:1–4. doi: 10.1111/nph.14920. [DOI] [PubMed] [Google Scholar]
- 2.Besson-Bard A, Wendehenne D.. New insights into nitric oxide signaling in plants. Annu Rev Plant Biol. 2008;59(1):21–39. doi: 10.1146/annurev.arplant.59.032607.092830. [DOI] [PubMed] [Google Scholar]
- 3.Yu M, Lamattina L, Spoel SH, Loake GJ. Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytol. 2014;202(4):1142–1156. doi: 10.1111/nph.12739. [DOI] [PubMed] [Google Scholar]
- 4.Wilson ID, Neill SJ, Hancock JT. Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 2008;31(5):622–631. doi: 10.1111/j.1365-3040.2007.01761.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 6.Kaur H, Bhatla SC. Melatonin and nitric oxide modulate glutathione content and glutathione reductase activity in sunflower seedling cotyledons accompanying salt stress. Nitric Oxide. 2016;59:42–53. doi: 10.1016/j.niox.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Jain P, Bhatla SC. Molecular mechanisms accompanying nitric oxide signalling through tyrosine nitration and S-nitrosylation of proteins in plants. Funct Plant Biol. 2018;45(2):70–82. doi: 10.1071/FP16279. [DOI] [PubMed] [Google Scholar]
- 8.Jain P, Von Toerne C, Lindermayr C, Bhatla SC. S-nitrosylation/denitrosylation as a regulatory mechanism of salt stress sensing in sunflower seedlings. Physiol Plant. 2018;162(1):49–72. doi: 10.1111/ppl.12641. [DOI] [PubMed] [Google Scholar]
- 9.Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, et al. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell. 2018;71(1):142–154. doi: 10.1016/j.molcel.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang J-G, Kwon E, Spoel SH, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478:264–268. doi: 10.1038/nature10427. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou J-M, Zuo J. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015;167:1604–1615. doi: 10.1104/pp.114.255216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, et al. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 13.Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Nat Acad Sci USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermos-tolerance and plant growth in Arabidopsis. Plant Cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S, Jia L, Chu H, Wu D, Peng X, Liu X, Zhang JJ, Zhao JF, Chen KM, Zhao LQ. Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by inhibiting S-nitrosoglutathione reductase via direct binding. PLoS Genet. 2016;12((9)–e1006255):1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, Zhang F, Guo J, Yang Y, Li B, Zhang L. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 2004;134:849–857. doi: 10.1104/pp.103.030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Wang L, Liu Y, Zhang Q, Zhang WW. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta. 2006;224:545–555. doi: 10.1007/s00425-006-0242-z. [DOI] [PubMed] [Google Scholar]
- 18.Xie Y, Mao Y, Lai D, Zhang W, Zheng T, Shen W. Roles of NIA/NR/NOA1-dependent nitric oxide production and HY1 expression in the modulation of Arabidopsis salt tolerance. J Exp Bot. 2013;64:3045–3060. doi: 10.1093/jxb/ert149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao MG, Tian QY, Zhang WH. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007;144:206–217. doi: 10.1104/pp.107.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterman TK, Ohol YM, Mcreynolds L, Luna E. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 2004;136(2):3080–3094. doi: 10.1104/pp.104.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Duan H, Liu Y, Sun X, Zhao J, Lin H. Patellin protein family functions in plant development and stress response. J Plant Physiol. 2019;234–235:94–97. doi: 10.1016/j.jplph.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H, Wang C, Tan T, Cai J, He J, Lin H. Patellin1 negatively modulates salt tolerance by regulating PM Na+/H+ antiport activity and cellular redox homeostasis in Arabidopsis. Plant Cell Physiol. 2018;59(8):1630–1642. doi: 10.1093/pcp/pcy081. [DOI] [PubMed] [Google Scholar]
- 23.Chu M, Li J, Zhang J, Shen S, Li C, Gao Y, Zhang S. AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J Exp Bot. 2018;69(21):5241–5253. doi: 10.1093/jxb/ery278. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee S, Bhatla SC. A novel fluorescence imaging approach to monitor salt stress-induced modulation of ouabain-sensitive ATPase activity in sunflower seedling roots. Physiol Plant. 2014;150(4):540–549. doi: 10.1111/ppl.12101. [DOI] [PubMed] [Google Scholar]


